Abstract
Critical to the mitigation of parasitic vector-borne diseases is the development of accurate spatial predictions that integrate environmental conditions conducive to pathogen proliferation. Species of Plasmodium and Trypanosoma readily infect humans, and are also common in birds. Here, we develop predictive spatial models for the prevalence of these blood parasites in the olive sunbird (Cyanomitra olivacea). Since this species exhibits high natural parasite prevalence and occupies diverse habitats in tropical Africa, it represents a distinctive ecological model system for studying vector-borne pathogens. We used PCR and microscopy to screen for haematozoa from 28 sites in Central and West Africa. Species distribution models were constructed to associate ground-based and remotely sensed environmental variables with parasite presence. We then used machine-learning algorithm models to identify relationships between parasite prevalence and environmental predictors. Finally, predictive maps were generated by projecting model outputs to geographically unsampled areas. Results indicate that for Plasmodium spp., the maximum temperature of the warmest month was most important in predicting prevalence. For Trypanosoma spp., seasonal canopy moisture variability was the most important predictor. The models presented here visualize gradients of disease prevalence, identify pathogen hotspots and will be instrumental in studying the effects of ecological change on these and other pathogens.
Keywords: avian malaria, Plasmodium, Trypanosoma, Maxent, random forest, predictive maps
1. Introduction
An improved understanding of the environmental correlates that give rise to the spread of infectious diseases constitutes one of the key challenges in disease ecology. With recent technological advances in satellite imagery, computer modelling and molecular biology, we are now capable of studying infectious diseases in an integrated fashion over relatively large spatial scales. It is believed that many infectious diseases originated in Africa [1], yet relatively little is known about the environmental factors that affect the evolution and spread of its endemic pathogens. Addressing the ecology of disease has taken on a new urgency with both the current and predicted rapid changes in climate and land use that are affecting the African continent and the entire planet [2,3]. The use of birds as a model system can provide important data regarding the ecology of infectious diseases because they represent natural ubiquitous populations, and can be studied in both human-impacted environments, and in pristine unaltered habitats where humans are largely absent. In addition, they are infected by several vector-borne pathogens, including malaria and trypanosomes. Predictive spatial maps for infectious diseases of birds can thus help visualize how natural ecological landscapes and climatic variables are associated with parasite transmission, and provide insight regarding the impacts of environmental change on the patterns of pathogen distribution.
Avian malaria is caused by species of Plasmodium that use a variety of mosquito species as vectors [4]. Trypanosomes, typically transmitted by simuliid and hippoboscid flies, are also common blood parasites found in African rainforest birds [5]. There is no evidence for the natural transmission of these parasites to mammals, but the study of avian malaria in particular has provided valuable information on the basic life-history strategies of human malaria parasites [6] and more recently on the evolutionary history of the disease (e.g. [7]). Infections of avian hosts with malaria are generally not fatal in natural ecosystems, but numerous exceptions have been documented [4], and infections certainly can affect host fitness [8,9]. Less is known about the impact of avian trypanosomes on natural populations [5,10–12]. Recent work has advanced the understanding of the evolution and ecology of these parasites [13–18]. But which ecological factors may contribute to the widespread distribution of avian blood parasites remain poorly understood. Prevalence of these parasites can vary greatly depending on location and host, but they are commonly found in birds throughout Africa [19–22]. In West Africa, several of these avian species are generalists that can be found in a variety of habitats, including primary rainforest, secondary forest and fragmented deforested areas of the ecotone. One of these species, the olive sunbird (Cyanomitra olivacea; [23,24]), is non-migratory, readily sampled and ubiquitous across the landscape, therefore acting as an excellent model to study the ecology of large-scale infectious disease patterns in Africa [19,20,23,25]. It is clear that distribution of avian haemosporidian parasites can vary on the local scale [14,26], but little information is available regarding the broad-scale environmental factors that can affect and help predict their transmission.
A number of studies have mapped the prevalence of human malaria using satellite-based techniques. Detailed risk maps based on environmental predictors of vector density and parasite incidence have been developed to estimate disease burden, and to aid in the control and prevention of human malaria (e.g. [27–29]). These predictive maps based on environmental correlates are valuable in understanding the ecology of the disease, and can inform studies concerning avian pathogens. Unlike birds, however, the prevalence of malaria in humans may be affected by socio-economic conditions [30]. In addition, little is known about the transmission of malaria parasites in human uninhabited rainforest regions. Thus, studying avian malaria can provide a baseline for the prevalence of Plasmodium parasites in general, and it is valuable to determine the environmental variables associated with parasite prevalence to provide unbiased and ecologically grounded models of disease distribution.
In order to better understand the ecology and transmission risks of infectious diseases, we sought to identify the environmental correlates of parasites of two important genera of avian haematozoa, Plasmodium and Trypanosoma, in Central and West Africa. Our objectives were to (i) empirically determine the parasites' prevalence at numerous rainforest sites, (ii) determine, using remote sensing, the ecological correlates associated with parasite prevalence, and (iii) use these correlates to develop flexible models and maps that accurately predict prevalence over a large scale. To accomplish these objectives, we first estimated parasite prevalence through analyses of blood samples using molecular methods and microscopy from an African bird species, the olive sunbird, across 28 sampling locations in Cameroon, Equatorial Guinea, Ghana and Côte d'Ivoire. This is a unique and unprecedented sample base from one rainforest bird species collected over 20 years. We then used a comprehensive environmental dataset comprising satellite-based and ground-based measurements of climate, land surface and vegetation characteristics and a random forest modelling framework to identify the underlying ecological factors that explain parasite prevalence and predict their broad-scale spatial distributions.
2. Material and methods
(a). Field sampling and parasite screening
The olive sunbird is a resident solitary sunbird that occurs across Central Africa, and feeds on both nectar and insects [31]. Using this species as a representative of generalist avian species within Central Africa, we captured and took blood samples from 622 individual olive sunbirds (C. olivacea, family: Nectariniidae) across 28 sites in Cameroon, Equatorial Guinea, Côte d'Ivoire and Ghana, and recorded site coordinates, dates and habitat type (primary forest, secondary forest and ecotone) in these sampling efforts (electronic supplementary material, figure S1 and table S1).
At each site, mist nets (12 m, 30 × 30 mm mesh) were erected to capture birds. Blood (20–30 µl) was collected from the brachial vein and stored in lysis buffer (10 mm Tris–HCL pH 8.0, 100 mm EDTA, 2% SDS). After sampling, all birds were released unharmed. Parasite screening was performed using molecular and microscopy approaches (see electronic supplementary material, methods). We used traditional estimates of parasite prevalence, defined as the percentage of individuals infected by a given parasite as compared with the total number of samples tested. We isolated 18 lineages of Plasmodium from the olive sunbird but were not able to develop statistically significant analyses with any of the individual lineages; as a result, we do not differentiate among the many possible species of Plasmodium or Trypanosoma in this study.
(b). Environmental variables
We compiled a set of ground-based climate data and satellite-based measurements and derived products to describe the ecology of two host–parasite systems across West and Central Africa. The satellite observations stem from both optical passive as well as microwave active sensors, and capture a diverse range of surface parameters including vegetation density, canopy moisture and roughness and topography (see electronic supplementary material for a complete description). The target spatial resolution is 1 km, close to the native grain size of a number of the used satellite data.
(c). Modelling approach
Our modelling approach consisted of three steps, used to (i) define the geographical distribution of the host–parasite system, (ii) identify and interpret the relationships between parasite prevalence and ecological predictor variables, and (iii) use these relationships to build a statistical model that can be used to project disease prevalence across the defined host/parasite distribution. In the first step, we used the Maxent species distribution algorithm [32] to predict the potential geographical distribution of the host-parasite system across West Africa based on presence of localities of infected birds and the corresponding environmental variables. The spatial output of the Maxent distribution model consists of a continuous range indicating relative probability of presence of, in the case of this study, the host–parasite system. A presence/absence map of the host–parasite system was obtained by applying an optimized threshold (balance threshold [32]) on the continuous Maxent probability distribution. This distribution was then used to define the spatial limits for all prevalence predictions.
In the second step, we used tree regression (as implemented in Tree v. 1.0–26; [33]) in the R statistical framework [34] to determine the ecological variables associated with variation in both Plasmodium and Trypanosoma prevalence across the host–parasite distribution. While tree regressions are straightforward to interpret (they include indications of directionality, strength and location along a response where predictors are important), they are poor predictors, as they make only a single statement about the relationships between response and predictors.
To make more robust predictions, in the third step, we grew a series of tree regressions (n = 2000), also known collectively as a random forest (as implemented in RandomForest v. 4.5–30 [35] in the R statistical framework; [34]), to predict the prevalence of each blood parasite genus in the olive sunbird within its geographical distribution determined from the first step. This non-parametric algorithm method was used instead of a more traditional data modelling technique (such as linear regression or ANOVA) because random forest procedures (i) do not require the use of any particular model (which might be difficult to assign given a complex response such as disease prevalence), (ii) do not require normalized data, and (iii) have consistently outperformed traditional regression procedures on a number of datasets [36–38]. The advantage of random forest models is their ability to predict a continuous (in this case, prevalence) rather than categorical (presence/absence) variable across a landscape, and their ability to model complex interactions among predictor variables [37]. Further details on the modelling approach are provided in the electronic supplementary material.
3. Results
(a). Parasite prevalence and environmental correlates
The prevalence of malaria parasites and trypanosomes in olive sunbird populations was estimated across 28 sampling sites in West Africa. Across regions, prevalence varied substantially, ranging from 0 to 83 per cent for parasites of the genus Plasmodium and 10 to 100 per cent for Trypanosoma. Significantly higher prevalence of each parasite genus was observed in West Africa (Ghana and Côte d'Ivoire) as compared with Cameroon (two-sided, two-sample test for equality of proportions, Plasmodium, χ2 = 39.54, p = 3.22e−10; Trypanosoma, χ2 = 22.01, p = 2.71e−6; electronic supplementary material, figure S3).
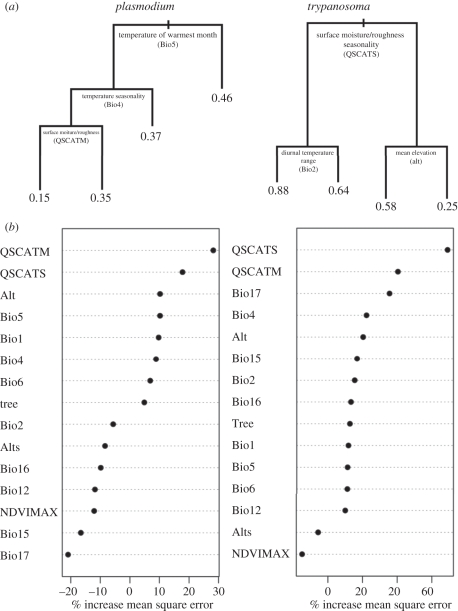
Tree regressions performed using all continuous environmental predictor variables for both Plasmodium and Trypanosoma spp. indicate that variance in parasite prevalence can be explained using relatively few predictors (figure 1a). For Plasmodium spp., the maximum temperature of the warmest month (as measured by Bio5) at a given site best delineates levels of prevalence, with higher temperatures associated with higher prevalence (figure 1a). At sites with relatively low Plasmodium spp. prevalence (and lower temperatures), temperature seasonality (Bio4) and annual mean surface moisture/roughness (QSCATM) are important in further characterizing prevalence. For Trypanosoma spp., surface moisture seasonality (QSCATS) is the key ecological variable distinguishing low and high prevalence, with higher prevalence occurring at sites with lower seasonality (figure 1a). At sites with relatively high trypanosome prevalence, diurnal temperature range also appears to affect prevalence levels, with highest prevalence levels recorded at lower diurnal temperature ranges. At those sites with relatively low trypanosome prevalence, mean elevation plays a role in further separating different levels of prevalence, with higher trypanosome prevalence occurring at lower elevation sites (figure 1a).
Figure 1.
(a) Tree regressions describing the relationships between environmental variables and prevalence of parasites of two genera (Plasmodium and Trypanosoma) in West Africa. Variables used to split prevalence values are indicated at each node, with higher values of each variable represented towards the right-side branch. Branch lengths correspond to the amount of deviance in prevalence explained by the variable at that node. Numbers at terminal nodes represent the average parasite prevalence within that group. (b) Importance scores for each environmental variable used as input in random forest algorithm models. Increase in mean square error is calculated as the average increase in squared residuals of the test set when the variable is permuted. A higher increase in mean square error when a variable is permuted is indicative of a more important variable [35]. Negative values indicate that with the removal of that particular variable, the per cent increase in mean square error decreases, and identifies that variable as a poor predictor in the model; one that adds error without contributing to explaining variation in prevalence.
When multiple randomly permuted tree regressions were performed, the resulting random forest model indicated that each of the two parasite genera had a unique set of environmental predictors identified as most important in explaining prevalence (figure 1b). Satellite-based radar backscatter variables that relate to canopy moisture/roughness (QSCATM) and canopy moisture seasonality (QSCATS) were consistently important predictors for these parasites. In terms of bioclimatic variables, temperature variables were repeatedly important in the case of Plasmodium spp., whereas precipitation during the driest quarter (Bio17) was a strong predictor for Trypanosoma spp. (lower precipitation in the driest quarter associated with lower prevalence). Although this variable was not identified using tree regressions, this probably points to cross-correlation with other predictor variables, such as QSCATS (there is a significant inverse relationship between QSCATS and Bio17, and together, these variables capture the seasonality in canopy moisture) revealed under random forest criteria, and suggests the importance of comparing predictor variables under particular regression frameworks when attempting to understand the relationship of prevalence to environmental conditions.
(b). Spatial predictions of parasite prevalence
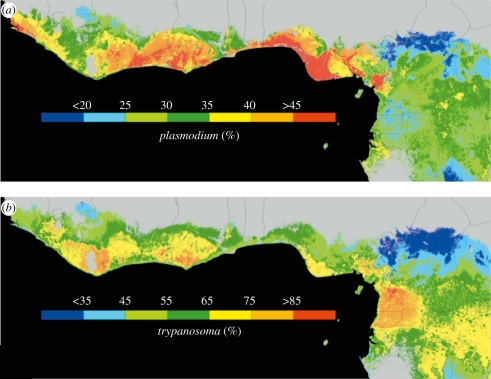
Using the point localities of infected birds, a set of environmental predictors and the Maxent algorithm, we first estimated the geographical distribution of the host–parasite system. Inspection of the Maxent predictions shows that infected birds generally reside in the humid forests and ecotones of Western Africa, consistent with the habitat requirements of the olive sunbird (figure 2). The most important environmental variables in the Maxent predictions were temperature and rainfall variables, as well as the percentage of tree cover (electronic supplementary material, table S2). Since at most sampling sites both parasite groups (Plasmodium and Trypanosoma) were detected (electronic supplementary material, table S1), the distribution of the host–parasite system was nearly identical for the two blood parasites.
Figure 2.
Random forest predictions of prevalence of avian blood parasites in the olive sunbird across West Africa and Cameroon: (a) Plasmodium spp.; (b) Trypanosoma spp. Regions shaded in grey correspond to absence of the host–parasite system based on the Maxent predictions.
We then projected the parasite prevalence–environment relationships (identified in the random forest models) onto unsampled regions defined by the Maxent distribution of the host–parasite system (see above). The resulting two predictive maps for blood parasites of two genera in the olive sunbird, however, showed a number of marked differences (figure 2a,b). For Plasmodium spp., regions of high prevalence were typically found along the coastal forested regions of Côte d'Ivoire, Ghana and Nigeria, with lower predicted prevalence in the dense humid forests of Cameroon, Gabon and Equatorial Guinea. For Trypanosoma spp., this trend was nearly the opposite, with higher prevalence in the dense humid forests of Equatorial Guinea, southeastern Cameroon and northeastern Gabon. For both parasite groups, prevalence exhibited decreases across the south-to-north rainforest–ecotone gradients, one of which is particularly prominent in Cameroon [39].
(c). Model spatial accuracy
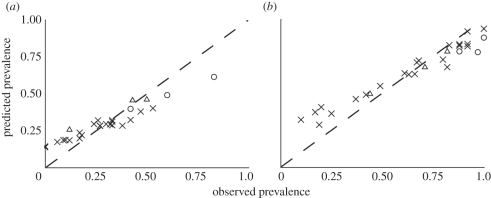
In order to assess the spatial accuracy for our predictions, prevalence levels at all sample sites were predicted and compared with actual observed prevalences at these sites (figure 3). Predictions generally closely match observed prevalence, despite the fact that the number of field-sampling sites was relatively small (n = 28). When the training data were also used for cross-validation under random forest models (figure 3), more than 90 per cent of the variance in each parasite's prevalence was explained, with a root mean square error of 0.091 for Plasmodium spp. and 0.101 for Trypanosoma spp. Predictions from random forest algorithm models were consistently accurate for sites with intermediate prevalence levels, while for sites with either extreme high or low detected prevalence, the predictions were characterized by higher variances (figure 3 and electronic supplementary material, figure S4). This is largely due to the fact that only a limited number of sites with prevalence levels within these lower and upper ranges were available to ‘train’ the algorithm.
Figure 3.
Predicted versus observed prevalence for two blood parasites in West Africa. Predicted values were significantly correlated to observed values for both (a) Plasmodium spp. (r = 0.968, d.f. = 26, p < 0.01) and (b) Trypanosoma spp. (r = 0.975, d.f. = 26, p < 0.01). Each point on the figure represents a single sample location (n = 28), all of which were used in random forest algorithm models. Observed prevalence for each parasite was significantly higher in Ghana (open circles) and Côte d'Ivoire (open triangles) when compared with prevalence observed in Cameroon (crosses; electronic supplementary material, figure S3).
4. Discussion
In this study, we present the first macroecological study predicting the prevalence of avian blood parasites. While much attention has been given to the ecological correlates of human malaria and the impact that future conditions might have on this disease [40,41], the blood pathogens of avian species in West and Central Africa represent an informative ecological model system to study vector-borne diseases for several reasons. Olive sunbirds are non-migratory, relatively sedentary [23] and occupy a range of habitats including humid dense rainforests and ecotone habitats, thus providing an accurate indication of parasite prevalence under a broad range of ecological conditions. This model system also allows the simultaneous detection and analysis of multiple parasites, in this case, species of Plasmodium and Trypanosoma. Finally, we capitalize on a unique blood sample base of the olive sunbird obtained from over two decades of fieldwork in geographically distant and often relatively inaccessible areas [42]. Our results prioritize the importance of climate and satellite-based data on habitat structure in explaining the observed prevalence of parasites and, in particular, reveal that environmental predictors differ between different parasite genera of the same host species in explaining distribution patterns.
In recent studies, environmental factors have been shown to be strong determinants of the dynamics of human vector-borne parasitic diseases (e.g. [43,44]). A key result of studies on human malaria is that temperature strongly influences malaria transmission [27,45]. For example, it has been shown to increase with a rise in the maximum monthly temperature [28]. Recent research has shown that the variation in diurnal temperatures is also a critical factor that affects the period of Plasmodium sporogony in mosquito hosts [46]. Similar results have been documented in studies of the effects of temperature on the development of avian malaria in mosquitoes [47]. Our results corroborate the importance of temperature on the prevalence of infection for Plasmodium parasites in the olive sunbird, as the maximum temperature of the warmest month was the most important indicator for elevated malaria prevalence. However, we did not examine parasite development or sporogony, and therefore could not determine whether this relationship between maximum temperature and disease prevalence is reflective of the same ecological drivers responsible for the link between diurnal temperature and disease development found in previous studies.
In our analyses, we found that annual mean radar backscatter (QSCATM) measurements are also strong predictors for the prevalence of avian malaria. Since the annual mean radar backscatter is sensitive to both canopy moisture and roughness (see above), the interpretation of these satellite microwave measurements and how they relate to prevalence is not straightforward. However, we found that the two radar metrics that we constructed (annual mean backscatter and backscatter seasonality) are correlated throughout the West African study region, and depict differences in habitat structure across the region (electronic supplementary material, figure S1). Since the seasonal metric is linked predominantly to moisture variability in evergreen rainforest systems, we are confident in our interpretation that the annual mean metric is also largely a moisture signal.
Although relatively little is known about the ecology of the mosquito vectors that transmit avian malaria parasites [15,48–50], it is certain that mosquito larval stages would require standing water [51]. In a recent study in Tanzania, precipitation seasonality and precipitation of the coldest quarter were the best predictors of mosquito prevalence for Anopheles arabiensis and Anopheles gambiae, respectively [52]. In the present study, the predicted prevalence of Plasmodium spp. was highest in coastal wet rainforest regions, especially of Nigeria (figure 2). While we have limited confidence in our predictions at high parasite prevalence levels in general, and for coastal Nigeria in particular (since no sampling efforts took place in that region, see also electronic supplementary material, figure S4), these results do differ from the predicted transmission intensity of human malaria by Gemperli et al. [28], where coastal Nigeria showed relatively low prevalence of malaria. On the other hand, our predictions closely concur with the human malaria study in Cameroon [28], where we performed more extensive sampling. In addition, it is clear that different mosquito species have very different distributions, as is evident among species of Anopheles mosquitoes in Cameroon [53]. With avian malaria, mosquitoes of eight genera transmit the parasites [4], and there is recent evidence that mosquitoes of the genus Coquillettidia can act as vectors in Africa [49]. Thus, it is probable that the mosquitoes of different genera that transmit avian malaria may also have varied distributions. It will be imperative to initiate an extensive survey of the vectors of avian malaria in these regions of Africa to disentangle the relationships among Plasmodium parasites and their insect vectors.
Almost nothing is known about the identity and ecology of the vector species that transmit avian trypanosomes in Africa [5]. Whereas mammalian trypanosomes are transmitted by Glossina tsetse flies, the vectors for avian Trypanosoma are typically louseflies of the family Hippoboscidae. However, avian trypanosomes may also be spread by black flies or mosquitoes, and it is believed that the parasite can be transmitted by ingestion of the insect [10,12]. In this study, the seasonal variation in canopy moisture, as inferred from satellite radar backscatter, was the primary predictor for the prevalence of trypanosomes infecting C. olivacea, with higher prevalence in areas exhibiting comparably low seasonality in canopy moisture. These areas also coincide with regions experiencing high rainfall totals during the driest quarter, suggesting that year-round wet conditions are a prerequisite for high prevalence. Such conditions are typically found along the coastal humid dense rainforests. The fact that rainfall appears to affect the prevalence of avian trypanosomes more significantly than temperature is not entirely unexpected: there is evidence that avian trypanosome development can occur in insect vectors at temperatures as low as 12–13°C [10]. Future research will be vital to better understand the vector ecology of avian trypanosomes, but the data presented here provide a first analysis of the macroecological dynamics of avian trypanosomes in Africa.
Previous studies have shown that changes in microclimate and the degree of disturbance can also significantly affect the fitness and vectorial capacity of Anopheles mosquitoes [54] as well as the prevalence of avian malaria [19,25]. In addition, there may also be substantial differences in distributions among the various species and strains of Plasmodium and Trypanosoma [5,19,55]. In our initial analysis, we did not find an influence of disturbance on prevalence of the two groups of blood parasites in the olive sunbird. Since our study is conducted at macroecological (regional to continental) scales with a spatial grain size at the kilometre level, it is likely that a more local disturbance effect is not captured. One noteworthy result in this context, however, is that the observed prevalence of Plasmodium spp. in our target species in Ghana and Côte d'Ivoire was higher than prevalences found in Cameroon (electronic supplementary material, figure S3) and higher than predicted by our models (figure 3). Several factors may explain this result, including differences in vector communities and overall biodiversity, but most strikingly, both countries are subjected to generally much higher levels of disturbance and deforestation when compared with Cameroon [2].
Patterns of parasite prevalence are also concordant with the forest–ecotone gradients, which have been shown to contribute to avian divergence and potentially speciation in Western and Central Africa [39,56]. Although future work is needed, the gradient in prevalence suggests that resistance and fitness may also vary, adding another potential source of differential selection on populations that occur across the forest–ecotone gradient [39,42].
We expect our approach to be of significant value and suitable for implementation in future studies. The approach makes use of host/vector distributions and tree regression algorithms that do not require a priori functions or assumptions (which may be difficult to derive given the complex nature of infectious disease systems). In this study, we present the first large-scale spatial predictions of the prevalence of avian blood parasites. As such, these maps still require further validation to assess their overall robustness, particularly in largely unsampled regions. Future sampling efforts should include the very humid Nigerian coastal rainforests, where we predicted high prevalence of Plasmodium spp. (figure 2), as well as more sites in West Africa where the sampling density was relatively low. In addition, research should also concentrate on insect vectors of avian parasites in Africa that are largely unstudied [49], and more thoroughly assess factors such as deforestation, and other anthropogenic changes that may critically impact pathogen prevalence in these systems.
Impending global climate change may impact the prevalence of avian blood parasites. Several reports have suggested that the incidence of malaria and other infectious diseases will increase with increased global average temperatures (e.g. [3,27,45]). However, there is considerable debate regarding a direct link between malaria prevalence and increasing temperatures in Africa (e.g. [41,57,58]). Studies focused on the effects of climate change on avian malaria, which would not be subject to the confounding patterns of human movement and economics, would greatly contribute to our understanding of the impacts of changing ecological conditions on natural disease systems. In addition, it will be important to investigate whether areas of high prevalence are indicators of an increased abundance of vectors, increased transmission capacity or decreased host resistance/immunity. Now with a baseline prevalence of blood parasites in the olive sunbird, projections can be made to include possible changes in temperature associated with climate change. Our study also highlights the importance of canopy moisture in this context, and further emphasizes the importance of identifying the complete set of environmental conditions that lead to proliferation of vector-borne parasitic diseases. Such information will be invaluable in assessing future effects of global change on these and other pathogens.
Acknowledgements
We thank the Governments of the Republic of Cameroon, Ghana, Equatorial Guinea and Côte d'Ivoire for permission to conduct the field research. We are grateful to Dennis Anye-Ndeh, Augustus Asamoah, Blaise Kadjo and Bertin Akpatou for expert assistance in the field, and Henri Thomassen, John Novembre, the editors of Proc. R. Soc. B and two anonymous reviewers for comments on the manuscript. This research was supported by grants from the National Geographic Society, National Environmental Research Council and the National Science Foundation DEB-9726425 and IRCEB9977072, by the joint NSF-NIH Ecology of Infectious Diseases Programme EF-0430146 and the Lithuanian State Science and Studies Foundation.
References
- 1.Wolfe N. D., Dunavan C. P., Diamond J. 2007. Origins of major human infectious diseases. Nature 447, 279–283 10.1038/nature05775 (doi:10.1038/nature05775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.FAO 2007. State of the world's forests, Africa. Rome, Italy: Food and Agriculture Organization [Google Scholar]
- 3.Patz J. A., Olson S. H., Uejio C. K., Gibbs H. K. 2008. Disease emergence from global climate and land use change. Med. Clin. North Am. 92, 1473–1491 10.1016/j.mcna.2008.07.007 (doi:10.1016/j.mcna.2008.07.007) [DOI] [PubMed] [Google Scholar]
- 4.Valkiūnas G. 2005. Avian malaria parasites and other haemosporidia. Boca Raton, FL: CRC Press [Google Scholar]
- 5.Sehgal R. N., Jones H. I., Smith T. B. 2001. Host specificity and incidence of Trypanosoma in some African rainforest birds: a molecular approach. Mol. Ecol. 10, 2319–2327 10.1046/j.1365-294X.2001.01339.x (doi:10.1046/j.1365-294X.2001.01339.x) [DOI] [PubMed] [Google Scholar]
- 6.Garnham P. C. C. 1966. Malaria parasites and other haemosporidia. Oxford, UK: Blackwell Scientific [Google Scholar]
- 7.Ricklefs R. E., Outlaw D. C. 2010. A molecular clock for malaria parasites. Science 329, 226–229 10.1126/science.1188954 (doi:10.1126/science.1188954) [DOI] [PubMed] [Google Scholar]
- 8.Palinauskas V., Valkiūnas G., Križanauskienė A., Bensch S., Bolshakov C. V. 2009. Plasmodium relictum (lineage P-SGS1): further observation of effects on experimentally infected passeriform birds, with remarks on treatment with Malarone. Exp. Parasitol. 123, 134–139 10.1016/j.exppara.2009.06.012 (doi:10.1016/j.exppara.2009.06.012) [DOI] [PubMed] [Google Scholar]
- 9.Knowles S. C., Palinauskas V., Sheldon B. C. 2010. Chronic malaria infections increase family inequalities and reduce parental fitness: experimental evidence from a wild bird population. J. Evol. Biol 23, 557–569 10.1111/j.1420-9101.2009.01920.x (doi:10.1111/j.1420-9101.2009.01920.x) [DOI] [PubMed] [Google Scholar]
- 10.Baker J. R. 1976. Biology of the trypanosomes of birds. In Biology of the kinetoplastida (eds Lumsden W. H. R., Evans D. A.), pp. 131–174 London, UK: Academic Press [Google Scholar]
- 11.Votypka J., Obornik M., Volf P., Svobodova M., Lukes J. 2002. Trypanosoma avium of raptors (Falconiformes): phylogeny and identification of vectors. Parasitology 125, 253–263 10.1017/S0031182002002093 (doi:10.1017/S0031182002002093) [DOI] [PubMed] [Google Scholar]
- 12.Votypka J., Svobodova M. 2004. Trypanosoma avium: experimental transmission from black flies to canaries. Parasitol. Res. 92, 147–151 10.1007/s00436-003-1034-z (doi:10.1007/s00436-003-1034-z) [DOI] [PubMed] [Google Scholar]
- 13.Bensch S., Stjernman M., Hasselquist D., Östman Ö., Hansson B., Westerdahl H., Pinheiro R. T. 2000. Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc. R. Soc. Lond. B 267, 1583–1589 10.1098/rspb.2000.1181 (doi:10.1098/rspb.2000.1181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cosgrove C. L., Wood M. J., Day K. P., Sheldon B. C. 2008. Seasonal variation in Plasmodium prevalence in a population of blue tits Cyanistes caeruleus. J. Anim. Ecol. 77, 540–548 10.1111/j.1365-2656.2008.01370.x (doi:10.1111/j.1365-2656.2008.01370.x) [DOI] [PubMed] [Google Scholar]
- 15.Gager A. B., Del Rosario Loaiza J., Dearborn D. C., Bermingham E. 2008. Do mosquitoes filter the access of Plasmodium cytochrome b lineages to an avian host? Mol. Ecol. 17, 2552–2561 10.1111/j.1365-294X.2008.03764.x (doi:10.1111/j.1365-294X.2008.03764.x) [DOI] [PubMed] [Google Scholar]
- 16.Hellgren O., Waldenström J., Perez-Tris J., Szollosi E., Hasselquist D., Križanauskienė A., Ottosson U., Bensch S. 2007. Detecting shifts of transmission areas in avian blood parasites—a phylogenetic approach. Mol. Ecol. 16, 1281–1290 10.1111/j.1365-294X.2007.03227.x (doi:10.1111/j.1365-294X.2007.03227.x) [DOI] [PubMed] [Google Scholar]
- 17.Martinsen E. S., Perkins S. L., Schall J. J. 2008. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol. Phylogenet. Evol. 47, 261–273 10.1016/j.ympev.2007.11.012 (doi:10.1016/j.ympev.2007.11.012) [DOI] [PubMed] [Google Scholar]
- 18.Ricklefs R. E. 2007. Estimating diversification rates from phylogenetic information. Trends Ecol. Evol. 22, 601–610 10.1016/j.tree.2007.06.013 (doi:10.1016/j.tree.2007.06.013) [DOI] [PubMed] [Google Scholar]
- 19.Chasar A., Loiseau C., Valkiūnas G., Iezhova T., Smith T. B., Sehgal R. N. 2009. Prevalence and diversity patterns of avian blood parasites in degraded African rainforest habitats. Mol. Ecol. 18, 4121–4133 10.1111/j.1365-294X.2009.04346.x (doi:10.1111/j.1365-294X.2009.04346.x) [DOI] [PubMed] [Google Scholar]
- 20.Sehgal R. N., Jones H. I., Smith T. B. 2005. Blood parasites of some West African rainforest birds. J. Vet. Med. Sci. 67, 295–301 10.1292/jvms.67.295 (doi:10.1292/jvms.67.295) [DOI] [PubMed] [Google Scholar]
- 21.Sehgal R. N., Valkiūnas G., Iezhova T. A., Smith T. B. 2006. Blood parasites of chickens in Uganda and Cameroon with molecular descriptions of Leucocytozoon schoutedeni and Trypanosoma gallinarum. J. Parasitol. 92, 1336–1343 [DOI] [PubMed] [Google Scholar]
- 22.Valkiūnas G., Sehgal R. N., Iezhova T. A., Smith T. B. 2005. Further observations on the blood parasites of birds in Uganda. J. Wildl. Dis. 41, 580–587 [DOI] [PubMed] [Google Scholar]
- 23.Borrow N., Demey R. 2001. A guide to the birds of western Africa. Princeton, NJ: Princeton University Press [Google Scholar]
- 24.Bowie R. C., Fjeldså J., Hackett S. J., Crowe T. M. 2004. Molecular evolution in space and through time: mtDNA phylogeography of the olive sunbird (Nectarinia olivacea/obscura) throughout continental Africa. Mol. Phylogenet. Evol. 33, 56–74 10.1016/j.ympev.2004.04.013 (doi:10.1016/j.ympev.2004.04.013) [DOI] [PubMed] [Google Scholar]
- 25.Bonneaud C., et al. 2009. The prevalence of avian Plasmodium is higher in undisturbed tropical forests of Cameroon. J. Trop. Ecol. 25, 439–447 10.1017/S0266467409006178 (doi:10.1017/S0266467409006178) [DOI] [Google Scholar]
- 26.Wood M. J., Cosgrove C. L., Wilkin T. A., Knowles S. C., Day K. P., Sheldon B. C. 2007. Within-population variation in prevalence and lineage distribution of avian malaria in blue tits, Cyanistes caeruleus. Mol. Ecol. 16, 3263–3273 10.1111/j.1365-294X.2007.03362.x (doi:10.1111/j.1365-294X.2007.03362.x) [DOI] [PubMed] [Google Scholar]
- 27.Craig M. H., Snow R. W., le Sueur D. 1999. A climate-based distribution model of malaria transmission in sub-Saharan Africa. Parasitol. Today 15, 105–111 10.1016/S0169-4758(99)01396-4 (doi:10.1016/S0169-4758(99)01396-4) [DOI] [PubMed] [Google Scholar]
- 28.Gemperli A., et al. 2006. Mapping malaria transmission in West and Central Africa. Trop. Med. Int. Health 11, 1032–1046 10.1111/j.1365-3156.2006.01640.x (doi:10.1111/j.1365-3156.2006.01640.x) [DOI] [PubMed] [Google Scholar]
- 29.Rogers D. J., Randolph S. E., Snow R. W., Hay S. I. 2002. Satellite imagery in the study and forecast of malaria. Nature 415, 710–715 10.1038/415710a (doi:10.1038/415710a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Worrall E., Basu S., Hanson K. 2005. Is malaria a disease of poverty? A review of the literature. Trop. Med. Int. Health 10, 1047–1059 10.1111/j.1365-3156.2005.01476.x (doi:10.1111/j.1365-3156.2005.01476.x) [DOI] [PubMed] [Google Scholar]
- 31.Cheke R. A., Mann C. F., Allen R. 2001. Sunbirds: a guide to the sunbirds, flowerpeckers, spiderhunters, and sugarbirds of the world. New Haven, CT: Yale University Press [Google Scholar]
- 32.Phillips S. J., Anderson R. P., Schapire R. E. 2006. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259 10.1016/j.ecolmodel.2005.03.026 (doi:10.1016/j.ecolmodel.2005.03.026) [DOI] [Google Scholar]
- 33.Ripley B. D. 1996. Pattern recognition and neural networks. Cambridge; New York: Cambridge University Press [Google Scholar]
- 34.R Development Core Team. 2004. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 35.Liaw A., Wiener M. 2002. Classification and regression by randomForest. R News 2, 18–22 [Google Scholar]
- 36.Breiman L. 2001. Random forests. Mach. Learn. 45, 5–32 10.1023/A:1010933404324 (doi:10.1023/A:1010933404324) [DOI] [Google Scholar]
- 37.Cutler D. R., Edwards T. C., Jr, Beard K. H., Cutler A., Hess K. T., Gibson J., Lawler J. J. 2007. Random forests for classification in ecology. Ecology 88, 2783–2792 10.1890/07-0539.1 (doi:10.1890/07-0539.1) [DOI] [PubMed] [Google Scholar]
- 38.Prasad A. M., Iverson L. R., Liaw A. 2006. Newer classification and regression tree techniques: bagging and random forests for ecological prediction. Ecosystems 9, 181–199 10.1007/s10021-005-0054-1 (doi:10.1007/s10021-005-0054-1) [DOI] [Google Scholar]
- 39.Smith T. B., Wayne R. K., Girman D. J., Bruford M. W. 1997. A role for ecotones in generating rainforest biodiversity. Science 276, 1855–1857 10.1126/science.276.5320.1855 (doi:10.1126/science.276.5320.1855) [DOI] [Google Scholar]
- 40.Lafferty K. D. 2009. The ecology of climate change and infectious diseases. Ecology 90, 888–900 10.1890/08-0079.1 (doi:10.1890/08-0079.1) [DOI] [PubMed] [Google Scholar]
- 41.Gething P. W., Smith D. L., Patil A. P., Tatem A. J., Snow R. W., Hay S. I. 2010. Climate change and the global malaria recession. Nature 465, 342–345 10.1038/nature09098 (doi:10.1038/nature09098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith T. B., Wayne R. K., Girman D., Bruford M. W. 2005. Evaluating the divergence-with-gene-flow model in natural population: the importance of ecotones in rainforest speciation. In Tropical rainforests: past, present and future (eds Bermingham E., Dick C. W., Moritz C.), pp. 148–165 Chicago, IL: University of Chicago Press [Google Scholar]
- 43.Rogers D. J., Randolph S. E. 2006. Climate change and vector-borne diseases. Adv. Parasitol. 62, 345–381 10.1016/S0065-308X(05)62010-6 (doi:10.1016/S0065-308X(05)62010-6) [DOI] [PubMed] [Google Scholar]
- 44.Kalluri S., Gilruth P., Rogers D., Szczur M. 2007. Surveillance of arthropod vector-borne infectious diseases using remote sensing techniques: a review. PLoS Pathog. 3, 1361–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers D. J., Randolph S. E. 2000. The global spread of malaria in a future, warmer world. Science 289, 1763–1766 10.1126/science.289.5485.1763 (doi:10.1126/science.289.5485.1763) [DOI] [PubMed] [Google Scholar]
- 46.Paaijmans K. P., Read A. F., Thomas M. B. 2009. Understanding the link between malaria risk and climate. Proc. Natl Acad. Sci. USA 106, 13 844–13 849 10.1073/pnas.0903423106 (doi:10.1073/pnas.0903423106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.LaPointe D. A., Goff M. L., Atkinson C. T. 2010. Thermal constraints to the sporogonic development and altitudinal distribution of avian malaria Plasmodium relictum in Hawai'i. J. Parasitol. 96, 318–324 10.1645/GE-2290.1 (doi:10.1645/GE-2290.1) [DOI] [PubMed] [Google Scholar]
- 48.Ishtiaq F., Guillaumot L., Clegg S. M., Phillimore A. B., Black R. A., Owens I. P., Mundy N. I., Sheldon B. C. 2008. Avian haematozoan parasites and their associations with mosquitoes across Southwest Pacific Islands. Mol. Ecol. 17, 4545–4555 10.1111/j.1365-294X.2008.03935.x (doi:10.1111/j.1365-294X.2008.03935.x) [DOI] [PubMed] [Google Scholar]
- 49.Njabo K. Y., Cornel A. J., Sehgal R. N., Loiseau C., Buermann W., Harrigan R. J., Pollinger J., Valkiūnas G., Smith T. B. 2009. Coquillettidia (Culicidae, Diptera) mosquitoes are natural vectors of avian malaria in Africa. Malar. J. 8, 193. 10.1186/1475-2875-8-193 (doi:10.1186/1475-2875-8-193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimura M., Darbro J. M., Harrington L. C. 2010. Avian malaria parasites share congeneric mosquito vectors. J. Parasitol. 96, 144–151 10.1645/GE-2060.1 (doi:10.1645/GE-2060.1) [DOI] [PubMed] [Google Scholar]
- 51.Kettle D. S. 1995. Medical and veterinary entomology. Wallingford, UK: CAB International [Google Scholar]
- 52.Kulkarni M. A., Desrochers R. E., Kerr J. T. 2010. High resolution niche models of malaria vectors in northern Tanzania: a new capacity to predict malaria risk? PLoS ONE 5, e9396. 10.1371/journal.pone.0009396 (doi:10.1371/journal.pone.0009396) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ayala D., Costantini C., Ose K., Kamdem G. C., Antonio-Nkondjio C., Agbor J. P., Awono-Ambene P., Fontenille D., Simard F. 2009. Habitat suitability and ecological niche profile of major malaria vectors in Cameroon. Malar. J. 8, 307. 10.1186/1475-2875-8-307 (doi:10.1186/1475-2875-8-307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afrane Y. A., Zhou G., Lawson B. W., Githeko A. K., Yan G. 2006. Effects of microclimatic changes caused by deforestation on the survivorship and reproductive fitness of Anopheles gambiae in western Kenya highlands. Am. J. Trop. Med. Hyg. 74, 772–778 [PubMed] [Google Scholar]
- 55.Loiseau C., Iezhova T. A., Valkiūnas G., Chasar A., Hutchinson A., Buermann W., Smith T. B., Sehgal R. N. 2009. Spatial variation of haemosporidian parasite infection in African rainforest bird species. J. Parasitol. 96, 21–29 10.1645/GE-2123.1 (doi:10.1645/GE-2123.1) [DOI] [PubMed] [Google Scholar]
- 56.Smith T. B., Calsbeek R., Wayne R. K., Holder K. H., Pires D., Bardeleben C. 2005. Testing alternative mechanisms of evolutionary divergence in an African rainforest passerine bird. J. Evol. Biol. 18, 257–268 10.1111/j.1420-9101.2004.00825.x (doi:10.1111/j.1420-9101.2004.00825.x) [DOI] [PubMed] [Google Scholar]
- 57.Hay S. I., Rogers D. J., Randolph S. E., Stern D. I., Cox J., Shanks G. D., Snow R. W. 2002. Hot topic or hot air? Climate change and malaria resurgence in East African highlands. Trends Parasitol. 18, 530–534 10.1016/S1471-4922(02)02374-7 (doi:10.1016/S1471-4922(02)02374-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ikemoto T. 2008. Tropical malaria does not mean hot environments. J. Med. Entomol. 45, 963–969 10.1603/0022-2585(2008)45[963:TMDNMH]2.0.CO;2 (doi:10.1603/0022-2585(2008)45[963:TMDNMH]2.0.CO;2) [DOI] [PubMed] [Google Scholar]