Abstract
There are numerous anthropological analyses concerning the importance of diet during human evolution. Diet is thought to have had a profound influence on the human phenotype, and dietary differences have been hypothesized to contribute to the dramatic morphological changes seen in modern humans as compared with non-human primates. Here, we attempt to integrate the results of new genomic studies within this well-developed anthropological context. We then review the current evidence for adaptation related to diet, both at the level of sequence changes and gene expression. Finally, we propose some ways in which new technologies can help identify specific genomic adaptations that have resulted in metabolic and morphological differences between humans and non-human primates.
Keywords: human evolution, natural selection, adaptation owing to diet
1. Introduction
Several recent studies have focused on signatures of natural selection in humans and non-human primates, both at the level of genome sequence as well as in gene expression data. Additionally, a number of studies have been very successful in identifying specific signatures of adaptation to dietary changes in the genome of modern humans (e.g. [1–4]). These case studies have shown that it is possible to identify specific coding, regulatory and copy number adaptive changes in the human genome related to changes in diet. However, the next challenge is understanding the larger sets of genes or pathways that have been under selection owing to shifts in the human diet, as compared with other primates, and in particular great apes. Gene set enrichment analyses, which focus on broader categories of genes showing signatures of positive selection, reveal a striking pattern across studies where many selective differences appear to be related to diet and metabolism. This consistent result suggests that selection acted to optimize differential metabolic requirements in humans and non-human primates. The challenge now is how to locate and make biological sense of the particular genes or pathways that underlie these metabolic adaptations. We propose that two sources of information need to be integrated to address this challenge: anthropological and comparative genomic analyses.
There are a number of prominent hypotheses in the anthropological literature concerning the importance of diet in human evolution. Comparisons with extant great apes as well as the fossil and archaeological record suggest that among the most important changes in diet was an increase in animal products (meat and fat) and starchy plant products during human evolution (reviewed in [5]). Diet and foraging patterns are thought to have had a profound influence on the human phenotype, and dietary differences have been hypothesized to contribute to the dramatic morphological changes seen in modern humans [6–8]. These dietary changes can inform our understanding of sequence and gene expression differences found in metabolic genes between primates.
Here, we attempt to integrate the results of genomic studies within this well-developed anthropological context. We then review the current evidence for adaptation related to diet, both at the level of sequence and gene expression. Finally, we propose some ways in which genomic technologies can help identify specific adaptations owing to metabolic differences between humans and non-human primates.
2. The importance of metabolic changes during human evolution
(a). Shifts in the diets of modern human populations as compared with ape diets
First we will compare the changes in diet between apes and modern human populations, and then review the changes that occurred in the time since the human lineage diverged from our most recent common ancestor with chimpanzees and bonobos. There is an enormous amount of geographical and temporal variation in the modern human diet, but the underlying strategy remains omnivory. The most dramatic change in the recent diet of humans is the domestication of animals and plants [9]. Nutrient intake varies widely among modern human populations, where some have a heavy reliance on carbohydrates in the form of cereals, roots and tubers from agriculture and gathering, while other populations have an emphasis on fat and protein extracted from animal husbandry, hunting and fishing [10]. These diets distinguish human populations from members of the larger family of living Hominidae, which includes the gorilla, chimpanzee and bonobo. All of the African apes have a diet that includes large quantities of fruit and/or structural plant parts. This is not to say these animals are exclusively vegetarian, as we know chimpanzees, bonobos and gorillas sometimes eat invertebrates [11], and chimpanzees [12,13] as well as bonobos hunt vertebrates [14,15].
(b). Dietary shifts in hominin evolution
The connection between diet and the appearance of possibly adaptive traits in hominins is of great importance for understanding human evolutionary history as well as the health and disease consequences of these adaptations for modern humans. Hominins, as a group, include humans and all of our ancestors arising after the human–chimpanzee divergence approximately 4.6–6.2 Ma [16]. In this review, we consider dietary traits acting at several scales, from molecular to organismal, associated with the intake and processing of food. In order to interpret the signatures of diet-related molecular changes, it is important to revisit the evolutionary history of humans at the organismal level. The phylogenetic affinities and the accompanying diets of many early hominin species remain unclear; however, ecological studies of extant primates and functional analyses of fossil remains suggest that hominins in general occupied an omnivore trophic niche [17] and provide us with several clear indicators of their dietary components. The fossil remains of several australopithecine and paranthropine species show that diet varied between 4.5 and 1.2 Ma, but that overall these hominins had large molars lacking well-developed shearing crests, thick enamel and powerful jaws [18–22]. These dental traits indicate crushing of hard food items during mastication and a diet that included seeds, rich in protein and fat, but do not preclude a diet including underground storage organs (USOs), such as roots and tubers, covered with abrasive soil and rich in carbohydrates [23–25]. The relative contribution of seeds versus USOs to the early hominin diet is currently an area of active discussion and research (reviewed in [26]). The genus Homo is first recognized in Africa approximately 2.5 Ma. The evolution of the genus Homo between 2.5 and 1.9 Ma is poorly understood because of the difficulties in assigning fossil specimens to distinct taxa. However, there appears to be a trend of gradually increasing brain size during this period [27–29]. Dental and skeletal traits of early Homo are difficult to interpret, though increased occlusal relief suggests an emphasis on shearing of food items during mastication [30]. It is notable that both stable isotope and dental microwear studies suggest that it is difficult to demonstrate a highly specialized diet for early hominins [31–35].
It is with the origin of Homo erectus approximately 1.9 Ma and the appearance of the Acheulean tool industry approximately 1.6 Ma that we see several traits that signify a clear shift towards the modern human form and a change in diet as compared with their more robust predecessors. With H. erectus, there is an increase in body size, skeletal indicators of a striding bipedal gait, a reduction in the size of the teeth and jaws and a substantial jump in relative brain size, which together with the evidence from the archaeological record suggest a dietary strategy that included bulk processing of a significant proportion of high-quality, calorie-rich food items [6,28,36–38] (figure 1). There is evidence at this time for extraction of marrow and flesh from large mammals using stone tools [39–41], although recent evidence argues that this may have occurred much earlier in Australopithecus afarensis [42]. Early representatives of the genus Homo probably used tools for the processing of both animal and plant materials and for wooden tool production [36,43]; however, hunting weapons do not show up unequivocally in the fossil record until about 400 thousand years ago [44]. While the archaeological record clearly shows that scavenging occurred at carnivore kill sites, there is little consensus as to what constitutes evidence for distinguishing passive scavenging, ‘power scavenging’, and hunting [45], where power scavenging is defined as actively driving another animal away from a carcase, as opposed to passive scavenging which involves harvesting food from an abandoned carcase [39]. The evidence from the feeding apparatus, particularly the reduction in post-canine tooth size, the increased occlusal relief and the gracilization of the jaws, indicates continued emphasis on shearing food items during mastication with a reduction in the hardness of foods consumed and/or the use of technology to process foods prior to ingestion [19,25,30,46]. The fossil and archaeological evidence suggests there was not only an increase in access to animal products during this period, but also the continued importance of plant material. Taken together, the hallmark of the early Homo diet is its great versatility and consistent access to high-quality foods [31,47]. Regardless of the predominate meat procurement mode, the increased availability of protein and fat in the diet of H. erectus via oil-rich seeds, USOs and meat [17] would provide consistently available, high-quality, calorie-rich fuels for such energetically expensive adaptations as a large brain.
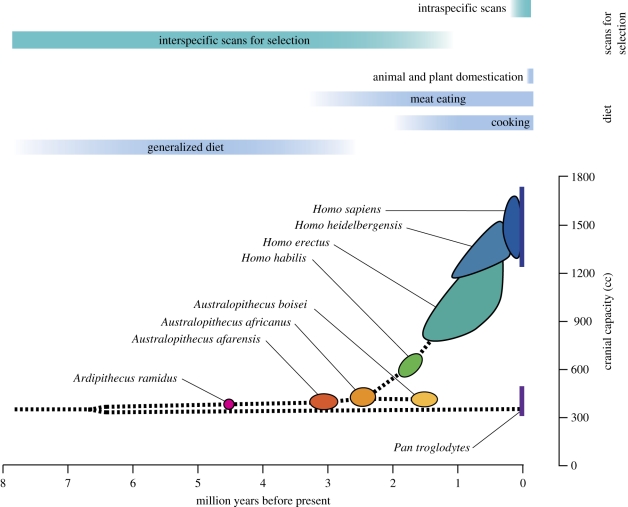
Figure 1.
A timeline showing some of the temporal intersections of diet, natural selection and one of the many changes in human morphology. Green bars indicate the temporal range on which different methods for scanning for selection are optimized to identify relevant changes in the genome (reviewed in [111]). Blue bars indicate the times in which there is evidence for shifts in human dietary intake [6,17,28,36,112,113]. The coloured bubbles are a general schematic of the time and range in size of cranial capacity found in various hominin species adapted from Schoenemann [27] with additional data from White et al. [114].
(c). Trade-off hypotheses regarding diet
As discussed above, a hallmark of the evolution of human diet is the inclusion of a high percentage of nutrient-rich foods (including animal products). Several hypotheses have been put forth to connect changes in nutrition with evidence of adaptation, specifically the increase in brain size over the past 2 Myr, and many of them focus on tissue mass changes. The first, and most prominent, of these hypotheses is the expensive tissue hypothesis proposed by Aiello & Wheeler [6]. They noticed that the total mass-specific basal metabolic rate of humans is well within the range of other primates, but that we have a larger brain, which results in greater energy requirements. Taken together, they predicted that some other structure or structures had to decrease in mass in order to reduce energy consumption and allow for the expansion of our metabolically demanding human brain. Aiello & Wheeler [6] hypothesized that the increasing quality and digestibility of the hominin diet during evolution allowed for the reduction of the energetically expensive gut tissue. Then, the net gain in energy could be allocated to the human brain. The differences in gut size among mammal species are consistent with empirical observations relating digestive organs to diet quality [48]; herbivores usually have large guts to better extract nutrients from plant tissue, whereas the simpler gut of humans is more typically found in carnivores. Another trade-off prediction comes from Leonard et al. [7], who suggest that a decrease in muscle mass and an increase in adiposity provided a potential source of energy to fuel the evolution of the human brain in two main ways. First, an energetically expensive tissue, skeletal muscle, was reduced. Second, a tissue known for its ability to store energy, fat, was increased.
Together these hypotheses illustrate that diet may have acted as a ‘releaser’ and a ‘challenger’ in human evolution. Examples of a release of energetic constraint would be the re-allocation of energy-expensive tissues, allowing for the development and maintenance of the human brain, or an increase in diet quality in terms of energy value [6,49–51]. Diet can also act as a challenger as foraging for foods high in quality often provides both an energetic challenge as well as a cognitive challenge [52–56]. When considering diet as a ‘releaser’ and a ‘challenger’ of metabolism, we are in the position to identify and characterize interesting genetic and molecular candidates that could be responsible for adaptive traits.
3. Genomic analyses allow us to identify trends across sets of genes
There has been an exponential increase in genomic information available for humans and non-human primates in the last decade (reviewed in [57–59]). This includes genomic variation between human populations as well as our closest primate relatives. Using these data, a number of studies looked for signatures of natural selection in gene sequence and/or differential gene expression within many primate lineages, most with the goal of determining human-specific adaptations [60–71]. Many of these studies have performed categorical enrichment analysis, which tests whether larger, pre-defined, classes of genes, such as ‘immunity and defence’ or ‘lipid metabolism’ are statistically over-represented among the genes showing signatures of selection or differential expression. Even if there is only a moderate signal for the individual genes with the category, there might be an important biological signal if many genes involved in a process are changing function in concert along a lineage. The predefined categories for human genes come from databases such as the Gene Ontology (GO) [72], or the Protein ANalysis THrough Evolutionary Relationships (PANTHER) [73] databases, which group genes in categories based on experimental evidence or predicted function. It is important to note that these annotations are partially hand-curated and so their interpretation presents some caveats: genes can be placed in multiple categories, some of the categories are not independent and some category labels can be difficult to interpret biologically. Nevertheless, by grouping genes into larger categories, we can begin to discern patterns of change at a genomic, rather than at a single gene, level. However, a change in expression or signature for positive selection in DNA sequence does not always imply a change in diet; even with a conserved diet, a change in gene expression may simply modify how the nutrients are processed and used. Therefore, it is critical to understand the biological context for all of these new datasets, and so it is essential to integrate them within the extensive anthropological framework concerning changes in diet during human evolutionary history.
4. Signatures of metabolic changes based on genome-wide scans for natural selection
In the search to uncover the genotypic changes underlying shifts in human phenotypes, there have been many in-depth accounts of polymorphisms segregating within human populations that confer some adaptive advantage in diet and digestion. The most prominent example is the ability to digest milk solids (lactose) into adulthood (lactase persistence) in some human populations [2,3,74,75]. Multiple regulatory polymorphisms in these different populations help drive lactose-phlorizin hydrolase (LPH) gene expression in adults. This regulatory locus shows one of the strongest signals of positive selection in the human genome [3,76–79]. There are now many examples of recent adaptations in genes as well as in gene families that appear to be shaped by more recent diet-related pressures (e.g. taste [80,81] and olfactory [82] receptor genes). However, as reviewed above, as hominoids diverged from our last common ancestors with the Pan lineage, there were significant changes in their diet—indicating that older adaptive changes might be found throughout the genome.
(a). Comparisons of metabolic categories changing between protein-coding and regulatory genomic regions between species
Comparative genomic studies looking for signatures of positive selection have taken two general approaches: the first is to scan the genome looking for regions where there is an overabundance of nucleotide substitutions as compared with nucleotides that are thought to be evolving neutrally [60–62,65,66]; the second is to look for regions that are extremely evolutionarily conserved, but show an accelerated number of changes in the human genome [63,64]. These studies have also differed in their focus on selection working on either protein-coding [60,62,65,66] or putative regulatory regions near genes [61,63,64]. Indeed, there appear to be different signatures coming from coding and regulatory regions [83] (i.e. coding and non-coding). Overall, however, many of these studies have found signatures of selection in genes that may have played a role in adaptations to dietary changes.
In coding regions, genes involved in sensory perception (i.e. processes such as taste and smell) have been under positive selection [60,65,66], as well as genes related to immune responses, which are expected to be under lineage-specific selection. Looking between mammalian clades, many genes involved in conveying sensory perception to the brain also appear to be under positive selection in the primate lineage, but not in the rodent lineage, correlating with the increased brain size and complexity in the primate lineage [62]. Other studies of more recent signals of positive selection within and between human populations also show some enrichments for metabolism-related genes, such as protein, carbohydrate and phosphate metabolism (for an extensive review, see [84]).
Regulatory regions in humans also show signatures of positive selection in genes related to metabolism, especially glucose metabolism [61]. Glucose metabolism categories include carbohydrate metabolism, glycolysis, other polysaccharide metabolism and anion transport. Glucose metabolism-related genes scoring high in humans include HK1, GCK and GPI, which are all involved in steps of glycolysis. One test to see whether these diet-related signals are biologically meaningful is to perform branch-specific enrichments, where only regions under positive selection in each species are used in the enrichments. These analyses show that the metabolic categories for each species can be quite different, and that even for the same category the specific genes showing evidence of selection are usually distinct subsets on different species' branches [61,62,71]. A weaker signal for glucose metabolism-related categories is seen on the chimpanzee branch, but the specific genes involved differ on the human and chimpanzee branches. On the chimpanzee branch, metabolic categories that are not enriched on the human branch include glycogen metabolism, sulphur redox metabolism and acyl-CoA metabolism [61].
A formal comparison of the different categories showing evidence for positive selection in coding and putative regulatory regions on the human branch revealed that very different categories of genes are evolving through changes in these regions [83]. Specifically, selection in coding regions is more prevalent on genes involved in olfaction, immunity and male reproduction; whereas selection in regulatory regions was associated with genes involved in neural development.
(b). Comparisons of metabolic categories changing between protein-coding and regulatory genomic regions within humans
In a scan for more recent adaptation in the DNA sequence owing to diet between human populations, Hancock et al. [85] show that there have been shifts in allele frequencies between populations in different ecoregions and with different diets. The strongest correlations between SNPs and environmental variables were seen in genic and non-synonymous SNPs. The strongest signal related to diet is seen in populations where the main dietary components are roots and tubers. Roots and tubers are mainly composed of starch and have low levels of other essential nutrients. Populations with diets rich in roots and tubers show significant shifts in allele frequency in the ‘starch and sucrose metabolism’ and ‘folate biosynthesis’ categories [85]. This study illustrates that there are numerous genetic changes of small effect scattered through the genome related to more recent dietary shifts. In contrast, the functional impact of regulatory SNPs throughout the genome is currently not well understood. Specific examples such as the regulatory variants that confer lactose tolerance (the ability to drink milk after weaning) in multiple populations [2,3,74,75] are known, suggesting that more are present and may be found in future functional genomic studies.
5. Evidence from the evolution of gene expression between humans and non-human primates
(a). Evidence for changes in metabolic pathways
One way to identify gene expression patterns that are consistent with positive selection is to look for lineage-specific changes in expression levels as compared with changes in multiple lineages. Cases where a lineage-specific shift in gene expression levels is seen in conjunction with low variance within the species are suggestive of directional selection driving that change [68,86]. There are clear changes in gene expression in humans, compared with non-human primates, that are centred in metabolic pathways in a number of different tissues [67–71], and some of these may be changes owing to directional selection. The metabolic pathways, again, differ on the human and chimpanzee branches [68]. For example, Blekhman et al. [68] found that fatty acid metabolism showed evidence of directional selection in gene expression in human heart tissue, whereas vitamin B6 metabolism and folate biosynthesis show evidence of directional selection on the chimpanzee branch. Additionally, Khaitovich et al. [67] found an enrichment for diet-related genes that both are differentially expressed between species and are in a large region of linkage disequilibrium (evidence of a selective sweep on that genomic region) in multiple human populations. Thus, it is likely that positive selection acted on gene expression changes before these populations diverged approximately 100 000 years ago [87–90], although many of these changes could have been selected for in morphologically modern human populations before moving out of Africa. Other changes in transcriptional regulation could also be important contributors to diet-related adaptations. For example, a recent study on alternative splicing within expressed genes in humans and non-human primates [70] found an enrichment for genes involved in metabolic processes in differentially spliced exons in humans as compared with chimpanzees and macaques.
(b). Evidence for changes in energy transport
In brain tissues, there is a consistent pattern of changes in expression of genes critical to aerobic energy metabolism [67,69,71,91]. This includes categories such as oxidative phosphorylation, electron transport and other nuclear-encoded genes that function in the mitochondria. This differential expression of aerobic energy metabolism-related genes could be due to the increased neural activity and metabolic requirements of the human brain. This trend is seen in adult human brain tissue, but not in foetal tissue, where selected enrichment categories are related to neuronal signalling and connectivity [71]. Some aerobic energy metabolism genes also show evidence of positive selection in their amino acid sequence during anthropoid primate evolution [92–94]; these changes correlate with changes in brain size and lifespan in primate life histories.
Unsurprisingly, different categorical enrichments are seen from different tissues. For example, the differential expression of genes related to aerobic energy metabolism has only been found in studies examining expression in brain regions between species [67,69,91]. To get a more complete picture of the important functional shifts in gene expression between tissues and species, future studies will need to examine more tissues throughout the body.
6. Other shifts in the genomic landscape owing to dietary shifts
Shifts in diet will also have effects on other functional levels, such as chromatin structure or protein processing. For instance, epigenetic changes may also play a large role in differential gene expression between species, and recent work in mouse models has shown that diet can play a role in changes in chromatin modification [95–97]. These epigenetic changes can also have a lasting impact since changes in maternal diet can affect methylation status for multiple generations (reviewed in [98]). Two preliminary studies of methylation change between humans and chimpanzees noted significant differences in methylation patterns, with a higher amount of methylation seen in humans [99,100]. Dietary changes incorporating more sources of methyl groups (e.g. methionine or choline) and folate would change the methylation states for many genes, and these changes could be important in development and later disease susceptibility (reviewed in [101,102]).
A recent study has also measured shifts in the concentration of 21 metabolites between humans and non-human primates [103], and found that a statistically significant number of them differ in relative concentration. Specifically, the relative concentration of metabolites related to energy metabolism, such as lactate and creatine, appear to have changed rapidly during human evolution. Genes involved in these metabolic processes also show greater sequence and gene expression divergence than expected. The authors suggest that the human brain may be working at the edge of its metabolic capabilities, as suggested through additional comparisons of metabolites between schizophrenic and control individuals [103].
7. Next steps in understanding the impact of diet on phenotype
In order to empirically test whether these patterns of genome-wide metabolism and energy metabolism are biologically relevant, the next steps will be to take different experimental approaches for a better understanding of the intersection of genotype, phenotype and diet in primates.
What does the anthropological evidence suggest to test questions surrounding the genetic impact of dietary shifts? One possibility would be to look for genetic and genomic signatures of adaptations related to digestion of meats, fats, and marrows, possibly even scavenged meats with attached immune challenges (rotting or parasite infested, for instance). This will be critical for distinguishing how during human evolution adaptations in gut, brain, muscle and fat and reproductive adaptations arose even though all are related to diet. A parallel investigation could be undertaken to see whether there is genetic evidence for more ancient adaptations to the adoption of cooking habits [104]. For instance, Blekhman et al. [68] hypothesized that signatures of selection in gene expression in the liver reflected the beginning of cooking meat during human evolution. Since it is challenging to detect specific dietary influences on musculoskeletal anatomy during human evolution, we contend that combining morphological approaches with genomic approaches is a next step in addressing these questions by looking at the evidence for natural selection at the molecular and tissue levels.
Experimentally, this could include exploring changes in pathways between species using other molecular approaches, such as in vitro cell culture assays. For example, one could treat a relevant cell type with varying levels of metabolites or oxygen, and then measure subsequent changes in gene expression or other metabolite concentrations between species. This would allow for a dissection of the genome-wide influence of a single factor between species, possibly helping to elucidate networks of genes that have changed.
Alternatively, detailed investigations of changes over large networks or pathways of genes would be informative. Like many complex traits, if changes in diet have been important in human evolution, we might expect many small changes at multiple loci. Likewise, genomic sequence and expression from other populations will increase our power to understand these adaptations. The recently published Neanderthal genome [105] will also be valuable in understanding the timing of certain specific mutations.
By comparing gene expression across tissues as well as between species, we may start to understand the genetic underpinnings of phenotypic changes related to dietary changes. For instance, paralogues within a gene family could be differentially ‘tuned’ to function in specific tissues. Phylogenetic histories of gene duplication, and gene family expansion, would help to illuminate this type of pattern, as seen in the olfactory receptors in humans [106]. Alternatively, if there are tissue changes (e.g. a reduced gut or enlarged brain) between species, analyses looking for shifting patterns of gene expression, protein function or methylation state in these tissues would be valuable. A similar effect might be expected at the level of natural selection on DNA sequence, showing an enrichment of selection on tissue-specific genes in tissues that have changed dramatically in size or energy consumption between species, whereas ubiquitously expressed genes may not show those enrichments.
The pattern we describe would also predict that other studies should see similar shifts in phenotypes (and the underlying genotypic shifts) in other organisms where diet has changed dramatically within a clade. With genome sequencing technologies rapidly advancing as costs decrease, it is now possible to create resources for new ‘model’ organisms to address specific questions. For example, measurements of brain and gut volume in addition to gene expression studies in Onychomys, a small (approx. 30 g), highly carnivorous cricetid rodent [107], would be an interesting natural experiment in the morphological and genetic patterns that occur when a carnivore evolves from a seed-eating ancestor. Another system to investigate is the elephant-nose fish Gnathonemus petersii. Gnathonemus petersii has an extremely large brain (particularly the cerebellum) that is exceptionally expensive for an ectotherm, with 60 per cent of total oxygen consumption being used by the brain compared with 2–8% in most vertebrates and 20 per cent in humans [108]. The enlarged cerebellum of G. petersii may be due to an energetic trade-off with the digestive tract [109], and the size of the brain varies widely within closely related species [110]. Experimental taxa such as these would be a powerful source of detailed information on the interplay between the genetic, physiological and morphological changes involved in energetic re-allocations.
A difficult gap between genotype and phenotype remains, and so next steps need to look at physiological, developmental and morphological differences—challenging in human and non-human primate species and populations. A combination of the experimental data reviewed here may assist in gaining a comprehensive understanding of how dietary changes have moulded the modern human phenotype.
Acknowledgements
We would like to thank Julie Horvath for critical comments on the manuscript and Nicholas J. Matzke for information concerning brain size changes. The authors were funded by National Science Foundation Grant NSF-BCS-08-27552 (HOMINID) and the Institute for Genome Sciences and Policy at Duke University.
References
- 1.Perry G. H., et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39, 1256–1260 10.1038/ng2123 (doi:10.1038/ng2123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enattah N. S., Sahi T., Savilahti E., Terwilliger J. D., Peltonen L., Jarvela I. 2002. Identification of a variant associated with adult-type hypolactasia. Nat. Genet. 30, 233–237 10.1038/ng826 (doi:10.1038/ng826) [DOI] [PubMed] [Google Scholar]
- 3.Tishkoff S. A., et al. 2007. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39, 31–40 10.1038/ng1946 (doi:10.1038/ng1946) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelley J. L., Swanson W. J. 2008. Dietary change and adaptive evolution of enamelin in humans and among primates. Genetics 178, 1595–1603 10.1534/genetics.107.077123 (doi:10.1534/genetics.107.077123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aiello L. C., Wells J. C. K. 2002. Energetics and the evolution of the genus Homo. Annu. Rev. Anthropol. 31, 323–338 10.1146/annurev.anthro.31.040402.085403 (doi:10.1146/annurev.anthro.31.040402.085403) [DOI] [Google Scholar]
- 6.Aiello L. C., Wheeler P. 1995. The expensive-tissue hypothesis—the brain and the digestive-system in human and primate evolution. Curr. Anthropol. 36, 199–221 10.1086/204350 (doi:10.1086/204350) [DOI] [Google Scholar]
- 7.Leonard W. R., Robertson M. L., Snodgrass J. J., Kuzawa C. W. 2003. Metabolic correlates of hominid brain evolution. Comp. Biochem. Physiol. Mol. Integr. Physiol. 136, 5–15 10.1016/S1095-6433(03)00132-6 (doi:10.1016/S1095-6433(03)00132-6) [DOI] [PubMed] [Google Scholar]
- 8.Martin R. D. 1996. Scaling of the mammalian brain: the maternal energy hypothesis. News Physiol. Sci. 11, 149–156 [Google Scholar]
- 9.Zeder M. A. 2006. Documenting domestication: new genetic and archaeological paradigms. Berkeley, MA: University of California Press [Google Scholar]
- 10.Murdock G. P. 1967. Ethnographic atlas: a summary. Ethnology 6, 109–236 10.2307/3772751 (doi:10.2307/3772751) [DOI] [Google Scholar]
- 11.McGrew W. C. 2001. The other faunivory: primate insectivory and early human diet. In Meat-eating and human evolution (eds Stanford C. B., Bunn H. T.), pp. 160–178 New York, NY: Oxford University Press [Google Scholar]
- 12.Stanford C. B., Wallis J., Matama H., Goodall J. 1994. Patterns of predation by chimpanzees on red colobus monkeys in Gombe National Park, 1982–1991. Am. J. Phys. Anthropol. 94, 213–228 10.1002/ajpa.1330940206 (doi:10.1002/ajpa.1330940206) [DOI] [PubMed] [Google Scholar]
- 13.Boesch C., Boesch-Achermann H. 2000. The chimpanzees of the Taï forest. New York, NY: Oxford University Press [Google Scholar]
- 14.Surbeck M., Hohmann G. 2008. Primate hunting by bonobos at LuiKotale, Salonga National Park. Curr. Biol. 18, R906–R907 10.1016/j.cub.2008.08.040 (doi:10.1016/j.cub.2008.08.040) [DOI] [PubMed] [Google Scholar]
- 15.Hohmann G., Fruth B. 2007. New records on prey capture and meat eating by Bonobos at Lui Kotale, Salonga National Park, Democratic Republic of Congo. Folia Primatol. 79, 103–110 10.1159/000110679 (doi:10.1159/000110679) [DOI] [PubMed] [Google Scholar]
- 16.Chen F.-C., Li H. 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Nat. Genet. 68, 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters C. R. 2007. Theoretical and actualistic ecobotanical perspectives on early hominin diets. In Evolution of the human diet: the known, the unknown, and the unknowable (ed. Ungar P. S.), pp. 233–261 Oxford, UK: Oxford University Press [Google Scholar]
- 18.Daegling D. J., Grine F. E. 1991. Compact-bone distribution and biomechanics of early hominid mandibles. Am. J. Phys. Anthropol. 86, 321–339 10.1002/ajpa.1330860302 (doi:10.1002/ajpa.1330860302) [DOI] [PubMed] [Google Scholar]
- 19.Robinson J. T. 1954. Prehominid dentition and hominid evolution. Evolution 8, 324–334 10.2307/2405779 (doi:10.2307/2405779) [DOI] [Google Scholar]
- 20.Demes B., Creel N. 1988. Bite force, diet, and cranial morphology of fossil hominids. J. Hum. Evol. 17, 657–670 10.1016/0047-2484(88)90023-1 (doi:10.1016/0047-2484(88)90023-1) [DOI] [Google Scholar]
- 21.Grine F. E., Martin L. B. 1988. Enamel thickness and development in Australopithecus and Paranthropus. In Evolutionary history of the ‘robust’ australopithecines (ed. Grine F. E.), pp. 3–42 New Brunswick, NJ: Transaction Publishers [Google Scholar]
- 22.Kay R. F., Grine F. E. 1988. Tooth morphology, wear and diet in Australopithecus and Paranthropus from Southern Africa. In Evolutionary history of the robustaustralopithecines (ed. Grine F. E.), pp. 427–447 Hawthorne, NY: Aldine De Gruyter [Google Scholar]
- 23.Peters C. R., O'Brian E. M. 1981. The early hominid plant-food niche: insights from an analysis of plant exploitation by Homo, Pan, and Papio in eastern and southern Africa. Curr. Anthropol. 22, 127–140 10.1086/202631 (doi:10.1086/202631) [DOI] [Google Scholar]
- 24.Teaford M. F., Ungar P. S. 2000. Diet and the evolution of the earliest human ancestors. Proc. Natl Acad. Sci. USA 97, 13 506–13 511 10.1073/pnas.260368897 (doi:10.1073/pnas.260368897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jolly C. J. 1970. The seed-eaters: a new model of hominid differentiation based on a baboon analogy. Man 5, 1–26 [Google Scholar]
- 26.Luca F., Perry G. H., Di Rienzo A. 2010. Evolutionary adaptations to dietary changes. Annu. Rev. Nutr. 30, 291–314 10.1146/annurev-nutr-080508-141048 (doi:10.1146/annurev-nutr-080508-141048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoenemann P. T. 2006. Evolution of the size and functional areas of the human brain. Annu. Rev. Anthropol. 35, 379–406 10.1146/annurev.anthro.35.081705.123210 (doi:10.1146/annurev.anthro.35.081705.123210) [DOI] [Google Scholar]
- 28.McHenry H. M. 1994. Tempo and mode in human evolution. Proc. Natl Acad. Sci. USA 91, 6780–6786 10.1073/pnas.91.15.6780 (doi:10.1073/pnas.91.15.6780) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Miguel C., Henneberg M. 2001. Variation in hominid brain size: how much is due to method? Homo 52, 3–58 10.1078/0018-442X-00019 (doi:10.1078/0018-442X-00019) [DOI] [PubMed] [Google Scholar]
- 30.Ungar P. 2004. Dental topography and diets of Australopithecus afarensis and early Homo. J. Hum. Evol. 46, 605–622 10.1016/j.jhevol.2004.03.004 (doi:10.1016/j.jhevol.2004.03.004) [DOI] [PubMed] [Google Scholar]
- 31.Ungar P. S., Grine F. E., Teaford M. F. 2006. Diet in early Homo: a review of the evidence and a new model of adaptive versatility. Annu. Rev. Anthropol. 35, 209–228 10.1146/annurev.anthro.35.081705.123153 (doi:10.1146/annurev.anthro.35.081705.123153) [DOI] [Google Scholar]
- 32.Ungar P. S., Grine F. E., Teaford M. F. 2008. Dental microwear and diet of the Plio-Pleistocene hominin Paranthropus boisei. PLoS ONE 3, e2044. 10.1371/journal.pone.0002044 (doi:10.1371/journal.pone.0002044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van der Merwe N. J., Masao F. T., Bamford M. K. 2008. Isotopic evidence for contrasting diets of early hominins Homo habilis and Australopithecus boisei of Tanzania. S. Afr. J. Sci. 104, 153–155 [Google Scholar]
- 34.Yeakel J. D., Bennett N. C., Koch P. L., Dominy N. J. 2007. The isotopic ecology of African mole rats informs hypotheses on the evolution of human diet. Proc. R. Soc. B 274, 1723–1730 10.1098/rspb.2007.0330 (doi:10.1098/rspb.2007.0330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sponnheimer M., Lee-Thorp J. A. 1999. Oxygen isotope ratios in enamel carbonate and their ecological significance. J. Archaeol. Sci. 26, 723–728 10.1006/jasc.1998.0388 (doi:10.1006/jasc.1998.0388) [DOI] [Google Scholar]
- 36.Shea J. J. 2007. Lithic archaeology, or, what stone tools can (and can't) tell us about early hominin diets. In Evolution of the human diet. The known, the unknown, and the unknowable (ed. Ungar P. S.), pp. 212–232 Oxford, UK: Oxford University Press [Google Scholar]
- 37.Brown F., Harris J., Leakey R., Walker A. 1985. Early Homo erectus skeleton from West Lake Turkana, Kenya. Nature 316, 788–792 10.1038/316788a0 (doi:10.1038/316788a0) [DOI] [PubMed] [Google Scholar]
- 38.McHenry H. M. 1992. Body size and proportions in early hominids. Am. J. Phys. Anthropol. 87, 407–431 10.1002/ajpa.1330870404 (doi:10.1002/ajpa.1330870404) [DOI] [PubMed] [Google Scholar]
- 39.Bunn H. T. 2001. Hunting, power scavenging, and butchering by Hadza foragers and by Plio-Pleistocene Homo. In Meat eating and human evolution (eds Stanford C. B., Bunn H. T.), pp. 199–281 Oxford, UK: Oxford University Press [Google Scholar]
- 40.Bunn H. T., Ezzo J. A. 1993. Hunting and scavenging by Plio-Pleistocene hominids: nutritional constraints, archaeological patterns, and behavioral implications. J. Archaeol. Sci. 20, 365–398 10.1006/jasc.1993.1023 (doi:10.1006/jasc.1993.1023) [DOI] [Google Scholar]
- 41.Blumenschine R. J., et al. 2003. Late Pliocene Homo and hominid land use from western Olduvai Gorge, Tanzania. Science 299, 1217–1221 10.1126/science.1075374 (doi:10.1126/science.1075374) [DOI] [PubMed] [Google Scholar]
- 42.McPherron S. P., Alemseged Z., Marean C. W., Wynn J. G., Reed D., Geraads D., Bobe R., Béarat H. A. 2010. Evidence for stone-tool-assisted consumption of animal tissues before 3.39 million years ago at Dikika, Ethiopia. Nature 466, 857–860 10.1038/nature09248 (doi:10.1038/nature09248) [DOI] [PubMed] [Google Scholar]
- 43.Keeley L. H., Toth N. 1981. Microwear polishes on early stone tools from Koobi Fora, Kenya. Nature 293, 464–465 10.1038/293464a0 (doi:10.1038/293464a0) [DOI] [Google Scholar]
- 44.Thieme H. 1997. Lower Paleolithic hunting spears from Germany. Nature 385, 807–810 10.1038/385807a0 (doi:10.1038/385807a0) [DOI] [PubMed] [Google Scholar]
- 45.Blumenschine R. J., Pobiner B. L. 2007. Zooarchaeology and the ecology of Oldowan hominin carnivory. In Evolution of the human diet: the known, the unknown and the unknowable (ed. Ungar P. S.), pp. 167–190 Oxford, UK: Oxford University Press [Google Scholar]
- 46.Teaford M. F., Ungar P. S., Grine F. E. 2002. Paleontological evidence for the diets of African Plio-Pleistocene hominins with special reference to early Homo. In Human diet: its origin and evolution (eds Ungar P. S., Teaford M. F.), pp. 143–166 Westport, CT: Bergin & Garvey [Google Scholar]
- 47.Schoeninger M. J., Bunn H. T., Murray S., Pickering T., Moore J. 2001. Meat-eating by the fourth African ape. In Meat-eating and human evolution (eds Stanford C. B., Bunn H. T.), pp. 179–198 New York, NY: Oxford University Press [Google Scholar]
- 48.Chivers D. J., Hladik C. M. 1980. Morphology of the gastrointestinal tract in primates: comparisons with other mammals in relation to diet. J. Morphol. 166, 337–386 10.1002/jmor.1051660306 (doi:10.1002/jmor.1051660306) [DOI] [PubMed] [Google Scholar]
- 49.Fish J. L., Lockwood C. A. 2003. Dietary constraints on encephalization in primates. Am. J. Phys. Anthropol. 120, 171–181 10.1002/ajpa.10136 (doi:10.1002/ajpa.10136) [DOI] [PubMed] [Google Scholar]
- 50.Gaulin S. J. C., Kurland J. A. 1976. Primate predation and bioenergetics. Science 191, 314–317 10.1126/science.1246618 (doi:10.1126/science.1246618) [DOI] [PubMed] [Google Scholar]
- 51.Leonard W. R., Robertson M. L. 1994. Evolutionary perspectives on human nutrition—the influence of brain and body-size on diet and metabolism. Am. J. Hum. Biol. 6, 77–88 10.1002/ajhb.1310060111 (doi:10.1002/ajhb.1310060111) [DOI] [PubMed] [Google Scholar]
- 52.Kaplan H., Hill K., Lancaster J., Hurtado A. M. 2000. A theory of human life history evolution: diet, intelligence, and longevity. Evol. Anthropol. 9, 156–185 (doi:10.1002/1520-6505(2000)9:4<156::AID-EVAN5>3.0.CO;2-7) [DOI] [Google Scholar]
- 53.McLean A. N. 2001. Cognitive abilities—the result of selective pressures on food acquisition? Appl. Anim. Behav. Sci. 71, 241–258 10.1016/S0168-1591(00)00181-7 (doi:10.1016/S0168-1591(00)00181-7) [DOI] [PubMed] [Google Scholar]
- 54.Hamilton W. J., III, Busse C. D. 1978. Primate carnivory and its significance to human diets. BioScience 28, 761–766 10.2307/1307249 (doi:10.2307/1307249) [DOI] [Google Scholar]
- 55.Milton K., May M. L. 1976. Body weight, diet and home range area in primates. Nature 259, 459–462 10.1038/259459a0 (doi:10.1038/259459a0) [DOI] [PubMed] [Google Scholar]
- 56.Ruxton G. D., Houston D. C. 2004. Obligate vertebrate scavengers must be large soaring fliers. J. Theor. Biol. 228, 431–436 10.1016/j.jtbi.2004.02.005 (doi:10.1016/j.jtbi.2004.02.005) [DOI] [PubMed] [Google Scholar]
- 57.Marques-Bonet T., Girirajan S., Eichler E. E. 2009. The origins and impact of primate segmental duplications. Trends Genet. 25, 443–454 10.1016/j.tig.2009.08.002 (doi:10.1016/j.tig.2009.08.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siepel A. 2009. Phylogenomics of primates and their ancestral populations. Genome Res. 19, 1929–1941 10.1101/gr.084228.108 (doi:10.1101/gr.084228.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tung J., Alberts S. C., Wray G. A. 2010. Evolutionary genetics in wild primates: combining genetic approaches with field studies of natural populations. Trends Genet. 26, 353–362 10.1016/j.tig.2010.05.005 (doi:10.1016/j.tig.2010.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bustamante C. D., et al. 2005. Natural selection on protein-coding genes in the human genome. Nature 437, 1153–1157 10.1038/nature04240 (doi:10.1038/nature04240) [DOI] [PubMed] [Google Scholar]
- 61.Haygood R., Fedrigo O., Hanson B., Yokoyama K. D., Wray G. A. 2007. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat. Genet. 39, 1140–1144 10.1038/ng2104 (doi:10.1038/ng2104) [DOI] [PubMed] [Google Scholar]
- 62.Kosiol C., Vinar T., da Fonseca R. R., Hubisz M. J., Bustamante C. D., Nielsen R., Siepel A. 2008. Patterns of positive selection in six mammalian genomes. PLoS Genet. 4, e1000144. 10.1371/journal.pgen.1000144 (doi:10.1371/journal.pgen.1000144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pollard K. S., et al. 2006. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2, e168. 10.1371/journal.pgen.0020168 (doi:10.1371/journal.pgen.0020168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prabhakar S., Noonan J. P., Paabo S., Rubin E. M. 2006. Accelerated evolution of conserved noncoding sequences in humans. Science 314, 786–786 10.1126/science.1130738 (doi:10.1126/science.1130738) [DOI] [PubMed] [Google Scholar]
- 65.Nielsen R., et al. 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3, 976–985 10.1371/journal.pbio.0030170 (doi:10.1371/journal.pbio.0030170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clark A. G., et al. 2003. Inferring nonneutral evolution from human–chimp–mouse orthologous gene trios. Science 302, 1960–1963 10.1126/science.1088821 (doi:10.1126/science.1088821) [DOI] [PubMed] [Google Scholar]
- 67.Khaitovich P., Tang K., Franz H., Kelso J., Hellmann I., Enard W., Lachmann M., Paabo S. 2006. Positive selection on gene expression in the human brain. Curr. Biol. 16, R356–R358 10.1016/j.cub.2006.03.082 (doi:10.1016/j.cub.2006.03.082) [DOI] [PubMed] [Google Scholar]
- 68.Blekhman R., Oshlack A., Chabot A. E., Smyth G. K., Gilad Y. 2008. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 4, e1000271. 10.1371/journal.pgen.1000271 (doi:10.1371/journal.pgen.1000271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Babbitt C. C., Fedrigo O., Pfefferle A. D., Horvath J., Furey T. S., Wray G. A. 2010. Both noncoding and protein-coding RNAs contribute to gene expression evolution in the primate brain. Genome Biol. Evol. 2010, 67–79 10.1093/gbe/evq002 (doi:10.1093/gbe/evq002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blekhman R., Marioni J. C., Zumbo P., Stevens M., Gilad Y. 2010. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 20, 180–189 10.1101/gr.099226.109 (doi:10.1101/gr.099226.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uddin M., et al. 2008. Distinct genomic signatures of adaptation in pre- and postnatal environments during human evolution. Proc. Natl Acad. Sci. USA 105, 3215–3220 10.1073/pnas.0712400105 (doi:10.1073/pnas.0712400105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ashburner M., et al. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 10.1038/75556 (doi:10.1038/75556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mi H., Dong Q., Muruganujan A., Gaudet P., Lewis S., Thomas P. D. 2010. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38, D204–D210 10.1093/nar/gkp1019 (doi:10.1093/nar/gkp1019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Enattah N. S., et al. 2008. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am. J. Hum. Genet. 82, 57–72 10.1016/j.ajhg.2007.09.012 (doi:10.1016/j.ajhg.2007.09.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Enattah N. S., et al. 2007. Evidence of still-ongoing convergence evolution of the lactase persistence T-13910 alleles in humans. Am. J. Hum. Genet. 81, 615–625 10.1086/520705 (doi:10.1086/520705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Voight B. F., Kudaravalli S., Wen X. Q., Pritchard J. K. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4, 446–458 10.1371/journal.pbio.0040446 (doi:10.1371/journal.pbio.0040446) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.The International HapMap Consortium 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 499, 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poulter M., Hollox E., Harvey C. B., Mulcare C., Peuhkuri K., Kajander K., Sarner M., Korpela R., Swallow D. M. 2003. The causal element for the lactase persistence/non-persistence polymorphism is located in a 1 Mb region of linkage disequilibrium in Europeans. Ann. Hum. Genet. 67, 298–311 10.1046/j.1469-1809.2003.00048.x (doi:10.1046/j.1469-1809.2003.00048.x) [DOI] [PubMed] [Google Scholar]
- 79.Bersaglieri T., Sabeti P. C., Patterson N., Vanderploeg T., Schaffner S. F., Drake J. A., Rhodes M., Reich D. E., Hirschhorn J. N. 2004. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 74, 1111–1120 10.1086/421051 (doi:10.1086/421051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fischer A., Gilad Y., Man O., Paabo S. 2005. Evolution of bitter taste receptors in humans and apes. Mol. Biol. Evol. 22, 432–436 10.1093/molbev/msi027 (doi:10.1093/molbev/msi027) [DOI] [PubMed] [Google Scholar]
- 81.Wang X., Thomas S. D., Zhang J. 2004. Relaxation of selective constraint and loss of function in the evolution of human bitter taste receptor genes. Hum. Mol. Genet. 13, 2671–2678 10.1093/hmg/ddh289 (doi:10.1093/hmg/ddh289) [DOI] [PubMed] [Google Scholar]
- 82.Gilad Y., Bustamante C. D., Lancet D., Paabo S. 2003. Natural selection on the olfactory receptor gene family in humans and chimpanzees. Am. J. Hum. Genet. 73, 489–501 10.1086/378132 (doi:10.1086/378132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haygood R., Babbitt C. C., Fedrigo O., Wray G. A. 2010. Contrasts between adaptive coding and non-coding changes during human evolution. Proc. Natl Acad. Sci. USA 107, 7853–7857 10.1073/pnas.0911249107 (doi:10.1073/pnas.0911249107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Akey J. M. 2009. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res. 19, 711–722 10.1101/gr.086652.108 (doi:10.1101/gr.086652.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hancock A. M., et al. 2010. Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc. Natl Acad. Sci. USA 107, 8924–8930 10.1073/pnas.0914625107 (doi:10.1073/pnas.0914625107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gilad Y., Rifkin S. A., Bertone P., Gerstein M., White K. P. 2005. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 15, 674–680 10.1101/gr.3335705 (doi:10.1101/gr.3335705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stringer C. B., Andrews P. 1988. Genetic and fossil evidence for the origin of modern humans. Science 239, 1263–1268 10.1126/science.3125610 (doi:10.1126/science.3125610) [DOI] [PubMed] [Google Scholar]
- 88.Fagundes N. J., Ray N., Beaumont M., Neuenschwander S., Salzano F. M., Bonatto S. L., Excoffier L. 2007. Statistical evaluation of alternative models of human evolution. Proc. Natl Acad. Sci. USA 104, 17 614–17 619 10.1073/pnas.0708280104 (doi:10.1073/pnas.0708280104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schaffner S. F., Foo C., Gabriel S., Reich D., Daly M. J., Altshuler D. 2005. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 15, 1576–1583 10.1101/gr.3709305 (doi:10.1101/gr.3709305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jobling M., Hurles M., Tyler-Smith C. 2004. Human evolutionary genetics: origins, peoples and disease. New York, NY: Garland Science [Google Scholar]
- 91.Uddin M., Wildman D. E., Liu G. Z., Xu W. B., Johnson R. M., Hof P. R., Kapatos G., Grossman L. I., Goodman M. 2004. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc. Natl Acad. Sci. USA 101, 2957–2962 10.1073/pnas.0308725100 (doi:10.1073/pnas.0308725100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang H. Y., et al. 2007. Rate of evolution in brain-expressed genes in humans and other primates. PLoS Biol. 5, e13. 10.1371/journal.pbio.0050013 (doi:10.1371/journal.pbio.0050013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grossman L. I., Schmidt T. R., Wildman D. E., Goodman M. 2001. Molecular evolution of aerobic energy metabolism in primates. Mol. Phylogenet. Evol. 18, 26–36 10.1006/mpev.2000.0890 (doi:10.1006/mpev.2000.0890) [DOI] [PubMed] [Google Scholar]
- 94.Grossman L. I., Wildman D. E., Schmidt T. R., Goodman M. 2004. Accelerated evolution of the electron transport chain in anthropoid primates. Trends Genet. 20, 578–585 10.1016/j.tig.2004.09.002 (doi:10.1016/j.tig.2004.09.002) [DOI] [PubMed] [Google Scholar]
- 95.Wolff G. L., Kodell R. L., Moore S. R., Cooney C. A. 1998. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 12, 949–957 [PubMed] [Google Scholar]
- 96.Dolinoy D. C., Weidman J. R., Waterland R. A., Jirtle R. L. 2006. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ. Health Perspect. 114, 567–572 10.1289/ehp.8700 (doi:10.1289/ehp.8700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Waterland R. A., Jirtle R. L. 2003. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23, 5293–5300 10.1128/MCB.23.15.5293-5300.2003 (doi:10.1128/MCB.23.15.5293-5300.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jirtle R. L., Skinner M. K. 2007. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 8, 253–262 10.1038/nrg2045 (doi:10.1038/nrg2045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Farcas R., et al. 2009. Differences in DNA methylation patterns and expression of the CCRK gene in human and nonhuman primate cortices. Mol. Biol. Evol. 26, 1379–1389 10.1093/molbev/msp046 (doi:10.1093/molbev/msp046) [DOI] [PubMed] [Google Scholar]
- 100.Enard W., Fassbender A., Model F., Adorjan P., Paabo S., Olek A. 2004. Differences in DNA methylation patterns between humans and chimpanzees. Curr. Biol. 14, R148–R149 [PubMed] [Google Scholar]
- 101.Choi S. W., Mason J. B. 2002. Folate status: effects on pathways of colorectal carcinogenesis. J. Nutr. 132, S2413–S2418 [DOI] [PubMed] [Google Scholar]
- 102.Lucock M. 2000. Folic acid: nutritional biochemistry, molecular biology, and role in disease processes. Mol. Genet. Metab. 71, 121–138 10.1006/mgme.2000.3027 (doi:10.1006/mgme.2000.3027) [DOI] [PubMed] [Google Scholar]
- 103.Khaitovich P., et al. 2008. Metabolic changes in schizophrenia and human brain evolution. Genome Biol. 9, R124. 10.1186/gb-2008-9-8-r124 (doi:10.1186/gb-2008-9-8-r124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wrangham R., Conklin-Brittain N. 2003. Cooking as a biological trait. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 136, 35–46 10.1016/S1095-6433(03)00020-5 (doi:10.1016/S1095-6433(03)00020-5) [DOI] [PubMed] [Google Scholar]
- 105.Green R. E., et al. 2010. A draft sequence of the Neanderthal genome. Science 328, 710–722 10.1126/science.1188021 (doi:10.1126/science.1188021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gilad Y., Segre D., Skorecki K., Nachman M. W., Lancet D., Sharon D. 2000. Dichotomy of single-nucleotide polymorphism haplotypes in olfactory receptor genes and pseudogenes. Nat. Genet. 26, 221–224 10.1038/79957 (doi:10.1038/79957) [DOI] [PubMed] [Google Scholar]
- 107.D'Elia G. 2003. Phylogenetics of Sigmodontinae (Rodentia, Muroidea, Cricetidae), with special reference to the akodont group, and with additional comments on historical biogeography. Cladistics 19, 307–323 10.1016/S0748-3007(03)00071-9 (doi:10.1016/S0748-3007(03)00071-9) [DOI] [Google Scholar]
- 108.Nilsson G. 1996. Brain and body oxygen requirements of Gnathonemus petersii, a fish with an exceptionally large brain. J. Exp. Biol. 199, 603–607 [DOI] [PubMed] [Google Scholar]
- 109.Kaufman J. 2003. On the expensive-tissue hypothesis: independent support from highly encephalized fish. Curr. Anthropol. 44, 705–706 10.1086/379258 (doi:10.1086/379258) [DOI] [Google Scholar]
- 110.Chapman L. J., Hulen K. G. 2001. Implications of hypoxia for the brain size and gill morphometry of mormyrid fishes. J. Zool. 254, 461–472 10.1017/S0952836901000966 (doi:10.1017/S0952836901000966) [DOI] [Google Scholar]
- 111.Sabeti P. C., et al. 2006. Positive natural selection in the human lineage. Science 312, 1614–1620 10.1126/science.1124309 (doi:10.1126/science.1124309) [DOI] [PubMed] [Google Scholar]
- 112.Goren-Inbar N., Alperson N., Kislev M. E., Simchoni O., Melamed Y., Ben-Nun A., Werker E. 2004. Evidence of hominin control of fire at Gesher Benot Ya'aqov, Israel. Science 304, 725–727 10.1126/science.1095443 (doi:10.1126/science.1095443) [DOI] [PubMed] [Google Scholar]
- 113.Jobling M. A., Hurles M. E., Tyler-Smith C. 2004. Human evolutionary genetics: origins, people, and disease. New York, NY: Garland Science [Google Scholar]
- 114.White T. D., Asfaw B., Beyene Y., Haile-Selassie Y., Lovejoy C. O., Suwa G., WoldeGabriel G. 2009. Ardipithecus ramidus and the paleobiology of early hominids. Science 326, 75–86 10.1126/science.1175802 (doi:10.1126/science.1175802) [DOI] [PubMed] [Google Scholar]