Abstract
It has long been maintained that the majority of terrestrial Antarctic species are relatively recent, post last glacial maximum, arrivals with perhaps a few microbial or protozoan taxa being substantially older. Recent studies have questioned this ‘recolonization hypothesis’, though the range of taxa examined has been limited. Here, we present the first large-scale study for mites, one of two dominant terrestrial arthropod groups in the region. Specifically, we provide a broad-scale molecular phylogeny of a biologically significant group of ameronothroid mites from across the maritime and sub-Antarctic regions. Applying different dating approaches, we show that divergences among the ameronothroid mite genera Podacarus, Alaskozetes and Halozetes significantly predate the Pleistocene and provide evidence of independent dispersals across the Antarctic Polar Front. Our data add to a growing body of evidence demonstrating that many taxa have survived glaciation of the Antarctic continent and the sub-Antarctic islands. Moreover, they also provide evidence of a relatively uncommon trend of dispersals from islands to continental mainlands. Within the ameronothroid mites, two distinct clades with specific habitat preferences (marine intertidal versus terrestrial/supralittoral) exist, supporting a model of within-habitat speciation rather than colonization from marine refugia to terrestrial habitats. The present results provide additional impetus for a search for terrestrial refugia in an area previously thought to have lacked ice-free ground during glacial maxima.
Keywords: ameronothroid mites, biogeography, dispersal, glacial refugia, vicariance
1. Introduction
The biogeography of Antarctica and its surrounding islands is both complex and contentious. The complexity is a consequence of the compound geological history of the region. Continental Antarctica is an amalgam of a large, eastern Antarctic block and a western complex of accreted terranes [1–3]. Likewise, its surrounding islands have a variety of geological origins and histories, ranging from relatively young volcanic islands such as the Prince Edwards [4], to more complex older islands such as South Georgia and the Kerguelen archipelago [5–8]. The contention stems largely from a long-standing debate about the origins of terrestrial Antarctic biotas. In particular, much controversy swirls around whether the biotas of the continent are relatively recent colonists or older palaeoendemics, and the extent of the relationships between the continental biota and those found on the more northerly maritime Antarctic and sub-Antarctic islands [9–15].
Apparently low species endemism in some groups, particularly the mosses and some microbiota, combined with the obvious and substantially greater glaciation of Antarctica and many of its surrounding islands during the last glacial maximum than at present [16,17], encouraged the early view [18] that the majority of terrestrial Antarctic species are relatively recent (post last glacial maximum) arrivals, with perhaps a few microbial or protozoan taxa being substantially older (reviews in [3,19]). While this view elicited some early controversy (e.g. [9,20]), conventional, morphology-based analyses of other taxa, such as algae [21] and crustacea [22], and renewed systematic and biogeographic work on the bryophytes (e.g. [23–25]) continue to support the idea of low endemism and, implicitly, recent arrival. Similarly, conventional, non-molecular biogeographic analyses of a variety of taxa have suggested that the assemblages of the sub-Antarctic islands and maritime Antarctic are clearly differentiated from those of continental Antarctica (e.g. [15,26,27]), and show only distant relationships among the main island groupings in the Pacific, Atlantic and Indian components of the Southern Ocean (e.g. [13,28,29]).
By contrast, a small number of recent studies and syntheses have questioned the ‘recolonization hypothesis’ [3,30–32]. Several of these studies have been based on patterns of diversity and species distributions that seem highly indicative of either an ancient Gondwana origin for the taxa concerned (e.g. [12,33]), or regional glacial refugia [34,35]. More compelling are molecular phylogenetic investigations suggesting that cyanobacteria, algae, midges and springtails display high diversity and show a range of colonization times, varying from ca 300–1 Ma, and almost certainly prior to the last glacial maximum [32,36–39]. However, to date these investigations have represented a relatively small set of taxa for strictly terrestrial species. In consequence, the timing of arrival in Antarctica and relationships among the continental and southern island terrestrial biotas remains poorly understood. This situation contrasts strongly with marine systems, where phylogenetic knowledge of the timing of events and relationships among the continent and its surrounding islands is growing rapidly (e.g. [40–50]).
Because mites are the most diverse group of terrestrial animals in the region [51], this key group is used here to further current understanding of the origins and biogeography of the terrestrial fauna of Antarctica and its surrounding islands. In particular, the ameronothroids are investigated for two reasons. First, it has been proposed, on the grounds of species distributional and habitat-use information, that colonization by these mites in the Antarctic has proceeded from marine habitats to terrestrial areas following glacial retreat [52] and, second, the group is also iconic in the sense that two members thereof, Alaskozetes antarcticus Michael, 1903 and Halozetes belgicae (Michael), are among the most well-studied terrestrial organisms in the region (e.g. [53–61]).
2. Material and methods
(a). Sampling
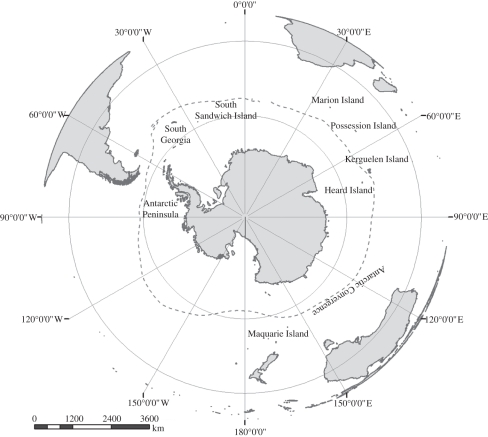
Following [52], the current analysis concerns the three major ameronothroid genera endemic to the Antarctic region, Podacarus (Grandjean 1955), Alaskozetes (Hammer 1955) and Halozetes Berlese 1916 (family Ameronothridae); [62]. These genera are closely related, endemic to the region and considered to belong to a monophyletic group within the Ameronothroidea [62–66]. Sampling included eight Halozetes species (H. fulvus, H. crozetensis, H. belgicae, H. marinus, H. marionensis, H. intermedius, H. macquariensis and H. capensis) incorporating four subspecies (H. m. devilliersi, H. m. marinus, H. b. micki and H. b. brevipilis), and Podacarus auberti and A. antarcticus (including A. a. intermedius). While there are no other known valid species of Podacarus, six other known Halozetes species, which are rare in the Antarctic/sub-Antarctic region or in New Zealand were not included in the present analysis, especially because in the Antarctic case recent collections at localities where these species are supposed to occur have not yielded material of them (see [52] for geographical information). Two other species of Alaskozetes, apparently found on Bouvetøya (A. bouvetoyaensis) and in Alaska (A. coriaceus), require further systematic investigation [52]. For outgroups, Aquanothrus montanus (Ameronothroidea; a Gondwanan species; [52]), Magellozetes antarcticus (Ceratozetoidea) and Platynothrus skottsbergi (Camisiidae) were used. These species were sampled from South Africa in the former case and in the latter case from both sub-Antarctic and maritime Antarctic localities across the region (figure 1; see also the electronic supplementary material, table S1). Reference specimens are currently housed at the Center for Invasion Biology by S.L.C., at the British Antarctic Survey by P.C. and by D.M. at the University of Brunei.
Figure 1.
Map of Antarctica and surrounding Southern Ocean Islands. The current position of the Antarctic Convergence is indicated.
(b). DNA extraction, PCR amplification and sequencing
Total genomic DNA was isolated using a phenol/chloroform extraction method [67] with slight modifications [68]. Conventional DNA extraction procedures sometimes failed to yield DNA of useable quality. In these cases, DNA was extracted with the DNeasy Tissue Kit (QIAGEN, Hilden, Germany) following the manufacturer's recommendations.
The widely used mitochondrial cytochrome oxidase subunit I gene (COI) was amplified using primers LCO1490 and HCO2198 [69] resulting in a fragment of 494 bp. The nuclear histone-3 gene (H3) was also amplified, but the H3 primers H3AF and H3AR [70] showed limited success with the amplification of Halozetes DNA. Hence, mite-specific primers were designed from the sequence alignment of the few successfully amplified specimens. The sense primer (HIST3F: CGTAAGTCGGCGCCCAGC) and the antisense primer (HIST3R: GACCCGTTTGGCGTGAATTGC) successfully amplified a 320 bp fragment. This fragment corresponded to the 2nd exon (partial), 2nd intron and the 3rd exon (partial) of the H3 gene of Drosophila hydei [71].
Mitochondrial and nuclear PCR reactions were performed in a final volume of 30 µl and included 2 µl of unquantified genomic DNA, 1X reaction buffer, 1.5 mM MgCl2, 200 µM dNTP solution, 2 µM of each primer and 1 unit of Taq polymerase (Super-Therm; JMR Holdings, London, UK). All PCR reactions were conducted with similar cycling parameters, which included an initial denaturation step at 94°C for 1 min followed by 35 cycles of 94°C for 30 s, 40°C (COI) or 50°C (H3) for 30 s, 72°C for 45 s. A final extension step at 72°C for 5 min completed the reactions.
Nucleotide sequencing was carried out using BigDye Terminator 3.1 mix (Applied Biosystems, Warrington, UK). Sequencing cocktails were cleaned using Centrisep spin columns (Applied Biosystems, Warrington, UK) and the products were analysed on an ABI3170 automated sequencer (Applied Biosystems, Warrington, UK). Electropherograms of the raw data were manually checked and edited with Sequence Editor (Applied Biosystems, Warrington, UK).
(c). Sequence and phylogenetic analyses
Sequences were aligned with Clustal X 1.81 [72] using the multiple alignment mode and all alignments were checked by eye. All sequences generated in the present study were submitted to GenBank under accession numbers HQ264 190-HQ264 403. In the nuclear H3 gene the exons were conserved across all taxa. The intron region of both the ingroup and outgroup species was highly variable and could not be unambiguously aligned. Because of this, the intron region was removed from further analyses.
Phylogenetic analyses included parsimony searches and Bayesian inference. All analyses were performed on (i) the mitochondrial COI gene, (ii) the H3 exon, and (ii) the combined COI and H3 datasets after the partition homogeneity test indicated no conflicting signal in the data (p = 1.0). Bootstrap values exceeding 70 per cent and posterior probabilities higher than 0.70 were considered somewhat supported with bootstrap values exceeding 80 per cent and posterior probability values exceeding 0.90 considered well supported. Parsimony searches employed the heuristic search criterion with TBR branch swapping as implemented in PAUP* 4.0b10 [73]. Nodal support was assessed through 1000 bootstrap replicates.
MrModeltest 2.3 [74] was used to determine the model of evolution that best fitted the data. The Akaike Information Criterion was preferentially used because it minimizes those parameters that do not contribute phylogenetic information [75,76]. Posterior probabilities for the Bayesian analyses were determined by running one cold and four heated chains for 5 000 000 generations in MrBayes 3.1.2 [77]. The combined data were analysed in a partitioned manner to allow the selection of different optimal models for each partition [77]. The optimal model was specified as prior for each partition. Each analysis was repeated and trees were sampled every 100 generations. Stability was determined using the ‘sump’ command in MrBayes and the first 5000 (or 10%) trees were excluded as burn-in. Posterior probabilities for nodes were calculated from the remaining topologies.
(d). Molecular divergence times
Two approaches were implemented to estimate the divergence times of the ameronothroid mites. First, an uncorrelated relaxed Bayesian clock (BEAST 1.5.4; [78]) was employed incorporating both geological and fossil calibration points. Representative COI sequences for the major groups within the Tardigrada (water bears; n = 8), Onychophora (velvet worms; n = 9) and Arthropoda (arthropods including two classes (Arachnida and Pycnogonida) and eight orders (Acari, Amblypygi, Araneae, Opiliones, Pseudoscorpiones, Scorpiones, Solifugae, Uropygi; n = 96)) were downloaded from public databases and retrospectively aligned based on their amino-acid composition (494 bp; dataset available upon request). We also included all COI sequences generated for the present study (n = 108). Panarthropod fossil calibration points [79–83] included the split between the Tardigrada and Onychophora/Arthropoda (between 850 and 400 Ma), the origin for the Arthropoda (530 Ma), the origin of the Arachnida (420 Ma), the origin for the Araneae (299 Ma) and the origin for the oribatid mites (117 Ma). As divergence times based on such ancient calibration points could have a notable impact on uncalibrated nodes, including the overestimation of more recent divergences (see [84]), we concurrently constrained the age of H. fulvus (including H. capensis) with the age of Marion Island given that H. fulvus is endemic to the Prince Edwards. The age of Marion Island is generally believed to be 0.5 Ma old [4], and as previous investigations have found support for using these bounds to delimit age of the taxa endemic to the island [85,86], we also calculated divergence times based on this single geological calibration point. The monophyly of the major panarthropod lineages was constrained a priori and an uncorrelated lognormal relaxed clock and the Yule speciation prior specified. Two independent Markov Chain Monte-Carlo runs were conducted of 50 000 000 generations (sampled every 500 generations) each with the first 10 per cent discarded as burn-in. Run convergences were verified using Tracer.
Second, we estimated divergence times in the absence of calibration points following [87]. Various arthropod COI rate estimates have been postulated ranging from 1.5 to 2.3 per cent per million years (e.g. [88–90]). To apply the most appropriate rate for our ingroup, rate specific mutation rates were estimated for the ingroup lineages. For this, we used the panarthropod data without calibration points. Parameters in BEAST followed those outlined above. Lineage-specific rates for Halozetes, Alaskozetes and Podacarus were calculated as the average of the (internal and single terminal) branch specific rates.
(e). Biogeographic analysis
The biogeographic history of these ameronothroid mites were reconstructed using diva 1.2 [91] and Lagrange 1.0.1 [92]. Diva is an event-based method that considers vicariance, dispersals and extinctions (see [93–95] for critical discussion). Lagrange is a likelihood approach that incorporates a parametric inference which takes both anagenetic and cladogenetic changes into account. Because dispersal across the Antarctic Convergence is our central concern, for the analyses, two geographical units were defined, viz. the sub-Antarctic which lies close to or north of the Antarctic Convergence (Marion, Kerguelen, Macquarie, Heard, Possession Islands and South Georgia) and Maritime Antarctic to the south of the Convergence (Antarctic Peninsula and Scotia Arc archipelagos of the South Shetland, South Orkney and South Sandwich Islands). The analyses were based on a simplified version of the combined (COI and H3) tree (all duplicate taxa were trimmed from the tree and we retained single representative specimens per clade; see the electronic supplementary material, figure S1). The current distribution of each representative was coded as present or absent in each of these areas.
3. Results
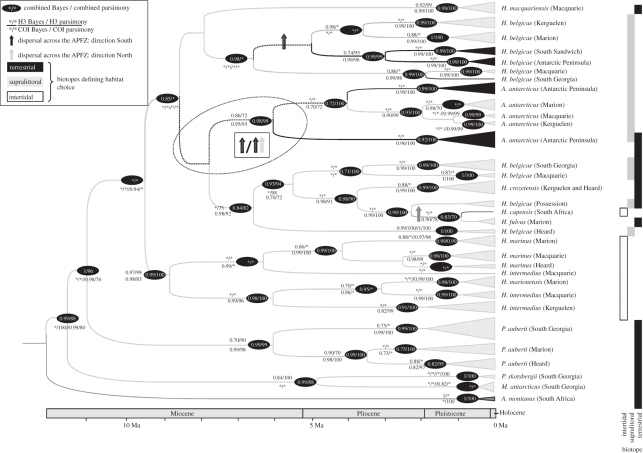
The combined ameronothroid dataset comprised 763 bp for 108 taxa (H3: 269 bp; COI: 494 bp; see the electronic supplementary material, table S2 for combined and single gene statistics). The combined Bayesian topology is shown in figure 2 (see also the electronic supplementary material, figure S2). Posterior probabilities and bootstrap support for the combined analyses as well as for the single gene fragments are indicated on the tree. With the exception of the analyses based on the H3 fragment alone that was mostly unresolved, largely congruent topologies (exceptions are discussed below) were retrieved. Five clades were found namely (i) Alaskozetes, (ii) Podacarus, (iii) H. belgicae from the Antarctic Peninsula, Kerguelen, Marion, South Sandwich, Macquarie and South Georgia Islands, (iv) H. crozetensis, H. fulvus, H. capensis and H. belgicae from South Georgia, Macquarie and Possession Islands and (v) H. marinus, H. intermedius and H. marionensis. Averaged uncorrected sequence divergence values among these groups for the COI gene varied between 15.9 per cent (GTR corrected value = 18.6%) between (i) Alaskozetes and (iv) H. crozetensis, H. fulvus, H. capensis and H. belgicae to 21.4 per cent (GTR corrected value = 25.9%) between (i) Alaskozetes and (v) H. marinus, H. intermedius and H. marionensis (sequence divergence values are presented in the electronic supplementary material, table S3). All of the evolutionary relationships among these five groups were not consistently supported by the different gene fragments and methods of analyses: specifically the evolutionary placement of H. macquariensis and the relationship of Alaskozetes and Podacarus to Halozetes. In spite of the uncertain placement, some support, most noticeably from the COI gene, was found for the clustering of Alaskozetes within Halozetes. The combined analyses placed Podacarus basal to the Halozetes group but in contrast, Podacarus clustered within the ingroup considering only COI. In addition, not all the species included in the analyses proved to be monophyletic. Halozetes belgicae is paraphyletic with membership in two of the larger clades. Halozetes intermedius was also paraphyletic.
Figure 2.
Bayesian topology inferred from the combined COI and H3 gene fragments for 108 taxa. Terminal groups have been collapsed, and the width of the triangle represents the number of distinct taxa included therein. Numbers at nodes indicate statistical support for the combined data as well as single gene fragments. Bayesian posterior probabilities (more than 0.7) and parsimony bootstrap support (more than 70%) are shown. Taxa occurring on maritime Antarctic localities (black), sub-Antarctic localities (light grey) and the taxon found in South Africa (dark grey) are indicated. Inferred ancestral area cladograms and biotopes (habitat choice) are shown. Ambiguous area cladograms are indicated by dashed lines (see text for further explanation). Dispersal events across the Antarctic Convergence are indicated by horizontal arrows. The scale bar follows the lineage-specific mutation rate divergence estimates.
Within the Halozetes group two main clades based on habitat preference were discernable (see figure 2). The first comprised the intertidal mite species (H. marinus, H. intermedius and H. marionensis), while the second clade included the supralittoral and terrestrial mite species (H. macquariensis, H. belgicae, H. crozetensis and H. fulvus).
With regard to the outcomes of the age calculations (see the electronic supplementary material, table S4), the lineage-specific mutation rates for the three ingroup genera (Halozetes, Alaskozetes and Podacarus) fall within the rates estimated across panarthropod lineages (see the electronic supplementary material, figure S3). As the rates estimated for Halozetes (0.98, s.d. = 0.23), Alaskozetes (0.98, s.d. = 0.34) and Podacarus (0.91, s.d. = 0.07) fall close to the median value for arthropods, we applied the average rate of 1.9 per cent to the ingroup. The origin of the ingroup ranged from 4.32 Ma when the tree was constrained using the single Marion Island geological calibration point to 35.90 Ma when the panarthropod fossil calibrations were included. In the absence of calibration points (and following [87]), the age of the ingroup was estimated at 10.86 Ma. Divergence times estimated using the very distant fossil calibration points are likely to be overestimates, whereas the dates calculated using the single recent geological calibration point probably underestimate true divergence times. As the divergence times estimated using the lineage-specific mutation rates yielded intermediate times (and bearing in mind that the standard errors associated with these age estimates are typically large), we chose to adopt these uncalibrated estimates based on the lineage-specific mutation rates. These dates are also in line with those reported for other arthropod taxa in the Antarctic (e.g. [37]).
Multiple dispersal events (diva estimated at least two events) across the Antarctic Convergence were found (see figure 2 and the electronic supplementary material, figure S1) in addition to the single dispersal of H. capensis to South Africa from Marion Island (Lagrange [92] confirmed multiple dispersal events). Ambiguous ancestral area cladograms were inferred for two internal branches resulting in either a single or two dispersal events. Many of the inferred dispersal events occurred across well supported areas of the topology and as such, the effects of the weakly supported deeper nodes are negligible here.
4. Discussion
Before proceeding to a discussion of the biogeographic implications of the current analyses, it should be noted that our analyses do not support the retention of the species A. antarcticus within the genus Alaskozetes (Hammer 1955) and that this species should be transferred to the genus Halozetes (Berlese 1916). Because Alaskozetes contains three species (A. antarcticus, Michael 1903; A. bouvetoyaensis, van Pletzen & Kok 1971; A. coriaceus, Hammer 1955), it is not possible to determine the current taxonomic status of the remaining two species and the genus, though it is clear that they require further systematic investigation. Moreover, the analyses also indicate that H. belgicae and H. intermedius each represent more than a single species. Further investigation of all individuals referred to these taxa is clearly required, although such complexity within a given, previously recognized species in the region is not unusual. For example, Stevens et al. [37] argued that the widespread springtail Cryptopygus antarcticus probably represents a complex of geographically separated species. Because both H. belgicae and the springtail C. antarcticus are among the most intensively studied Antarctic arthropods (see §1), and especially in the context of climate change impacts in the maritime and sub-Antarctic [96–99] the paraphyly of these taxa should be recognized in the interpretation of differences in responses among sites. Fortunately, this problem does not extend to the other, heavily studied species.
From a biogeographic perspective, and adopting the age estimates based on the lineage-specific mutation rates, the analyses suggest that this group of ameronothroid mites colonized the Antarctic region ca 10 Ma. Similar or older estimates for colonization times of the region have been made for other terrestrial invertebrate taxa such as springtails and midges [36,37], as well as for some marine groups (e.g. [50]). The independent dispersals across the Antarctic Convergence in Alaskozetes and H. belgicae between ca 10 and 6 Ma are also in keeping with data from other groups, most notably the genus Cryptopygus [37].
If these dates generated based on lineage-specific mutation rates are accepted, this suggests colonizations, across the Antarctic Convergence, of the Antarctic Peninsula, sometime during the late Miocene or early Pliocene. Although evidence exists for exposure from glaciation of some regions in East Antarctica since 12–5 Ma, available geological evidence, along with ice sheet reconstruction through modelling, suggests that the Antarctic Peninsula region remained heavily glaciated until the end of the last glacial maximum (reviewed in [3,100]). The current phylogenetic analyses therefore suggest, irrespective of how the divergence dates for the lineages are constrained, that despite the geological evidence, some refugia must have been present in the Peninsula region. In so doing, they support a growing body of molecular information showing that recent, post-glacial colonization is not a plausible explanation for the origins of much of the Antarctic terrestrial and freshwater invertebrate biota [32,36–39]. In consequence, not only does the geological interpretation require further scrutiny, but a search for refugia, in keeping with at least some level of geological plausibility is also required (e.g. [35]), particularly for the northern Peninsula region. The multiple colonizations of Antarctica from the surrounding islands also make this continent unusual by comparison with others, where colonization from the continent to surrounding areas is the norm, and where exceptions are considered worthy of comment [101]. However, perhaps the group is unusual, given that it appears that H. capensis also represents a recent colonization (less than 0.5 Ma) of South Africa from the sub-Antarctic [102].
In the case of the sub-Antarctic islands it is clear that successful dispersal among them has not only been common, but often has not required a prolonged period to occur after the islands have emerged (see also [37,85]). Thus, despite the apparent level of biogeographic separation among the islands identified using conventional assemblage (or species-identity)-based analyses (e.g. [15,26,28,29]), it is clear that considerable movement among them does take place (see also [103]). Although the differences in analytical outcome may partly be a consequence of variation in dispersal ability among the groups concerned [14], the current phylogeographic evidence suggests that a much greater role for dispersal across the region, among what are often considered distinct biogeographic regions, will be found in all groups. This conclusion is in keeping with molecular studies from elsewhere in the Southern Hemisphere emphasizing the role of dispersal processes in its biogeography (reviewed in [104]). However, it does not mean that the biogeographic regions identified to date (e.g. [13,26]) are meaningless, but rather that the boundaries among them are more permeable than might previously have been supposed, and that the biogeographic history of the region is consequently more complicated (see also [19]).
The latter is readily illustrated by examining the proposal that the species within the Halozetes group (sensu lato) survived glaciation in marine refuges and subsequently colonized terrestrial areas of the sub-Antarctic and Antarctic [105]. While the proposal appears compelling, given evidence for weevils derived from biogeographic studies of the group (e.g. [106]), it is not supported by the current molecular data. Rather, Halozetes clearly comprises two distinct clades, one associated with marine intertidal environments and the other with terrestrial and supralittoral ones, which diverged early in the history of the group in the region. No evidence exists of speciation from a marine to a terrestrial group (or vice versa). Rather, it seems probable that substantial ecological flexibility has enabled species in the terrestrial and supralittoral clade to inhabit a relatively diverse range of environments. Recent molecular work at the population level in a weevil species from Marion Island likewise suggests that inland rather than coastal refugia have been more significant in its history [86], in keeping with new geological interpretations for the island [107].
In conclusion, this work has demonstrated the significant role for colonization of Antarctica and its surrounding islands prior to the last glacial maximum as an important component of their fauna. By doing so, it has highlighted the need for additional investigations of taxa across the continent and on its surrounding islands to clarify discrepancies among the biological and geological data, and to provide insights into a region that has long been the subject of biogeographic controversy.
Acknowledgements
We thank Anne Ropiquet, Brent Emerson, Savel Daniels and an anonymous reviewer for critical comments on a previous version of the ms. This work was supported by SANAP Grant 2069543 and a Center for Invasion Biology stipend to BvV. PC is supported through the ‘Polar Sciences for Planet Earth’ core programme of BAS. We thank several researchers for making collections of mites, particularly J. Deere, C. Hänel, D. Bergstrom and L. Coetzee. This paper also contributes to the SCAR ‘Evolution and Biodiversity in Antarctica’ research programme.
References
- 1.Cande S. C., Stock J., Müller R. D., Ishihara T. 2000. Cenozoic motion between East and West Antarctica. Nature 404, 145–150 10.1038/35004501 (doi:10.1038/35004501) [DOI] [PubMed] [Google Scholar]
- 2.Vaughan A. P. M., Storey B. C. 2000. The eastern palmer land shear zone: a new terrane accretion model for the Mesozoic development of the Antarctic Peninsula. J. Geol. Soc. Lond. 157, 1243–1256 10.1144/jgs.157.6.1243 (doi:10.1144/jgs.157.6.1243) [DOI] [Google Scholar]
- 3.Convey P., Gibson J. A. E., Hillenbrand C. D., Hodgson D. A., Pugh P. J. A., Smellie J. L., Stevens M. I. 2008. Antarctic terrestrial life—challenging the history of the frozen continent? Biol. Rev. 83, 103–117 10.1111/j.1469-185X.2008.00034.x (doi:10.1111/j.1469-185X.2008.00034.x) [DOI] [PubMed] [Google Scholar]
- 4.McDougall I., Verwoerd W., Chevallier L. 2001. K-Ar geochronology of Marion Island, Southern Ocean. Geol. Mag. 138, 1–17 10.1017/S0016756801005039 (doi:10.1017/S0016756801005039) [DOI] [Google Scholar]
- 5.Chown S. L., Gremmen N. J. M., Gaston K. J. 1998. Ecological biogeography of southern ocean islands: species–area relationships, human impacts, and conservation. Am. Nat. 152, 562–575 10.1086/286190 (doi:10.1086/286190) [DOI] [PubMed] [Google Scholar]
- 6.Wallace P. J., Frey F. A., Weis D., Coffin M. F. 2002. Origin and evolution of the Kerguelen Plateau, Broken Ridge and Kerguelen Archipelago: editorial. J. Petrol. 43, 1105–1108 10.1093/petrology/43.7.1105 (doi:10.1093/petrology/43.7.1105) [DOI] [Google Scholar]
- 7.Craig D. A. 2003. Deconstructing Gondwana—words of warning from the Crozet Island Simuliidae (Diptera). Cimbebasia 19, 157–164 [Google Scholar]
- 8.Ali J. R., Aitchison J. C. 2009. Kerguelen Plateau and the Late Cretaceous southern-continent bioconnection hypothesis: tales from a topographic ocean. J. Biogeogr. 36, 1778–1784 10.1111/j.1365-2699.2009.02105.x (doi:10.1111/j.1365-2699.2009.02105.x) [DOI] [Google Scholar]
- 9.Brundin L. Z. 1966. Transantarctic relationships and their significance as evidenced by chironomid midges with a monograph of the sub-families Podonominae and Aphroteniinae and the austral Heptagyiae. K. Svenska. Vetensk. Handl. 11, 1–472 [Google Scholar]
- 10.Darlington P. J. 1970. A practical criticism of Hennig-Brundin ‘phylogenetic systematics’ and Antarctic biogeography. Syst. Zool. 19, 1–18 10.2307/2412024 (doi:10.2307/2412024) [DOI] [Google Scholar]
- 11.Udvardy M. D. F. 1987. The biogeographical realm Antarctica: a proposal. J. R. Soc. N. Z. 17, 187–194 [Google Scholar]
- 12.Marshall D. J., Pugh P. J. A. 1996. Origin of the inland Acari of Continental Antarctica, with particular reference to Dronning Maud Land. Zool. J. Linn. Soc. 118, 101–118 10.111/j.1096-3642.1996.tb00221.x (doi:10.111/j.1096-3642.1996.tb00221.x) [DOI] [Google Scholar]
- 13.Morrone J. J. 1998. On Udvardy's Insulantarctica province: a test from the weevils (Coleoptera: Curculionoidea). J. Biogeogr. 25, 947–955 10.1046/j.1365-2699.1998.00240.x (doi:10.1046/j.1365-2699.1998.00240.x) [DOI] [Google Scholar]
- 14.Greve M., Gremmen N. J. M., Gaston K. J., Chown S. L. 2005. Nestedness of South Ocean island biotas: ecological perspectives on a biogeographical conundrum. J. Biogeogr. 32, 155–168 10.1111/j.1365-2699.2004.01169.x (doi:10.1111/j.1365-2699.2004.01169.x) [DOI] [Google Scholar]
- 15.Pugh P. J. A., Convey P. 2008. Surviving out in the cold: Antarctic endemic invertebrates and their refugia. J. Biogeogr. 35, 2176–2186 10.1111/j.1365-2699.2008.01953.x (doi:10.1111/j.1365-2699.2008.01953.x) [DOI] [Google Scholar]
- 16.Hall K. 2002. Review of Present and Quaternary periglacial processes and landforms of the maritime and sub-Antarctic region. SouthAfr. J. Sci. 98, 71–81 [Google Scholar]
- 17.Peck L. S., Convey P., Barnes D. K. A. 2006. Environmental constraints on life histories in Antarctic ecosystems: tempos, timings and predictability. Biol. Rev. 81, 75–109 10.1017/S1464793105006871 (doi:10.1017/S1464793105006871) [DOI] [PubMed] [Google Scholar]
- 18.Gressitt J. L. 1967. Entomology of Antarctica. Antarct. Res. Ser. 10, 1–395 [Google Scholar]
- 19.Barnes D. K. A., Hodgson D. A., Convey P., Allen C. S., Clarke A. 2006. Incursion and excursion of Antarctic biota: past, present and future. Global Ecol. Biogeogr. 15, 121–142 10.1111/j.1466-822X.2006.00216.x (doi:10.1111/j.1466-822X.2006.00216.x) [DOI] [Google Scholar]
- 20.Wallwork J. A. 1973. Zoogeography of some terrestrial micro-arthropoda in Antarctica. Biol. Rev. 48, 233–259 10.1111/j.1469-185X.1973.tb00981.x (doi:10.1111/j.1469-185X.1973.tb00981.x) [DOI] [Google Scholar]
- 21.Broady P. A. 1996. Diversity, distribution and dispersal of Antarctic terrestrial algae. Biodivers. Conserv. 5, 1307–1335 10.1007/BF00051981 (doi:10.1007/BF00051981) [DOI] [Google Scholar]
- 22.Pugh P. J. A., Dartnall H. J. G., McInnes S. J. 2002. The non-marine Crustacea of Antarctica and the islands of the Southern Ocean: biodiversity and biogeography. J. Nat. Hist. 36, 1047–1103 10.1080/00222930110039602 (doi:10.1080/00222930110039602) [DOI] [Google Scholar]
- 23.Bednarek-Ochyra H., Väňa J., Ochyra R., Lewis Smith R. I. 2000. The liverwort flora of Antarctica. Cracow, Poland: W. Szafer Institute of Botany, Polish Academy of Sciences [Google Scholar]
- 24.Peat H. J., Clarke A., Convey P. 2007. Diversity and biogeography of the Antarctic flora. J. Biogeogr. 34, 132–146 10.1046/j.1365-2699.1998.00240.x (doi:10.1046/j.1365-2699.1998.00240.x) [DOI] [Google Scholar]
- 25.Ochyra R., Bednarek-Ochyra H., Lewis Smith R. I. 2008. The illustrated moss flora of Antarctica. Cambridge, UK: Cambridge University Press [Google Scholar]
- 26.McInnes S. J., Pugh P. J. A. 1998. Biogeography of limno-terrestrial Tardigrada, with particular reference to the Antarctic fauna. J. Biogeogr. 25, 31–36 10.1046/j.1365-2699.1998.251176.x (doi:10.1046/j.1365-2699.1998.251176.x) [DOI] [Google Scholar]
- 27.Pugh P. J. A., Convey P. 2000. Scotia Arc Acari: antiquity and origin. Zool. J. Linn. Soc 130, 309–328 10.1111/j.1096-3642.2000.tb01633.x (doi:10.1111/j.1096-3642.2000.tb01633.x) [DOI] [Google Scholar]
- 28.Pugh P. J. A., Scott B. 2002. Biodiversity and biogeography of non-marine Mollusca on the islands of the Southern Ocean. J. Nat. Hist. 36, 927–952 10.1080/00222930110034562 (doi:10.1080/00222930110034562) [DOI] [Google Scholar]
- 29.Pugh P. J. A. 2004. Biogeography of spiders (Araneae: Arachnida) on the islands of the Southern Ocean. J. Nat. Hist. 38, 1461–1487 10.1080/0022293031000155403 (doi:10.1080/0022293031000155403) [DOI] [Google Scholar]
- 30.Convey P., Stevens M. I. 2007. Antarctic Biodiversity. Science 317, 1877–1878 10.1126/science.1147261 (doi:10.1126/science.1147261) [DOI] [PubMed] [Google Scholar]
- 31.Convey P., et al. 2009. Exploring biological constraints on the glacial history of Antarctica. Quaternary Sci. Rev. 28, 3035–3048 10.1016/j.quascirev.2009.08.015 (doi:10.1016/j.quascirev.2009.08.015) [DOI] [Google Scholar]
- 32.De Wever A., Leliaert F., Verleyen E., Vanormelingen P., Van der Gucht K., Hodgson D. A., Sabbe K., Vyverman W. 2009. Hidden levels of phylodiversity in Antarctic green algae: further evidence of glacial refugia. Proc. R. Soc. B 276, 3591–3599 10.1098/rspb.2009.0994 (doi:10.1098/rspb.2009.0994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayly I. A. E., Gibson J. A. E., Wagner B., Swadling K. M. 2003. Taxonomy, ecology and zoogeography of two East Antarctic freshwater calanoid copepod species: Boeckella poppei and Gladioferens antarcticus. Antarct. Sci. 15, 439–448 10.1017/S0954102003001548 (doi:10.1017/S0954102003001548) [DOI] [Google Scholar]
- 34.Lawley B., Ripley S., Bridge P., Convey P. 2004. Molecular analysis of geographic patterns of eukaryotic diversity in Antarctic soils. Appl. Environ. Microb. 70, 5963–5972 10.1128/aem.70.10.5963-5972.2004 (doi:10.1128/aem.70.10.5963-5972.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maslen N. R., Convey P. 2006. Nematode diversity and distribution in the southern maritime Antarctic—clues to history? Soil Biol. Biochem. 38, 3141–3151 10.1016/j.soilbio.2005.12.007 (doi:10.1016/j.soilbio.2005.12.007) [DOI] [Google Scholar]
- 36.Allegrucci G., Carchini G., Todisco V., Convey P., Sbordoni V. 2006. A molecular phylogeny of Antarctic Chironomidae and its implications for biogeographical history. Polar Biol. 29, 320–326 10.1007/s00300-005-0056-7 (doi:10.1007/s00300-005-0056-7) [DOI] [Google Scholar]
- 37.Stevens M. I., Greenslade P., Hogg I. D., Sunnucks P. 2006. Southern hemisphere springtails: could any have survived glaciation of Antarctica? Mol. Biol. Evol. 23, 874–882 10.1093/molbev/msj073 (doi:10.1093/molbev/msj073) [DOI] [PubMed] [Google Scholar]
- 38.Taton A., Grubisic S., Balthasart P., Hodgson D. A., Laybourn-Parry J., Wilmotte A. 2006. Biogeographical distribution and ecological ranges of benthic cyanobacteria in East Antarctic lakes. FEMS Microbiol. Ecol. 57, 272–289 10.1111/j.1574-6941.2006.0110.x (doi:10.1111/j.1574-6941.2006.0110.x) [DOI] [PubMed] [Google Scholar]
- 39.McGaughran A., Torricelli G., Carapelli A., Frati F., Stevens M. I., Convey P., Hogg I. D. 2010. Contrasting phylogeographical patterns for springtails reflect different evolutionary histories between Antarctic Peninsula and continental Antarctica. J. Biogeogr. 37, 103–119 10.1111/j.1365-2699.2009.02178.x (doi:10.1111/j.1365-2699.2009.02178.x) [DOI] [Google Scholar]
- 40.Patarnello T., Bargelloni L., Varotto V., Battaglia B. 1996. Krill evolution and the Antarctic ocean currents: evidence of vicariant speciation as inferred by molecular data. Mar. Biol. 126, 603–608 10.1007/BF00351327 (doi:10.1007/BF00351327) [DOI] [Google Scholar]
- 41.Bargelloni L., Marcato S., Zane L., Patarnello T. 2000. Mitochondrial phylogeny of notothenioids: a molecular approach to Antarctic fish evolution and biogeography. Syst. Biol. 49, 114–129 10.1080/10635150050207429 (doi:10.1080/10635150050207429) [DOI] [PubMed] [Google Scholar]
- 42.Jarman S. N., Elliot N. G., McMinn A. 2002. Genetic differentiation in the Antarctice coastal krill Euphausia crystallorophias. Heredity 88, 280–287 10.1038/sj.hdy.6800041 (doi:10.1038/sj.hdy.6800041) [DOI] [PubMed] [Google Scholar]
- 43.Page T. J., Linse K. 2002. More evidence of speciation and dispersal across the Antarctic Polar Front through molecular systematics of Southern Ocean Limatula (Bivalvia: Limidae). Polar Biol. 25, 818–826 10.1007/s00300-002-0414-7 (doi:10.1007/s00300-002-0414-7) [DOI] [Google Scholar]
- 44.Ritchie P. A., Millar C. D., Gibb G. C., Baroni C., Lambert D. M. 2004. Ancient DNA enables timing of the Pleistocene origin and Holocene expansion of two Adélie penguin lineages in Antarctica. Mol. Biol. Evol. 21, 240–248 10.1093/molbev/msh012 (doi:10.1093/molbev/msh012) [DOI] [PubMed] [Google Scholar]
- 45.Baker A. J., Pereira S. L., Haddrath O. P., Edge K. E. 2006. Multiple gene evidence for expansion of extant penguins out of Antarctica due to global cooling. Proc. R. Soc. B 273, 11–17 10.1098/rspb.2005.3260 (doi:10.1098/rspb.2005.3260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogers A. D. 2007. Evolution and biodiversity of Antarctic organisms: a molecular perspective. Phil. Trans. R. Soc. B 362, 2191–2214 10.1098/rstb.2006.1948 (doi:10.1098/rstb.2006.1948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunter R. L., Halanych K. M. 2008. Evaluating connectivity in the brooding brittle star Astrotoma agassizii across the Drake Passage in the Southern Ocean. J. Hered. 99, 137–148 10.1093/jhered/esm119 (doi:10.1093/jhered/esm119) [DOI] [PubMed] [Google Scholar]
- 48.Thornhill D. J., Mahon A. R., Norenburg J. L., Halanych K. M. 2008. Open-ocean barriers to dispersal: a test case with the Antarctic Polar Front and the ribbon worm Parborlasia corrugatus (Nermetea: Lineidae). Mol. Ecol. 17, 5104–5117 10.1111/j.1365-294X.2008.03970.x (doi:10.1111/j.1365-294X.2008.03970.x) [DOI] [PubMed] [Google Scholar]
- 49.Fraser C. I., Nikula R., Spencer H. G., Waters J. M. 2009. Kelp genes reveal effects of subAntarctic sea ice during the Last Glacial Maximum. Proc. Natl Acad. Sci. USA 106, 3249–3253 10.1073/pnas.0810635106 (doi:10.1073/pnas.0810635106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.González-Wevar C. A., Nakano T., Cañete J. I., Poulin E. 2010. Molecular phylogeny and historical biogeography of Nacella (Patellogastropoda: Nacellidae) in the Southern Ocean. Mol. Phylogen. Evol. 56, 115–124 10.1016/j.ympev.2010.02.001 (doi:10.1016/j.ympev.2010.02.001) [DOI] [PubMed] [Google Scholar]
- 51.Pugh P. J. A. 1993. A synonymic catalogue of the Acari from Antarctica, the sub-Antarctic islands and the Southern Ocean. J. Nat. Hist. 27, 323–421 10.1080/00222939300770171 (doi:10.1080/00222939300770171) [DOI] [Google Scholar]
- 52.Marshall D. J., Convey P. 2004. Latitudinal variation in habitat specificity of ameronothrid mites (Oribatida). Exp. Appl. Acarol. 34, 21–35 10.1023/b:appa.0000044437.17333.82 (doi:10.1023/b:appa.0000044437.17333.82) [DOI] [PubMed] [Google Scholar]
- 53.Minto L. B., Shepherd G. J., Usher M. B. 1991. The cryptostigmatid mite Halozetes belgicae (Michael) in the maritime Antarctic. Antarct. Sci. 3, 53–59 [Google Scholar]
- 54.Young S. R., Block W. 1980. Experimental studies on the cold tolerance of Alaskozetes antarcticus. J. Insect Physiol. 26, 189–200 10.1016/0022-1910(80)90080-3 (doi:10.1016/0022-1910(80)90080-3) [DOI] [Google Scholar]
- 55.Convey P. 1994. Growth survival strategy of the Antarctic mite Alaskozetes antarcticus. Ecography 17, 97–107 10.1111/j.1600-0587.1994.tb00081.x (doi:10.1111/j.1600-0587.1994.tb00081.x) [DOI] [Google Scholar]
- 56.Hayward S. A. L., Bale J. S., Worland M. R., Convey P. 2001. Influence of temperature on the hygropreference of the collembolan, Cryptopygus antarcticus, and the mite, Alaskozetes antarcticus from the maritime Antarctic. J. Insect Physiol. 47, 11–18 10.1016/S0022-1910(00)00088-3 (doi:10.1016/S0022-1910(00)00088-3) [DOI] [PubMed] [Google Scholar]
- 57.Worland M. R., Convey P. 2001. Rapid cold hardening in Antarctic microarthropods. Funct. Ecol. 15, 515–524 10.1046/j.0269-8463.2001.00547.x (doi:10.1046/j.0269-8463.2001.00547.x) [DOI] [Google Scholar]
- 58.Deere J. A., Chown S. L. 2006. Testing the beneficial acclimation hypothesis and its alternatives for locomotor performance. Am. Nat. 168, 630–644 10.1086/508026 (doi:10.1086/508026) [DOI] [PubMed] [Google Scholar]
- 59.Deere J. A., Sinclair B. J., Marshall D. J., Chown S. L. 2006. Phenotypic plasticity of thermal tolerances in five oribatid mite species from sub-Antarctic Marion Island. J. Insect Physiol. 52, 693–700 10.1016/j.jinsphys.2006.03.009 (doi:10.1016/j.jinsphys.2006.03.009) [DOI] [PubMed] [Google Scholar]
- 60.Hawes T. C., Bale J. S., Worland M. R., Convey P. 2007. Plasticity and superplasticity in the acclimation potential of the Antarctic mite Halozetes belgicae (Michael). J. Exp. Biol. 210, 593–601 10.1242/jeb.02691 (doi:10.1242/jeb.02691) [DOI] [PubMed] [Google Scholar]
- 61.Benoit J. B., Yoder J. A., Lopez-Martinez G., Elnitsky M. A., Lee R. E., Denlinger D. L. 2008. Adaptations for the maintenance of water balance by three species of Antarctic mites. Polar Biol. 31, 539–547 10.1007/s00300-007-0385-9 (doi:10.1007/s00300-007-0385-9) [DOI] [Google Scholar]
- 62.Subias L. S. 2008. Listado sistematico, sinonimico y biogeografico de los acaros oribatidos (Acariformes: Oribatida) del Mundo (Excepto fosiles). Graellsia 60, 3–305 See http://www.ucm.es/info/zoo/Artropodos/Catalogo.(2004) [Google Scholar]
- 63.Wallwork J. A. 1962. A redescription of Notaspis antarctica Michael, 1903 (Acari: Oribatei). Pac. Insects 4, 869–890 [Google Scholar]
- 64.Wallwork J. A. 1967. Cryptostigmata (Oribatid mites). In Entomology of Antarctica (ed. Gressitt J. L.), pp. 105–122 University of Antarctic Research Series, vol. 10 Washington, DC: American Geophysical Society [Google Scholar]
- 65.Starý J., Block W. 1998. Distribution and biogeography of oribatid mites (Acari: Oribatida) in Antarctica, the sub-Antarctic islands and nearby land areas. J. Nat. Hist. 32, 861–894 10.1080/00222939800770451 (doi:10.1080/00222939800770451) [DOI] [Google Scholar]
- 66.Marshall D. J., Procheş Ş. 2007. The origins of marine mites: interpreting geographical and ecological patterns. In Acarology XI: proceedings of the international congress (eds Morales-Malacara J. B., Behan-Pelletier V., Ueckermann E., Perez T. M., Estrada-Venegas E. G., Badii M.), pp. 97–103 Mexico city, Mexico: Instituto de Biologia and Facultad de Ciencias, Universidad Nacional Autonoma de Mexico [Google Scholar]
- 67.Maniatis T., Fritsch E., Sambrook J. 1982. Molecular cloning. New York, NY: Cold Spring Harbor [Google Scholar]
- 68.Mortimer E., Jansen van Vuuren B. 2007. Phylogeography of Eupodes minutus (Acari: Prostigmata) on sub-Antarctic Marion Island reflects the impact of historical events. Polar Biol. 30, 471–476 10.1007/s00300-006-0205-7 (doi:10.1007/s00300-006-0205-7) [DOI] [Google Scholar]
- 69.Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299 [PubMed] [Google Scholar]
- 70.Colgar D. J., McLauchlan A., Wilson G. D. F., Livingston S. P., Edgecombe G. D., Macaranas J., Cassis G., Gray M. R. 1998. Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Aust. J. Zool. 46, 419–437 10.1071/ZO98048 (doi:10.1071/ZO98048) [DOI] [Google Scholar]
- 71.Akhmanova A. S., Bindels P. C., Xu J., Miedema K., Kremer H., Hennig W. 1995. Structure and expression of histone H3.3 genes in Drosophila melanogaster and Drosophila hydei. Genome 38, 586–600 10.1139/g95-075 (doi:10.1139/g95-075) [DOI] [PubMed] [Google Scholar]
- 72.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 10.1093/nar/25.24.4876 (doi:10.1093/nar/25.24.4876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Swofford D. 2000. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland, MA: Sinauer Associates [Google Scholar]
- 74.Nylander J. A. A. 2004. MrModeltest version 2. Program distributed by the author. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University [Google Scholar]
- 75.Burnham K. P., Anderson D. R. 2002. Model selection and multimodel inference: a practical-theoretic approach, 2nd edn. Berlin, Germany: Springer [Google Scholar]
- 76.Nylander J. A. A., Ronquist F., Huelsenbeck J. P., Nieves-Aldrey J. L. 2004. Bayesian phylogenetic analysis of combined data. Syst. Biol. 53, 47–67 10.1080/10635150490264699 (doi:10.1080/10635150490264699) [DOI] [PubMed] [Google Scholar]
- 77.Ronquist F., Huelsenbeck J. 2003. MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 78.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dunlop J. A. 1996. A trigonotarbid arachnid from the upper Silurian of Shropshire. Palaeontology 39, 605–614 [Google Scholar]
- 80.Selden P. A., Baker A. S., Phipps K. J. 2008. An oribatid mite (Arachnida: Acari) from the Oxford clay (Jurassic: upper Callovian) of South Cave Station Quarry, Yorkshire, UK. Palaeontology 51, 623–633 10.1111/j.1475-4983.2008.00769.x (doi:10.1111/j.1475-4983.2008.00769.x) [DOI] [Google Scholar]
- 81.Edgecombe G. D. 2010. Arthropod phylogeny: an overview from the perspectives of morphology, molecular data and the fossil record. Arthropod Struct. Dev. 39, 74–87 10.1016/j.asd.2009.10.002 (doi:10.1016/j.asd.2009.10.002) [DOI] [PubMed] [Google Scholar]
- 82.Sanders K. L., Lee M. S. Y. 2010. Arthropod molecular divergence times and the Cambrian origin of pentastomids. Syst. Biodivers. 8, 63–74 10.1080/14772000903562012 (doi:10.1080/14772000903562012) [DOI] [Google Scholar]
- 83.Schaefer I., Norton R. A., Scheu S., Maraun M. 2010. Arthropod colonization of land—linking molecules and fossils in oribatid mites (Acari, Oribatida). Mol. Phylogen. Evol. 57, 113–121 10.1016/j.ympev.2010.04.015 (doi:10.1016/j.ympev.2010.04.015) [DOI] [PubMed] [Google Scholar]
- 84.Near T. J., Sanderson M. J. 2004. Assessing the quality of molecular divergence time estimates by fossil calibrations and fossil-based model selection. Phil. Trans. R. Soc. Lond. B 359, 1477–1483 10.1098/rstb.2004.1523 (doi:10.1098/rstb.2004.1523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grobler G. C., Janse van Rensburg L., Bastos A. D. S., Chimimba C. T., Chown S. L. 2006. Molecular and morphometric assessment of the taxonomic status of Ectemnorhinus weevil species (Coleoptera: Curculionidae, Entiminae) from the sub-Antarctic Prince Edward Islands. J. Zool. Syst. Evol. Res. 44, 200–211 10.1111/j.1439-0469.2006.00358.x (doi:10.1111/j.1439-0469.2006.00358.x) [DOI] [Google Scholar]
- 86.Grobler G. C., Bastos A. D. S., Chimimba C. T., Chown S. L. Submitted Inter-island dynamics in a flightless weevil, Bothrometopus parvulus (Coleoptera: Curculionidae) from the sub-Antarctic Prince Edward Islands. [Google Scholar]
- 87.Cicconardi F., Nardi F., Emerson B. C., Frati F., Fanciulli P. P. 2010. Deep phylogeographic divisions and long-term persistence of forest invertebrates (Hexapoda: Collembola) in the north-western Mediterranean basin. Mol. Ecol. 19, 386–400 10.1111/j.1365-294X.2009.04457.x (doi:10.1111/j.1365-294X.2009.04457.x) [DOI] [PubMed] [Google Scholar]
- 88.Salomone N., Emerson B. C., Hewitt G. M., Bernini F. 2002. Phylogenetic relationships among the Canary Island Steganacaridae (Acari, Oribatida) inferred from mitochondrial DNA sequence data. Mol. Ecol 11, 79–89 10.1046/j.0962-1083.2001.01421.x (doi:10.1046/j.0962-1083.2001.01421.x) [DOI] [PubMed] [Google Scholar]
- 89.Heethoff M., Domes K., Laumann M., Maraun M., Norton R. A., Scheu S. 2007. High genetic divergences indicate ancient separation of parthenogenetic lineages of the oribatid mite Platynothrus peltifer (Acari, Oribatida). Eur. Soc. Evol. Biol. 20, 392–402 10.1111/j.1420-9101.2006.01183.x (doi:10.1111/j.1420-9101.2006.01183.x) [DOI] [PubMed] [Google Scholar]
- 90.McGaughran A., Hogg I. D., Stevens M. I. 2008. Patterns of population genetic structure for springtails and mites in southern Victoria Land, Antarctica. Mol. Phylogen. Evol. 46, 606–618 10.1016/j.ympev.2007.0.003 (doi:10.1016/j.ympev.2007.0.003) [DOI] [PubMed] [Google Scholar]
- 91.Ronquist F. 1997. Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Syst. Biol. 46, 195–203 10.1093/sysbio/46.1.195 (doi:10.1093/sysbio/46.1.195) [DOI] [Google Scholar]
- 92.Ree R. H., Moore B. R., Webb C. O., Donoghue M. J. 2005. A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution 59, 2299–2311 10.1111/j.0014-3820.2005.tb00940.x (doi:10.1111/j.0014-3820.2005.tb00940.x) [DOI] [PubMed] [Google Scholar]
- 93.Brooks D. R., McLennan D. A. 2001. A comparison of a discovery-based and an event-based method of historical biogeography. J. Biogeogr. 28, 757–768 10.1046/j.1365-2699.2001.00598.x (doi:10.1046/j.1365-2699.2001.00598.x) [DOI] [Google Scholar]
- 94.Cook L. G., Crisp M. D. 2005. Directional asymmetry of long-distance dispersal and colonization could mislead reconstructions of biogeography. J. Biogeogr. 32, 741–754 10.1111/j.1365-2699.2005.01261.x (doi:10.1111/j.1365-2699.2005.01261.x) [DOI] [Google Scholar]
- 95.Yuan Y., Wohlhauser S., Moeller M., Klackenberg J., Callmander M. W., Kuepfer P. 2005. Phylogeny and biogeography of Exacum (Gentianaceae): a disjunctive distribution in the Indian Ocean basin resulted from long distance dispersal and extensive radiation. Syst. Biol. 54, 21–34 10.1080/10635150590905867 (doi:10.1080/10635150590905867) [DOI] [PubMed] [Google Scholar]
- 96.Convey P., Pugh P. J. A., Jackson C., Murray A. W., Ruhland C. T., Xiong F. S., Day T. A. 2002. Response of Antarctic terrestrial arthropods to multifactorial climate manipulation over a four year period. Ecology 83, 3130–3140 10.1890/0012-9658(2002)083[3130:roatmt]2.0.co;2 (doi:10.1890/0012-9658(2002)083[3130:roatmt]2.0.co;2) [DOI] [Google Scholar]
- 97.Convey P., Block W., Peat H. J. 2003. Soil arthropods as indicators of water stress in Antarctic terrestrial habitats? Global Change Biol. 9, 1718–1730 10.1046/j.1365-2486.2003.00691.x (doi:10.1046/j.1365-2486.2003.00691.x) [DOI] [Google Scholar]
- 98.Chown S. L., Slabber S., McGeoch M. A., Janion C., Leinaas H. P. 2007. Phenotypic plasticity mediates climate change responses among invasive and indigenous arthropods. Proc. R. Soc. B 274, 2531–2537 10.1098/rspb.2007.0772 (doi:10.1098/rspb.2007.0772) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bokhorst S., Huiskes A., Convey P., Van Bodegom P. M., Aerts R. 2008. Climate change effects on soil arthropod communities from the Falkland Islands and the Maritime Antarctic. Soil Biol. Biochem. 40, 1547–1556 10.1016/j.soilbio.2008.01.017 (doi:10.1016/j.soilbio.2008.01.017) [DOI] [Google Scholar]
- 100.Johnson J. S., Smellie J. L., Nelson A. E., Stuart F. M. 2009. History of the Antarctic Peninsula Ice Sheet since the early Pliocene—evidence from cosmogenic dating of Pliocene lavas on James Ross Island, Antarctica. Global Planet. Change 69, 205–213 10.1016/j.gloplacha.2009.09.001 (doi:10.1016/j.gloplacha.2009.09.001) [DOI] [Google Scholar]
- 101.Bellemain E., Ricklefs R. E. 2008. Are islands the end of the colonization road? Trends Ecol. Evol. 23, 461–468 10.1016/j.tree.2008.05.001 (doi:10.1016/j.tree.2008.05.001) [DOI] [PubMed] [Google Scholar]
- 102.Coetzee L., Marshall D. J. 2003. A new Halozetes species (Acari: Oribatida: Ameronothridae) from the marine littoral of southern Africa. Afr. Zool. 38, 327–331 [Google Scholar]
- 103.Muñoz J., Felicísimo A. M., Cabezas F., Burgaz A. R., Martínez I. 2004. Wind as a long-distance dispersal vehicle in the Southern Hemisphere. Science 304, 1144–1147 10.1126/science.1095210 (doi:10.1126/science.1095210) [DOI] [PubMed] [Google Scholar]
- 104.McGlone M. S. 2005. Goodbye Gondwana. J. Biogeogr. 32, 739–740 10.1111/j.1365-2699.2005.01278x (doi:10.1111/j.1365-2699.2005.01278x) [DOI] [Google Scholar]
- 105.Wallwork J. A. 1965. The Cryptostigmata (Acari) of Antarctica with special reference to the Antarctic Peninsula and South Shetland Islands. Pacific Insects 7, 453–468 [Google Scholar]
- 106.Chown S. L. 1994. Historical ecology of sub-Antarctic weevils: patterns and processes on isolated islands. J. Nat. Hist. 28, 411–433 10.1080/00222939400770191 (doi:10.1080/00222939400770191) [DOI] [Google Scholar]
- 107.Boelhouwers J. C., Meiklejohn I. K., Holness S. D., Hedding D. W. 2008. Geology, geomorphology and change. In The Prince Edward Islands: land–sea interactions in a changing ecosystem (eds Chown S. L., Froneman P. W.), pp. 65–96 Stellenbosch, South Africa: Sun Press [Google Scholar]