Abstract
Technological developments allow increasing numbers of markers to be deployed in case-control studies searching for genetic factors that influence disease susceptibility. However, with vast numbers of markers, true ‘hits’ may become lost in a sea of false positives. This problem may be particularly acute for infectious diseases, where the control group may contain unexposed individuals with susceptible genotypes. To explore this effect, we used a series of stochastic simulations to model a scenario based loosely on bovine tuberculosis. We find that a candidate gene approach tends to have greater statistical power than studies that use large numbers of single nucleotide polymorphisms (SNPs) in genome-wide association tests, almost regardless of the number of SNPs deployed. Both approaches struggle to detect genetic effects when these are either weak or if an appreciable proportion of individuals are unexposed to the disease when modest sample sizes (250 each of cases and controls) are used, but these issues are largely mitigated if sample sizes can be increased to 2000 or more of each class. We conclude that the power of any genotype–phenotype association test will be improved if the sampling strategy takes account of exposure heterogeneity, though this is not necessarily easy to do.
Keywords: tuberculosis, association study, power, infectious disease, heterozygosity, population simulation
1. Introduction
Understanding the genetic basis of susceptibility to disease has become an increasingly important target for research. Moreover, with the publication of complete genome sequences for humans and other species, the development of literally millions of polymorphic markers [1,2] and emerging technologies that make it feasible to collect genetic data at unprecedented rates, we appear to be faced with an embarrassment of riches. However, progress remains slower than many might predict [3], raising the question of why this power is not more effective.
Studies aimed at uncovering genetic predispositions to disease usually attempt to demonstrate an association between the genotype at one or more polymorphic markers and a phenotype related to disease susceptibility [4]. There are two main approaches, one based on candidate genes (CG) [5,6], the other based on testing the entire genome (genome-wide association (GWA)). Both approaches enjoy a combination of benefits and drawbacks. Broadly, CG studies tend to have rather high statistical power but are incapable of discovering new genes or gene combinations, while GWA studies can pinpoint genes regardless of whether their function was known before [7] but have low power owing to the number of independent tests performed [3,8,9]. Indeed, the problem of false positives, already an issue in early studies deploying a few hundred microsatellite markers, is becoming acute as we move into the era when single nucleotide polymorphisms (SNPs) are replacing microsatellites and more than a million markers may be used. With one to two million SNPs it is estimated that an alpha level of 5 × 10−8 is required [3], though this number may be reduced by using a subset of ‘tag SNPs’ whose genotypes correlate strongly with those of neighbouring loci [10,11].
Recent reviews have reached contrasting conclusions about the effectiveness of CG compared with GWA approaches. One analysis asks whether GWA studies ‘are a waste of time’ since even with thousands of samples the approach can be underpowered [12]. However, SNP-based GWA studies in humans have had considerable success in identifying regions that are important in disease development in such conditions as diabetes [13], Crohn's disease [14] and other autoimmune and genetic conditions. By contrast, GWA studies have had relatively limited success in finding novel genes involved in susceptibility to infectious diseases. One reason for this may lie with a number of added complexities that infectious diseases imply, including the key element of exposure. Only exposed individuals can contract the disease, meaning that even highly susceptible genotypes can be found in unaffected individuals. Moreover, exposure is often correlated with other factors such as age and behaviour.
Issues such as exposure imply studies of genetic susceptibility to infectious disease will have modest effect sizes and hence that statistical power will be at a premium. If so, then the greater inherent power of CG may make it preferable over GWA. This expectation seems partially fulfilled, in that small-scale studies in both human [15] and non-human systems have proved surprisingly effective in identifying genomic regions associated with disease susceptibility [16–18]. The non-human studies are particularly interesting because as few as around 10 markers have revealed rather convincing links to immune-related genes such as methylmalonyl-CoA [19,20], despite often not being initially selected for proximity to CGs. This apparent success probably reflects more than just the reduced need for multiple testing. Thus, microsatellites have higher variability, increasing the chance that one allele shows strong linkage disequilibrium with a causative gene allele; microsatellites are selected for high polymorphism, increasing the chance that such markers lie near genes experiencing balancing selection [21]; genetic effects in natural populations may be much stronger than in humans owing to the absence of medical intervention.
The problems associated with finding genes that influence susceptibility to infectious diseases are nicely illustrated by tuberculosis, an important disease caused by various species of Mycobacteria and affecting many species including humans and cattle [22,23]. In the UK cattle herd, bovine tuberculosis (bTB) is monitored using a skin test that is less than 100 per cent effective [24]. Cows testing positive (‘reactors’) may variously have a current infection, a latent infection, have previously resisted infection or were infected but are now recovered. Exposure rates also vary widely with factors such as farm management practice [25], proximity to infected badgers [26] and geography. Untangling the influence of genetics versus exposure is difficult. Thus, a farm may lack disease because it has no exposure or because its (related) cattle mostly carry an allele that confers sufficient resistance. Conversely, farms may have high incidence because of high exposure or high genetic susceptibility.
Attempts to calculate statistical power in association studies in order to determine the optimum sample size for individuals and markers have generated a large literature [11,27,28]. However, analytical calculations of power necessarily have to ignore a range of real, stochastic complexities such as variation in exposure rate, variation in recombination rate, the impact of local selection pressure, population structure and the genetic background of the original causative mutation. Consequently, for human studies, the tendency is to assume the use of SNPs and then to exploit the wealth of HapMap data to estimate parameters such as average levels of linkage disequilibrium [10,29,30]. However, for non-human systems and microsatellite data, these calculations have little relevance. Moreover, most power calculations assume that the most resistant genotype is homozygous [28,31], as do methods used to correct for population structure [32], when for infectious diseases it may well be the heterozygote that is most resistant. Thus, current methods for calculating power appear poorly suited to studies of infectious disease, particularly in non-human systems.
In view of the above, we decided to conduct a simulation-based study to assess the relative power of GWA and CG approaches to detect genetic susceptibility to infectious disease in the presence of varying exposure rates. We consider both microsatellite and SNP markers and explore the relative chance that an informative marker lies near to a susceptibility factor or is, in the case of a SNP, the functional mutation itself. Although inspired by studies focused on bTB, our approach has implications for any case–control studies where factors such as variable disease exposure cause individuals with susceptible genotypes to be included in the control group.
2. Rationale and methods
We consider a single gene affecting the probability that an individual exposed to a pathogen contracts the disease. Individuals are deemed either ‘susceptible’ or ‘resistant’ and may be either ‘exposed’ or ‘unexposed’ to the disease. Unexposed individuals do not get the disease while exposed susceptible individuals always do. Exposed resistant individuals become infected with a probability that can be varied to simulate different levels of genetic benefit. We refer to this as the resistant infected fraction (RIF). Naturally, susceptible individuals may not be 100 per cent susceptible, but in our model, such individuals are treated as unexposed: effectively, exposure is normalized to the point where all susceptible individuals become infected.
Genetic susceptibility is assumed to be dictated by a single gene with two alleles where heterozygotes are resistant and homozygotes are susceptible. Other scenarios are possible, but are difficult to simulate because any directional selection (i.e. the disease influences survival/reproduction) tends to lead to the elimination of susceptible alleles. Near the gene is a single locus, designated the marker, that mutates according to a strict stepwise mutation model. By varying the mutation rate, it is possible to select scenarios where the population carries three genotypes, taken as equivalent in informativeness to a SNP, or more than 10 genotypes, taken as representing a microsatellite. In each simulation, a population of 1000 diploid individuals is initialized with a single marker allele and random gene alleles (0 and 1, each with p = 0.5). Evolution progresses under random mating, selection (heterozygote fitness = 1, homozygote fitness = 0.75), mutation at the marker and recombination (range examined 10−3–10−5 per generation).
At the end of each simulation, the frequencies of each compound genotype (gene and marker) are determined and, within each, the expected frequency of diseased animals determined according to the risk factors of gene genotype, exposure and RIF. Based on these frequencies, a sample of N cases and N controls is generated deterministically and a simple χ2-test used to test for a significant difference in genotype frequencies between cases and controls. Using the same terminal genotypes, this process is then repeated across a range of exposures (range tested = 0.1 − 0.9 in steps of 0.1) and a range of RIF values (range tested = 0.1 − 0.9 in steps of 0.1). Simulations are repeated 100 times for each recombination rate, yielding a per cent score for how often the χ2-test yielded a significant association. For significance, we use an alpha level equivalent to p = 0.05 after correction for the number of markers tested, i.e. with X markers, we use α = 1/20X. Finally, to simulate a causative SNP in the gene itself, the same process is repeated, but this time using the genotype of the gene instead of the marker. For GWA we assumed 50 000 SNPs, while for the CG study we assume 10 candidate markers. The stochastic simulation program was coded in C++, using Mersenne Twister random number generation.
Using many SNPs invokes an important trade-off. Using more SNPs increases the chance that the functional SNP is included, but reduces power owing to an increase in false positives. To obtain a measure of whether there is an optimal number of SNPs, we simulated this scenario. We assumed a maximum of five million SNPs in the genome and that a study uses somewhere between 2000 and 2 000 000 randomly selected SNPs. The probability of detecting an effect is then taken as 1−(1 − PQ)(1 − RS), where P is the probability that the functional SNP is included in the marker set, Q is the probability that the functional SNP itself detects a significant association at alpha (see above), R is the probability that at least one SNP is close enough to the gene to be linked by a recombination rate of 10−5, and S is the probability that a linked SNP detects a significant association. Since cattle exhibit high levels of linkage disequilibrium owing to their small effective population size [33], we conservatively equate a recombination rate of 10−5 with a distance of 0.1 Mb.
3. Results
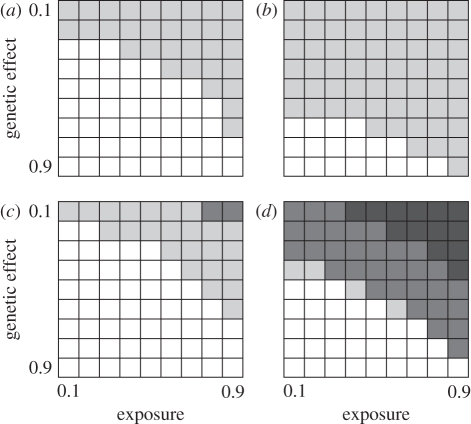
Figure 1 summarizes the results of our simulations for the two extreme recombination rates, 10−3 and 10−5, and the two marker categories, SNPs (= three genotypes) and microsatellites (10+ genotypes). Several trends are apparent. First, as expected, higher exposure rates and higher effect sizes (difference in susceptibility between susceptible and resistant genotypes) both act to increase the chance that a significant association is detected. Noticeably, at exposure rates below around 30 per cent only very strong genetic effects (twofold or greater risk) are likely to be detected. Second, low recombination rates promote detection, with the 10−3 level requiring almost deterministic infection where genetic resistance is higher than 70 per cent. Third, where associations are detected, microsatellites tend to be around twice as likely as a SNP to detect an association, the exception being around the borderline of detection, where SNPs perform somewhat better. We interpret this as reflecting the χ2-test; for medium-strength associations, the high genotype diversity helps increases power, but when the effect is weak, the loss of degrees of freedom becomes critical.
Figure 1.
How the probability of finding a significant genotype–phenotype association for an infectious disease varies with marker type, exposure rate, recombination rate and size of genetic effect. Each cell contains the proportion of approximately 100 replicate simulations that yielded an experiment-wide significant association at α = 5%. Greyscale represents: white = 0–5% significant; light grey = 5–50% significant; dark grey = 50–90% significant; black = 90–100% significant. Exposure is the proportion of individuals exposed to the disease, genetic effect is the probability of an exposed, genetically resistant individual catching the disease relative to an exposed, susceptible individual (p = 1). (a) ‘SNPs’ (=marker carries three genotypes), recombination rate = 10−3; (b) ‘SNPs’, recombination rate = 10−5; (c) ‘microsatellites’ (=marker carries 10+ genotypes), recombination rate = 10−3; (d) ‘microsatellites’, recombination rate = 10−5.
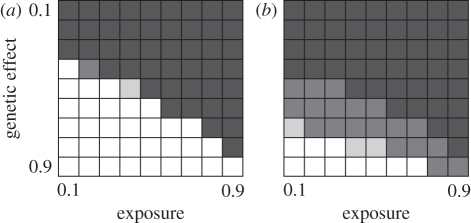
Figure 2a summarizes the results of a simulated GWA study in which we assume that 50 000 SNPs are deployed and include the functional mutation itself. Here, a rather similar region of the parameter space is filled with zeros compared with the 10−5 recombination rate simulations. By implication, when the genetic effect is weak and/or the exposure rate is low, throwing more SNPs at the problem has relatively little impact on the chance of finding a significant association. However, almost all the non-zero cells contain ‘1’s, indicating that where the approach can detect an association, it almost invariably does so. Compared with linked markers, there are few grey areas where some studies will find an effect and others not.
Figure 2.
How the probability of finding a significant genotype–phenotype association for an infectious disease varies with exposure rate and size of genetic effect. (a) Results for a panel of 50 000 SNPs that includes the functional mutation. (b) Results for a CG approach using a sample size of 4000 (equal numbers of cases and controls). For details of simulations and greyscale, see legend of figure 1.
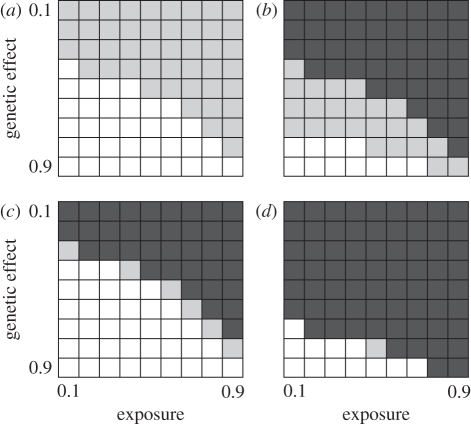
Finally, we considered the impact of using larger sample sizes and the way the joint probability that a SNP is either in strong linkage disequilibrium with the functional gene or is the gene mutation itself varies with SNP number. Figure 2b summarizes results for a CG approach with 10 microsatellites and a sample size increased from 500 to 4000. Only the weakest genetic effects are not detected with high probability and exposure rates have reduced influence. Figure 3 summarizes the results for simulations based either on 50 000 or two million SNPs for the two different sample sizes. Increasing the number of SNPs to two million brings some benefit, though arguably not commensurate with the added empirical effort. As with the CG approach, increasing sample sizes brings much greater benefit, with a much larger proportion of scenarios yielding good to high power. Noticeably, for an equivalent sample size, the CG approach generally offers higher power.
Figure 3.
Effectiveness of a genome-wide association (GWA) study to reveal a genotype–phenotype association with sample sizes of 500 and 4000. In each set of simulations, we assume a large, SNP-based study deploying either 50 000 or two million markers. The probability of finding an association is calculated by combining the probability of one marker lying close enough to the functional gene for the recombination rate to be 10−5 with the probability that the functional mutation itself is included in the panel. For other details, see legend of figure 1. (a) 50 000 SNPs, sample size = 500 (250 case plus 250 controls); (b) 50 000 SNPs, sample size = 4000; (c) two million SNPs, sample size = 500; (d) two million SNPs, sample size = 4000.
4. Discussion
We have used stochastic simulations to explore how the power of genetic association studies to detect genes that influence susceptibility to an infectious disease vary with factors such as exposure rates and the strength of genetic resistance/susceptibility. We find that when either exposure rates are very high or the genetic effects are strong, an association can be detected, but that many plausible scenarios are unlikely to be detected regardless of the experimental effort deployed. In our simulations, a CG approach tends to outperform a GWA approach, even when this involves literally millions of SNPs.
GWA studies are expensive and are currently outside the likely budget for most non-model species study systems. However, rapidly improving technology and falling prices mean that we are just reaching the point where such analyses can be contemplated and are indeed starting [34]. Consequently, it seems a good time to ask whether such investment is likely to yield results commensurate with the outlay, a question whose importance is emphasized by recent papers that suggest many GWA studies in humans may be underpowered, even with thousands of samples and excellent study design [12]. In non-human and in non-laboratory-based systems, the situation is likely to be worse because factors that would ideally be controlled often cannot be. This may be particularly true for infectious diseases, where both exposure and diagnosis potentially contribute statistical noise.
To illustrate this, we explore a system based loosely on bTB. Here, the ideal would be to control for exposure by using experimental infections, but this is unlikely to yield large-enough sample sizes for an association study. In reality, therefore, comparisons are made between cows that are unaffected and those that respond positively to an antigenic challenge. Exposure rates are unknown and appear to vary greatly between farms [25] and even individuals [26]. However, the true amount of variation in exposure rate is difficult to estimate because to do so currently requires an assumption that genetic factors exert a negligible impact, which seems likely to be false [34]. A further complication, mentioned above, is that the most frequently used test for infection is ambiguous, potentially indicating anything from disease exposure without properly contracting the disease, through contraction and full recovery to a current infection. Clearly, any genetically resistant cows that react in this test, either because they resisted infection or recovered from it, will undermine statistical power.
We show that the chance of detecting an association is profoundly affected by the proportion of individuals exposed to the pathogen. This makes intuitive sense because unexposed individuals cannot become infected regardless of their genotype, thereby adding noise to any underlying signal. Put more generally, for any given sample size and disease, power will be greatest when there is maximum correlation between genotype and disease status: sampling should aim to minimize individuals who have resistant genotypes but contract the disease through high exposure just as much as individuals with susceptible genotypes who are unaffected because they were never exposed. In the specific case of bTB, the optimal strategy is unclear because the critical interaction between exposure rates and genetics has yet to be resolved. We do not yet know whether high-incidence farms occur because of high exposure rates or because they carry many cattle with susceptible genotypes. For this reason, sampling strategies that strive to increase power by making a priori assumptions, for example, that zero-incidence farms should be excluded because they have not been exposed, may achieve their aims but run the risk of producing a false negative result (in this example, if most of the genetic effect is manifest as resistant genotypes carried mainly by cattle on low- or zero-incidence farms).
The above contrast between high and low breakdown farms illustrates two important points. First, resistance and susceptibility are not simply the opposite sides of the same coin. From a genetic perspective, what matters is which genotypes are common and which are rare: for bTB, if most cows are resistant, the key farms are those with high disease incidence, but if most cows are susceptible, the key farms are those where disease is rare or absent, even though some of these may be clean, with a genuine absence of the bacterium. Second, although good experimental design should avoid unexposed cattle, there is also a need to sample both relatively resistant and relatively susceptible genotypes. This introduces a quandary because the purest design would focus on a single farm with a single breed, but if these cows are all related [33] to the extent that they are all genetically either resistant or susceptible, genotype-fitness associations may be much reduced or even absent.
Comparing the CG and GWA approaches suggests a general advantage to CG, regardless of the number of SNPs being used. Several effects contribute to this overall picture. With a few thousand SNPs, the problem of false positives is small but so is the chance that one of the SNPs used is close to a particular gene. Moreover, even if a SNP is close enough, the higher diversity of microsatellites appears, under most circumstances, to increase the chance of finding an association, presumably because more alleles make it more likely that one is strongly indicative of a key allele at the gene. With medium numbers of SNPs, say 50 000, strong linkage disequilibrium with any given gene becomes likely, but correction for multiple testing reduces the power of the test, handing the advantage to a method that chooses the gene a priori. Finally, with a million SNPs or more, it becomes increasingly likely that one SNP is the actual functional mutation. When this happens, the effect size leaps and extremely small p-values tend to result. However, the probability of finding an experiment-wide significant association remains well below one because, unless all possible SNPs are deployed, every time the functional SNP is not included, the signal from the gene itself gets lost in a veritable sea of false positives.
To say that the CG approach is superior would overstate the case. If the list of target genes identified a priori fails to include the gene(s) that are important, this method will also fail. For example, an individual might become susceptible indirectly, through being in a poor nutritional state owing to a gene involved in metabolism, and such genes are unlikely to be included as candidates. Second, we have assumed a modest sample size of 500 samples. Increasing sample size increases statistical confidence, reducing the impact of false positives and making any given association more detectable. We tested this by repeating the simulations with 2000 cases and 2000 controls, finding an increase in the parameter space in which associations could be detected, with exposure rates having a reduced impact and only the weakest genetic effects (less than 30% fitness advantage) likely to pass undetected. Despite this, the relative performances of CG and GWA remain rather unchanged, with any given combination of parameter values being more likely to be detected by CG. This presumably reflects the fact that, if the primary functional gene is included as a candidate for CG, GWA studies only ever have an advantage when they include the functional mutation itself, and this depends on SNP number not sample size.
In conclusion, variable exposure rates present an important challenge to the experimental design of association studies. Greatest statistical power is gained by sampling exposed individuals across a range of genotypes, including both those that are relatively resistant and those that are susceptible. However, it is potentially dangerous to base a sampling strategy primarily on disease incidence assuming this to be a proxy for exposure, because the more this assumption is true, the weaker, and hence less detectable, the genetic effects will be. Our simulations suggest that CG approaches tend to offer some extra power compared with genome-wide tests, though this assumes prior knowledge of genes likely to play a role. This power gain is most likely to be important in smaller studies, since when thousands of samples can be analysed, all but weak genetic effects are likely to be detected by both approaches. Our work thus supports the handful of recent papers that question whether very large SNP-based studies offer the only viable approach [12], and instead suggests that much cheaper analyses based on microsatellites can be highly effective.
Acknowledgements
We thank the Editor and two anonymous referees for comments that helped improve the text.
References
- 1.Hinds D. A., Stuve L. L., Nilsen G. B., Halperin E., Eskin E., Balinger D. G., Frazer K. A., Cox D. R. 2005. Whole-genome patterns of common DNA variation in three human populations. Science 307, 1072–1079 10.1126/science.1105436 (doi:10.1126/science.1105436) [DOI] [PubMed] [Google Scholar]
- 2.The International HapMap Consortium 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–853 10.1038/nature06258 (doi:10.1038/nature06258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarthy M. I., Abecasis G. R., Cardon L. R., Goldstein D. B., Little J., Ionnidis J. P. A., Hirschhorn J. N. 2008. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9, 356–369 10.1038/nrg2344 (doi:10.1038/nrg2344) [DOI] [PubMed] [Google Scholar]
- 4.Schulze T. G., McMahon F. J. 2002. Genetic association mapping at the crossroads: which test and why? Overview and practical guidelines. Am. J. Med. Genet. (Neuropsych. Genet.) 114, 1–11 10.1002/ajmg.10042 (doi:10.1002/ajmg.10042) [DOI] [PubMed] [Google Scholar]
- 5.Altet L., Francimo O., Solano-Gallego L., Renier C., Sánchez A. 2002. Mapping and sequencing of the canine NRAMP1 gene and identification of mutations in Leishmaniasis-susceptible dogs. Infect. Immun. 70, 2763–2771 10.1128/IAI.70.6.2763-2771.2002 (doi:10.1128/IAI.70.6.2763-2771.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ridruechai C., et al. 2010. Association analysis of susceptibility candidate region on chromosome 5q31 for tuberculosis. Genes Immun. 11, 1–7 [DOI] [PubMed] [Google Scholar]
- 7.Cooke G. S., et al. 2008. Mapping of a novel susceptibility locus suggests a role for MC3R and CTSZ in human tuberculosis. Am. J. Respir. Crit. Care Med. 178, 203–207 10.1164/rccm.200710-1554OC (doi:10.1164/rccm.200710-1554OC) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 [Google Scholar]
- 9.Wu M. C., Kraft P., Epstein M. P., Taylor D. M., Chanock S. J., Hunter D. J., Lin X. 2010. Powerful SNP-set analysis for case–control genome-wide association studies. Am. J. Hum. Genet. 86, 929–942 10.1016/j.ajhg.2010.05.002 (doi:10.1016/j.ajhg.2010.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pe'er I., de Bakker P. I. W., Maller J., Yelensky R., Altschuler D., Daly M. J. 2006. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat. Genet. 38, 663–667 10.1038/ng1816 (doi:10.1038/ng1816) [DOI] [PubMed] [Google Scholar]
- 11.Schaid D. J. 2005. Power and sample size for testing associations of haplotypes with complex traits. Ann. Hum. Genet. 70, 116–130 10.1111/j.1529-8817.2005.00215.x (doi:10.1111/j.1529-8817.2005.00215.x) [DOI] [PubMed] [Google Scholar]
- 12.Yoo Y. J., Bull S. B., Paterson A. D., Waggott D., Sun L. & The Diabetes Control and Complications Trial/Edidemiology of Diabetes Interventions and Complications Research Group 2010. Were genome-wide linkage studies a waste of time? Exploiting candidate regions within genome-wide association studies. Genet. Epidemiol. 34, 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sladek R., et al. 2007. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445, 881–885 10.1038/nature05616 (doi:10.1038/nature05616) [DOI] [PubMed] [Google Scholar]
- 14.Barrett J. C., et al. 2008. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 40, 955–962 10.1038/ng.175 (doi:10.1038/ng.175) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyons E. J., et al. 2009. Homozygosity and risk of childhood death due to invasive bacterial disease. BMC Med. Genet. 10, 55. 10.1186/1471-2350-10-55 (doi:10.1186/1471-2350-10-55) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acevedo-Whitehouse K., Spraker T. R., Lyons E., Melin S. R., Gulland F., DeLong R. L., Amos W. 2006. Contrasting effects of heterozygosity on survival and hookworm resistance in California sealion pups. Mol. Ecol. 15, 1973–1982 10.1111/j.1365-294X.2006.02903.x (doi:10.1111/j.1365-294X.2006.02903.x) [DOI] [PubMed] [Google Scholar]
- 17.Amos W., Acevedo-Whitehouse K. A. 2009. A new test for genotype—fitness associations reveals a single microsatellite allele that strongly predicts the nature of tuberculosis infections in wild boar. Mol. Ecol. Res. 9, 1102–1111 10.1111/j.1755-0998.2009.02560.x (doi:10.1111/j.1755-0998.2009.02560.x) [DOI] [PubMed] [Google Scholar]
- 18.Luikart G., Pilgrim K., Visty J., Ezenwa V. O., Schwartz M. K. 2008. Candidate gene microsatellite variation is associated with parasitism in bighorn sheep. Biol. Lett. 4, 228–231 10.1098/rsbl.2007.0633 (doi:10.1098/rsbl.2007.0633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naranjo V., Acevedo-Whitehouse K. A., Vicente J., Gortazar C., de la Fuente J. 2008. Influence of methylmalonyl-CoA mutase alleles on resistance to bovine tuberculosis in the European wild boar (Sus scrofa). Anim. Genet. 39, 316–320 10.1111/j.1365-2052.2008.01725.x (doi:10.1111/j.1365-2052.2008.01725.x) [DOI] [PubMed] [Google Scholar]
- 20.Naranjo V., Villar M., Martín-Hernando M. P., Vidal D., Höfle U., Gortazar C., Kocan K. M., Vázquez J., de la Fuente J. 2007. Proteomic and transcriptome analyses of differential stress/inflammatory responses in mandibular lymph nodes and oropharyngeal tonsils of European wild boars infected with Mycobacterium bovis. Proteomics 7, 220–231 10.1002/pmic.200600527 (doi:10.1002/pmic.200600527) [DOI] [PubMed] [Google Scholar]
- 21.Hoffman J. I., Forcada J., Amos W. 2010. Exploring the mechanisms underlying a heterozygosity-fitness correlation for canine size in the Antarctic fur seal Arctocephalus gazella. J. Hered. 101, 539–552 10.1093/jhered/esq046 (doi:10.1093/jhered/esq046) [DOI] [PubMed] [Google Scholar]
- 22.Allen A. R., Minozzi G., Glass E. J., Skuce R. A., McDowell S. W. J., Woolliams J. A., Bishop S. C. 2010. Bovine tuberculosis: the genetic basis of host susceptibility. Proc. R. Soc. B 277, 2737–2745 10.1098/rspb.2010.0830 (doi:10.1098/rspb.2010.0830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellamy R., Ruwende C., Corrah T., McAdam K. P., Whittle H. C., Hill A. V. 1998. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N. Engl. J. Med. 338, 640–644 10.1056/NEJM199803053381002 (doi:10.1056/NEJM199803053381002) [DOI] [PubMed] [Google Scholar]
- 24.Doherty M. L., Cassidy J. P. 2002. New perspectives on bovine tuberculosis. Vet. J. 163, 109–110 10.1053/tvjl.2002.0654 (doi:10.1053/tvjl.2002.0654) [DOI] [PubMed] [Google Scholar]
- 25.Ramírez-Villaescusa G. F., Medley G. F., Mason S., Green L. E. 2010. Risk factors for herd breakdown with bovine tuberculosis in 148 cattle herds in the south west of England. Prevent. Vet. Med. 95, 224–230 10.1016/j.prevetmed.2010.03.009 (doi:10.1016/j.prevetmed.2010.03.009) [DOI] [PubMed] [Google Scholar]
- 26.Böhm M., Hutchings M. R., White P. C. L. 2009. Contact networks in a wildlife-livestock host community: identifying high-risk individuals in the transmission of bovine TB among badgers and cattle. PLoS ONE 4, e5016. 10.1371/journal.pone.0005016 (doi:10.1371/journal.pone.0005016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardon L. R., Bell J. I. 2001. Association study designs for complex diseases. Nat. Rev. Genet. 2, 91–98 10.1038/35052543 (doi:10.1038/35052543) [DOI] [PubMed] [Google Scholar]
- 28.Ohashi J., Tokunaga K. 2001. The power of genome-wide association studies of complex disease genes: statistical limitations of indirect approaches using SNP markers. J. Hum. Genet. 46, 478–482 10.1007/s100380170048 (doi:10.1007/s100380170048) [DOI] [PubMed] [Google Scholar]
- 29.Klein R. J. 2007. Power analysis for genome-wide association studies. BMC Genet. 8, 58. 10.1186/1471-2156-8-58 (doi:10.1186/1471-2156-8-58) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nannya Y., Taura K., Kurokawa M., Chiba S., Ogawa S. 2007. Evaluation of genome-wide power of genetic association studies based on empirical data from the HapMap project. Hum. Mol. Genet. 16, 2494–2505 10.1093/hmg/ddm205 (doi:10.1093/hmg/ddm205) [DOI] [PubMed] [Google Scholar]
- 31.Menashe I., Rosenberg P. S., Chen B. E. 2008. PGA: power calculation for case-control genetic association analyses. BMC Genet. 9, 36. 10.1186/1471-2156-9-36 (doi:10.1186/1471-2156-9-36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price A. L., Patterson N. J., Plenge R. M., Wieinblatt M. E., Shadick N. A., Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 10.1038/ng1847 (doi:10.1038/ng1847) [DOI] [PubMed] [Google Scholar]
- 33.MacLeod I. M., Hayes B. J., Savin K. W., Chamberlain A. J., McPartlan H. C., Goddard M. E. 2010. Power of a genome scan to detect and locate quantitative trait loci in cattle using single nucleotide polymorphisms. J. Anim. Breed. Genet. 127, 133–142 10.1111/j.1439-0388.2009.00831.x (doi:10.1111/j.1439-0388.2009.00831.x) [DOI] [PubMed] [Google Scholar]
- 34.Settles M., et al. 2009. A whole genome association analysis identifies loci associated with Mycobacterium avium subsp. paratuberculosis infection status in US holstein cattle. Anim. Genet. 40, 655–662 10.1111/j.1365-2052.2009.01896.x (doi:10.1111/j.1365-2052.2009.01896.x) [DOI] [PubMed] [Google Scholar]