Abstract
SNF5, a core component of the SWI/SNF chromatin remodeling complex, is expressed as two isoforms, SNF5a and SNF5b. SNF5 is a tumor suppressor as mutation of SNF5 leads to tumor formation and cooperates with p53 deficiency to enhance cancer susceptibility. Interestingly, lack of SNF5 inhibits cell survival and embryonic development potentially via abnormal activation of p53. To further examine this, we generated cell lines in that SNF5a, SNF5b, or both can be inducibly knocked down. We found that SNF5 knockdown leads to cell cycle arrest in G1, and SNF5a and SNF5b are functionally redundant. We also showed that SNF5 knockdown impairs p53-dependent transcription of p21 and MDM2. However, contrary to earlier reports that p53 is activated by SNF5 knockout in murine cells, SNF5 knockdown leads to decreased, but not increased, expression of both basal and stress-induced p53 in multiple human cell lines. In addition, we showed that SNF5 knockdown induces AMPK activation and inhibits eIF4E expression. Finally, we demonstrated that SNF5 knockdown inhibits p53 translation via eIF4E and replacement of eIF4E in SNF5-knockdown cells restores p53 expression and cell survival. Together, our results suggest that the p53 pathway is regulated by, and mediates the activity of, SNF5 in tumor suppression and pro-survival.
Keywords: p53, SNF5, chromatin remodeling, p21, Mdm2, eIF4E, AMPK
Introduction
The SWI/SNF complex is an evolutionarily conserved multi-subunit chromatin remodeling complex, which uses ATP to mobilize nucleosomes and remodel chromatin (Kingston and Narlikar 1999). As a core subunit, human SNF5 is required for both the in vivo and in vitro remodeling activity of SWI/SNF (Carlson and Laurent 1994; Peterson 1996). The SNF5 gene is expressed as two alternative spliced forms, SNF5a and SNF5b. SNF5 is found to be mutated or deleted in various tumors including rhabdoid, brain and lung cancers, suggesting that SNF5 functions as a potential tumor suppressor (Roberts and Orkin 2004; Sevenet et al. 1999a; Sevenet et al. 1999b; Versteege et al. 1998). Indeed, loss of heterozygosity of SNF5 predisposes mice to cancer development at a high frequency (Guidi et al. 2001; Klochendler-Yeivin et al. 2000; Roberts et al. 2000). In addition, conditional inactivation of SNF5 in hematopoietic tissues resulted in an extremely rapid onset of CD8+ lymphomas (Roberts et al. 2002), potentially via transcriptional activation of p16 and inhibition of cyclin D1 (Chai et al. 2007; Oruetxebarria et al. 2004; Vries et al. 2005; Zhang et al. 2002). Interestingly, SNF5 is also required for cell survival and differentiation (Caramel et al. 2008; Gresh et al. 2005). Homozygous deletion of SNF5 in mice causes embryonic lethality and conditional inactivation of SNF5 in mice leads to rapid bone marrow failure and death (Klochendler-Yeivin et al. 2000; Roberts et al. 2000; Roberts et al. 2002). In addition, targeted inactivation of SNF5 in liver impairs glucose metabolism and cell differentiation (Gresh et al. 2005).
P53 is a transcription factor and regulates target genes involved in cell death and survival in response to stresses (K. Harms et al. 2004; Ko and Prives 1996). One study showed that SNF5 is capable of regulating p53 transcriptional activity in vitro (Lee et al. 2002). Recently, we demonstrated that Brg1- and Brm-containing SWI/SNF complexes differentially regulate p53 transcriptional activity (Xu et al. 2007). In addition, loss of p53 accelerates tumor formation in mice bearing conditional inactivation of Snf5, suggesting that SNF5 and p53 cooperate to prevent cancer development (Isakoff et al. 2005; Klochendler-Yeivin et al. 2006). However, whether endogenous SNF5 has an effect on p53-dependent transcription and whether p53 target genes play a role in SNF5 pro-survival function remain to be determined. Here, we showed that knockdown of SNF5 in tumor cells leads to cell cycle arrest in G1. We also showed that p53 translation is inhibited upon SNF5 knockdown and replacement of eIF4E in SNF5-knockdown cells restores p53 expression and cell survival. Thus, we hypothesized that the p53 pathway is regulated by, mediates the activity of, SNF5 in tumor suppression and pro-survival.
Results
Generation of cell lines in which total SNF5 or an individual SNF5 isoform can be inducibly knocked down
SNF5a, a polypeptide with 385 amino acids and SNF5b, a polypeptide with 376 amino acids, are identical except that SNF5b lacks nine amino acids between codons 69–77 (Fig. 1A). To investigate a potential role of SNF5 in the p53 pathway, we generated MCF7 and RKO cell lines which inducibly express siRNAs targeting both SNF5 isoforms under the control of the tetracycline-regulated promoter. To reduce non-specific effect of RNA interference, two siRNAs, siSNF5-1049 and siSNF5-1063, were used to knock down total SNF5. siSNF5-1049 was used to generate SNF5-KD in both MCF7 and RKO cells. Two representative SNF5-KD MCF7 and RKO cell lines, MCF7-SNF5-KD#5/#73 and RKO-SNF5-KD#10/#11, were shown in Fig. 1B–C. siSNF5-1063 was used to generate SNF5-KD MCF7 cell lines. Two representative cell lines, MCF7-SNF5-KD#18/#24 were shown in Fig. 1B. Western blot analysis showed that both isoforms of SNF5 were expressed in MCF7 and RKO cells and the levels of SNF5 proteins were significantly decreased upon expression of either siSNF5-1049 or siSNF5-1063 (Fig. 1B–C). We would like to note that in MCF7-SNF5-KD#5 cells, both SNF5a and SNF5b were expressed but overlapped due to insufficient separation. We would also like to mention that the relative basal level of SNF5a is lower in MCF7 cells than that in RKO cells (Fig. 1B–C). Next, to investigate the physiological relevance of each isoform, we generated multiple MCF7 and RKO cell lines which inducibly express siRNAs targeting each individual SNF5 isoform. Western blot analysis showed that the levels of SNF5a and SNF5b in MCF7 and RKO cells were significantly decreased upon expression of siRNA targeting SNF5a and SNF5b, respectively (Fig. 1D). Interestingly, we found that knockdown of one isoform led to an increased expression of other isoform, especially in MCF7 cell lines in which SNF5b was knocked down (Fig. 1D, compare lanes 5 and 7 with 6 and 8, respectively). This suggests that SNF5a and SNF5b are compensatory in their expression. Previously, expression of the SNF5 alleles was found to be compensatory. Upon knockout of one allele, expression of the remaining allele was found to be increased to compensate for the lost expression from the deleted allele (Guidi et al. 2004). Here, for the first time, we demonstrated that a compensatory effect exists between SNF5 isoforms, possibly to maintain a proper level of SNF5 proteins, suggesting that SNF5 is necessary for cell survival.
Fig. 1. Generation of inducible SNF5-KD cell lines.
(A) Schematic presentation of SNF5 functional domains. Residues 69–77 is absent in SNF5b. (B, C) Generations of MCF7 and RKO cell lines in that both SNF5 isoforms can be inducibly knocked down. Western blots were prepared using extracts from MCF7 and RKO cells that were uninduced (−) or induced (+) to express SNF5 siRNAs for 72 h. The blots were analyzed by anti-SNF5 and anti-actin, respectively. (D) Generation of MCF7 and RKO cell lines in that an individual SNF5 isoform can be inducibly knocked down. The experiments were performed as in B–C.
Knockdown of total SNF5, but not an individual SNF5 isoform, inhibits cell proliferation
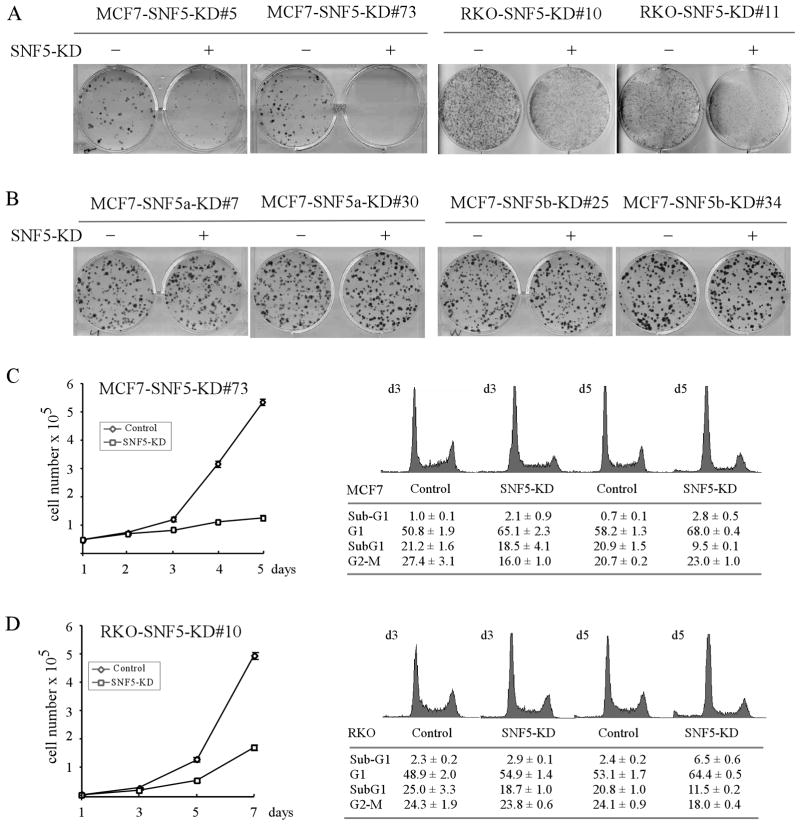
To determine whether endogenous SNF5, especially an individual SNF5 isoform, has a long-term impact on cell survival, colony formation assay was performed and showed that upon knockdown of total SNF5, colony formation by MCF7 and RKO cells was significantly inhibited (Fig. 2A), consistent with previous report that SNF5 is necessary for embryonic development (Klochendler-Yeivin et al. 2000; Roberts et al. 2000; Roberts et al. 2002). Interestingly, knockdown of either SNF5a or SNF5b had no significant effect on colony formation in MCF7 cells (Fig. 2B). As a control, tetracycline had no effect on cell proliferation in parental MCF7 or RKO cell lines (data not shown). We also performed cell proliferation assay and showed that cell growth was suppressed by knockdown of total SNF5 over a 5- or 7-day testing period in MCF7 and RKO cells (Fig. 2C–D, left panels). To determine whether the growth suppressive effect was due to cell cycle arrest, apoptosis, or both, DNA histogram analysis was performed. We found that upon knockdown of total SNF5 for 3–5 days, the growth suppression was primarily due to G1 arrest and to a less extent, apoptosis in MCF7 and RKO cells (Fig. 2C–D, right panels). Together, we showed that SNF5a and SNF5b can functionally compensate each other and that lack of both isoforms impairs cell proliferation.
Fig. 2. Knockdown of total SNF5, but not an individual SNF5 isoform, inhibits cell proliferation.
(A) SNF5 is required for colony formation. SNF5-KD MCF7 and RKO cells were cultured in the absence (−) or presence (+, SNF5-KD) of tetracycline for 14 d and then fixed and stained with crystal violet. (B) SNF5a and SNF5b are functionally compensatory. SNF5a-or SNF5b-KD MCF7 cells were cultured in the absence (−) or presence (+) of tetracycline for 14 d. The experiments were performed as in (A). (C–D) Left panel: The growth rate of MCF7 (C) and RKO (D) cells that were uninduced or induced to knock down total SNF5 was measured over a 5- or 7-day period. Right panel: The cell cycle profile of MCF7 (C) and RKO (D) cells that were uninduced or induced to knock down total SNF5 for 3 and 5 d. The percentage of cells in each phase of the cell cycle was quantified by fluorescence-activated cell sorter.
Knockdown of total SNF5, but not an individual SNF5 isoform, sensitizes cancer cells to DNA damage-induced apoptosis
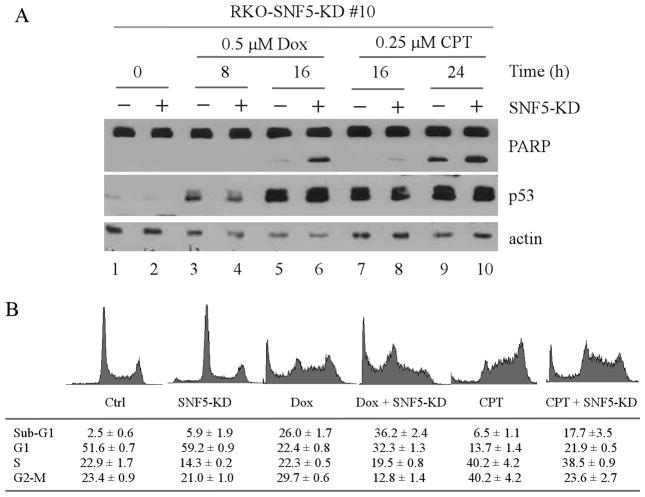
Although loss of SNF5 has been shown to increase cell sensitivity to DNA damage via activation of p53 in mouse embryo fibroblasts (MEFs) (Isakoff et al. 2005; Klochendler-Yeivin et al. 2006), it is not clear whether knockdown of SNF5 or an individual isoform has similar effect in human tumor cells. Thus, SNF5-KD RKO cells were treated with various DNA damage agents, and the extent of PARP cleavage, an indicative of apoptosis, was measured. We showed that upon treatment with doxorubicin or camptothecin, PARP cleavage was significantly increased by total SNF5 knockdown in a time-dependent manner (Fig. 3A, PARP panel). It is well established that p53 is a critical mediator of the DNA damage response and that p53 stabilization enhances apoptosis induced by DNA damage. Therefore, we wanted to examine whether increased apoptosis was due to increased p53 stabilization. Surprisingly, in contrast to early report that loss of SNF5 in MEF cells leads to p53 stabilization, we found that the level of p53 was slightly decreased, but not increased, in RKO cells by total SNF5 knockdown although p53 was efficiently stabilized by DNA damage (Fig. 3A, p53 panel). Next, DNA histogram analysis was performed to measure the number of cells with sub-G1 DNA content. We showed that upon total SNF5 knockdown, DNA damage-induced apoptosis was increased from 26.0% to 36.2% following treatment with doxorubicin and from 6.5% to 17.7% following treatment with camptothecin (Fig. 3B). Similar results were obtained with SNF5 knockdown MCF7 cell lines (data not shown). Furthermore, we showed that knockdown of SNF5a or SNF5b had no effect on the DNA damage-induced apoptotic response (data not shown), which is consistent with the above results that knockdown of one isoform had little if any effect on cell proliferation (Fig. 2B). Together, these data indicate that SNF5 plays a protective role in the DNA damage response.
Fig. 3. SNF5 knockdown sensitizes cancer cells to DNA damage-induced apoptosis.
(A) Western blots were prepared using extracts from RKO-SNF5-KD#10 cells that were uninduced (−) or induced (+) to express siRNA to knock down SNF5 for 72 h and then untreated or treated with 0.5μM doxorubicin for 8–16 h or 0.25μM camptothecin for 16–24 h. The blots were analyzed with antibodies against PARP, p53, and actin, respectively. (B) RKO cells were uninduced or induced to knock down SNF5 for 48 h along with treatment of 0.5μM doxorubicin for 18 h or 0.25μM camptothecin for 22 h, and then used for DNA histogram analysis.
SNF5 knockdown impairs p53-dependent transcription of p21 and MDM2
The p53 protein functions as a sequence-specific transcription factor that activates or inhibits transcription of various target genes. We sought to determine whether endogenous SNF5 is required for p53 to induce its target genes. To explore this, we examined induction of p21 and MDM2 in MCF7 cells in which total SNF5 or one isoform was inducibly knocked down. We found that upon treatment with doxorubicin or Nutlin-3, an inhibitor of p53-MDM2 interaction, p53 was stabilized and subsequently p21 and MDM2 were induced (Fig. 4A, compare lanes 1 and 9 with 3, 5, 7, 11, and 13, respectively). However, upon knockdown of SNF5, the level of DNA damage-induced p53 was decreased in MCF7 cells (Fig. 4A, compare lanes 5 and 7 with 6 and 8, respectively), consistent with the above result (Fig. 3A). Interestingly, Nutlin-3-mediated stabilization of p53 was increased upon SNF5 knockdown (Fig. 4A, compare lanes 11 and 13 with 12 and 14, respectively), suggesting that inhibition of p53 degradation via Mdm2 inhibitor is able to mitigate the effect of SNF5 on p53 expression. Nevertheless, the extent of p21 and MDM2 expression was still decreased in a time-dependent manner when total SNF5 was inducibly knocked down (Fig. 4A), suggesting that in addition to its effect on p53 stabilization, SNF5 is necessary for p53-dependent activation of p21 and MDM2. To rule out potential cell type-specific effect, we examined p53 transcriptional activity upon SNF5 knockdown in RKO cells (Fig. 4A, lanes 15–20). We found that p53 induction of p21 and MDM2 upon DNA damage was significantly inhibited by total SNF5 knockdown (Fig. 4A, compare lanes 17 and 19 with 18 and 20, respectively). In addition, we found that p53 stabilization was slightly decreased by SNF5 knockdown (Fig. 4A, compare lanes 17 and 19 with 18 and 20, respectively), consistent with the above result (Fig. 3A). Next, we examined whether each individual SNF5 has an effect on p53-dependent transcription and showed that the expression of p53 and p21 was not consistently decreased upon knockdown of either SNF5a or SNF5b along with or without treatment of doxorubicin or camptothecin in MCF7 cells (Fig. 4B).
Fig. 4. SNF5 knockdown impairs p53-dependent p21 and MDM2 transcription.
(A) Western blots were prepared using extracts from MCF7-SNF5-KD#73 and RKO-SNF5-KD#10 cells that were uninduced (−) or induced (+) to express SNF5 siRNAs for 72 h and then treated with 0.5μM doxorubicin, 0.25μM camptothecin, or 2.5μM Nutlin for 0–8 h. The blots were analyzed with antibodies against p53, p21, MDM2, and actin, respectively. (B) Western blots were prepared using extracts from MCF7-SNF5a-KD#30 or MCF7-SNF5b-KD#25 cells that were uninduced (−) or induced (+) to knock down SNF5a or SNF5b for 72 h and then treated with 0.5 μM doxorubicin or 0.25μM camptothecin for 6 h. The blots were analyzed with antibodies against p53, p21 and actin, respectively. (C) Northern blots were prepared with total RNAs isolated from MCF7-SNF5-KD#73 cells that were uninduced (−) or induced (+) to express SNF5 siRNAs for 72 h along with treatment of 0.5 μM doxorubicin or 0.25μM camptothecin for 6 h. The blots were probed with 32P-labeled cDNAs derived from MDM2, p21, and GAPDH genes, respectively. (D) Top panel: schematic presentation of the p21 (left) and MDM2 (right) promoters with the locations of the p53 responsive elements and PCR primers used for ChIP assay. Bottom panel: knockdown of SNF5 decreases the ability of p53 to bind to the p53 responsive element in the p21 (left) and MDM2 (right) promoters. MCF7-SNF5-KD#73 cells were uninduced (−) or induced (+) to knock down SNF5 for 72 h followed by mock-treatment or treatment with 0.5 μM of doxorubicin or 0.25 μM of camptothecin for 8 h. ChIP assay were performed as described in Materials and Methods.
To determine whether the impaired induction of p21 and MDM2 protein is due to decreased expression of their transcripts, we performed Northern blot analysis and found that the level of p21 and MDM2 transcripts, although up-regulated upon treatment with doxorubicin or camptothecin, was markedly decreased upon total SNF5 knockdown (Fig. 4C, compare lanes 3 and 5 with 4 and 6, respectively). We would like to mention that Mdm2 transcripts were induced in MCF7 cells treated with camptothecin, which was decreased by SNF5 knockdown (longer exposure, data not shown). As a key component of chromatin remodeling complex, Snf5 may regulate p53 transcriptional activity by altering the DNA-binding activity of p53 to its target genes, including p21 and Mdm2. To test this, the binding of p53 to the p21 and Mdm2 promoters was examined by ChIP assay with MCF7 cells uninduced or induced to knock down SNF5 along with or without treatment with doxorubicin or camptothecin for 8 h. The p53-DNA complexes were immunoprecipitated with anti-p53 as well as mouse IgG as a control. PCR was performed to amplify the region spanning the p53 responsive elements in the p21 and MDM2 genes with primers shown in Fig. 4D (top panel). We showed that the extent of p53 binding to the p21 and MDM2 promoters was increased in MCF7 cells upon treatment with doxorubicin or camptothecin (Fig. 4D, bottom panels, compare lanes 1 and 7 with lanes 3, 5, 10, and 12, respectively). No DNA fragment was enriched by control antibody (Fig. 4D, IgG panels). Interestingly, we found that SNF5 knockdown markedly inhibited p53 to bind to the p21 and MDM2 promoters (Fig. 4D, bottom panels, compare lanes 3, 5, 9, and 11 with 4, 6, 10, and 12, respectively), consistent with the requirement of SNF5 for p53 induction of p21 and Mdm2 in MCF7 cells (Fig. 4A and 4C). Taken together, these data suggest that SNF5 is required for p53 transcriptional activity and induction of pro-survival p53 targets, including p21 and MDM2, may play a role in SNF5 pro-survival activity.
SNF5 knockdown inhibits p53 translation but not protein stability
It is well established that p53 protein stability is regulated by post-translational modifications, leading to accumulation of p53 upon DNA damage (Scoumanne and Chen 2008). Since p53 expression was found to be decreased in cells upon knockdown of SNF5, we reasoned that p53 protein stability may be regulated by SNF5. To test this, we measured p53 protein half life and found that upon inhibition of new protein synthesis with cycloheximide, p53 protein was found to be rapidly degraded in both MCF7 and RKO cells (Fig. 5A–B, lanes 1–7). However, there was no substantial difference in the stability of p53 protein between control and SNF5-knockdown cells (Fig. 5A–B, compare lanes 1–7 with 8–14, respectively). Next, we examined the rate of p53 translation since translation is found to regulate p53 expression (Maltzman and Czyzyk 1984; Takagi et al. 2005). To test this, newly synthesized p53 protein was labeled with 35S-methionine for a short 30 min in MCF7 cells uninduced or induced to knock down SNF5. We showed that upon knockdown of SNF5, the level of newly synthesized p53 protein was markedly decreased in MCF7 cells under both the control and stress-induced conditions (Fig. 5C, compare lanes 5 and 11 with 6 and 12, respectively).
Fig. 5. SNF5 knockdown inhibits p53 translation but not protein stability.
(A, B) The stability of p53 protein was not affected by knockdown of SNF5. Western blots were prepared with extracts from MCF7 (A) and RKO (B) cells, which were uninduced (−) or induced (+) to knock down SNF5 for 3 days, treated with cycloheximide (CHX, 1mg/ml) for 0–180 minutes, and then probed with antibodies against p53 or actin. (C) p53 protein translation was inhibited by knockdown of SNF5. Cell extracts were prepared from MCF7 cells, which were uninduced (−) or induced (+) to knock down SNF5 for 3 days, mock-treated (lanes 1–6) or treated with 0.5 μM DOX for 1 h (lanes 7–12), and then labeled with [35S]methionine for 30 min. The levels of 35S-labeled proteins were measured by scintillation counter and an equal amount of 35S-labeled proteins were used for immunoprecipitation. 35S-labeled p53 protein was immunoprecipitated with rabbit anti-p53 polyclonal antibodies or a control IgG and visualized by autoradiography. Input was used as a loading control.
SNF5 knockdown induces AMPK phosphorylation and inhibits eIF4E expression
Previous report showed that mice with liver-specific inactivation of SNF5 died of severe impaired energy metabolism, which was due to down regulation of a plethora of genes involved in gluconeogenesis as well as glucogen storage (Gresh et al. 2005). As a energy sensor, AMPK becomes activated under energy stress to coordinate glucose metabolism with cell proliferation. To test this, the levels of activated AMPK were measured in SNF5-KD MCF7 and RKO cells (MCF7-SNF5-KD#73 and RKO-SNF5-KD#10) as well as the control MCF7 and RKO cells (MCF7-pcDNA6-TR#7 and RKO-pcDNA6-TR#13). We found that upon SNF5 knockdown, activated AMPK was markedly increased (Fig. 6A, compare lanes 3 and 7 with 4 and 8, respectively), suggesting that AMPK plays a role in the SNF5 pro-survival pathway.
Fig. 6. SNF5 knockdown inhibits p53 translation and cell survival via enhanced AMPK phosphorylation and decreased eIF4E expression.
(A) SNF5 knockdown enhanced AMPK phosphorylation. Western blots were prepared using extracts from MCF7-pcDNA6-TR#7, MCF7-SNF5-KD#73, RKO-pcDNA6-TR#13 and RKO-SNF5-KD#10 that were cultured with (+) or without (−) tetracycline for 72 h. The blots were analyzed with antibodies against phospho-AMPK, SNF5 and actin, respectively. (B) SNF5 knockdown inhibits eIF4E expression. Western blots were prepared using extracts from RKO-SNF5-KD#10 that were uninduced (−) or induced (+) to knock down SNF5 for 72 h and then treated with 0.5μM doxorubicin for 6 h. The blots were analyzed with antibodies against SNF5, eIF4E, p53 and actin, respectively. (C) Left panel: overexpression of myc-tagged eIF4E restores p53 expression in RKO cells in which SNF5 is inducibly knocked down. Western blots were prepared with extracts from eIF4E-producing RKO cells, which were uninduced (−) or induced (+) to knock down SNF5 for 3 days and then probed with antibodies against SNF5, myc-tag, eIF4E, p53 and actin, respectively. Right panel: overexpression of myc-tagged eIF4E rescues the proliferation defect of RKO cells in which SNF5 is inducibly knocked down. (D) A model for the role of SNF5 in the p53 pathway.
To further demonstrate how SNF5 regulates cell survival, we wanted to identify potential SNF5 targets, which are not directly regulated by p53. Thus, we performed microarray analysis with Affymetrix GeneChip with RNAs purified from MCF7 cells uninduced or induced to knock down SNF5 for 3 days. We found that many genes were down-regulated (Table 1), including p21, S100a8, and S100a9, which were also found to be altered in mice upon liver-specific inactivation of SNF5 (Gresh et al. 2005). Among these was eIF4E, an initiation factor for protein translation. eIF4E is found to be over-expressed in many tumors and have transforming and antiapoptotic activities (Graff et al. 2008). Thus, Western blot analysis was performed to confirm the microarray study and showed that the expression level of eIF4E along with p53 was inhibited by knockdown of SNF5 regardless of treatment with doxorubicin (Fig 6B, compare lanes 1 and 3 with 2 and 4, respectively).
Table 1.
Effect of SNF5 knockdown on gene expression
| SNF5-KD Vs. Control (fold) |
||
|---|---|---|
| Genes | Mock-treated | Treated with 0.5 μM Dox |
| P53 target Genes | ||
| p21 | −1.3 | |
| Mdm2 | −1.4 | |
| Gadd45a | −1.6 | |
| DRAM | −1.8 | −1.9 |
| p52R2 | −1.3 | |
| Cathepsin D | −1.3 | −1.7 |
| DR5 | −1.3 | |
| Trail | −2.3 | −4.3 |
| Other genes | ||
| eIF4E | −1.3 | −1.74 |
| S100a8 | −3.25 | −2.3 |
| S100a9 | −4.59 | −4.0 |
| IFIT1 | −2.46 | −2.83 |
| DUSP6 | −2.14 | −1.8 |
| KIAA1430 | −2.0 | −2.0 |
| PTP4A2 | −1.87 | −1.87 |
| VAV3 | −1.74 | −2.14 |
| E2IG4 | −1.62 | −1.74 |
| CCNG2 | −2.3 | −1.74 |
| PDK4 | −4.92 | −5.66 |
We showed that p53 translation but not stability is controlled by SNF5 (Fig. 5C). We also showed that eIF4E expression is decreased by SNF5 knockdown (Fig. 6B). As a key factor for translation, we reasoned that SNF5 regulation of p53 may be mediated through eIF4E. To test this, eIF4E was constitutively expressed in RKO cells in which endogenous SNF5 can be inducibly knocked. We showed that upon knockdown of SNF5, the level of endogenous eIF4E was decreased whereas exogenous myc-tagged eIF4E was constitutively expressed (Fig. 6C, left panel, compare lanes 1 and 3 with 2 and 4, respectively). Interestingly, we observed that the decreased expression of p53 due to knockdown of SNF5 was abrogated regardless of treatment with doxorubicin (Fig. 6C, left panel). Furthermore, we found that upon restoration of eIF4E, SNF5-knockdown cells regained the ability to proliferate as measured by colony formation assay (Fig. 6C, right panel). Taken together, these data suggest that eIF4E is a mediator of SNF5 in p53 expression and cell survival.
Discussion
Through inducible knockdown of one or both SNF5 isoforms in MCF7 and RKO cells, several novel observations were made: (1) SNF5a and SNF5b are functionally redundant and compensatory in their expression, and knockdown of either one alone has no effect on cell survival; (2) knockdown of SNF5 inhibits cell survival by inducing G1 arrest and to a lesser extent, apoptosis; (3) SNF5 is a co-activator required for p53 transcriptional activity; and (4) p53 translation was inhibited via reduced eIF4E expression and possibly enhanced activity of AMPK (Fig. 6D).
We showed that knockdown of SNF5 leads to growth inhibition and loss of survival, which is consistent with earlier reports (Isakoff et al. 2005; Klochendler-Yeivin et al. 2006). However, the mechanisms by which lack of SNF5 inhibits cell survival are different. In MEFs with SNF5 knockout, p53 is activated and subsequently induces cell cycle arrest and apoptosis (Isakoff et al. 2005; Klochendler-Yeivin et al. 2006). Here, in human carcinoma cells with SNF5 knockdown, p53 expression is decreased along with decreased expression of eIF4E (Fig. 6A–B). When the decreased expression of endogenous eIF4E is compensated by exogenous eIF4E in SNF5-KD cells, p53 expression is restored (Fig. 6C). We also found that SNF5 knockdown leads to increased AMPK activation (Fig. 6A). AMPK is an inhibitor of the AKT pro-survival pathway and AMPK activation also inhibits mTOR kinase and subsequently translation (Gwinn et al. 2008; Inoki et al. 2003b; Inoki et al. 2003a). Thus, we hypothesize that inhibition of eIF4E and activation of AMPK are one of the mechanisms by which SNF5 controls cell survival and p53 expression via translation. Thus, future study is needed to address how activation of AMPK and decreased expression of eIF4E coordinately mediate SNF5 in regulating p53 expression and cell survival.
The defective p53 pathway caused by SNF5 knockdown is consistent with the in vivo observation that p53 knockout cooperates with SNF5 knockout to promote cancer susceptibility in mice, including early onset and high penetrance (Isakoff et al. 2005; Klochendler-Yeivin et al. 2006). Thus, how to explain lack of SNF5 in MEF cells increases, whereas knockdown of SNF5 in human carcinoma cells decreases, p53 expression? One explanation is simply due to the difference between species: humans vs. rodents. The other explanation is that in MEF cells, persistent lack of SNF5 generates severe stresses, potentially including DNA damage, which then activate and stabilize p53. However, in this study, SNF5 was inducibly knocked down. In addition, siRNA knockdown of SNF5 was transient and incomplete. Together, inducible and incomplete knockdown of SNF5 may not be sufficient to generate stresses, such as DNA damage, to activate p53 albeit SNF5 knockdown is sufficient to inhibit cell survival.
It is well established that SNF5 is a tumor suppressor and loss of SNF5 predisposes humans and mice to spontaneous tumor formation (Roberts and Orkin 2004). However, SNF5 deficiency in MEF cells and in human tumor cells leads to growth suppression, which cannot be rescued by inactivation of p53 (Isakoff et al. 2005; Klochendler-Yeivin et al. 2006). This suggests that SNF5 is required for proper expression of pro-survival genes. Consistent with this notion, we and others (Gresh et al. 2005) showed that expression of many pro-survival genes, including p53, p21, and eIF4E, was decreased upon SNF5 knockdown or knockout. Conversely, in order for transform cells in the absence of SNF5, activation of other pro-survival pathways must be made to counter SNF5 deficiency-induced growth suppression. Indeed, expression of cyclin D1 and c-Myc was markedly increased in SNF-deficiency mouse and human tumors (McKenna et al. 2008). However, although lack of p53 promotes SNF5 deficiency-induced tumor formation (Isakoff et al. 2005; Klochendler-Yeivin et al. 2006), there is no evidence that SNF5 deficiency leads to increased p53 mutation in mouse and human tumors. In contrast, a recent study showed that loss of SNF5 does not lead to genomic instability (McKenna et al. 2008). Since genomic instability is correlated with a high frequency of p53 mutation in human cancers (Brosh and Rotter 2009), it is unlikely that lack of genomic instability in SNF5-deficient tumors would increase the frequency of p53 mutation. In addition, considering that knockdown of SNF5 leads to decreased expression and activity of the p53 pathway (this study) and lack of genomic instability in SNF5-knockout mice (McKenna et al. 2008), it suggest that there is no selective pressure to mutate the p53 gene.
SNF5 functions as co-activator and co-repressor (Martens and Winston 2003; Sif et al. 2001), and many genes are regulated by SNF5, including approximately 6% of genes in the whole genome of yeast (Holstege et al. 1998; Sudarsanam et al. 2000) and up to 70% of genes in mouse liver (Gresh et al. 2005). Consistent with this, the transcriptional activity of p53 was found to be enhanced by co-transfection of SNF5 (Lee et al. 2002). Here, we found that SNF5 is required for p53 to induce expression of endogenous p21 and Mdm2. We would like to note that in MCF7 cells treated with nutlin-3, an inhibitor of Mdm2, p53 was accumulated but its ability to induce p21 and Mdm2 was still inhibited by SNF5 knockdown (Fig. 4A). It should also be noted that in SNF5-knockdout mouse hepatic cells, p21 expression was found to be markedly inhibited (Gresh et al. 2005), consistent with the effect of SNF5 knockdown in human carcinoma cells (this study).
Materials and Methods
Plasmids
To generate inducible small interfering RNA (siRNA) constructs against SNF5, two individual siRNAs were designed to target total SNF5 and one each was designed to target SNF5a or SNF5b. These siRNAs were separately cloned into pBabe-H1 vector as previously described (Xu et al. 2007). The constructs that target total SNF5 were designated pBabe-H1-siSNF5-1049 and pBabe-H1-siSNF5-1063. The constructs that targets SNF5a and SNF5b were designated pBabe-H1-siSNF5a and pBabe-H1-siSNF5b, respectively. The sense oligonucleotide in pBabe-H1-siSNF5-1049 (targeting region shown in upper case) is gatcccc GGACATGTCAGAGAAGGAGAAC ttcaagaga GTTCTCC TTCTCTGACATGTCC tttttggaaa and the antisense oligonucleotide is agcttttccaaaaa GGACATGTCAGAGAAGGAGAAC tctcttgaa GTTCTCCTTCTCTGACATGTCC ggg. The sense oligonucleotide in pBabe-H1-siSNF5-1063 is gatcccc GGAGAACTCACCAGAGAAGTT ttcaagaga AACTTCTCTGGTGAGTTCTCC tttttggaaa and the antisense oligonucleotide is agcttttccaaaaa GGAGAACTCACCAGAGAAGTT tctcttgaa AACTTCTCTGGTGAGTTCTCC ggg. The sense oligonucleotide in pBabe-H1-siSNF5a is gatcccc AACACTAAGGATCACGGATAC ttcaagaga GTATCCGTGATCCTTAGTGTT tttttggaaaand the antisense oligonucleotide is agcttttccaaaaa AACACTAAGGATCACGGATAC tctcttgaa GTATCCGTGATCCTTAGTGTT ggg. The sense oligonucleotide in pBabe-H1-siSNF5b is gatcccc GTCACATGATCACGGATACACG ttcaagaga CGTGTATCCGTGATCATGTGAC tttttggaaa and the antisense oligonucleotide is agcttttccaaaaa GTCACATGATCACGGATACACG tctcttgaa CGTGTATCCGTGATCATGTGAC ggg.
Cell culture and generation of stable cell lines
MCF7 and RKO cells were cultured in DMEM supplement with 8% tetracycline-free fetal bovine serum and antibiotics. To generate MCF7 and RKO cell lines in which SNF5, SNF5a, or SNF5b can be inducibly knocked down under the control of the tetracycline inducible promoter, each siRNA expression vector was transfected into MCF7-pcDNA6-TR-7 and RKO-pcDNA6-TR-13 cells, both of which express a tetracycline repressor by pcDNA6 (Liu and Chen 2006). SNF5 knockdown cell lines were selected with puromycin and confirmed by Western blot analysis. To generate cell lines in which myc-tagged eIF4E is stably overexpressed and SNF5 is inducibly knocked down, pcDNA3-myc-eIF4E (Scoumanne et al. 2009) was transfected into RKO cells in which SNF5 can be inducibly knocked down and cell lines were then selected with G418.
Growth rate analysis, colony formation assay, and DNA histogram analysis
Growth rate was measured as described (Xu et al. 2007). Briefly, 5×104 of MCF7 cells or 3×104 of RKO cells were seeded with or without tetracycline in triplicate. Attached cells were counted at the indicated times. For colony formation assay, 500 MCF7 cells and 300 RKO cells seeded in 6-well plates were cultured in the absence or presence of tetracycline (1.0 μg/ml) for 11–14 days. DNA histogram analysis was done as described (Xu et al. 2007). Briefly, Cells seeded at 2×105 per well were cultured with or without tetracycline along with treatment of a DNA damage agent for various times. The percentages of cells in the sub-G1, G1, S, and G2-M phases were determined using the CELLQuest program (BD Biosciences).
Western blot analysis
Whole cell extracts were prepared by lysing cells with 2×SDS sample buffer. Proteins were separated on 7–10 % SDS-PAGE, transferred onto a nitrocellulose membrane, and probed with indicated antibodies followed by ECL detection. Antibodies against SNF5 (BAF47), eIF4E, and PARP were purchased from BD Pharmingen. Phospho-AMPK-alpha (Thr172) antibody was purchased from Cell Signaling. p21 (C19) and MDM2 (SMP14) antibodies were purchased from Santa Cruz Biotechnology. Antibodies against p53, HA and actin were as described (Xu et al. 2007).
Affymetrix GeneChip assay and Northern blot Analysis
Total RNAs were isolated from MCF7-SNF5-KD#73 cells using Trizol reagent (Invitrogen). U133 plus GeneChip was purchased from Affymetrix (Santa Clara, CA, USA), which contains oligos representing 37,000 unique human transcripts. GeneChip analysis was performed according to the manufacturer’s instruction. Northern blot analysis and preparation of p21, MDM2 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probes were as described previously (Chen et al. 1995).
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed as previously described (K. L. Harms and Chen 2005). After induction (+) or no induction (−) of SNF5 siRNA for 72 h, cells were treated with doxorubicin or camptothecin for 8h and cross-linked with 1% formaldehyde for 10 min at room temperature, which were then sonicated to generate 500- to 1000-bp DNA fragments and immunoprecipitated with anti-p53 (DO-1) and control mouse IgG. After reverse cross-linking and phenol-chloroform extraction, the bound DNA fragments were purified by a QIAGEN column. PCR was performed to visualize the enriched DNA fragments. Primers designed to amplify the region from nt −2312 to −2131 in the p21 promoter were forward primer, 5′-CAGGCTGTGGCTCT GATTGG-3′, and the reverse primer, 5′-TTCAGAGTAACAGGCTAAGG-3′. Primers designed to amplify the region from nt +3779 to +3969 in the MDM2 intron 1 were forward primer, 5′-GGATTGGGCCGGTTCAGTGG-3′, and the reverse primer, 5′-GGTCTACCCTCCAATCGCCAC-3′.
Protein stability assay
Western blots were prepared with extracts from MCF7 and RKO cells uninduced (′) or induced (+) to knock down SNF5 for 3 days along with treatment of cycloheximide (1mg/ml) for 0–180 minutes, and then probed with antibodies against p53 or actin.
Measurement of newly synthesized p53
Immunoprecipitation was performed to measure newly synthesized p53 protein in MCF7 cells, which were uninduced (−) or induced (+) to knock down SNF5 for 3 days along with or without treatment of 0.5 μM DOX for 1 h and then labeled with [35S]-methionine for 30 min.
Acknowledgments
Thiswork is supported in part by NIH grants CA081237, CA076069 and CA102188.
Footnotes
Conflicts of Interest
No conflicts of interest were disclosed.
References
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- Caramel J, et al. The requirement for SNF5/INI1 in adipocyte differentiation highlights new features of malignant rhabdoid tumors. Oncogene. 2008;27:2035–2044. doi: 10.1038/sj.onc.1210847. [DOI] [PubMed] [Google Scholar]
- Carlson M, Laurent BC. The SNF/SWI family of global transcriptional activators. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Chai J, et al. Tumor-specific cooperation of retinoblastoma protein family and Snf5 inactivation. Cancer Res. 2007;67:3002–3009. doi: 10.1158/0008-5472.CAN-06-4207. [DOI] [PubMed] [Google Scholar]
- Chen X, Bargonetti J, Prives C. p53, through p21 (WAF1/CIP1), induces cyclin D1 synthesis. Cancer Res. 1995;55:4257–4263. [PubMed] [Google Scholar]
- Graff JR, et al. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008;68:631–4. doi: 10.1158/0008-5472.CAN-07-5635. [DOI] [PubMed] [Google Scholar]
- Gresh L, et al. The SWI/SNF chromatin-remodeling complex subunit SNF5 is essential for hepatocyte differentiation. EMBO J. 2005;24:3313–3324. doi: 10.1038/sj.emboj.7600802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi CJ, et al. Transcriptional compensation for loss of an allele of the Ini1 tumor suppressor. J Biol Chem. 2004;279:4180–4185. doi: 10.1074/jbc.M312043200. [DOI] [PubMed] [Google Scholar]
- Guidi CJ, et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol Cell Biol. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms K, Nozell S, Chen X. The common and distinct target genes of the p53 family transcription factors. Cell Mol Life Sci. 2004;61:822–842. doi: 10.1007/s00018-003-3304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms KL, Chen X. The C terminus of p53 family proteins is a cell fate determinant. Mol Cell Bio. 2005;25:2014–2030. doi: 10.1128/MCB.25.5.2014-2030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003a;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Inoki K, et al. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003b;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff MS, et al. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc Natl Acad Sci U S A. 2005;102:17745–17750. doi: 10.1073/pnas.0509014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, Picarsky E, Yaniv M. Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex. Mol Cell Biol. 2006;26:2661–2674. doi: 10.1128/MCB.26.7.2661-2674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, et al. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Lee D, et al. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J Biol Chem. 2002;277:22330–22337. doi: 10.1074/jbc.M111987200. [DOI] [PubMed] [Google Scholar]
- Liu G, Chen X. DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol Cell Biol. 2006;26:1398–1413. doi: 10.1128/MCB.26.4.1398-1413.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltzman W, Czyzyk L. UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- McKenna ES, et al. Loss of the epigenetic tumor suppressor SNF5 leads to cancer without genomic instability. Mol Cell Biol. 2008;28:6223–6233. doi: 10.1128/MCB.00658-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oruetxebarria I, et al. P16INK4a is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhabdoid tumor cells. J Biol Chem. 2004;279:3807–3816. doi: 10.1074/jbc.M309333200. [DOI] [PubMed] [Google Scholar]
- Peterson CL. Multiple SWItches to turn on chromatin? Curr Opin Genet Dev. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH. The SWI/SNF complex--chromatin and cancer. Nat Rev Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- Roberts CW, et al. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell. 2002;2:415–425. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- Roberts CW, et al. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci U S A. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoumanne A, Chen X. Protein methylation: a new mechanism of p53 tumor suppressor regulation. Histol Histopathol. 2008;23:1143–1149. doi: 10.14670/hh-23.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoumanne A, Zhang J, Chen X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 2009;37:4965–4976. doi: 10.1093/nar/gkp516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenet N, et al. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J Hum Genet. 1999a;65:1342–1348. doi: 10.1086/302639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenet N, et al. Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum Mol Genet. 1999b;8:2359–2368. doi: 10.1093/hmg/8.13.2359. [DOI] [PubMed] [Google Scholar]
- Sif S, et al. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsanam P, et al. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, et al. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Versteege I, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Vries RG, et al. Cancer-associated mutations in chromatin remodeler hSNF5 promote chromosomal instability by compromising the mitotic checkpoint. Genes Dev. 2005;19:665–670. doi: 10.1101/gad.335805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang J, Chen X. The activity of p53 is differentially regulated by Brm- and Brg1-containing SWI/SNF chromatin remodeling complexes. J Biol Chem. 2007;282:37429–37435. doi: 10.1074/jbc.M706039200. [DOI] [PubMed] [Google Scholar]
- Zhang ZK, et al. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol Cell Biol. 2002;22:5975–5988. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]