Abstract
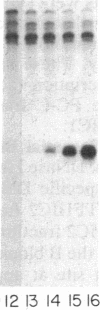
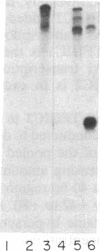
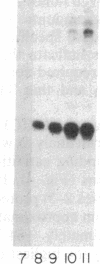
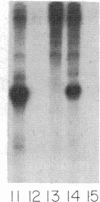
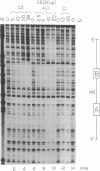
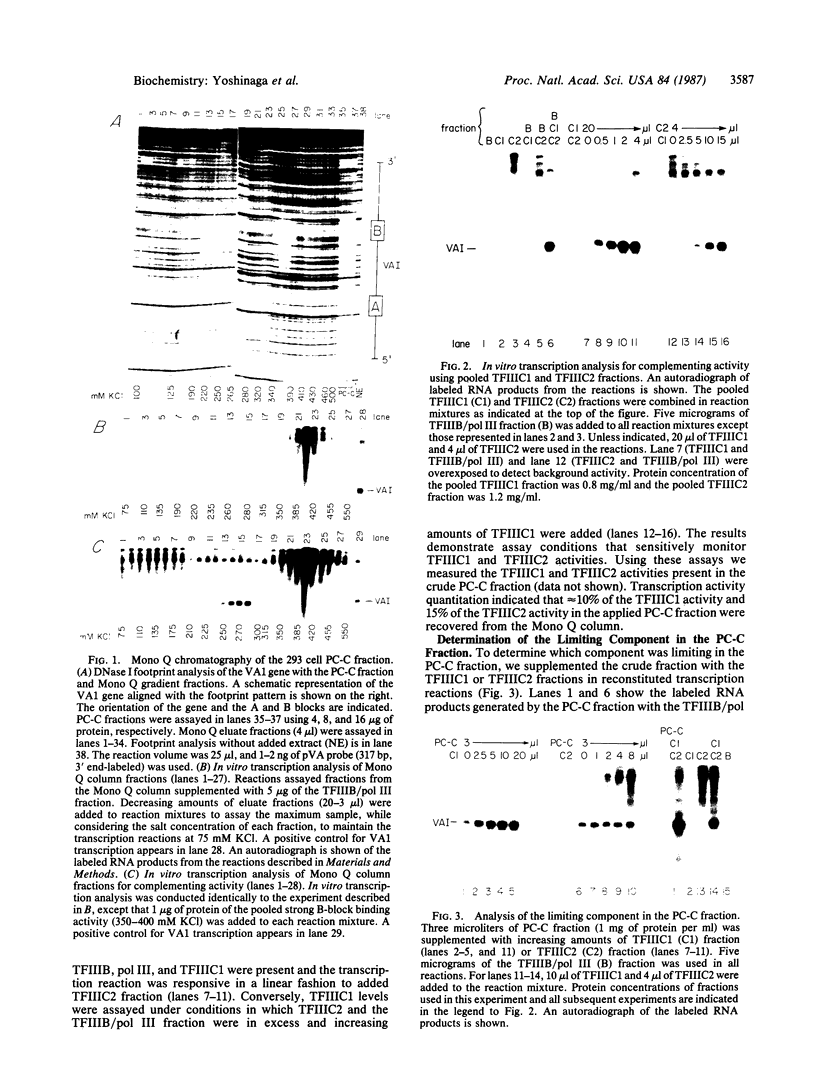
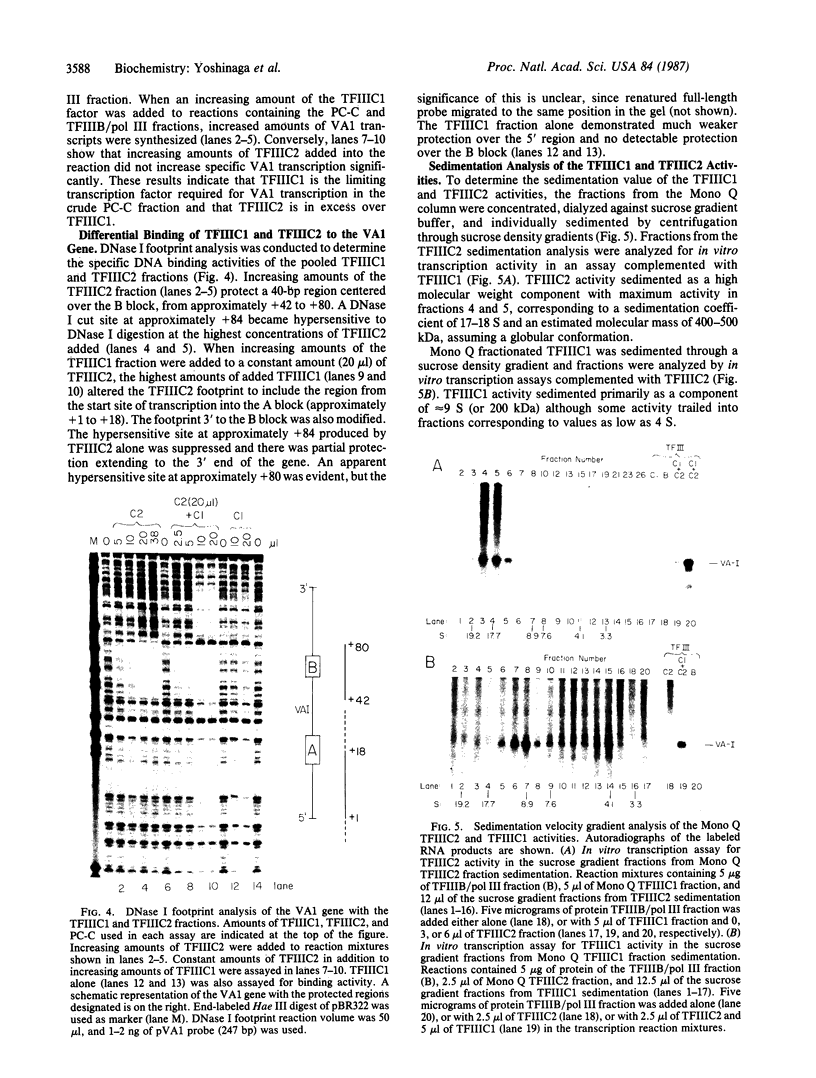
tRNA genes and adenovirus viral-associated (VA) genes are transcribed by RNA polymerase III. Transcription of these genes in vitro requires two protein fractions containing transcription factors designated TFIIIB and TFIIIC, in addition to RNA polymerase III. We report that the TFIIIC fraction derived from human cells in culture can be separated into two functional components, which we call TFIIIC1 and TFIIIC2. Both TFIIIC1 and TFIIIC2 fractions are required for in vitro transcription of the VA1 gene. In DNase I "footprinting" experiments, the TFIIIC2 fraction protects the internal control region termed the B block. Addition of the TFIIIC1 fraction extends the footprint over the internal control region called the A block. TFIIIC1 activity is the limiting transcription factor activity required for VA1 transcription in the crude extract. TFIIIC2 activity sediments as a large component of approximately 18 S, while TFIIIC1 activity sediments at approximately 9 S. These data indicate that the two activities are unique components and when added together reconstitute TFIIIC activity.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger S. L., Folk W. R. Differential activation of RNA polymerase III-transcribed genes by the polyomavirus enhancer and the adenovirus E1A gene products. Nucleic Acids Res. 1985 Feb 25;13(4):1413–1428. doi: 10.1093/nar/13.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J. Functions of adenovirus E1A. Cancer Surv. 1986;5(2):367–387. [PubMed] [Google Scholar]
- Burke D. J., Söll D. Functional analysis of fractionated Drosophila Kc cell tRNA gene transcription components. J Biol Chem. 1985 Jan 25;260(2):816–823. [PubMed] [Google Scholar]
- Camier S., Gabrielsen O., Baker R., Sentenac A. A split binding site for transcription factor tau on the tRNA3Glu gene. EMBO J. 1985 Feb;4(2):491–500. doi: 10.1002/j.1460-2075.1985.tb03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M. F., Gerrard S. P., Cozzarelli N. R. Analysis of RNA polymerase III transcription complexes by gel filtration. J Biol Chem. 1986 Mar 25;261(9):4309–4317. [PubMed] [Google Scholar]
- Ciliberto G., Castagnoli L., Cortese R. Transcription by RNA polymerase III. Curr Top Dev Biol. 1983;18:59–88. doi: 10.1016/s0070-2153(08)60579-7. [DOI] [PubMed] [Google Scholar]
- Cozzarelli N. R., Gerrard S. P., Schlissel M., Brown D. D., Bogenhagen D. F. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983 Oct;34(3):829–835. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- Fuhrman S. A., Engelke D. R., Geiduschek E. P. HeLa cell RNA polymerase III transcription factors. Functional characterization of a fraction identified by its activity in a second template rescue assay. J Biol Chem. 1984 Feb 10;259(3):1934–1943. [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor R. B., Feldman L. T., Berk A. J. Transcription of class III genes activated by viral immediate early proteins. Science. 1985 Oct 25;230(4724):447–450. doi: 10.1126/science.2996135. [DOI] [PubMed] [Google Scholar]
- Gonzalez N., Wiggs J., Chamberlin M. J. A simple procedure for resolution of Escherichia coli RNA polymerase holoenzyme from core polymerase. Arch Biochem Biophys. 1977 Aug;182(2):404–408. doi: 10.1016/0003-9861(77)90521-5. [DOI] [PubMed] [Google Scholar]
- Hall B. D., Clarkson S. G., Tocchini-Valentini G. Transcription initiation of eucaryotic transfer RNA genes. Cell. 1982 May;29(1):3–5. doi: 10.1016/0092-8674(82)90083-6. [DOI] [PubMed] [Google Scholar]
- Hoeffler W. K., Roeder R. G. Enhancement of RNA polymerase III transcription by the E1A gene product of adenovirus. Cell. 1985 Jul;41(3):955–963. doi: 10.1016/s0092-8674(85)80076-3. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Marzouki N., Camier S., Ruet A., Moenne A., Sentenac A. Selective proteolysis defines two DNA binding domains in yeast transcription factor tau. Nature. 1986 Sep 11;323(6084):176–178. doi: 10.1038/323176a0. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Ogden R. C., Abelson J. tRNA gene transcription in yeast: effects of specified base substitutions in the intragenic promoter. Cell. 1983 Nov;35(1):117–125. doi: 10.1016/0092-8674(83)90214-3. [DOI] [PubMed] [Google Scholar]
- Ruet A., Camier S., Smagowicz W., Sentenac A., Fromageot P. Isolation of a class C transcription factor which forms a stable complex with tRNA genes. EMBO J. 1984 Feb;3(2):343–350. doi: 10.1002/j.1460-2075.1984.tb01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setzer D. R., Brown D. D. Formation and stability of the 5 S RNA transcription complex. J Biol Chem. 1985 Feb 25;260(4):2483–2492. [PubMed] [Google Scholar]
- Stillman D. J., Caspers P., Geiduschek E. P. Effects of temperature and single-stranded DNA on the interaction of an RNA polymerase III transcription factor with a tRNA gene. Cell. 1985 Feb;40(2):311–317. doi: 10.1016/0092-8674(85)90145-x. [DOI] [PubMed] [Google Scholar]
- Stillman D. J., Geiduschek E. P. Differential binding of a S. cerevisiae RNA polymerase III transcription factor to two promoter segments of a tRNA gene. EMBO J. 1984 Apr;3(4):847–853. doi: 10.1002/j.1460-2075.1984.tb01895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., Sivertsen A. L., Zentner P. G., Geiduschek E. P. Correlations between transcription of a yeast tRNA gene and transcription factor-DNA interactions. J Biol Chem. 1984 Jun 25;259(12):7955–7962. [PubMed] [Google Scholar]
- Wingender E., Shi X. P., Houpert A., Seifart K. H. Isolation of a transcription complex for ribosomal 5S RNA. EMBO J. 1984 Aug;3(8):1761–1768. doi: 10.1002/j.1460-2075.1984.tb02043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. J. Adenovirus DNA-directed transcription of 5.5S RNA in vitro. Proc Natl Acad Sci U S A. 1978 May;75(5):2175–2179. doi: 10.1073/pnas.75.5.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga S., Dean N., Han M., Berk A. J. Adenovirus stimulation of transcription by RNA polymerase III: evidence for an E1A-dependent increase in transcription factor IIIC concentration. EMBO J. 1986 Feb;5(2):343–354. doi: 10.1002/j.1460-2075.1986.tb04218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]