Abstract
We hypothesised that comparing the protein mixture in bronchoalveolar lavage fluid (BALF) between humans and mice may lead to mechanistic insights into common and divergent pathways that evolved in each species.
BALF from four humans and six mice was pooled separately and underwent identical shotgun proteomic analysis. Functional and network analysis was applied to identify overlapping and distinct pathways enriched in the BALF. Follow-up experiments using Western analysis in unpooled BALF samples were performed.
We identified 91 unique proteins in human and 117 unique proteins in mouse BALF samples. Functional analysis of the proteins revealed conservation of several key processes between the species, including defence response. Oxidative stress response, however, was selectively enriched only in mouse BALF. Differences in the expression of peroxiredoxin-1, a key member of the defence pathway against oxidative injury, were confirmed between normal human and mouse BALF and in models of lung injury.
A computational proteomics approach of mouse and human BALF confirms the conservation of immune and defence-mediated pathways while highlighting differences in response to oxidative stress. These observations suggest that the use of mice models to study human lung disorders should be undertaken with an appreciation of interspecies variability.
Keywords: Lung injury, network, oxidative stress, peroxiredoxin, proteomics
Mouse is the most common mammalian species for studying human diseases, including pulmonary disorders. However, promising findings based on murine models of lung disease often fail to translate into effective therapeutic targets in follow-up human studies. Since mice and humans diverged over 90 million yrs ago, significant interspecies differences may have accumulated between their respiratory system as each species adapted to diverse environmental challenges and exposures [1–3]. The functional and physiological divergence within compartments of the lung may account, in part, for the difficulties encountered when using the mouse as a model for human respiratory disorders.
The vast surface area of mammalian alveoli is lined by a thin layer of liquid that maintains the structural integrity of the airspaces, participates in gas exchange, and serves as a critical barrier in protecting the host from environmental and infectious insults. The alveolar epithelial lining fluid is a complex mixture of phospholipids, neutral lipids and proteins that is derived from resident cells (including type 1 and type 2 pneumocytes, alveolar macrophages and neutrophils) and through active and passive transport from plasma [4]. Profound alterations in the constituents of bronchoalveolar fluid occur in many human pulmonary disorders, prompting widespread analysis of bronchoalveolar lavage fluid (BALF) for diagnostic and prognostic purposes and to elucidate disease mechanisms. We and others have previously studied the human BALF proteome in various lung disorders [5–9]. Recently, we performed shotgun proteomic analysis of normal human BALF and observed that despite inter-individual differences in the detection of specific proteins, there was extensive overlap in the functional enrichment of these proteins across healthy subjects [10].
In this study we extend our previous work by initially assessing whether a global comparison of the complex protein mixture in BALF between humans and mice can lead to mechanistic insights into common and divergent pathways that have evolved in each species. Next, using Western analysis, we demonstrate that the expression level of a candidate member of an enriched pathway in mouse BALF is increased relative to normal human BALF levels, but follows species-specific expression pattern during acute lung injury.
Materials and Methods
BALF sample collection and processing
Normal human BALF
The protocol for collecting human BALF was approved by the institutional review board at the University of Washington, Seattle, WA, USA. Written informed consent was obtained from each healthy, nonsmoking subject prior to enrolment in the study. Samples from four male subjects were pooled and analysed. Bronchoalveolar lavage was performed as previously described [11]. Briefly, five separate 30-mL aliquots of 0.89% sterile saline were instilled into the right middle lobe or lingula. BALF was centrifuged immediately and cell-free supernatants were aliquoted and stored at -80°C.
Acute lung injury BALF
This study was approved by the institutional review board at the University of Washington. BALF was obtained from seven patients with acute lung injury defined as: 1) arterial oxygen tension (Pa,o2)/inspiratory oxygen fraction (Fi,o2) <300 mmHg; 2) diffuse parenchymal infiltrates; 3) pulmonary artery wedge pressure <18 mmHg or no clinical evidence of congestive heart failure; and 4) no other obvious explanation for these findings [12] in patients who were undergoing bronchoscopy for evaluation of clinically suspected ventilator-associated pneumonia based on the protocol of Fagon et al. [13]. Patient characteristics included: four males, three females, mean±sd age 55±10 yrs, number of days on ventilator prior to bronchoscopy 10±8; Pa,o2/Fi,o2 183±59 mmHg. Following bronchoscopy, the samples were immediately transported to the microbiology laboratory, where 5 mL was removed for microbiological analysis and the remaining sample was processed as described above.
Normal mouse BALF
All animal experiments were approved by the institutional animal care and use committee of University of Washington. Normal adult, male C57BL/6 mice (n=6) were lavaged with 1 mL of PBS containing 0.6 mM EDTA warmed to 37°C. BALF was centrifuged immediately after collection and cell-free supernatants were aliquoted into polypropylene tubes and stored at -80°C prior to proteomics analysis. Total protein measurements were made on aliquots of supernatants from all samples using the Bradford assay [14].
Shotgun proteomics analysis
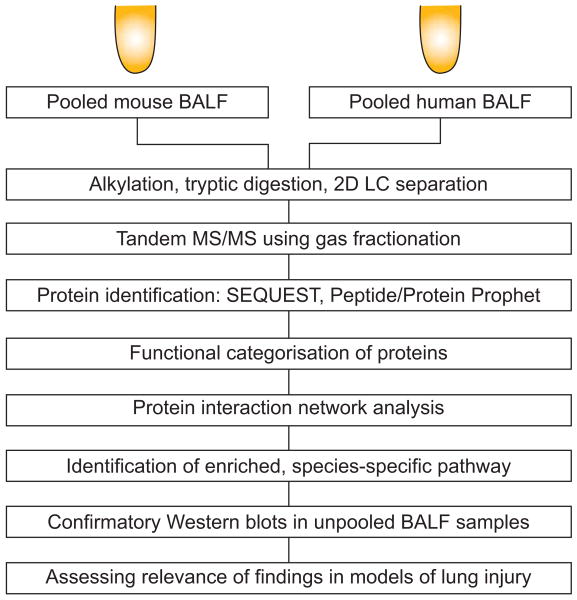
Pooled BALF samples from humans and mice were reduced, alkylated, tryptically digested and separated using two-dimensional liquid chromatography followed by tandem mass spectrometric analysis (LC-MS/MS) (fig. 1). BALF proteins were identified using SEQUEST search engine followed by PeptideProphet and ProteinProphet. Details of the shotgun proteomics methods are available in the supplementary material.
Figure 1.
Schematic overview of shotgun proteomic analysis of mouse and human bronchoalveolar lavage fluid (BALF). 2D LC: two-dimensional liquid chromatography; MS: mass spectrometry.
Sample preparation
Equal volumes of each cell-free BALF supernatant sample were denatured with 8 M urea and reduced by addition of 5 mM tris(2-carboxyethyl)phosphine hydrochloride. The solution underwent proteolysis for 16 h using a 1:20 w/w trypsin-to-protein ratio followed by desalting with micro-spin cartridge (Nest Group Inc., Southborough, MA, USA) and completely dried using a Speed-Vac (Thermo-Savant, Milford, MA, USA).
LC-MS/MS analysis
Equal sample concentrations of each sample was analysed by LC-MS/MS via electrospray ionisation online to a linear ion trap quadrupole (LTQ) Fourier transform (FT) ion cyclotron resonance mass spectrometer, as previously described [15]. The LTQ-FT mass spectrometer was operated in a data-dependent mode. The five most intense ions were sequentially isolated in the linear ion trap and subjected to collision-induced dissociation in series using a trap target value of 5,000 and 60 s of dynamic exclusion. Precursor ions were selected over the entire m/z range (350–2,000). Then, the experiment was repeated using gas phase fraction (e.g. precursor ions were selected over a limited m/z range during each injection). The following six m/z ranges were applied: 350–550, 500–700, 650– 850, 800–1,000, 950–1,500 and 1,450–2,000.
Protein identification
Tandem mass spectral RAW (ThermoFinnigan, Waltham, MA, USA) files were first converted to mzXML format [16]. The tandem mass spectra were then matched to protein sequence in the IPI Human and Mouse database (European Bioinformatics Institute, Hinxton, UK; www.ebi.ac.uk/IPI) using SEQUEST. Criteria for matching a peptide tandem mass spectrum to a peptide sequence were: Xcorr >1.9 with charge state 1+, Xcorr >2.2 with charge state 2+, or Xcorr >3.75 with charge state 3+, as well as ΔCn >0.1. Peptide tandem mass spectra passing these criteria were utilised for protein identifications. A protein was considered to be identified only if ProteinProphet probability >0.8 [17] and if more than one unique peptide was found for each protein.
Functional and network analyses
Enrichment of biological modules in the BALF proteome of each species was determined relative to its respective global proteome which was defined based on its known genome and annotated using Entrez Gene Identification (www.ncbi.nlm.nih.gov/Entrez/) [1, 10]. We used a web-based implementation of Database for Annotation, Visualization and Integrated Discovery (DAVID) [18] and GOTree Machine [19] to functionally annotate identified proteins in BALF based on the Gene Ontology (GO) database (www.geneontology.org) [20]. Enrichment p-values for functional categories were determined using a variant of the one-tailed Fisher exact probability of the hypergeometric distribution. Since many GO categories were evaluated simultaneously, a function was deemed significantly “enriched” based on a permutation-based false discovery rate cut-off <0.05.
A protein interaction network was created from combining all proteins mapped to catalytic and oxidative stress functional groups, and using several databases to obtain biologically verified protein interactions: 1) Ingenuity System's knowledge base, which has been manually curated from >200,000 full-text, peer-reviewed scientific articles and consists of protein relationships from over 15,000 human, mouse and rat genes [21]; 2) Human Protein Reference Database, consisting of ∼35,000 protein interactions [22]; 3) Biomolecular Interaction Network Database, comprising ∼200,000 interactions [23]; and 4) Database of Interacting Proteins, containing ∼55,000 interactions [24].
Animal models of acute lung injury
All animal experiments were performed using adult, male C57BL/6 mice from the same vendor (Jackson Laboratory, Bar Harbor, ME, USA). Additional details are provided in the online supplementary material.
Hyperoxia
Mice (n=6) were housed in a plexiglass chamber located in a biosafety hood. Oxygen flow through the cage was maintained at 5 L·min-1, which was sufficient to achieve an oxygen fraction of 95% within the plexiglass chamber. Mice were observed daily and oxygen fraction was checked daily. Animals were exposed to hyperoxia or normoxia (Fi,o2 0.21) for 72 h, and BALF collected, centrifuged and processed as described above. In some mice, the lungs were inflated and fixed with 4% paraformaldehyde at 25 cmH2O pressure, paraffin-embedded and sections were stained with haematoxylin and eosin for histology.
Ischaemia/reperfusion
Mechanically ventilated, isoflurane anaesthetised mice (n=3) were ventilated with a tidal volume of 10 mL·kg-1, a rate of 150 breaths·min-1, an end-expiratory pressure of 3 cmH2O and Fi,o2 0.4. Left lung warm ischaemia for 60 min was achieved via thoracotomy and placement of a vascular clamp across the left hilum. During left hilar clamping, tidal volume was decreased to 6 mL·kg-1 and respiratory rate increased as necessary to achieve a similar end-tidal CO2. Subsequently, the clamp was removed, tidal volumes and respiratory rates were returned to baseline, and left lung reperfusion was permitted for an additional 4 h prior to euthanasia and necropsy. After euthanasia, the right hilum was clamped and left lung was lavaged and BALF was processed as described.
Staphylococcus aureus pneumonia
S. aureus was originally isolated from a patient with catheter-related sepsis and cultured as previously described [25]. Mice (n=3) were inoculated with 2×107 CFU of S. aureus in 50 μL PBS via oropharyngeal aspiration [25]. Mice were euthanised 24 h post-infection and right lung was lavaged and BALF was collected and processed as above.
Influenza pneumonia
Mice (n=3) were inoculated with 4,000 PFU of A/PR8/34 (mouse-adapted H1N1 influenza strain) in 50 μL PBS via oropharyngeal aspiration. At 120 h post-infection, mice were euthanised and BALF obtained and processed as described above.
Western analysis
Equal amounts of BALF protein (20 mg) were separated by SDS-PAGE, and electrophoretically transferred to polyvinylidene fluoride membrane. Membranes were blocked with 5% nonfat dry milk/0.05% Tween-20/PBS for 2 h, incubated with rabbit polyclonal anti-PRDX1 antibody (1:10,000, Abcam, Cambridge, MA, USA) for 2 h at room temperature, washed with 0.05% Tween-20/PBS, incubated with horseradish peroxidase-conjugated secondary antibody (1:5,000) for 1 h, washed with 0.05% Tween-20/PBS and then developed with enhanced chemilumi-nescence technique (Amersham, Buckinghamshire, UK).
Results
The BALF proteome in human and mouse
We defined the BALF proteome from four human subjects and six mice using a shotgun approach. We identified 91 distinct proteins in humans and 117 distinct proteins in mice (supplementary table 1). 21 of these proteins were products of homologous genes in both species. Since we applied stringent criteria to increase our confidence in accurate protein identification, we may not have generated an exhaustive list of BALF proteins. Therefore, we approached the interspecies proteomics comparison via a pathway-centric focus.
Functional and network analysis of BALF proteome
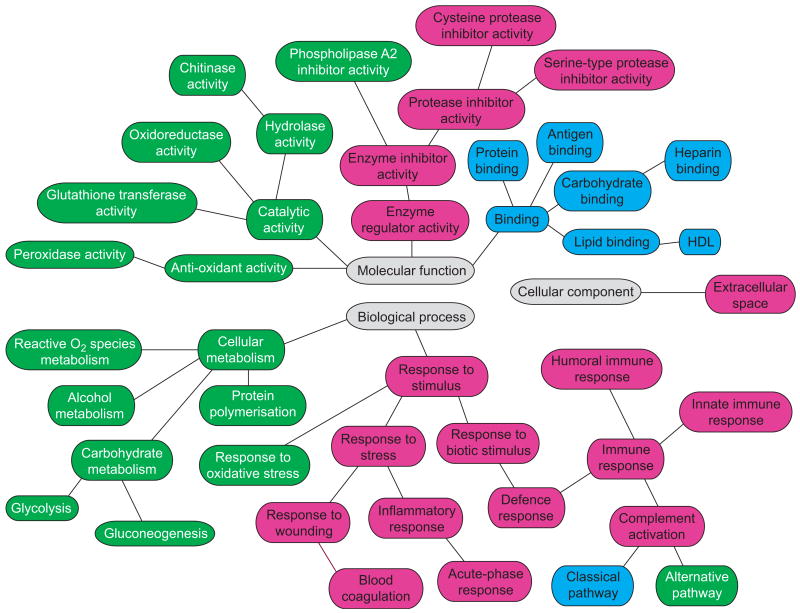
We categorised identified proteins in mouse and human BALF based on their GO annotation and determined enriched categories for each species relative to its respective global proteome. Despite differences in the actual protein identification between mice and human BALF, functional analysis revealed significant interspecies overlap in many processes, including inflammatory response, defence response, immunity and protease activity (fig. 2). In general, the over-represented categories common to both mouse and human were among the most enriched, suggesting that these pathways have been preserved across these species and encompass key functions of the lung's epithelial barrier. However, comparing the enrichment of functional categories side by side (fig. 3) revealed relative differences among processes over-represented in both species. For example, immune and defence pathways were highly enriched in human BALF (enrichment p-value ∼10-10 for human and ∼10-4 in mouse), while protease and enzyme inhibition was more over-represented in mouse BALF (enrichment p-value ∼10-12 for mouse and ∼10-8 in human). Please refer to supplementary table 2 for more details.
Figure 2.
Functional analysis of mouse and human bronchoalveolar lavage fluid proteome using Gene Ontology's hierarchical annotation tree. Categories representing biological processes, molecular functions and cellular components are colour-coded based on the species-specific enrichment (false discovery rate <0.05). Pink: modules enriched in mouse and human; green: modules enriched in mouse; blue: modules enriched in human. HDL: high-density lipoprotein.
Figure 3.
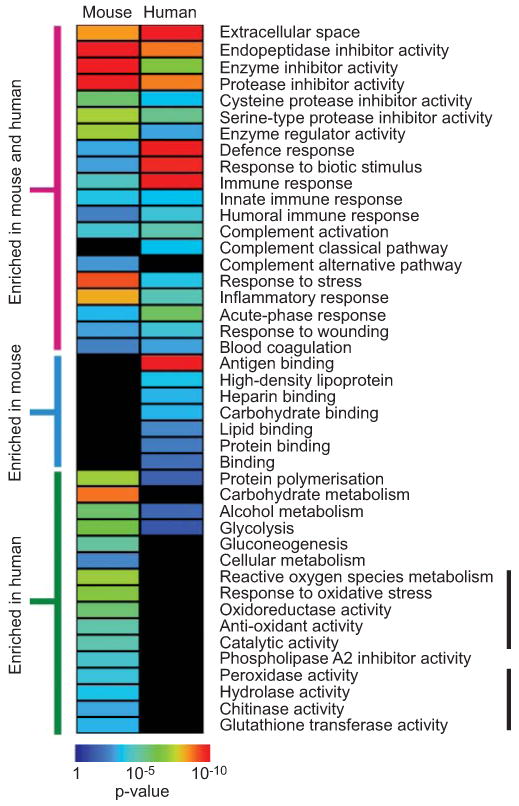
Heat map representation of over-represented functional categories in bronchoalveolar lavage fluid (BALF) proteome of mice and humans. This depiction of the same data from figure 2 allows side by side comparison of the relative enrichment of each category between the two species. Note the selective enrichment of functional modules involved in oxidative stress response in mouse BALF (indicated by vertical black bar). p-values indicate the probability of identifying a given category as enriched by random chance alone.
Importantly, we also identified processes that were significantly enriched in one species but not the other. Biological processes and molecular functions involved in cellular metabolism, catalytic activity, and antioxidant activity were differentially over-represented only in murine BALF, while a number of molecular functions involving antigen, heparin and carbohydrate binding were only enriched in human BALF.
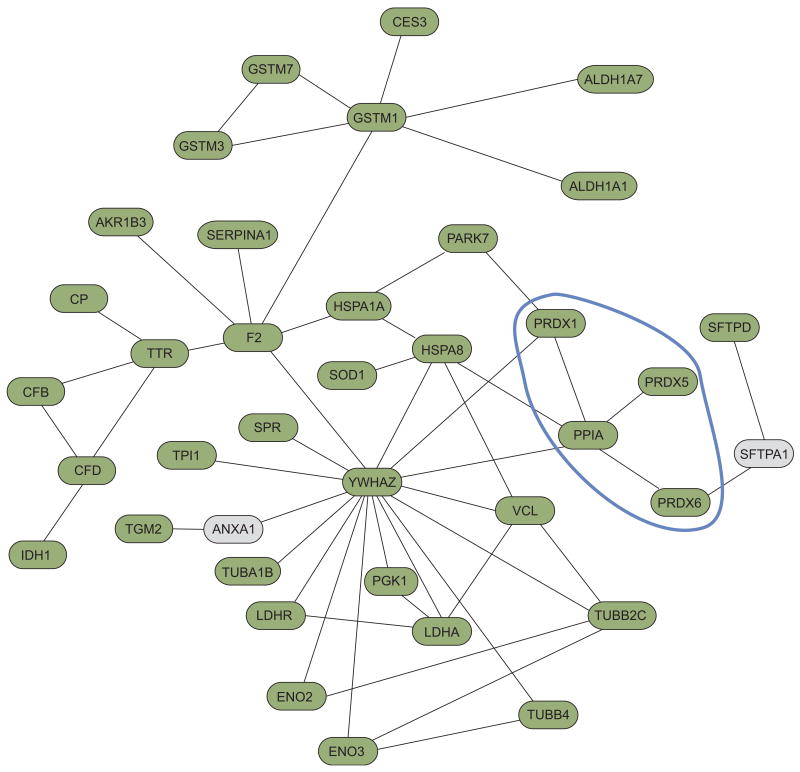
The most striking observation was the persistent and selective enrichment of processes involved in oxidative stress in mouse BALF (∼20-fold enrichment, p-value ∼10-6 to 10-7). To better elucidate the relationship among the proteins mapped to this module, we created a protein network based on previously published gene product interactions (fig. 4). This “interactome” mapped the relationships among key players in cellular response to oxidative challenge and included members of the glutathione pathway (GSTM1, GSTM3 and GSTM7), superoxide dismutase (SOD1), and heat shock proteins (HSPA1A and HSPA8). Furthermore, we identified a sub-network comprised of several members of the peroxiredoxin family (PRDX1, PRDX5 and PRDX6) interacting with peptidylprolyl isomerase A (PPIA; also known as cyclophilin A).
Figure 4.
Protein interaction network of the oxidative stress module. This interactome is comprised of proteins linked to processes selectively enriched in mouse bronchoalveolar lavage fluid. A few proteins not mapped to this module (shown in grey) were included to maximise the spanning topology of the network. The peroxiredoxin family, including its relationship with peptidylprolyl isomerase A, is marked by the blue boundary.
PRDX1 expression in human and mouse BALF
To verify interspecies differences in the oxidative stress module, we performed Western analysis to compare the relative expression of PRDX1 in independent, unpooled BALF samples from normal mice and human subjects (n=4 per group). We observed a prominent signal in all mice BALF samples but minimal signal in the human samples (fig. 5). Next, we examined how PRDX1 levels in BALF were altered during acute lung injury using independent human patient samples and in mouse samples during four different models of lung injury: 1) exposure to hyperoxia; 2) influenza pneumonia; 3) S. aureus pneumonia; and 4) ischaemia/reperfusion.
Figure 5.
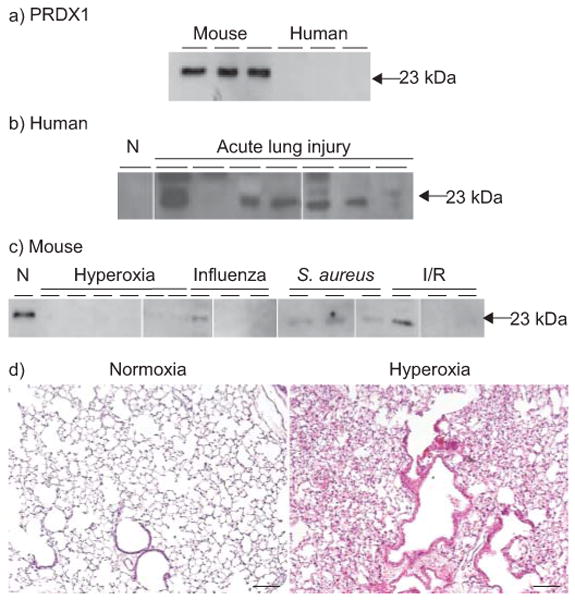
Western blot analysis of peroxiredoxin 1 (PRDX1) in bronchoalveolar lavage fluid (BALF). a) BALF from three separate normal human and three separate mouse samples confirms differential expression of PRDX1 in mouse. b) Increased BALF PRDX1 in patients with acute lung injury (n=7) compared with normal subjects (N). c) Decreased BALF PRDX1 levels in four mouse models of lung injury compared with control mice (N): hyperoxia (n=6); influenza pneumonia (n=3); Staphylococcus aureus pneumonia (n=3); and ischaemia/reperfusion (I/R; n=3). d) Representative histology of mouse lungs during hyperoxia (inspiratory oxygen fraction (Fi,o2) 0.95, 72 h) demonstrates diffuse lung injury compared with normoxia (Fi,o2 0.21).
In patients with acute lung injury, PRDX1 was significantly increased in six out of seven patients compared with healthy controls (fig. 5b). In mice, an opposite pattern emerged during lung injury, with PRDX1 levels dramatically reduced compared with normal controls, especially during hyperoxia exposure (fig. 5c). We further confirmed the profound susceptibility of mice to hyperoxia by examining lung histology of animals exposed to 72 h of 95% oxygen. We observed diffuse epithelial injury, with perivascular oedema and inflammatory cellular infiltrates within airspaces (fig. 5d).
Discussion
We performed a comparative shotgun proteomics analysis of bronchoalveolar fluid in mice and humans to assess common and divergent functional pathways (fig. 1). We found that many processes critical for host defence against a hostile environment, including immunity, defence response and protease inhibitor activity, are highly enriched in the BALF of both species. However, we also observed significant interspecies differences, particularly in oxidative stress pathways, implying that evolution under unique external and genetic pressures has led to distinct functional profiles in the bronchoalveolar fluid proteome of mice and humans. We confirmed the relevance of our findings using unpooled samples by demonstrating a distinctly different expression pattern of PRDX1, a representative oxidative stress protein, in normal human and mice BALF and during lung injury.
Although many key functional categories were highly enriched in both species, there were differences in their relative enrichment (fig. 3). For example, proteins mapping to defence response and immunity were more over-represented in human BALF compared with mice. A simple explanation may be that since the mice were raised under specific pathogen-free conditions and not exposed to the full spectrum of pathogenic micro-organisms, the proteomic profile of their epithelial lining fluid was less enriched in immune-related functions.
The most prominent interspecies difference in BALF proteome was the selective enrichment of processes involved in cellular defence against oxidative stress in mice. We further explored this finding by creating an interacting network comprised of proteins functionally mapped to these processes. Members of this interactome included well-established mediators of cellular defence against reactive oxygen species, such as the glutathione pathway and superoxide dismutase. In mice, isoenzymes of superoxide dismutase play a critical role in protecting the lung against hyperoxia-induced injury [26, 27].
Within this murine-enriched oxidative stress module, we identified a sub-network of interacting peroxiredoxins (PRDX1, PRDX5 and PRDX6). These proteins belong to family of nonseleno-peroxidases that catalyse the reduction of a broad spectrum of peroxides by electron transfer from thioredoxins or cyclophilins. Peroxiredoxins are emerging as key protectors of the lung from injury due to reactive oxygen metabolites. For example, transgenic mice lacking PRDX6 are much more susceptible to lung damage from hyperoxia, whereas over-expression of PRDX6 protects mice from hyperoxia-induced lung injury [28, 29]. We confirmed the differential expression of PRDX1 in mouse BALF samples relative to humans. Since a role for PRDX1 in lung injury has not been reported, we analysed PRDX1 expression in several mouse models of lung injury associated with prominent oxidative stress. We demonstrated that PRDX1, while abundantly expressed in the BALF of control mice, is dramatically decreased during lung injury due to hyperoxia, ischaemia/reperfusion and pneumonia. Interestingly, PRDX1 followed an opposite expression pattern in human BALF, becoming much more abundant after acute lung injury. These observations suggest that while PRDX1 may serve as an important peroxidase in lung defence against oxidative insults, there are distinct interspecies differences in its expression profile.
One advantage of integrating our proteomics approach with network analysis is its potential ability to provide novel insights into molecular mechanisms. For example, all of the identified peroxiredoxins in mouse BALF interacted with PPIA (also known as cyclophilin A). There is increasing evidence that hubs of high connectivity within biological networks are important for its functional stability [30–32]. Therefore, based on our computational analysis, we predict that PPIA is a putative target for modulating the lung's anti-oxidant defence programme. In support of this proposition, previous in vitro experiments have demonstrated that PPIA directly binds to peroxiredoxins and profoundly increases their enzymatic activity [33]. Interestingly, cyclosporine A can bind to PPIA and alter its activity, and mice treated with this drug demonstrate significant attenuation in lung injury during exposure to hyperoxia [34].
While our study provides insights into the mechanisms of susceptibility of C57BL/6 mice to hyperoxia-induced lung injury, there is significant strain-dependent variability in this response [35]. We chose C57BL/6 mice because they are the most commonly used strain in biomedical research and have an “intermediate” susceptibility to hyperoxia-induced lung injury [35]. Future work based on a similar proteomics approach may help elucidate pathway-specific mechanisms across phenotypically distinct strains.
Other investigators have also reported on proteomic analysis of BALF in mice and humans. For example, Guo et al. [36] combined a gel-based and shotgun approach to define the mouse BALF proteome, and identified a total of 297 unique proteins. Although this number is substantially larger than our finding of 117 unique proteins, many factors, ranging from sample preparation to instrumentation and database searching criteria, can lead to significant variability in shotgun proteomic experiments. Nevertheless, if the number of proteins identified by Guo et al. [36] is limited to those obtained using the shotgun method, and based on identifying more than one unique peptide, the number of unique proteins is reduced to 126 and is comparable to our report. Further comparison of their study with ours reveals that many common functional categories, including oxidative stress, complement activation, proteolysis and inflammatory response, were identified in the mouse BALF proteome by both groups. Interestingly, and consistent with our findings, Guo et al. [36] also reported the presence of PPIA and several peroxiredoxin proteins, including PRDX1, PRDX5 and PRDX6 in mouse BALF.
There are a number of limitations in this study. Since the primary aim of this work was to gain insights into common and divergent pathways in human and mouse BALF, we decided to pool the lavage samples from each species. Although pooling raises the possibility that an outlier sample could bias the findings, we previously demonstrated that despite inter-individual differences, most of the highly enriched processes in human BALF proteome are tightly preserved across subjects [10]. Our (unpublished) data show even tighter homogeneity of the BALF proteome in mice. We did not directly compare the relative abundance of proteins between mouse and human BALF samples. We refrained from performing quantitative comparisons, e.g. using spectral counting [37], because the majority of the identified proteins did not overlap between the species, making a direct comparison impossible. However, we verified the differential expression of PRDX1 in mice relative to humans using Western analysis. Finally, while detection of BALF proteins does not indicate their cellular sources, there is ample evidence that differential protein expression in BALF correlates with disease phenotype and severity. Furthermore, the relative ease in sampling the airspaces to search for mediators of lung disorders ensures its continued widespread application in research and clinical settings.
In summary, our work provides a framework for the comparative analysis of human and mouse BALF proteome. Despite the identification of relatively few common homologous proteins, key functional categories involved in immunity, defence response and protease inhibition were preserved in both species. However, we also identified significant functional differences in the BALF proteome, most prominently in the selective enrichment of oxidative stress pathways in mice. Thus, while the utilisation of murine models to study human lung disorders has been successful in bringing bench research to bedside practice, this approach should be undertaken with an appreciation of inherent interspecies differences.
Supplementary Material
Acknowledgments
The authors would like to thank Y-A. Goo (Dept of Medicinal Chemistry, University of Washington, Seattle, WA, USA) for assisting with the proteomics analysis, D. An and T. Lozon (Center for Lung Biology, University of Washington) for assisting with the animal experiments, and F. Turcer and M. Whang (Center for Lung Biology) for performing the Western analyses.
Support Statement: This work was supported by National Institutes of Health/National Heart, Lung, and Blood Institute (Bethesda, MD, USA) HLBI HL074223 (S.A. Gharib), HL083481 (L.M. Schnapp), HL086796 (L.M. Schnapp), and UL1RR025014 (S.A. Gharib).
Footnotes
This article has supplementary material available from www.erj.ersjournals.com
Statement of Interest: A statement of interest for C.E. Doneanu can be found at www.erj.ersjournals.com/misc/statements.dtl
References
- 1.Waterston RH, Lindblad-Toh K, Birney E, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 2.Church DM, Goodstadt L, Hillier LW, et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy WJ, Pringle TH, Crider TA, et al. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray JF, Nadel JA. Murray and Nadel's Textbook of Respiratory Medicine. 4th. Philadelphia: Saunders; 2005. [Google Scholar]
- 5.Chang DW, Hayashi S, Gharib SA, et al. Proteomic and computational analysis of bronchoalveolar proteins during the course of the acute respiratory distress syndrome. Am J Respir Crit Care Med. 2008;178:701–709. doi: 10.1164/rccm.200712-1895OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gharib SA, Vaisar T, Aitken ML, et al. Mapping the lung proteome in cystic fibrosis. J Proteome Res. 2009;8:3020–3028. doi: 10.1021/pr900093j. [DOI] [PubMed] [Google Scholar]
- 7.Schnapp LM, Donohoe S, Chen J, et al. Mining the acute respiratory distress syndrome proteome: identification of the insulin-like growth factor (IGF)/IGF-binding protein-3 pathway in acute lung injury. Am J Pathol. 2006;169:86–95. doi: 10.2353/ajpath.2006.050612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rottoli P, Magi B, Perari MG, et al. Cytokine profile and proteome analysis in bronchoalveolar lavage of patients with sarcoidosis, pulmonary fibrosis associated with systemic sclerosis and idiopathic pulmonary fibrosis. Proteomics. 2005;5:1423–1430. doi: 10.1002/pmic.200301007. [DOI] [PubMed] [Google Scholar]
- 9.Kriegova E, Melle C, Kolek V, et al. Protein profiles of bronchoalveolar lavage fluid from patients with pulmonary sarcoidosis. Am J Respir Crit Care Med. 2006;173:1145–1154. doi: 10.1164/rccm.200507-1126OC. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Ryu S, Gharib SA, et al. Exploration of the normal human bronchoalveolar lavage fluid proteome. Proteomics Clin Appl. 2008;2:585–595. doi: 10.1002/prca.200780006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinberg KP, Milberg JA, Martin TR, et al. Evolution of bronchoalveolar cell populations in the adult respiratory distress syndrome. Am J Respir Crit Care Med. 1994;150:113–122. doi: 10.1164/ajrccm.150.1.8025736. [DOI] [PubMed] [Google Scholar]
- 12.Bernard GR, Artigas A, Brigham KL, et al. Report of the American-European Consensus conference on acute respiratory distress syndrome: definitions, mechanisms, relevant outcomes, and clinical trial coordination. Consensus Committee. J Crit Care. 1994;9:72–81. doi: 10.1016/0883-9441(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 13.Fagon JY, Chastre J, Wolff M, et al. Invasive and noninvasive strategies for management of suspected ventilator-associated pneumonia. A randomized trial. Ann Intern Med. 2000;132:621–630. doi: 10.7326/0003-4819-132-8-200004180-00004. [DOI] [PubMed] [Google Scholar]
- 14.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 15.Yi EC, Lee H, Aebersold R, et al. A microcapillary trap cartridge-microcapillary high-performance liquid chromatography electrospray ionization emitter device capable of peptide tandem mass spectrometry at the attomole level on an ion trap mass spectrometer with automated routine operation. Rapid Commun Mass Spectrom. 2003;17:2093–2098. doi: 10.1002/rcm.1150. [DOI] [PubMed] [Google Scholar]
- 16.Pedrioli PG, Eng JK, Hubley R, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 17.Nesvizhskii AI, Keller A, Kolker E, et al. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 18.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 19.Zhang B, Schmoyer D, Kirov S, et al. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calvano SE, Xiao W, Richards DR, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 22.Peri S, Navarro JD, Amanchy R, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alfarano C, Andrade CE, Anthony K, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salwinski L, Miller CS, Smith AJ, et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhanireddy S, Altemeier WA, Matute-Bello G, et al. Mechanical ventilation induces inflammation, lung injury, and extra-pulmonary organ dysfunction in experimental pneumonia. Lab Invest. 2006;86:790–799. doi: 10.1038/labinvest.3700440. [DOI] [PubMed] [Google Scholar]
- 26.Carlsson LM, Jonsson J, Edlund T, et al. Mice lacking extracellular superoxide dismutase are more sensitive to hyperoxia. Proc Natl Acad Sci USA. 1995;92:6264–6268. doi: 10.1073/pnas.92.14.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wispe JR, Warner BB, Clark JC, et al. Human Mn-superoxide dismutase in pulmonary epithelial cells of transgenic mice confers protection from oxygen injury. J Biol Chem. 1992;267:23937–23941. [PubMed] [Google Scholar]
- 28.Wang Y, Feinstein SI, Manevich Y, et al. Lung injury and mortality with hyperoxia are increased in peroxiredoxin 6 gene-targeted mice. Free Radic Biol Med. 2004;37:1736–1743. doi: 10.1016/j.freeradbiomed.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Phelan SA, Manevich Y, et al. Transgenic mice over-expressing peroxiredoxin 6 show increased resistance to lung injury in hyperoxia. Am J Respir Cell Mol Biol. 2006;34:481–486. doi: 10.1165/rcmb.2005-0333OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gharib SA, Liles WC, Klaff LS, et al. Noninjurious mechanical ventilation activates a proinflammatory transcriptional program in the lung. Physiol Genomics. 2009;37:239–248. doi: 10.1152/physiolgenomics.00027.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovegrove FE, Gharib SA, Patel SN, et al. Expression microarray analysis implicates apoptosis and interferon-responsive mechanisms in susceptibility to experimental cerebral malaria. Am J Pathol. 2007;171:1894–1903. doi: 10.2353/ajpath.2007.070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luscombe NM, Babu MM, Yu H, et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 33.Lee SP, Hwang YS, Kim YJ, et al. Cyclophilin A binds to peroxiredoxins and activates its peroxidase activity. J Biol Chem. 2001;276:29826–29832. doi: 10.1074/jbc.M101822200. [DOI] [PubMed] [Google Scholar]
- 34.Matthew E, Pun R, Simonich M, et al. Cyclosporin A protects lung function from hyperoxic damage. Am J Physiol. 1999;276:L786–L795. doi: 10.1152/ajplung.1999.276.5.L786. [DOI] [PubMed] [Google Scholar]
- 35.Whitehead GS, Burch LH, Berman KG, et al. Genetic basis of murine responses to hyperoxia-induced lung injury. Immunogenetics. 2006;58:793–804. doi: 10.1007/s00251-006-0147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Y, Ma SF, Grigoryev D, et al. 1-DE MS and 2-D LC-MS analysis of the mouse bronchoalveolar lavage proteome. Proteomics. 2005;5:4608–4624. doi: 10.1002/pmic.200500052. [DOI] [PubMed] [Google Scholar]
- 37.Fu X, Gharib SA, Green PS, et al. Spectral index for assessment of differential protein expression in shotgun proteomics. J Proteome Res. 2008;7:845–854. doi: 10.1021/pr070271+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.