Abstract
Peroxisome proliferator-activated receptor γ (PPAR γ) is widely expressed in macrophages and has been identified as a putative target for the development of novel therapies against inflammatory bowel disease (IBD). Computational simulations identified macrophages as key targets for therapeutic interventions against IBD. This study aimed to characterize the mechanisms underlying the beneficial effects of macrophage PPAR γ in IBD. Macrophage-specific PPAR γ deletion significantly exacerbated clinical activity and colonic pathology, impaired the splenic and mesenteric lymph node regulatory T cell compartment, increased percentages of LP CD8+ T cells, increased surface expression of CD40, Ly6C, and TLR-4 in LP macrophages, and upregulated expression of colonic IFN-γ, CXCL9, CXCL10, IL-22, IL1RL1, CCR1, suppressor of cytokine signaling 3 and MCH class II in mice with IBD. Moreover, macrophage PPAR γ was required for accelerating pioglitazone-mediated recovery from DSS colitis, providing a cellular target for the anti-inflammatory effects of PPAR γ agonists in IBD.
INTRODUCTION
Inflammatory bowel disease (IBD), with its two clinical manifestations, Ulcerative Colitis (UC) and Crohn’s Disease (CD), is an immune-mediated disease characterized by widespread inflammation and immune cell infiltration of the gastrointestinal tract. The etiology of IBD is multifactorial, and entails interaction among genetic predisposition, environmental factors and the gut microbiota. Treatments targeted to down-modulate the immune and inflammatory responses, such as the corticosteroid prednisone or the anti-TNF-α antibody Remicade, have shown promise in reducing severity and reoccurrence of the disease. These treatments, however, are also associated with various adverse side effects, such as cushingoid appearance, weight gain, and systemic immunosuppression, thus stressing the need to develop safer alternatives for the long-term management of IBD 1.
Peroxisome proliferator-activated receptor γ (PPAR γ) agonists have also demonstrated efficacy in ameliorating intestinal inflammation associated with IBD 2–4. PPAR γ is a transcription factor expressed highly in all the major cell types involved in IBD pathogenesis, including intestinal epithelial cells (IECs), macrophages, dendritic cells, and lymphocytes. Chronic administration of synthetic PPAR γ ligands (i.e., thiazolidinedione, TZD, class of antidiabetic drugs) is also linked with adverse side-effects that led to a mandatory labeling with a black box warning 5. Nonetheless, PPAR γ can also become activated by safer compounds such as dietary lipids including conjugated linoleic acid (CLA), or endogenous lipid mediators produced during inflammation such as 15(S)-HETE, 13(S)-HODE, and other unsaturated fatty acids 6, 7 or the isoprenoid abscisic acid (ABA) 8, 9, thereby heightening the potential for developing safer and more efficacious therapies against gut inflammatory diseases.
We demonstrated that mice lacking PPAR γ broadly in all immune and epithelial cells are irresponsive to the beneficial affects of the naturally occurring ligand of PPAR γ CLA in experimental models of colitis 2, 10–12 and colorectal cancer 13, though the specific immune and/or epithelial cells required for the PPAR γ-induced anti-inflammatory effects of CLA were still unclear. By using the CD4+CD45RBhi adoptive transfer model of colitis we demonstrated that PPAR γ is required for the anti-inflammatory activity of Treg against effector CD4+ T cell-induced colitis 12. Recent examination of more narrow cell-specific PPAR γ knockout mice that underwent CD4-Cre- and Villin-Cre-mediated recombination revealed that PPAR γ expressed in T cells and IECs, respectively, contribute to protection against experimental IBD through immunoregulatory mechanisms involving both T cells and macrophages 14–16. For instance, in T cell-specific PPAR γ knockout (CD4-Cre+) mice, MLN and blood had less regulatory T (Treg) cells and inflammatory and cell adhesion molecules and suppressor of cytokine signaling 3 (SOCS-3) were significantly upregulated in the colonic mucosa as compared to CD4-Cre-with a wild-type phenotype 14. The IEC-specific PPAR γ knockout (Villin-Cre+) mice had significantly worsened disease severity and up-regulated lysosomal pathway and antigen presentation-related gene expression while modulating expression of genes in the p53 tumor suppressor pathway in comparison to their littermate PPAR γ -expressing (Villin-Cre−) controls 16.
Computational simulations using a mathematical model of the cellular interactions at colonic mucosa and MLN during colitis identified macrophages and their mechanisms of plasticity as key targets for therapeutic interventions against IBD 17. In this regard, the effect of the macrophage-specific PPAR γ deletion on experimental IBD was previously examined by Shah et al 18, who found that macrophage-specific PPAR γ knockout mice were more susceptible to DSS colitis than wild-type counterparts. In addition to showing significantly worsened disease activity, macrophage-specific PPAR γ knockout mice showed an up-regulation in colonic expression of chemokines and inflammatory cytokines 18. These findings suggest that PPAR γ expression in macrophages may act to limit colonic recruitment of blood-derived immune cells. However, Shah and colleagues did not examine whether deletion of PPAR γ in macrophages affected T cell populations in effector or inductive sites of the gut associated lymphoid tissue or the global colonic transcriptome. This is important since the balance between effector and regulatory immune cell populations can significantly modulate the inflammatory response and tissue damage in IBD. This study aimed to characterize the immunoregulatory mechanisms by which macrophage PPAR γ ameliorates experimental IBD. We demonstrate that the deficiency of PPAR γ in macrophages increases the expression of colonic inflammatory and fatty acid oxidation genes, impairs the peripheral Treg compartment, favors a pro-inflammatory macrophage phenotype in the colonic LP and an influx of effector T cells at the colonic mucosa during IBD; all of which implies a possible cross-talk between macrophages and T cells coordinated by PPAR γ.
RESULTS
The deficiency of PPAR γ in macrophages worsens colitis severity
To examine the effect of macrophage-specific PPAR γ deficiency on colitis severity LysM-Cre+ and LysM-Cre− mice were treated with 2.5% DSS for 7 days. Figure 1A illustrates the deletion of PPAR γ in the macrophages, spleen, MLN and colon of LysM-Cre+ mice as well as LysM-Cre− mice. The results show that the deletion is more efficient in purified macrophages in comparison to the other tissues containing mixed cell populations. These PCR results are in line with the western blot analyses showing that the amount of PPAR γ protein is lower in macrophages and colons from LC+ mice when compared to LC− mice (Figure 1B). In addition, the degree of deletion of PPAR γ is more effective in macrophages than in colonic samples. This can be explained due to the fact that the entire colon contains epithelial cells and other immune cells that express normal levels of PPAR γ since the Lysozyme M promoter is expressed in myeloid cells but not epithelial or lymphoid cells. Figure 1C illustrates the suppressive effect of macrophage-specific PPAR γ deficiency on colonic PPAR γ and CD36 mRNA expression in healthy mice. Similar suppression of PPAR γ and CD36 expression was found for the other tissues (data not shown).
Figure 1. Genotyping of PPAR γ flfl; Lysozyme M-Cre+ (LC+) and Lysozyme MCre- (LC−) control mice.
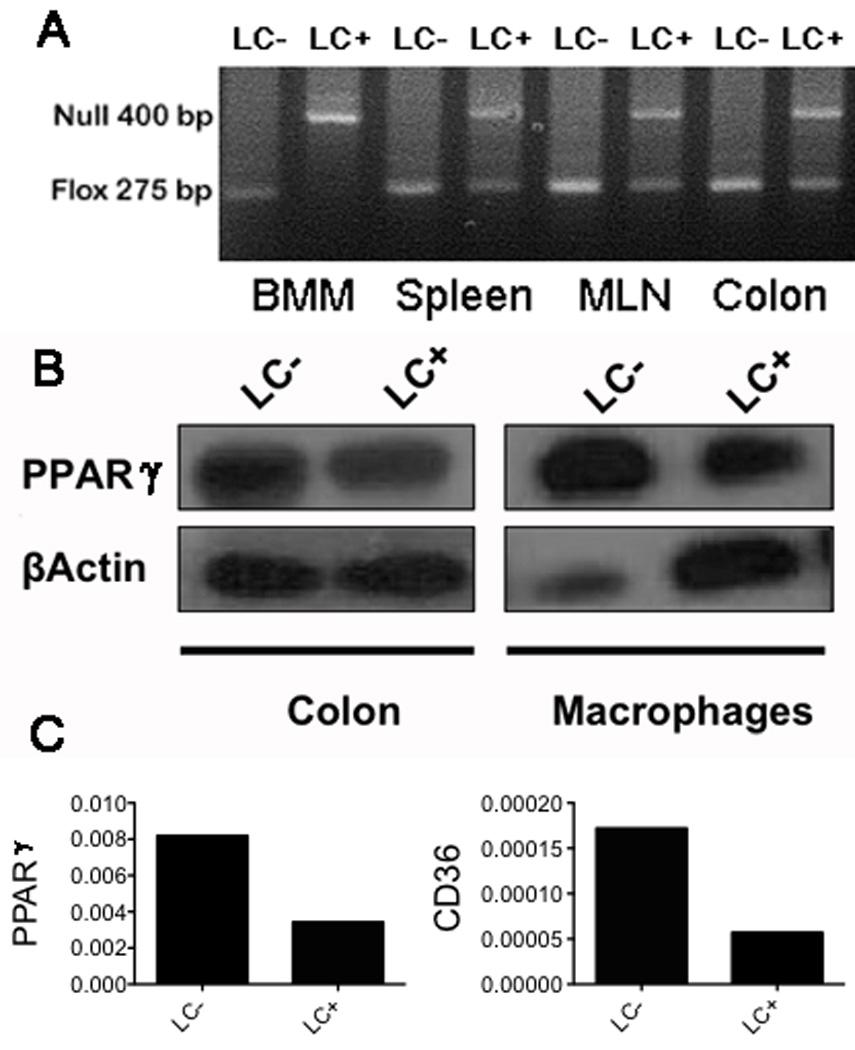
Conditional deletion of the PPAR γ gene via Lysozyme M-Cre-mediated recombination was examined in mouse tissues by PCR analysis. The flox (fl) allele at 275 bp and the null allele at 400 bp. (A) Left to right: depicts fl/fl in bone marrow-derived macrophages (BMM) without recombination (LC−) (lane 1) or with recombination (LC+) (lane 2). Homogenized whole spleen, mesenteric lymph node (MLN), and colon without recombination (LC−) (lanes 3, 5, 7) or with recombination (LC+) (lanes 4, 6, 8). (B) Left to right: depicts PPAR γ (top panel) and β actin (bottom panel) protein expression as measured by Western Blot in BMM and colon of LC− (lanes 1 and 3) and LC+ (lanes 2 and 4) mice. (C) Quantification of PPAR γ (left panel) and CD36 (right panel) colonic mRNA expression by real-time quantitative RT-PCR in LC− and LC+ mice.
To investigate the effect of macrophage-specific PPAR γ deficiency on the severity of experimental IBD, PPAR γ flfl Cre+ mice and control PPAR γ flfl Cre-littermates were treated with 2.5% DSS in the drinking water for 7 days, and disease activity was monitored daily. Macrophage-specific PPAR γ null mice had significantly worsened disease activity throughout the 7-day challenge as compared to PPAR γ-expressing littermates (Figure 2A). On day 7 colons, MLN and spleens from mice in each group were scored on gross anatomical signs of inflammation. In line with the DAI scores, both the colons and spleens were significantly more inflamed in PPAR γ flfl Cre+ mice than in PPAR γ-expressing littermate mice (Figures 2B and C).
Figure 2. Effect of macrophage-specific PPAR γ deletion on disease severity and macroscopic lesions in spleen and colon of mice with experimental IBD.
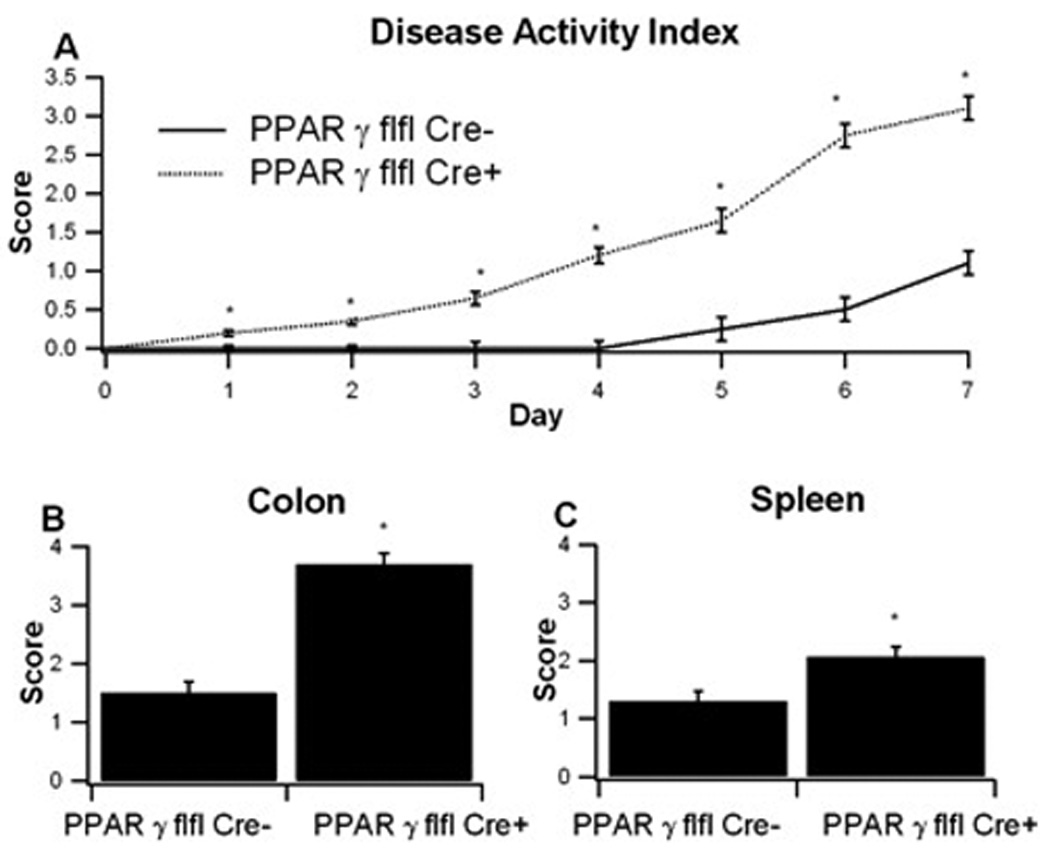
PPAR γ flfl; Lysozyme M-Cre+ and Lysozyme M-Cre− control mice were challenged with 2.5% dextran sodium sulfate (DSS) for 7 days. The disease activity index (DAI), a composite score reflecting clinical sings of the disease was assessed daily (A) and the macroscopic lesions in colon (B) and spleen (C) were determine on day 7. Data are represented as mean ± standard error of groups of 10 mice. Points with an asterisk are significantly different (P<0.05).
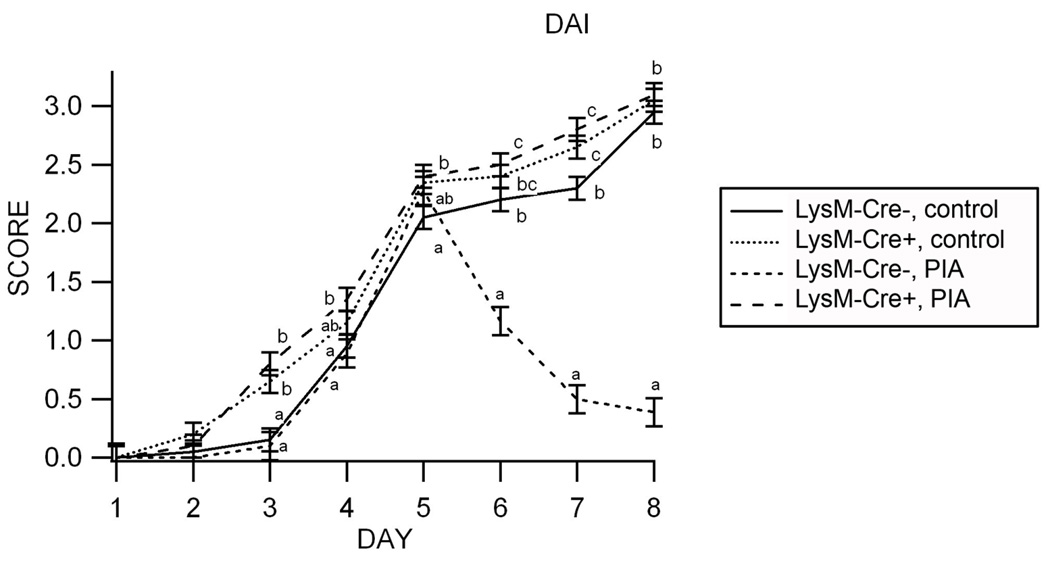
To determine whether exogenous administration of a synthetic PPAR γ agonist (pioglitazone, PIA) affects the clinical recovery from DSS colitis we challenged PPAR γ flfl Cre+ mice and control PPAR γ flfl Cre- mice with 2.5% DSS for 5 days followed by a 3-day recovery phase using a previously published protocol of colonic regeneration 19. We used selective instead of safer and equally efficacious compounds such as conjugated linoleic acid (CLA) 2, 10, 13, 20 and abscisic acid (ABA) 8, 9 because PIA is known to be a more selective activator of PPAR γ than these naturally occurring compounds. During the recovery phase mice were administered either control or PIA-supplemented diets. In the colonic mucosa by activating macrophage PPAR γ with PIA facilitated recovery from experimental IBD (Figure 3). Moreover, the deletion of PPAR γ in macrophages (but not in other immune or epithelial cells) abrogated the ability of PIA to induce recovery from colitis, suggesting that macrophages are important targets for the action of TZDs.
Figure 3. Effect of pioglitazone and macrophage-specific PPAR γ deletion in the clinical recovery from experimental IBD.
Treatment with the PPAR gamma agonist pioglitazone (PIA) accelerates recovery from gut inflammation in Lysozyme M-Cre− mice but not in macrophage-specific PPAR γ null mice (Lysozyme M-Cre+). Data are represented as mean ± standard error of groups of 10 mice. Means without a common letter superscript are significantly (P<0.05) different.
Colonic histopathology was significantly worsened in macrophage-specific PPAR γ-deficient mice
To further examine the affect of macrophage-specific PPAR γ deficiency on DSS colitis we examined the histopathological changes in the colon resulting from the DSS challenge of LysM-Cre− and LysM-Cre+ mice for the amount epithelial erosion, leukocyte infiltration, and mucosal thickening. Our data indicate that LysM-Cre+ mice had significantly higher scores for all three parameters tested, showing that observations of worse disease activity translate to more severe colonic immunopathology (Figure 4).
Figure 4. Effect of macrophage-specific PPAR γ deletion on colon histopathology and inflammation of mice with experimental IBD.
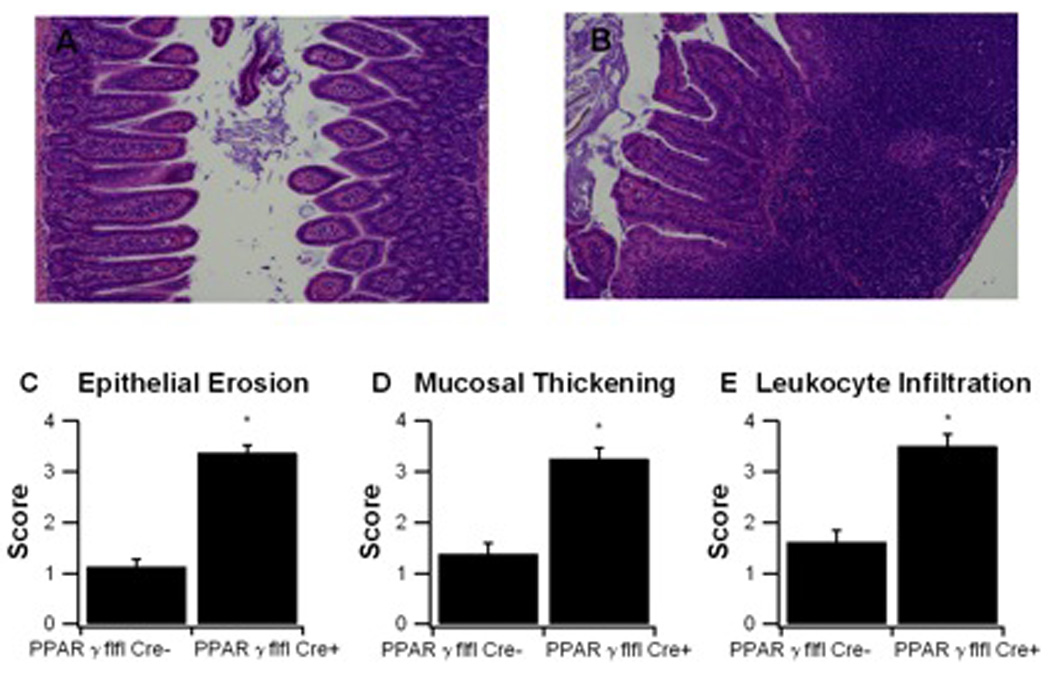
PPAR γ flfl; Lysozyme M-Cre+ and Lysozyme M-Cre− control mice were challenged with 2.5% dextran sodium sulfate (DSS) for 7 days. Representative photomicrographs of colonic samples from LC− (A) and LC+ (B) mice with DSS colitis (Original magnification, 40 ×). Colonic specimens underwent blinded histological examination and were scored (1–4) on epithelial erosion (C), mucosal wall thickening (D), and leukocyte infiltration (E) on day 7 of the challenge. Data are represented as mean ± standard error of groups of 10 mice. Points with an asterisk are significantly different (P<0.05).
Effect of macrophage-specific PPAR γ deficiency on immune cell populations in the blood, spleen, MLN, and LP
In blood, F4/80+CD11b+ monocytes from LysM-Cre+ mice expressed significantly less MCP-1 but greater levels MHC II than control mice, as indicated by median fluorescence intensity (MFI) (Figure 5A–C). Spleens from macrophage-specific PPAR γ deficient mice had reduced CD4+ T cells and CD4+CD25+FoxP3+ Treg cells on day 7 of DSS challenge (Figures 5D–F).
Figure 5. Effect of macrophage-specific PPAR γ deletion on immune cell subsets in blood and spleen from mice with experimental IBD.
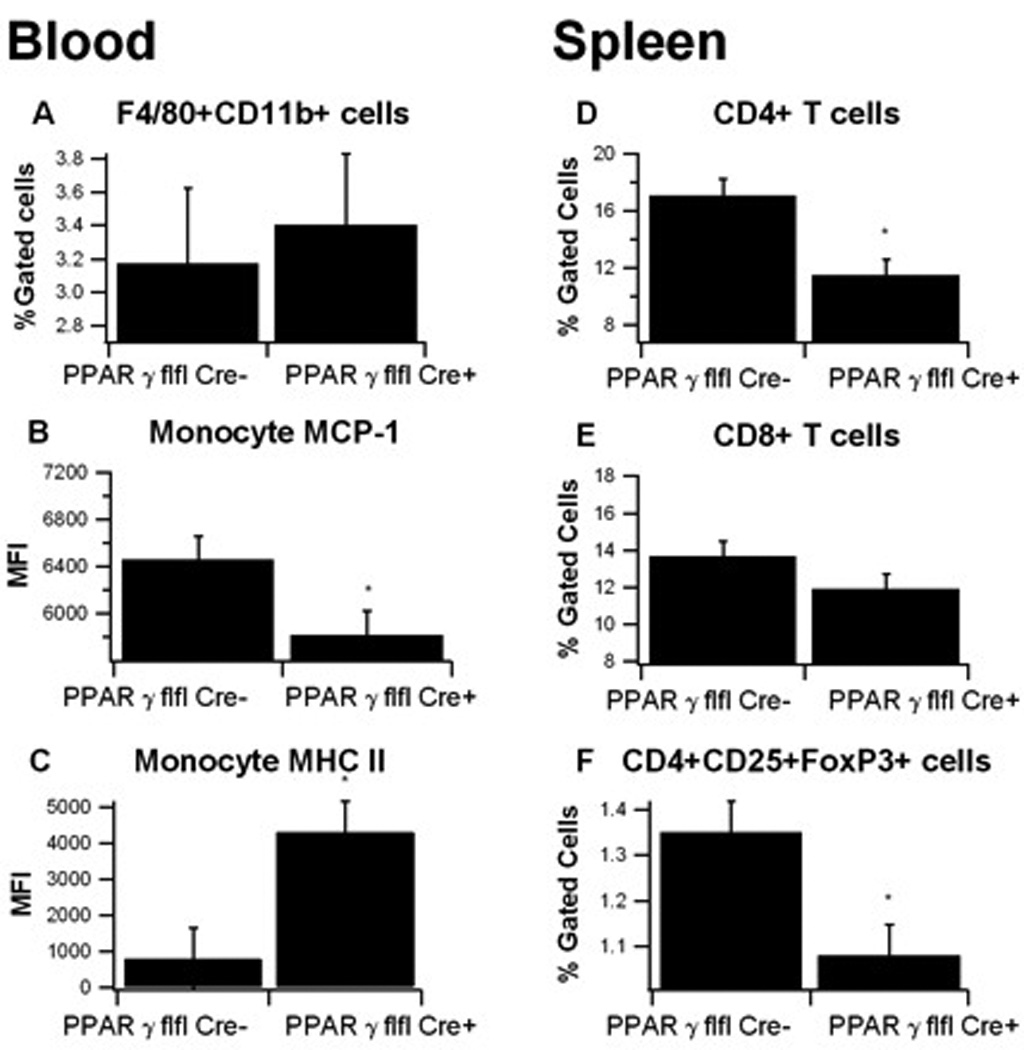
Blood (A–C) and spleen (D–F) from PPAR γ flfl; Lysozyme M-Cre+ and Lysozyme M-Cre− mice were immunophenotyped. Data were collected on day 7 of the dextran sodium sulfate (DSS) challenge and were analyzed with the FACS Diva software. Blood monocytes or spleen T cell subsets were selected by gating first on the viable cells based on FSC and SSC. Monocytes were then gated on F4/80+ CD11b+ and within this cell population, MCP-1 and MHC-II mean fluorescence intensity (MFI) were then examined. Splenocytes were gated on CD3+CD4+ and CD3+CD8+ or CD4+CD25+FOXP3+ to characterize the T cell subsets. Data are represented as mean ± standard error of groups of 10 mice. Points with an asterisk are significantly different (P<0.05).
In MLN, macrophage-specific PPAR γ deficient mice had significantly more F4/80+CD11b+ macrophages than in PPAR γ-expressing mice (Figure 6). Additionally, macrophages from macrophage-specific PPAR γ deficient mice expressed greater levels of CD40, a costimulatory molecule involved in the pathogenesis of DSS-induced intestinal inflammation 21. Similar to the spleen, MLN from macrophage-specific PPAR γ deficient mice had significantly fewer Treg than those from LysM-Cre− mice.
Figure 6. Effect of macrophage-specific PPAR γ deletion on immune cell subsets in mesenteric lymph nodes (MLN) from mice with experimental IBD.
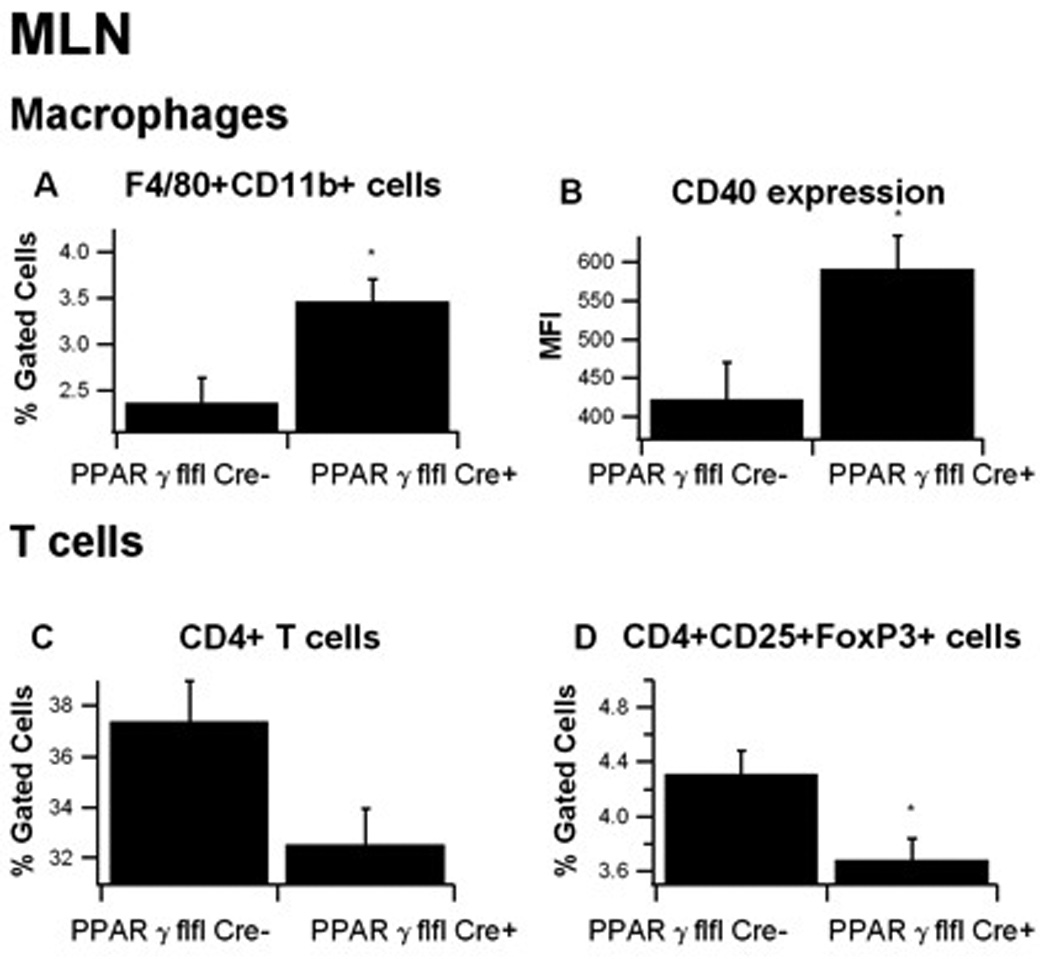
MLN-derived cells from PPAR γ flfl; Lysozyme M-Cre+ and Lysozyme M-Cre− mice were immunophenotyped to identify immune cell subsets by flow cytometry. Data were collected on day 7 of the dextran sodium sulfate (DSS) challenge and were analyzed with the FACS Diva software. MLN macrophages were selected based on F4/80 and CD11b expression in the cell viable gate and then CD40 expression within this population was determined by assessing mean fluorescence activity (MFI). For T-cells, all viable MLN cells were selected using FSC and SSC and the gated on CD3+CD4+ and CD3+CD8+ and CD4+CD25+FOXP3+. Data are represented as mean ± standard error of groups of 10 mice. Points with an asterisk are significantly different (P<0.05).
In the colonic LP, there were no differences in the overall percent of F4/80+CD11b+ macrophages between LysM-Cre+ and LysM-Cre− mice, though macrophages from LysM-Cre+ mice differed phenotypically than those of LysM-Cre-mice (Figure 7). Specifically, macrophage-specific PPAR γ deficient macrophages expressed significantly more Ly6C, and the percent of CD40+ and TLR-4+ macrophages were elevated. These findings suggest a more inflammatory, possibly M1, classically activated phenotype of macrophages in macrophage-specific PPAR γ deficient mice. We also observed a significant influx of CD8+ T cells in the colonic of LP macrophage-specific PPAR γ deficient mice, which is indicative of an enhanced secondary recruitment of effector T cells.
Figure 7. Effect of macrophage-specific PPAR γ deletion on immune cell subsets in the colonic lamina propria (LP) from mice with experimental IBD.
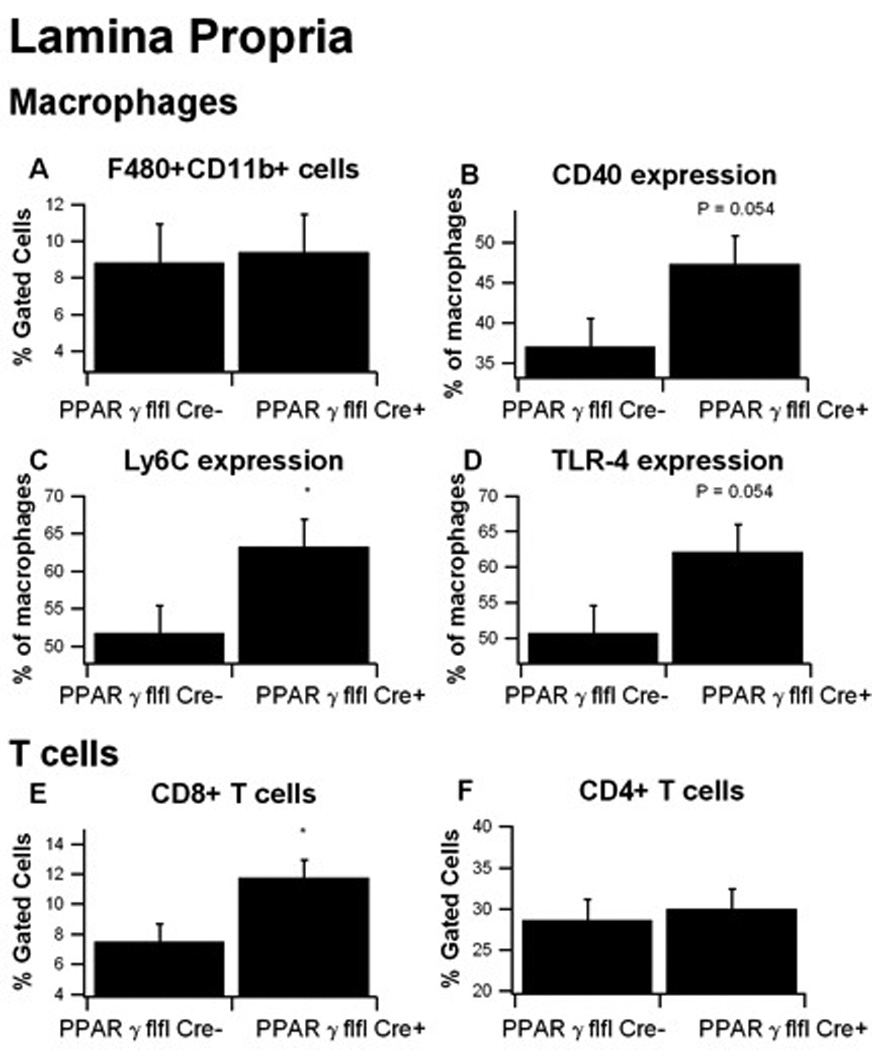
Colonic LP cells from PPAR γ flfl; Lysozyme M-Cre+ and Lysozyme M-Cre− mice were immunophenotyped to identify immune cell subsets by flow cytometry. Data were collected on day 7 of the dextran sodium sulfate (DSS) challenge and were analyzed with the FACS Diva software. LP macrophages were selected based on F4/80 and CD11b expression in the cell viable gate and then CD40, Ly6C and TLR-4 expression within this population was determined by assessing mean fluorescence activity (MFI). For T-cells, all viable MLN cells were selected using FSC and SSC and the gated on CD3+CD4+ and CD3+CD8+. Data are represented as mean ± standard error of groups of 10 mice. Points with an asterisk are significantly different (P<0.05).
Effect of macrophage-specific PPAR γ deficiency on colonic inflammation
To assess the effect of macrophage-specific PPAR γ deletion on colonic inflammation real time quantitative RT-PCR was performed on colonic samples of LysM-Cre+ and LysM-Cre− mice with DSS colitis. As anticipated, the expression of PPAR γ was greater in LysM-Cre− mice, though the data were not statistically significant (Figure 8), due to the fact that PPAR γ was deleted in macrophages but not in lymphocytes, IECs, endothelial cells, dendritic cells, fibroblasts and other colonic cell types that express normal PPAR γ levels. In contrast to the effects of T cell-specific PPAR γ deletion 14, IL-6 and IL-1β, did not differ significantly between groups. However, IFN γ levels were significantly greater in LysM-Cre+ mice, a finding in line with the greater abundance of CD8+ T cells in the LP and increased levels of macrophage inflammatory markers. The expressions of cellular adhesion molecule mucosal address in cellular adhesion molecule 1 (MAdCAM-1) and intracellular adhesion molecule 1 (ICAM-1) also showed a strong numerical elevation in LysM-Cre+ mice, though the data did not reach statistical significance. In their study using macrophage-specific PPAR γ -deficient mice, Shah et al reported a significant increase in colonic expression of MCP-1 and CCR2 following treatment with DSS. Here, we found that MCP-1 expression was greater in colons of LysM-Cre+ mice, though differences were not statistically significant.
Figure 8. Effect of macrophage-specific PPAR γ deletion on target gene expression in the colonic mucosa of mice with experimental IBD.
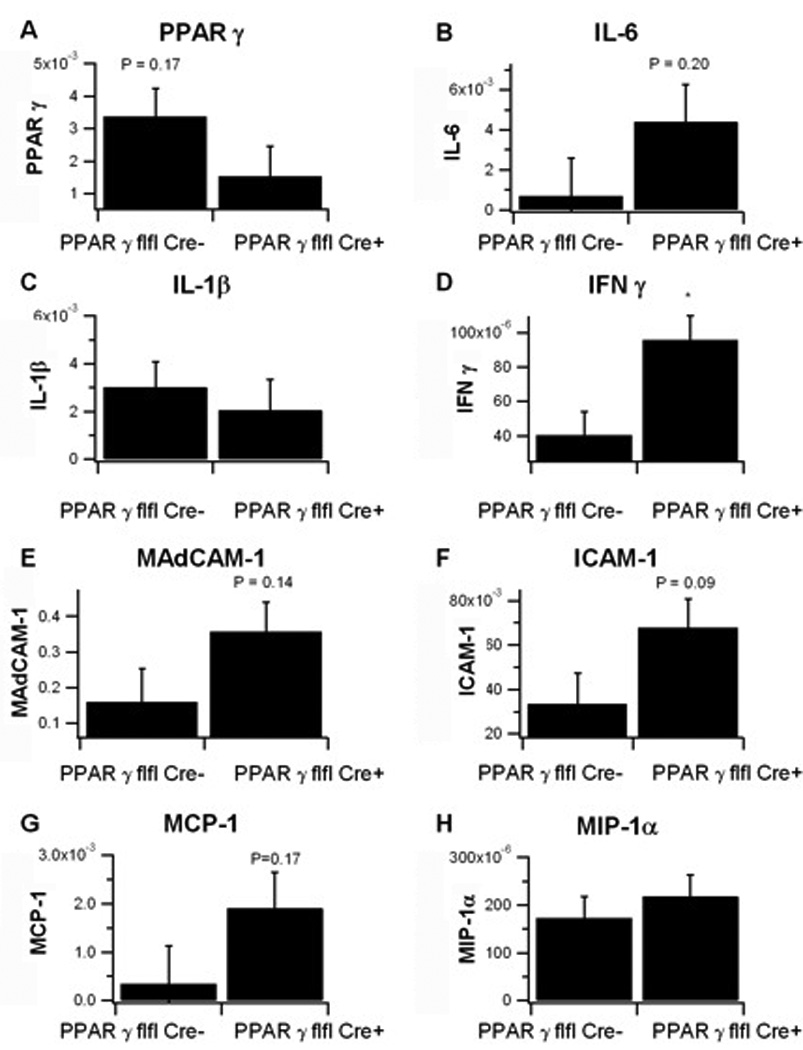
Expression of PPAR γ (A), IL-1β (B) MAdCAM-1 (C), MCP-1 (D), IL-6 (E), IFN-γ (F), ICAM-1 (G), MIP-1α (H) was assessed in colonic mucosal specimens recovered from PPAR γ flfl; Lysozyme M-Cre+ and Lysozyme M-Cre− mice. Data are represented as mean ± standard error of groups of 10 mice. Points with an asterisk are significantly different (P<0.05).
Global gene expression analyses in the colonic mucosa of macrophage-specific PPAR γ deficient mice and floxed littermates
A total of 124 genes had statistically significant differences in colonic expression (P <0.01 and two-fold change) between groups as described in the heat map (Supplementary Figure 1). Gene ontology (GO) analyses revealed that the deletion of PPAR γ in macrophages during IBD is significantly enriched for genes involved in response to stimulus, immune response, immune system process, cellular metabolic process and oxidation/reduction (Supplementary Table 1). These differentially expressed genes were submitted to ingenuity pathway for network analyses and the top two networks retrieved were carbohydrate metabolism (Supplementary Figure 2) and immunological disease (Supplementary Figure 3).
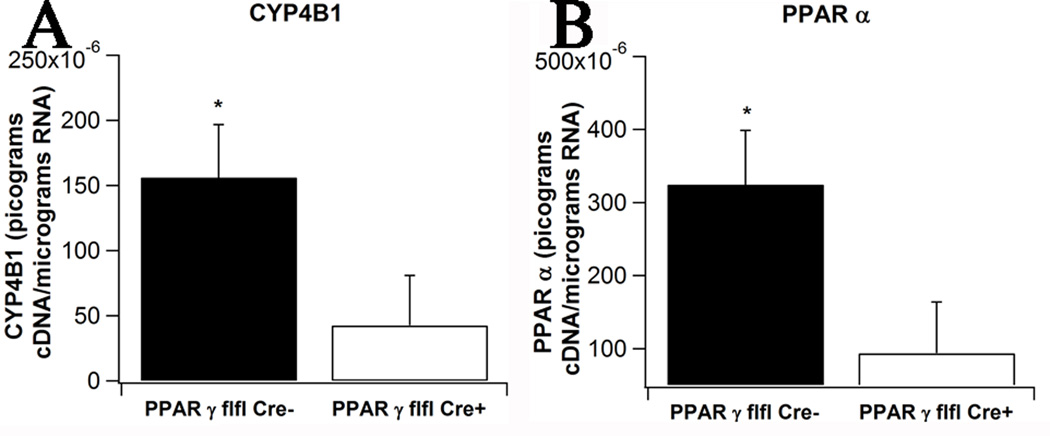
The carbohydrate metabolism network reveals that the deletion of PPAR γ in macrophages caused a down-regulation of oxidation genes stearoyl-coenzyme A desaturase (SCD), PPAR α, CYP4B1, a member of the cytochrome P450 monooxygenase family, casein kinase 1 (CK1), plasma membrane calcium-transporting ATPase 1 (ATP2B1) and immunoglobulin mu chain C region (IGHM). Additionally, the deletion of PPAR γ in macrophages caused an up-regulation of DNA primase large subunit 2 (PRIM2), macrophage scavenger receptor 1 (MSR1), nuclear factor, IL-3 regulated (NFIL3), the zinc finger transcription factor EGR2, the atypical member of the E2F transcription factor family E2F8, the anti-apoptotic molecule BCL2A1 and the GTP cyclohydrolase I (GCH1). The microarray results of two key metabolically active genes (i.e., PPAR α and CYP4B1) were validated by real-time RT-PCR (Figure 9), thereby confirming the microarray results.
Figure 9. Effect of macrophage-specific PPAR γ deletion on metabolically active gene expression in the colonic mucosa of mice with experimental IBD.
Expression of cytochrome P450 (CYP4B1, A) and PPAR α (B) was assessed in colonic mucosal specimens recovered from PPAR γ flfl; Lysozyme M-Cre+ and Lysozyme M-Cre− mice. Data are represented as mean ± standard error of groups of 10 mice. Points with an asterisk are significantly different (P<0.05).
The immunological disease network demonstrates for the first time that the deletion of PPAR γ in macrophages caused a down-regulation of retinoid acid orphan receptor γt (ROR γt, also described as RORC), Bmp and activin membrane-bound inhibitor (BAMBI, initially described as a pseudoreceptor antagonizing TGF-β receptor activation) and Staphylococcal complement inhibitor (SCIN) in the colon of mice with IBD. Additionally, the deletion of PPAR γ in macrophages caused an up-regulation of chemokine (C-X-C motif) ligand 9 (CXCL9), CXCL10, IL-22, IL1RL1, CCR1, SOCS3 (an gene responsive to IFN-γ), MCH class II, ADM, interferon induced tetratricopeptide repeat protein 2 (IFIT-2), which belongs to the type I interferon response genes, receptor interacting protein kinase 3 (RIPK3), C3a receptor gene (C3AR1) an important receptor of the complement system, IIGP is a member of the p47 GTPase family induced upon exposure to Escherichia coli LPS 22, cytochrome b-245, beta polypeptide (CYBB), receptor 3A for the Fc region of immunoglobulin G (FCGR3A).
DISCUSSION
The appeal of PPAR γ as a putative therapeutic target for human IBD lies in its high expression levels in the majority of cells found in mucosal barriers, including macrophages, lymphocytes, and IECs, although PPAR γ expression in the colonic epithelium is decreased in UC patients 23. In previous experiments we have shown that PPAR γ expressed in IECs and T cells protects mice from experimental IBD 2, 9, 12, 14, 16. In IECs, PPAR γ expression was shown to be important in the lysosomal and p53 tumor suppressor pathways and antigen presentation complex genes, while deletion of PPAR γ in T cells resulted in an increase in pro-inflammatory genes and cellular adhesion molecules 14, 16. Macrophage PPAR γ has also been reported to protect mice from experimental IBD 18, a finding consistent with its noted anti-inflammatory effects in this cell type 24–27 and the purported role of colonic macrophages as possible sites for therapeutic intervention against gut inflammation 17.
Rosiglitazone, a TZD also known as Avandia, has been proven to be efficacious in treating human UC 4, 28, which provides clinical validation in humans for the use of PPAR γ as target for new IBD treatments. Additionally, PPAR γ can become activated by naturally occurring, non-toxic compounds such as dietary lipids and phytochemicals (e.g., ABA and CLA) that are safer than Avandia (rosiglitazone) and Actos (pioglitazone). The long-term objective of this program was to investigate the immunoregulatory mechanisms by which macrophage PPAR γ prevents or ameliorates mucosal inflammation, thereby adding to the development of safer, more effective therapies against gut immunoinflammatory disorders.
Macrophage-specific PPAR γ deficiency significantly worsened disease activity, colonic histopathology and abrogated pioglitazone’s ability to accelerate recovery from DSS colitis. These clinical and pathological differences were consistent with the predictions of our computational and mathematical model of IBD, which suggested that macrophages and their mechanisms of plasticity are key targets for therapeutic intervention against IBD 17. Additionally, these clinical and pathological findings are associated with significant differences in peripheral and colonic immune cell populations. More specifically, macrophage-specific PPAR γ null mice overall had significantly fewer Treg cells in spleen and MLN than their littermates expressing PPAR γ in macrophages, suggesting a possible role of macrophage populations in defining T cell ontogeny and development; a process that may be regulated by macrophage PPAR γ. We have previously shown that Treg accumulation in mucosal inductive sites is associated with the prevention of chronic colitis caused by the adoptive transfer of CD4+CD45RBhi T cells in SCID mice 12. Further studies examining the role of PPAR γ in modulating the cross-talk between macrophages and T cells at the mucosal surfaces and the effects of this cross-talk on gastroenteric inflammation are warranted.
In the MLN, macrophage infiltration was significantly increased in macrophage-specific PPAR γ null mice in comparison to PPAR γ-expressing littermates. MLN macrophages also expressed significantly greater amounts of surface costimulatory molecule CD40 than PPAR γ-expressing littermate mice. PPAR γ activation has been shown to reduce CD40 expression in the hippocampus 29, atherosclerotic plaques 30, and renal tubular epithelial cells 31, though this is the first study to demonstrate a similar effect in colonic macrophages. Combined with our results showing a significantly decreased Treg compartment, these results indicate that macrophage PPAR γ deficiency may increase effector responses in mucosal inductive sites.
Similar to our findings in the MLN, macrophage CD40 expression was also enhanced in the colonic LP. Our findings are consistent with the elevated levels of CD40+ macrophages in colons of CD and UC patients 32–35 where they have been linked to activation of IECs 36, pro-inflammatory cytokine production 32, and costimulation of T cells 35; all of which can lead to effector and inflammatory responses at the gut mucosa. Combined with increases in Ly6C and TLR-4, our findings suggest that a greater percentage of colonic LP macrophages in macrophage-specific PPAR γ null mice were in a classically activated M1, pro-inflammatory state. These findings are consistent with studies showing that PPAR γ plays an important role in alternative M2 macrophage differentiation 24. While we did not find significant increases in pro-inflammatory cytokines derived from macrophages, such as IL-6 and IL-1β, or in chemokines MCP-1 or MIP-1α, there were consistent numerical increases in chemokines and cytokines in the colons of macrophage-specific PPAR γ null mice.
Colonic IFN-γ expression and the percentages of LP CD8+ T cells were significantly increased in macrophage-specific PPAR γ null mice, indicating the predominance of a Th1 phenotype and that macrophage PPAR γ may be an important component in cross-talk between macrophages and lymphocytes in the colon. Our results on colonic gene expression differ somewhat from those previously reported by Shah et al, who found no significant difference in IFN-γ and significant increases in IL-1β and MCP-1 in macrophage-specific PPAR γ knockout mice 18. The differences between our results and those reported may be due to the different genetic backgrounds of the mice. Whereas mice from our experiments are from a C57BL/6 background, Shah et al used mice from a mixed Sv129/C57BL/6/CB.20 background. Our group and others have demonstrated that the mouse genetic background plays an important role in delineating disease activity in and colonic inflammatory lesions during DSS colitis 16, 37. Since gut microbial populations vary across mouse vivaria, possible differences in the gut microbiota between our mice and those used by Shah et al may also have contributed to differential clinical and pathological findings reported.
In conclusion, our results show that macrophage PPAR γ deficiency worsens experimental IBD by modulating the expression of inflammatory/effector (CXCL9, CXCL10, IL-22, IFN-γ, CCR1, SOCS3, IL1RL1) and metabolic (SCD1, PPAR α, CYP4B1) genes, impairing the peripheral Treg compartment and increasing both the influx and activation of LP macrophages and T cells at the colonic mucosa. We also provide in vivo evidence demonstrating that colonic macrophages are important targets for TZDs in the gut mucosa during experimental IBD.
METHODS
Animal Procedures
Eight week old PPAR γ flfl Cre+ mice, with a Cre recombinase targeted to the Lysozyme M-Cre promoter (LysM-Cre+, n=20), and control Lysozyme M-Cre- (LysM-Cre−, n=20) littermates in a C57BL/6 background were housed at the animal facilities at Virginia Polytechnic Institute and State University in a room maintained at 23.9°C, with a 12:12 h light-dark cycle starting from 6:00 AM. All experimental procedures were approved by the Institutional Animal Care and Use Committee of Virginia Polytechnic Institute and State University and met or exceeded requirements of the Public Health Service/National Institutes of Health and the Animal Welfare Act. Colitis was induced with 2.5% dextran sodium sulfate (DSS), 36,000–44,000 mol wt (ICN Biomedicals, Aurora, OH) in the drinking water. After the DSS challenge mice were weighed on a daily basis and examined for clinical signs of disease associated with colitis (i.e., perianal soiling, rectal bleeding, diarrhea, and piloerection). For the DSS challenge, the disease activity indices and rectal bleeding scores were calculated using a modification of a previously published compounded clinical score 2. Briefly, disease activity index consisted of a scoring for diarrhea and lethargy (0–3), whereas rectal bleeding consisted of a visual observation of blood in feces and the perianal area (0–4). Mice in the DSS study were euthanized on day 7 of the DSS challenge by CO2 narcosis and blood was drawn from the heart. Colons and spleens were scored based on size and macroscopic inflammatory lesions (0–3).
Pioglitazone treatment during recovery from DSS colitis
To determine whether the activation of PPAR γ in macrophages using pioglitazone (Actos) would accelerate the recovery from DSS colitis we challenged PPAR γ flfl LysM-Cre+ mice (n=20) and LysM-Cre- (n=20) mice with 2.5% DSS for 5 days. On day 6 of the study, the DSS was withdrawn from the drinking water and half of the mice within group were given either control or pioglitazone (70 mg/Kg)-supplemented diets during the recovery phase for 3 additional days.
Histology
Colonic sections were fixed in 10% buffered neutral formalin, later embedded in paraffin, and then sectioned (5 µm) and stained with H&E stain for histologic examination. Colons were graded with a compounded histologic score including the extent of (1) epithelial cell erosion, (2) leukocyte infiltration, and (3) mucosal thickening. The sections were graded with a score of 0–4 for each of the previous categories and data were analyzed as a normalized compounded score.
Protein extraction and Western Blot
Macrophages were washed 3 times with ice-cold PBS, harvested and lysed with RIPA buffer (150 mM NaCl, 1.0% IGEPAL® CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0.) containing protease and phosphatase inhibitors and incubated 30 min on ice. Colons were collected, cleaned and weighed. The tissue was mixed with RIPA buffer (150 mM NaCl, 1.0% IGEPAL® CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0.) containing protease and phosphatase inhibitors. Cells and tissues were then homogenized and incubated on ice for 30 min. Whole lysates were cleared by centrifugation (10,000 g for 10 min) and protein concentration was measured using a DC protein assay kit (Bio-Rad Laboratories). Diluted samples containing equal amounts of protein were mixed with 2× Laemmli Sample Buffer (Bio-Rad Laboratories). Proteins were separated on a 10% SDS-PAGE gel and transferred to polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with 1× TBS-T (20 mM Tris-HCl pH 7.6, 8.5% NaCl, 0.1% Tween-20) containing 5% nonfat dry milk 30 min at room temperature. Anti-PPARγ (Santa Cruz Biotechnology, inc.) primary antibody at 1:250 dilution in 1× TBS-T 5% nonfat dry milk was incubated overnight at 4 °C. After 3 washes with 1× TBS-T, membrane was incubated for 45 min at room temperature with anti-mouse IgG conjugated to horseradish peroxidase (HRP) at 1:2000 dilution. The antigen detection was performed with the ECL (BioRad) chemiluminescent detection system.
Isolation of colonic lamina propria immune cells
Colons from mice were excised, placed in CMF (HBSS/10% FBS/0.251 EDTA)/HEPES (Mediatech), cleaned of contents, and sectioned into 5-mm pieces. To remove IEL, tissues were incubated for 45 min in 15 ml of CMF/EDTA. Medium was changed every 15 min. To release lamina propria cells (LPC), tissues were digested for 3 h with type VIII collagenase (Sigma-Aldrich) in HBSS. Medium was changed every hour and supernatants containing LPC were collected. After digestion, supernatants were pooled, spun at 400 × g and enumerated with a Coulter Counter (Beckman Coulter, Fullerton, CA).
Flow Cytometry
MLN and spleen-derived cells were excised, crushed with frosted slides, and were resuspended in PBS and enumerated with a Coulter Counter. LPC, MLN, and spleen-derived cells (2 × 105 cells/well) or whole blood (10 µL/well) were seeded onto 96-well plates, centrifuged at 4°C at 3000 rpm for 4 minutes, and washed with PBS containing 5% serum and 0.09% sodium azide (FACS buffer). To assess differential monocyte/macrophage subsets, the cells were then incubated in the dark at 4°C for 20 minutes in FcBlock (20 µg/ml; BD Pharmingen), and then for an additional 20 minutes with fluorochrome-conjugated primary antibodies anti-F4/80-PE-Cy5 (5 µg/mL, ebioscience), anti-CD11b-Alexa Fluor 700 (2 µg/mL, eBioscience), anti-CD40-APC (3 µg/mL, BD), anti-MHC II-PE (2 µg/mL, eBioscience), anti-Ly6C-PerCP-Cy5.5 (2 µg/mL, eBioscience), anti TLR-4-PE-Cy7 (2 µg/mL, eBioscience), and anti MCP-1-FITC (3 µg/mL, BD). For lymphocyte assessment, cells were incubated with anti-CD4-FITC (3 µg/mL; BD Pharmingen), anti-CD8-PerCp-Cy5.5 (2 µg/mL, eBioscience), anti-CD3 PE-Cy5 (2 µg/mL; BD Pharmingen), anti-CD25-APC (2 µg/mL, BD), anti-FoxP3-PE (2 µg/mL, eBioscience) as previously shown 38. Flow results were computed with a BD LSR II flow cytometer and data analyses was performed with FACS Diva software (BD).
Quantitative real time RT-PCR
Total RNA was isolated from colons using the RNA isolation Minikit (Qiagen) according to the manufacturer’s instructions. Total RNA (1 µg) was used to generate complementary DNA (cDNA) template using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). The total reaction volume was 20 µL with the reaction incubated as follows in an MJ MiniCycler: 5 minutes at 25°C, 30 minutes at 52°, 5 minutes at 85°C, and hold at 4°C. PCR was performed on the cDNA using Taq DNA polymerase (Invitrogen, Carlsbad, CA) and using previously described conditions 2. Each gene amplicon was purified with the MiniElute PCR Purification Kit (Qiagen) and quantitated on an agarose gel by using a DNA mass ladder (Promega). These purified amplicons were used to optimize real-time RT-PCR conditions and to generate standard curves in the real-time PCR assay. Primer concentrations and annealing temperatures were optimized for the iCycler iQ system (Bio-Rad) for each set of primers using the system’s gradient protocol. PCR efficiencies were maintained between 92 and 105% and correlation coefficients above 0.98 for each primer set during optimization and also during the real-time PCR of sample DNA.
Complementary DNA (cDNA) concentrations for genes of interest were examined by real-time quantitative PCR using an iCycler IQ System and the iQ SYBR green supermix (Bio-Rad). A standard curve was generated for each gene using 10-fold dilutions of purified amplicons starting at 5 pg of cDNA and used later to calculate the starting amount of target cDNA in the unknown samples. SYBR green I is a general double-stranded DNA intercalating dye and may therefore detect nonspecific products and primer/dimers in addition to the amplicon of interest. In order to determine the number of products synthesized during the real-time PCR, a melting curve analysis was performed on each product. Real-time PCR was used to measure the starting amount of nucleic acid of each unknown sample of cDNA on the same 96-well plate.
GeneChip hybridization
RNA from 3 of macrophage-specific PPAR γ-deficient mice (PPAR γ flfl Cre+) and 3 of control (Cre−) littermates was processed and labeled according to the standard target labeling protocols and the samples were hybridized, stained, and scanned per standard Affymetrix protocols at the Virginia Bioinformatics Institute (VBI) core laboratory on Mouse 430 2.0 expression arrays (Affymetrix Inc., Santa Clara, CA).
Microarray data analysis
Quality checking of array hybridizations was done by Bioconductor package "ArrayQualityMatrics". All of six sample results passed quality control. Data input and subsequent steps were performed using the Bioconductor package "affy" 39. Raw microarray data obtained from CEL files were pre-processed by the gcRMA algorithm (GC Robust Multiarray Average) and then log2 transformed by using Bioconductor package “gcrma” 40. Only probe sets with at least one Present call across all the compared hybridizations will be kept for downstream statistical analysis. All statistical analysis of the data was performed within R statistical environment - Version 2.9.0 41 using Bioconductor packages 40]. A cut-off of P < 0.01 and > = 2-fold change was used as the criterion for whether or not a gene was significantly modulated by a treatment. All the 124 significantly modulated genes were submitted to IPA (www.ingenuity.com) for pathway analysis and to GO:termfinder for GO analysis. The microarray data (both raw and normalized) have been submitted at the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/, Data set: GSE23421).
Statistics
Data were analyzed as a completely randomized design. To determine the statistical significance of the model, analysis of variance (ANOVA) was performed using the general linear model procedure of Statistical Analysis Software (SAS), and probability value (P) < 0.05 was considered to be significant. When the model was significant, ANOVA was followed by Fisher’s Protected Least Significant Difference multiple comparison method.
Supplementary Material
ACKNOWLEDGMENTS
Supported by award number 5R01AT004308 of the National Center for Complementary and Alternative Medicine at the National Institutes of Health awarded to J.B.-R., European Commission grant number 224836, the Ramon y Cajal Program, the Virginia Bioinformatics Institute-Fralin CRI grants program to J.B.-R., NIAID Contract No. HHSN272200900040C to B.W.S., and funds from the Nutritional Immunology and Molecular Medicine Laboratory.
Footnotes
DISCLOSURE
The authors declared no conflict of interest.
REFERENCES
- 1.Hanauer SB, Present DH. The state of the art in the management of inflammatory bowel disease. Rev Gastroenterol Disord. 2003;3(2):81–92. [PubMed] [Google Scholar]
- 2.Bassaganya-Riera J, Reynolds K, Martino-Catt S, Cui Y, Hennighausen L, Gonzalez F, et al. Activation of PPAR gamma and delta by conjugated linoleic acid mediates protection from experimental inflammatory bowel disease. Gastroenterology. 2004;127(3):777–791. doi: 10.1053/j.gastro.2004.06.049. [DOI] [PubMed] [Google Scholar]
- 3.Desreumaux P, Dubuquoy L. PPARgamma agonists as a new class of effective treatment for ulcerative colitis. Inflamm Bowel Dis. 2009;15(6):959–960. doi: 10.1002/ibd.20765. [DOI] [PubMed] [Google Scholar]
- 4.Lewis JD, Lichtenstein GR, Deren JJ, Sands BE, Hanauer SB, Katz JA, et al. Rosiglitazone for active ulcerative colitis: a randomized placebo-controlled trial. Gastroenterology. 2008;134(3):688–695. doi: 10.1053/j.gastro.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcy TR, Britton ML, Blevins SM. Second-generation thiazolidinediones and hepatotoxicity. Ann Pharmacother. 2004;38(9):1419–1423. doi: 10.1345/aph.1E072. [DOI] [PubMed] [Google Scholar]
- 6.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20(5):649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 7.Bassaganya-Riera J, Pogranichniy RM, Jobgen SC, Halbur PG, Yoon KJ, O'Shea M, et al. Conjugated linoleic acid ameliorates viral infectivity in a pig model of virally induced immunosuppression. J Nutr. 2003;133(10):3204–3214. doi: 10.1093/jn/133.10.3204. [DOI] [PubMed] [Google Scholar]
- 8.Guri AJ, Hontecillas R, Bassaganya-Riera J. Abscisic acid ameliorates experimental IBD by downregulating cellular adhesion molecule expression and suppressing immune cell infiltration. Clinical Nutrition. 2010 doi: 10.1016/j.clnu.2010.02.009. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guri AJ, Evans NP, Hontecillas R, Bassaganya-Riera J. T cell PPAR gamma is required for the anti-inflammatory efficacy of abscisic acid against experimental IBD. Journal of Nutritional Biochemistry. 2010 doi: 10.1016/j.jnutbio.2010.06.011. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hontecillas R, Wannemeulher MJ, Zimmerman DR, Hutto DL, Wilson JH, Ahn DU, et al. Nutritional regulation of porcine bacterial-induced colitis by conjugated linoleic acid. J Nutr. 2002;132(7):2019–2027. doi: 10.1093/jn/132.7.2019. [DOI] [PubMed] [Google Scholar]
- 11.Bassaganya-Riera J, Hontecillas R. CLA and n-3 PUFA differentially modulate clinical activity and colonic PPAR-responsive gene expression in a pig model of experimental IBD. Clin Nutr. 2006;25(3):454–465. doi: 10.1016/j.clnu.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Hontecillas R, Bassaganya-Riera J. Peroxisome proliferator-activated receptor gamma is required for regulatory CD4+ T cell-mediated protection against colitis. J Immunol. 2007;178(5):2940–2949. doi: 10.4049/jimmunol.178.5.2940. [DOI] [PubMed] [Google Scholar]
- 13.Evans NP, Misyak SA, Schmelz EM, Guri AJ, Hontecillas R, Bassaganya-Riera J. Conjugated linoleic acid ameliorates inflammation-induced colorectal cancer in mice through activation of PPARgamma. J Nutr. 2010;140(3):515–521. doi: 10.3945/jn.109.115642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guri AJ, Mohapatra SK, Horne WT, 2nd, Hontecillas R, Bassaganya-Riera J. The role of T cell PPAR gamma in mice with experimental inflammatory bowel disease. BMC Gastroenterol. 2010;10:60. doi: 10.1186/1471-230X-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adachi M, Kurotani R, Morimura K, Shah Y, Sanford M, Madison BB, et al. Peroxisome proliferator activated receptor gamma in colonic epithelial cells protects against experimental inflammatory bowel disease. Gut. 2006;55(8):1104–1113. doi: 10.1136/gut.2005.081745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohapatra SK, Guri AJ, Climent M, Vives C, Carbo A, Horne WT, et al. Immunoregulatory actions of epithelial cell PPAR gamma at the colonic mucosa of mice with experimental inflammatory bowel disease. PLoS One. 2010;5(4):e10215. doi: 10.1371/journal.pone.0010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wendelsdorf K, Bassaganya-Riera J, Hontecillas R, Eubank S. Model of colonic inflammation: immune modulatory mechanisms in inflammatory bowel disease. J Theor Biol. 2010;264(4):1225–1239. doi: 10.1016/j.jtbi.2010.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah YM, Morimura K, Gonzalez FJ. Expression of peroxisome proliferator-activated receptor-gamma in macrophage suppresses experimentally induced colitis. Am J Physiol Gastrointest Liver Physiol. 2007;292(2):G657–G666. doi: 10.1152/ajpgi.00381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boismenu R, Havran WL. Modulation of epithelial cell growth by intraepithelial gamma delta T cells. Science. 1994;266(5188):1253–1255. doi: 10.1126/science.7973709. [DOI] [PubMed] [Google Scholar]
- 20.Bassaganya-Riera J, Hontecillas R. Dietary conjugated linoleic acid and n-3 polyunsaturated fatty acids in inflammatory bowel disease. Curr Opin Clin Nutr Metab Care. 2010 doi: 10.1097/MCO.0b013e32833b648e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vowinkel T, Anthoni C, Wood KC, Stokes KY, Russell J, Gray L, et al. CD40-CD40 ligand mediates the recruitment of leukocytes and platelets in the inflamed murine colon. Gastroenterology. 2007;132(3):955–965. doi: 10.1053/j.gastro.2006.12.027. [DOI] [PubMed] [Google Scholar]
- 22.Lapaque N, Takeuchi O, Corrales F, Akira S, Moriyon I, Howard JC, et al. Differential inductions of TNF-alpha and IGTP, IIGP by structurally diverse classic and non-classic lipopolysaccharides. Cell Microbiol. 2006;8(3):401–413. doi: 10.1111/j.1462-5822.2005.00629.x. [DOI] [PubMed] [Google Scholar]
- 23.Dubuquoy L, Jansson EE, Deeb S, Rakotobe S, Karoui M, Colombel JF, et al. Impaired expression of peroxisome proliferator-activated receptor gamma in ulcerative colitis. Gastroenterology. 2003;124(5):1265–1276. doi: 10.1016/s0016-5085(03)00271-3. [DOI] [PubMed] [Google Scholar]
- 24.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447(7148):1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437(7059):759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ricote M, Huang J, Fajas L, Li A, Welch J, Najib J, et al. Expression of the peroxisome proliferator-activated receptor gamma (PPARgamma) in human atherosclerosis and regulation in macrophages by colony stimulating factors and oxidized low density lipoprotein. Proc Natl Acad Sci U S A. 1998;95(13):7614–7619. doi: 10.1073/pnas.95.13.7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature. 1998;391(6662):79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 28.Lewis JD, Lichtenstein GR, Stein RB, Deren JJ, Judge TA, Fogt F, et al. An open-label trial of the PPAR-gamma ligand rosiglitazone for active ulcerative colitis. Am J Gastroenterol. 2001;96(12):3323–3328. doi: 10.1111/j.1572-0241.2001.05333.x. [DOI] [PubMed] [Google Scholar]
- 29.Sun H, Huang Y, Yu X, Li Y, Yang J, Li R, et al. Peroxisome proliferator-activated receptor gamma agonist, rosiglitazone, suppresses CD40 expression and attenuates inflammatory responses after lithium pilocarpine-induced status epilepticus in rats. Int J Dev Neurosci. 2008;26(5):505–515. doi: 10.1016/j.ijdevneu.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Hu Q, Zhang XJ, Zhang C, Zhao YX, He H, Liu CX, et al. Peroxisome proliferator-activated receptor-gamma1 gene therapy attenuates atherosclerosis and stabilizes plaques in apolipoprotein E-deficient mice. Hum Gene Ther. 2008;19(3):287–299. doi: 10.1089/hum.2007.0142. [DOI] [PubMed] [Google Scholar]
- 31.Zhang YJ, Yang X, Kong QY, Zhang YF, Chen WY, Dong XQ, et al. Effect of 15d-PGJ2 on the expression of CD40 and RANTES induced by IFN-gamma and TNF-alpha on renal tubular epithelial cells (HK-2) Am J Nephrol. 2006;26(4):356–362. doi: 10.1159/000094735. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Colpaert S, D'Haens GR, Kasran A, de Boer M, Rutgeerts P, et al. Hyperexpression of CD40 ligand (CD154) in inflammatory bowel disease and its contribution to pathogenic cytokine production. J Immunol. 1999;163(7):4049–4057. [PubMed] [Google Scholar]
- 33.Battaglia E, Biancone L, Resegotti A, Emanuelli G, Fronda GR, Camussi G. Expression of CD40 and its ligand, CD40L, in intestinal lesions of Crohn's disease. Am J Gastroenterol. 1999;94(11):3279–3284. doi: 10.1111/j.1572-0241.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- 34.Polese L, Angriman I, Cecchetto A, Norberto L, Scarpa M, Ruffolo C, et al. The role of CD40 in ulcerative colitis: histochemical analysis and clinical correlation. Eur J Gastroenterol Hepatol. 2002;14(3):237–241. doi: 10.1097/00042737-200203000-00006. [DOI] [PubMed] [Google Scholar]
- 35.Carlsen HS, Yamanaka T, Scott H, Rugtveit J, Brandtzaeg P. The proportion of CD40+ mucosal macrophages is increased in inflammatory bowel disease whereas CD40 ligand (CD154)+ T cells are relatively decreased, suggesting differential modulation of these costimulatory molecules in human gut lamina propria. Inflamm Bowel Dis. 2006;12(11):1013–1024. doi: 10.1097/01.mib.0000234135.43336.72. [DOI] [PubMed] [Google Scholar]
- 36.Borcherding F, Nitschke M, Hundorfean G, Rupp J, von Smolinski D, Bieber K, et al. The CD40-CD40L pathway contributes to the proinflammatory function of intestinal epithelial cells in inflammatory bowel disease. Am J Pathol. 176(4):1816–1827. doi: 10.2353/ajpath.2010.090461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsutani T, Anantha Samy TS, Kang SC, Bland KI, Chaudry IH. Mouse genetic background influences severity of immune responses following trauma-hemorrhage. Cytokine. 2005;30(4):168–176. doi: 10.1016/j.cyto.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 38.Bassaganya-Riera J, Misyak S, Guri AJ, Hontecillas R. PPAR gamma is highly expressed in F4/80(hi) adipose tissue macrophages and dampens adipose-tissue inflammation. Cell Immunol. 2009;258(2):138–146. doi: 10.1016/j.cellimm.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 40.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willems F, Vollstedt S, Suter M. Phenotype and function of neonatal DC. Eur J Immunol. 2009;39(1):26–35. doi: 10.1002/eji.200838391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.