The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A
The Endopeptidase-like and Kinase associated to transcribed Chromatin (KEOPS)/Endopeptidase-like and Kinase associated to transcribed Chromatin (EKC) complex has been implicated in multiple biological processes including transcription, telomere maintenance and chromosome segregation. This study reveals a role in the biosynthesis of a universal tRNA modification threonyl carbamoyl adenosine (t6A).
Keywords: Kae1, KEOPS–EKC complex, Sua5, t6A, tRNA
Abstract
The highly conserved Kinase, Endopeptidase and Other Proteins of small Size (KEOPS)/Endopeptidase-like and Kinase associated to transcribed Chromatin (EKC) protein complex has been implicated in transcription, telomere maintenance and chromosome segregation, but its exact function remains unknown. The complex consists of five proteins, Kinase-Associated Endopeptidase (Kae1), a highly conserved protein present in bacteria, archaea and eukaryotes, a kinase (Bud32) and three additional small polypeptides. We showed that the complex is required for a universal tRNA modification, threonyl carbamoyl adenosine (t6A), found in all tRNAs that pair with ANN codons in mRNA. We also showed that the bacterial ortholog of Kae1, YgjD, is required for t6A modification of Escherichia coli tRNAs. The ATPase activity of Kae1 and the kinase activity of Bud32 are required for the modification. The yeast protein Sua5 has been reported previously to be required for t6A synthesis. Using yeast extracts, we established an in vitro system for the synthesis of t6A that requires Sua5, Kae1, threonine, bicarbonate and ATP. It remains to be determined whether all reported defects of KEOPS/EKC mutants can be attributed to the lack of t6A, or whether the complex has multiple functions.
Introduction
In 2006, two groups independently described a previously unknown conserved nuclear protein complex. They used very different yeast genetic screens as starting points and each proposed a different function for the complex. One group named this complex KEOPS (Kinase, Endopeptidase and Other Proteins of small Size) (Downey et al, 2006). Their starting point was a genome-wide search for mutations that improved the growth of a cdc13 mutant. Cdc13 protein is crucial for telomere protection and function. They found that deletion of a previously uncharacterized gene partially suppressed a cdc13-1 temperature-sensitive (ts) mutation. They called this yeast gene CGI121 because the protein was a homologue of a human protein, Cgi-121, that interacted with a p53 kinase called PRPK. Yeast also has a clear ortholog of human PRPK called Bud32. To identify interacting partners, yeast Cgi121 was TAP tagged and used to purify a protein complex that they named KEOPS. The complex consisted of Cgi121, the kinase Bud32, a putative endopeptidase called Kae1 (Kinase-Associated Endopeptidase) and a small protein called Pcc2/Gon7. Mutations in the genes for three of these proteins caused severe growth defects and shortened telomeres. Downey et al concluded that the KEOPS complex was in some way involved in telomere regulation and maintenance.
Another lab discovered the same complex while searching for multicopy suppressors of a splicing defect caused by a point mutation in U1 RNA (Kisseleva-Romanova et al, 2006). The screen yielded a gene encoding a small protein of 88 amino acids. Deletion of this gene, which they named PCC1, caused extremely poor growth. A pcc1 ts mutant had defects in transcription of many genes as well as genetic interactions with other genes involved in transcription. They TAP-tagged Pcc1 to discover proteins that bound to it and identified the same four proteins Kae1, Bud32, Cgi121 and Pcc2 that were identified by Downey et al. It is possible that the screen of Downey et al missed Pcc1 because of its small size. Kisseleva-Romanova et al named this complex EKC for ‘Endopeptidase-like and Kinase associated to transcribed Chromatin.' Chromatin immunoprecipitation experiments showed that the complex was associated with transcribed genes, and thus these authors concluded that it was involved in transcription. Deletions of four of the five genes encoding the complex led to extremely slow growth or lethality, the only exception being CGI121 whose deletion had very little effect on growth.
Four of the five proteins in the KEOPS/EKC complex show extraordinary sequence conservation from archaea to mammals. One of these proteins, Kae1, is even more conserved, with readily detectable orthologs in bacteria. Notably, Kae1 is one of the very few proteins that are nearly ubiquitous in cellular life forms, but its exact function(s) remains elusive; accordingly, it has been described as the top target for experimental study among functionally uncharacterized conserved proteins (Galperin and Koonin, 2004). Interestingly, genomes of all eukaryotes also code for a mitochondrial homologue of Kae1, called Qri7 in yeast. The sequence conservation and the severe pleiotropic phenotypes of mutants clearly suggest that Kae1 has a crucial role in all species. A recent study described phenotypes of a yeast kae1 ts mutant and concluded it had defects in chromosome segregation (Ben-Aroya et al, 2008). Thus, although three different roles have been attributed to this complex, namely telomere maintenance, transcription and chromosome segregation, its actual function(s) in these processes is unknown.
In this study, we show that the KEOPS/EKC complex is required for a universal tRNA modification called threonyl carbamoyl adenosine (t6A), found in all tRNAs that pair with ANN codons, where N is any of the four bases (the only exception being prokaryotic fMet tRNA). This modification consists of a carbonyl group and a threonine attached to the amino group of adenine at nucleotide 37 immediately 3′ of the anticodon (see Figure 3A). Structural work has demonstrated that t6A strengthens the A-U codon–anticodon interaction on the ribosome (Murphy et al, 2004). Very little is known about the enzymes responsible for the t6A modification. A recent paper reported that a yeast protein, Sua5, which in not part of the KEOPS/EKC complex, is required for t6A formation (El Yacoubi et al, 2009). In addition, early reports demonstrated that a partially purified Escherichia coli extract was able to carry out the modification reaction in vitro and that the reaction required threonine, bicarbonate and ATP (Elkins and Keller, 1974; Korner and Soll, 1974).
Results
kae1 Mutants are defective in translation
Given the extraordinary conservation of Kae1 among eukaryotes (Figure 1) and the various functions attributed to this protein and its associated complex, we sought to identify the essential role of Kae1. To this end, we tested the effects of a kae1 ts mutation at the non-permissive temperature on macromolecular synthesis. Although the rates of DNA and RNA synthesis were reduced in the mutant, as compared with a wild-type strain, protein synthesis was more severely affected (Supplementary Figure S1). This finding suggested that the primary function of Kae1 could be associated with translation, in accord with the prediction, based on genomic neighbourhood analysis, that the bacterial Kae1 ortholog performs a translation-associated function (Wolf et al, 2001). Further support for the functional association of the KEOPS/EKC complex with translation comes from a comparative genomic analysis of several Methanocaldococcus species in which the PCC1 gene is fused with the gene for an acetyltransferase, an apparent ortholog of RimI, known to acetylate alanine in a bacterial ribosomal protein (Yoshikawa et al, 1987). Given that the KEOPS/EKC complex was previously implicated in transcription (Kisseleva-Romanova et al, 2006), we considered the possibility that it was required for rRNA synthesis which would affect translation. We found that neither the steady-state levels nor the maturation of rRNA was affected in kae1 ts mutants.
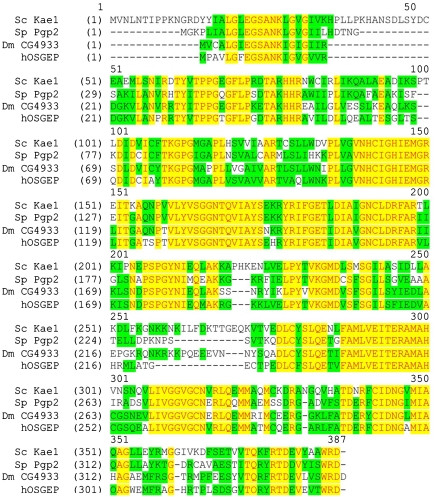
Figure 1.
ClustalW alignment of eukaryotic Kae1 orthologs. The alignment of the amino acid sequences was generated using Vector NTI software (Invitrogen). Identical residues are shaded in yellow and conservative substitutions are shaded in green. Dm, Drosophila melanogaster; h, Homo sapiens; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe.
t6A Modification of tRNAs requires Kae1
We became aware of the observation that some bacterial carbamoyl transferases contain a domain also found in Kae1, raising the possibility that Kae1 and its orthologs are involved in the t6A modification of tRNAs (El Yacoubi B and de Crécy-Lagard V, http://genomicscience.energy.gov/pubs/2009abstracts/2009GTLAbstractsFinal3.pdf). This finding prompted us to test whether Kae1 was required for this modification. KAE1 is annotated as an essential gene but we were able to obtain a viable but extremely slowly growing deletion strain by dissecting a diploid, heterozygous for the mutation. To examine the involvement of Kae1 in t6A modification of tRNAs, we analysed tRNAs isolated from the kae1 deletion strain by primer extension. As mentioned above, Sua5 has been reported to be required for the t6A modification (El Yacoubi et al, 2009), and thus we used a sua5 deletion strain as a control for the primer extension experiments. Bulk tRNAs were isolated from wild type, kae1 and sua5 deletion strains and subjected to extension with primers specific to either tRNA Ile (AAU) that normally contains the t6A modification, or tRNA Val(UAC) that lacks the modification. Strikingly, in case of the Ile tRNA, both kae1 and sua5 deletion strains showed an identical pattern of extension that differed from the wild-type pattern at nucleotides 37 and 38, near the position of the t6A modification (Figure 2). On the other hand, all three strains had an identical primer extension pattern for the Val tRNA. These results suggested that the difference observed at positions 37 and 38 of the Ile tRNA could be indicative of an effect on t6A modification of tRNA in both kae1 and sua5 mutant strains. We also noticed that the Ile RNA in the two mutants was extended beyond that of the wild type, suggesting that the mutants had a defect in 5′-end processing. To test if this was the case, we examined bulk tRNAs isolated from wild type, kae1 and sua5 mutants by northern blotting analysis using hybridization with specific probes. We observed that all the Ile tRNAs from the kae1 and sua5 mutants that were detected by this method migrated at the position of fully processed wild-type tRNA (Supplementary Figure S2), and thus the difference observed in primer extension was most likely due to a minor fraction of unprocessed tRNAs in the two mutants. Therefore, we focused our attention on the difference in primer extension seen at the position of the t6A modification.
Figure 2.
Primer extension analysis of tRNA Ile (AAU) and Val (UAC). tRNAs isolated from wild type (WT), kae1Δ and sua5Δ mutant strains were subjected to primer extension analysis as described in Materials and methods. End-labeled primers specific to either tRNA Ile (AAU) or Val (UAC) were used for the analysis. Cloverleaf structures of the Ile and Val tRNAs are shown. The specific primers used are complementary to the sequence shown in bold face in the cloverleaf structures. The position of the t6A-modified base (A37) in the Ile tRNA, marked by horizontal arrows, was confirmed by running length standards and sequence ladders.
Modification of cytoplasmic and mitochondrial tRNAs requires Kae1 and Qri7, respectively
To test directly whether Kae1 was involved in t6A modification of tRNAs, we analysed the nucleosides derived from total tRNA preparations of wild type, kae1Δ and sua5Δ strains by LC-MS/MS. The structure of t6A and its m/z spectrum are shown in Figure 3A. We determined the ratio of t6A/A in total tRNAs derived from the three strains. The analysis revealed that the ratio of t6A/A in the wild type was about 2.5% while the kae1Δ mutant had about 40-fold reduced levels of t6A (t6A/A=0.06%) (Figure 3B), demonstrating that Kae1 is essential for t6A modification of tRNAs. Interestingly, the sua5Δ strain had a more than 10-fold lower t6A/A ratio than the kae1 mutant (t6A/A=0.004%), effectively at background levels (Figure 3B). To explain why kae1 mutants had a higher than background level of t6A, we hypothesized that Qri7, the mitochondrial homologue of Kae1, might still be functional in a kae1Δ strain and thus would contribute to the total t6A. We, therefore, tested the amount of t6A in tRNA from qri7Δ and qri7Δ kae1Δ mutants. The qri7 mutant had a slightly reduced amount of t6A in comparison to wild type, whereas the amount of t6A in the qri7 kae1 double mutant was at the background level observed in the sua5 mutant (Figure 3B). Thus, it is likely that Kae1 is involved in t6A modification of cytoplasmic tRNAs while Qri7 is involved in modification of mitochondrial tRNAs. Additional evidence came from primer extension studies using a primer specific for a mitochondrial Ile tRNA. The qri7 mutant affected only the primer extension pattern for the mitochondrial tRNA whereas the kae1 mutant affected only the cytoplasmic pattern (Supplementary Figure S3). Given that the sua5Δ mutant completely abolished t6A, it seems likely that Sua5 is required for modification of both the cytoplasmic and mitochondrial tRNAs (although the Sua5 protein does not have an obvious mitochondrial import signal).
Figure 3.
(A) LC-MS/MS spectrum of t6A. Based on the fragmentation of an adenosine standard, t6A was analysed by LC-MS/MS in selected reaction monitoring (SRM) mode (m/z 413.2 → m/z 280.9). (B) t6A content of tRNA isolated from the indicated strains. tRNA isolated from wild type (WT), kae1Δ, sua5Δ, qri7Δ and kae1Δ qri7Δ mutant strains was hydrolysed to nucleosides as described in Materials and methods. The t6A content of each strain, normalized to the respective adenosine content, is plotted as the % t6A/A and the numerical values are also shown below the graph. (C) t6A content of an E. coli strain depleted of YgjD. An E. coli strain with the natural ygjD promoter replaced by the inducible arabinose promoter was grown with (YgjD+) and without arabinose (YgjD−). tRNA was isolated from the two cultures and processed for LC-MS/MS analysis as above. The % t6A/A for each condition is plotted and the numerical values are also shown below the graph.
The t6A-modifying activity of Kae1 is evolutionarily conserved
As mentioned above, the gene for Kae1 is represented in at least one copy per genome in (nearly) all organisms from all three domains of life. The gene is essential in all bacterial species tested, thus far including E. coli (Arigoni et al, 1998). To test whether the E. coli ortholog of Kae1, YgjD, is required for t6A modification, we used an E. coli strain that has the natural ygjD promoter replaced by the inducible arabinose promoter. Growing this strain in the absence of arabinose depletes the intracellular pool of YgjD (Handford et al, 2009). To determine whether YgjD is required for t6A modification, we used LC-MS/MS quantification of the t6A/A ratio in tRNAs isolated from E. coli cells under YgjD-depleted (−arabinose) or YgjD-repleted (+arabinose) conditions (Figure 3C). When the strain was grown in the presence of arabinose, the t6A/A ratio was ∼0.2%. In sharp contrast, depletion of YgjD by growing cells in the absence of arabinose reduced the t6A/A ratio by 40-fold (Figure 3C). This indicates that the E. coli Kae1 ortholog, YgjD, is required for t6A modification of tRNAs, and thus the function is conserved between yeast and bacteria.
The t6A modification requires Pcc1, the ATPase activity of Kae1 and the kinase activity of Bud32
As mentioned above, Pcc1, Bud32 and Cgi121 are the other highly conserved subunits of the KEOPS/EKC complex. Moreover, in many archaeal genomes the genes for the Kae1 and Bud32 orthologs are fused, supporting a tight functional linkage between these proteins. We tested whether the other subunits of the complex are required for t6A modification by primer extension of a t6A containing tRNA (Ile AAU) isolated from WT, pcc1Δ, kae1Δ, bud32Δ and cgi121Δ strains. The primer extension profiles of the pcc1 and bud32 mutants look exactly like that of the kae1 mutant, with a marked difference at position 37 in comparison to the wild type. In contrast, the profile from the cgi121 mutant was indistinguishable from that of the wild type (Figure 4). This result suggests that Pcc1 and Bud32 along with Kae1 are required for t6A modification, whereas Cgi121 is dispensable. It is noteworthy that the pcc1Δ and bud32Δ strains grow as poorly as the kae1Δ mutant whereas the cgi121Δ mutant grows at a close to normal rate (Supplementary Figure S4).
Figure 4.
Primer extension analysis of tRNA Ile (AAU) isolated from KEOPS/EKC mutants. tRNAs isolated from wild type (WT), pcc1Δ, kae1Δ, bud32Δ and cgi121Δ mutant strains were subjected to primer extension analysis as in Figure 1. The cloverleaf structure of the Ile tRNA is shown. The specific primer used for the analysis is complementary to the sequence shown in bold face in the cloverleaf structure. The position of the t6A-modified base (A37) in the Ile tRNA is marked by horizontal arrows.
Sequence and structure analyses show that Kae1 adopts an actin/HSP70/DnaK structural fold and retains all the conserved amino acid residues that are required for ATP binding and hydrolysis by the molecular chaperones of the HSP70/DnaK family (Aravind and Koonin, 1999; Koonin, 2010). Furthermore, a weak metal-dependent ATPase activity of Kae1 has been demonstrated (Mao et al, 2008). In order to test whether the ATPase activity is required for t6A formation, we used mass spectroscopy to quantify the t6A content of tRNAs isolated from a kae1Δ strain transformed with a low copy CEN plasmid containing the wild-type KAE1 gene, or a kae1 mutant that abolishes metal binding and ATPase activity (kae1H141,145A) (Mao et al, 2008), or the empty vector. The analysis revealed that the kae1H141,145A mutant grew as poorly as the kae1Δ strain and had a very low t6A/A ratio compared with wild type (Figure 5A). This finding strongly suggests that the ATPase activity of Kae1 is required for t6A formation.
Figure 5.
Kae1 ATPase activity and Bud32 kinase activity are required for t6A formation. (A) LC-MS/MS analysis of tRNA nucleosides from kae1 mutants. A kae1Δ strain was transformed with a low copy CEN plasmid containing the wild-type KAE1 gene (WT), a mutant of the ATP-binding site (kae1H141,145A) or the empty vector (kae1Δ). Ten-fold serial dilutions of the respective cultures were spotted onto a selective medium and assessed for growth at 30°C. tRNA isolated from the three strains was processed for LC-MS/MS quantification of t6A and A as described in Materials and methods. The t6A content of each sample, normalized to the respective adenosine content, is plotted as the % t6A/A and the numerical values are also shown below the graph. (B) A similar analysis was done with bud32 mutants. A bud32Δ strain was transformed with a low copy CEN plasmid containing the wild-type BUD32 gene (WT), a catalytic site mutant (bud32D161A) or the empty vector (bud32Δ). Ten-fold serial dilutions of the respective cultures were spotted onto a selective medium and assessed for growth at 30°C. The t6A content of each sample, normalized to the respective adenosine content, is plotted as the % t6A/A and the numerical values are also shown below the graph.
Bud32 is an atypical, Rio-like, protein kinase (Leonard et al, 1998; Facchin et al, 2002). To test whether the kinase activity of Bud32 is required for t6A modification, we used a strategy similar to that used for the Kae1 ATPase mutant. A bud32Δ strain was transformed with a low copy CEN plasmid containing the wild-type BUD32 gene, or a catalytic site mutant known to abolish kinase activity in vitro (bud32D161A) (Lopreiato et al, 2004) or the empty vector. The analysis by mass spectroscopy revealed that in the bud32Δ mutant, the t6A/A ratio was reduced 10-fold compared with wild type (t6A/A=0.023% as opposed to 0.23% in the wild type), while in the bud32D161A catalytic mutant, the t6A/A ratio was reduced to 30% of the wild type (t6A/A=0.069%, as opposed to 0.23 in the wild type) (Figure 5B). Thus, the kinase activity of Bud32 appears to be required for t6A modification. The t6A/A ratio of the wild-type strain in this experiment (bud32Δ with BUD32 on a CEN plasmid) was lower than that seen for a strain with a genomic copy of the wild-type gene (Figure 3B). This difference is most likely due to plasmid loss and the use of synthetic medium in this experiment. The fact that the catalytic site mutant exhibits better growth than the bud32 deletion (Figure 5B), an observation that has been reported previously (Lopreiato et al, 2004), correlates well with the data showing the catalytic site mutant has more t6A than the deletion mutant. Possibly, the catalytic mutant retains some kinase activity in vivo; alternatively, Bud32 contributes to t6A formation as a structural subunit of the complex, to some extent independent of its kinase activity.
In vitro reconstitution of the t6A-modification reaction
It has been reported previously that partially purified E. coli extracts can carry out t6A modification of tRNA in vitro (Elkins and Keller, 1974; Korner and Soll, 1974). The reaction required threonine, bicarbonate and ATP (Elkins and Keller, 1974). Under similar conditions, using a mixture of the KEOPS/EKC complex and Sua5 partially purified from yeast by TAP-tag affinity chromatography, we were unable to incorporate radioactive threonine into tRNA-lacking t6A (isolated from a sua5 mutant and thus lacking the modification). However, using a yeast whole-cell extract and the same tRNA substrate, we observed incorporation of radioactive threonine into TCA precipitable material in an ATP, bicarbonate and Kae1-dependent manner (Table I). The dependence on bicarbonate and Kae1 made it unlikely that we were simply observing charging of threonyl tRNA; instead the results suggested that threonine was being incorporated into t6A. Yeast extracts prepared from a kae1Δ mutant showed a modest 1.6-fold increase in incorporation over that seen without ATP and bicarbonate (Table I). This result was suggestive of a low level of t6A synthesis that might be due to the presence of Qri7 in the extract. In contrast, the sua5 mutant extracts were just as effective as the wild type in bicarbonate and ATP-dependent incorporation of threonine (Table I). This observation was surprising in light of the fact that Sua5 is necessary to modify tRNAs in vivo ((El Yacoubi et al, 2009) and our results). In an attempt to understand this puzzling result, and to confirm that the observed threonine incorporation was indicative of t6A biosynthesis, we replaced the radioactive threonine with heavy threonine (containing one N15 and four C13 atoms) and analysed the reaction product by LC-MS/MS. As shown in Figures 3A and 6A, t6A with natural isotopes has a retention time of 12.5 min under our conditions and m/z=413.2 fragmenting to 280.9. Using wild-type extracts, substrate tRNA and heavy threonine, we detected a peak with a retention time of 12.5 min and m/z=418.6 with a fragmentation product with m/z=286.0 (shifted by 5 Da because of the heavy isotopes). The 418 peak was not seen in reactions that lacked either bicarbonate or ATP (Figure 6A). This result provides strong evidence that the threonine was indeed being incorporated into t6A (Figure 6A). Under similar conditions, an extract from a kae1Δ mutant did not yield heavy t6A (Figure 6B). Interestingly, we did not detect the heavy t6A peak in reactions containing extracts from a sua5Δ mutant either. This observation contradicted the results obtained with the radioactive threonine assay (Table I). We speculate that the bicarbonate and ATP-dependent incorporation of the radioactive threonine observed in the sua5Δ mutant could be due to a reaction intermediate in t6A modification (see Discussion). In summary, the observations that wild-type yeast extracts could carry out the t6A modification in vitro, while neither kae1 nor sua5 mutant extracts could, confirms the requirement of Kae1 and Sua5 for t6A modification of tRNAs.
Table 1. t6A synthesis by yeast cell extractsa.
| Incorporation of 14C threonineb | |||
|---|---|---|---|
| Complete | −HCO3− | −ATP | |
| WT | 2380 | 474 | 301 |
| kae1Δ | 800 | 497 | 433 |
| sua5Δ | 2418 | 421 | 376 |
| −Protein | 121 | ||
| −RNA | 334 | ||
| aThe assay system contained 10 μg of t6A-deficient RNA and 100 μg of cell extract. | |||
| bc.p.m. In TCA-precipitable material. | |||
Figure 6.
(A) In vitro t6A biosynthesis requires bicarbonate and ATP. The top panel shows the retention time (RT=12.5 min) and m/z value (413.2) of t6A as measured by LC-MS/MS. The three panels below that show the results found when t6A-deficient tRNA was incubated with wild-type cell extract and heavy threonine (one 15N and four 13C atoms), in presence of bicarbonate and ATP (complete), lacking bicarbonate (−HCO3−), or lacking ATP (−ATP). The t6A heavy isotope-labeled reaction product was confirmed by LC-MS/MS in SRM mode (m/z 418.6 → m/z 286.0). (B) In vitro t6A biosynthesis requires Kae1 and Sua5. t6A-deficient tRNA was incubated with cell extracts prepared from wild type, sua5Δ or kae1Δ strains in the presence of heavy threonine, bicarbonate and ATP. The reaction products were processed and analysed as in (A). The mass spectrum of the nucleotide component with a RT of 12.5 min is shown.
Discussion
We presented evidence that Kae1 and the conserved KEOPS/EKC complex are required for the synthesis of t6A in yeast, and confirmed previous work indicating that Sua5 is also required for this reaction (El Yacoubi et al, 2009). Interestingly, three other tRNA-modified bases, pseudouracil, 5-methyl cytosine and 1-methyl guanine, are present in normal amounts in kae1 and sua5 mutants (S D'Silva and E Phizicky, unpublished data). We also showed that the bacterial ortholog of Kae1, YgjD, is required for t6A synthesis in E. coli, as is the yeast mitochondrial homologue, Qri7, for mitochondrial tRNAs. It is thus very likely that Kae1 and its orthologs perform this function in all species. Given that Kae1 is a subunit of the KEOPS/EKC complex, highly conserved from archaea to mammals, it seems likely that t6A formation is a primary function of this complex. In addition to Kae1, the Pcc1 and Bud32 subunits of the complex, but not Cgi121, are required for t6A synthesis. Furthermore, the kinase activity of Bud32 is required. Little is known about in vivo substrates of Bud32, except for the glutaredoxin Grx4 (Peggion et al, 2008). Biochemical experiments in vitro have demonstrated that Bud32 phosphorylates Kae1, and that binding of Kae1 to Bud32 inhibits its autophosphorylation activity (Hecker et al, 2008). In vitro, Cgi121 greatly stimulates the kinase activity of archaeal Bud32 (Mao et al, 2008). Perhaps, another protein can supply this function in yeast, thus making Cgi121 less important for normal growth.
Importantly, deletion mutations of the KAE1, BUD32 and PCC1 genes grow poorly and are defective in t6A synthesis whereas the cgi121Δ mutant grows reasonably well and is able to synthesize t6A. This finding highlights the importance of t6A for normal growth. As noted earlier, this tRNA modification strengthens the interaction of an A-U codon–anticodon base pair on the ribosome to allow proper decoding of ANN codons (Weissenbach and Grosjean, 1981; Stuart et al, 2000; Murphy et al, 2004). Mutations in SUA5 are known to increase the frequency of frameshifting during protein synthesis, most likely because of the lack of the t6A modification (Lin et al, 2010). Furthermore, the original isolation of a sua5 mutant in a genetic screen can be explained by abnormal frameshifting in the mutant (Na et al, 1992).
Biochemical activity of Kae1
Kae1 and its orthologs have been annotated as O-sialoglycoprotein endopeptidases based on the work of a single laboratory that purified this protein from the bacterium Pasteurella haemolytica and subsequently cloned the gene from that species (Abdullah et al, 1991). The gene was cloned from a genomic expression library by screening recombinant plasmids for protease activity on a sialylated protein, glycophorin. The authors sequenced the gene they had cloned and noticed how conserved it was; that is, they had cloned the P. haemolytica ortholog of Kae1. All subsequent annotations of the protein as a sialoglycoprotein endopeptidase are based on this initial report. However, a subsequent paper from the same group casts doubt that the gene they cloned actually encodes such an enzyme (Jiang and Mellors, 2004). Also, another group could not detect protease activity using the Kae1 ortholog from an archaeal species (Hecker et al, 2007). Additionally, a report that an archaeal Kae1 ortholog possessed AP lyase activity (Hecker et al, 2007) could not be confirmed with an ortholog purified from a different archaeal species (Mao et al, 2008). We expressed and purified yeast Kae1 in E. coli and initially detected AP lyase activity. However, subsequent control experiments showed that a low level contamination from the highly active E. coli enzyme, Fpg, was responsible for the observed AP lyase activity. Thus, it appears extremely unlikely that Kae1 has endopeptidase or AP lyase activity. As mentioned above, sequence analysis indicates that Kae1 is a member of the HSP70/DnaK family of molecular chaperones; in this study, we show that its ATPase activity is required for t6A formation (Figure 5A). These findings suggest that Kae1 could be an ATP-dependent chaperone for a protein or possibly for tRNA. For example, Kae1 could hold substrate tRNAs in the correct orientation for modification by Sua5 that is likely to be the actual catalyst for the t6A formation.
The telomere defect and possible other roles of the complex
It is significant that yeast KEOPS/EKC and SUA5 mutants have a very similar telomere maintenance defect (Downey et al, 2006; Meng et al, 2009). Given that the synthesis of full-length yeast Est3 protein, necessary for normal telomere function, requires a programmed frameshift (Morris and Lundblad, 1997), we considered the possibility that the telomere defect of the mutants was due to inefficient frameshifting and hence a low amount of Est3. Using reagents generously supplied by V Lundblad, we tested this possibility, but found no evidence that there was less full-length Est3 in the mutants. Another possibility that we are currently testing is that the telomerase RNA itself has a functionally important t6A modification. Finally, it seems possible that the putative chaperone activity of Kae1 and the KEOPS/EKC complex are important for proper folding of the telomerase RNA. Although the present results show that the complex is required for t6A synthesis, this might not be its only role. For instance, the convincing chromatin IP data that the complex is associated with transcribed genes (Kisseleva-Romanova et al, 2006) cannot be easily explained by our demonstration of its involvement in t6A biosynthesis. Therefore, the function of the KEOPS/EKC complex in telomere maintenance might be unrelated to t6A. More specifically, it is possible that the predicted chaperone activity of this complex contributes to several processes, t6A formation being one of them.
Enzymology of t6A synthesis
Using heavy isotope-labeled threonine and a tRNA preparation from a sua5 mutant (hence lacking t6A), we were able to obtain convincing evidence that a yeast extract could catalyse the synthesis of t6A in vitro. The reaction required Kae1, Sua5, Bud32, and threonine, bicarbonate and ATP (Figure 6). The requirement for those three small molecules for t6A synthesis was also seen many years ago using a partially purified E. coli extract (Elkins and Keller, 1974). The precise roles of the proteins involved are not understood; however. It is very likely that ATP is used to activate one or more of the molecules involved in the reaction. Reasoning that such activated intermediates might accumulate in either kae1 or sua5 mutants, we isolated tRNA from each of the mutants, hydrolysed the RNA to nucleosides as in the experiment shown in Figure 3, and used mass spectroscopy to look for activated adenosine intermediates with a carboxyl group, a carbonyl phosphate group or carbonyl AMP attached to the amino group of adenosine. We did not find evidence for any of these compounds, either because they do not exist or because they were hydrolysed during the isolation procedure. Another possibility is that threonine itself is modified and activated, followed by transfer of that intermediate to adenosine on tRNA, with ATP being required for one or both steps of the reaction.
The enzyme assays using radioactive threonine provided our initial evidence that t6A was being synthesized in vitro. Wild-type extracts were able to incorporate threonine into TCA-precipitable material while kae1 mutant extracts were not (Table I). On the other hand, we obtained the surprising result that sua5 mutant extracts incorporated just as much threonine as wild-type extracts, and the incorporation also depended on the presence of bicarbonate and ATP. However, the experiments with heavy threonine provided strong evidence that no t6A was synthesized in sua5 mutant extracts. We, therefore, assume that a TCA-precipitable threonine intermediate accumulates in the sua5 mutant, and that Sua5 protein is required to transfer it to adenosine on tRNA. An activated carbonyl threonine moiety might be assembled by and on the KEOPS/EKC complex and then transferred to tRNA by Sua5 protein. Perhaps another as yet unidentified protein is involved in the reaction. That could explain why we were unable to incorporate threonine into tRNA using a mixture of partially purified TAP-tagged Kae1 and Sua5. More experiments are needed to decipher the steps involved in this key tRNA modification and the enzymes that carry them out.
In conclusion, the involvement of the KEOPS complex in the formation of t6A would in itself suffice to account for its exceptional conservation, because this modification is essential for accurate decoding of mRNAs (Yarian et al, 2002). Loss of t6A modification, due to deletion of SUA5 causes severe translational errors such as frameshifting, nonsense suppression and incorrect initiation at internal AUG codons (Lin et al, 2010). These defects are also compatible with the severe pleiotropic effects observed in KEOPS complex mutants. Our findings highlight the importance of the KEOPS complex in maintaining the stringency of the genetic code and translational fidelity.
Materials and methods
Strains, plasmids and growth conditions
The strains and plasmids used in this study are listed in Supplementary Table SI. Sequences of primers and probes used are available upon request. Yeast strains were grown in YPD medium (1% yeast extract, 2% peptone, 2% dextrose) or in synthetic complete medium lacking uracil to select for the pRS316-based plasmids. Plasmid transformations were performed according to standard protocols (Ausubel et al, 1987). Deletions of KAE1, PCC1, BUD32, CGI121 and QRI7 were made by gene replacement using the Schizosaccharomyces pombe his5+, the E. coli kanMX6 or the Saccharomyces cerevisiae TRP1 gene in a W303 diploid strain (Longtine et al, 1998). The heterozygous diploid was sporulated and haploid strains carrying the deletions were selected using the specific marker. The deletions and the mating type of the haploid mutants were confirmed by PCR.
E.coli strains were cultured aerobically at 37°C in LB medium. The conditional E. coli ygjD strain was grown on LB agar plates supplemented with 0.2% arabinose. To maximally deplete intracellular YgjD, a single colony was inoculated into 1 ml LB and grown overnight in the absence of arabinose. The cells were subcultured into 100 ml of LB supplemented with 0.5% glucose, allowed to grow to OD600=0.2–0.3 and harvested by centrifugation. For YgjD+ growth, cells from the depleted culture were subcultured at 37°C in LB supplemented with 0.2% arabinose to OD600=0.2–0.3 and harvested by centrifugation. Harvested cells were washed in cold water and stored at −80°C.
The plasmid pYY128 containing the BUD32 gene was constructed by cloning the BUD32 promoter region (−296 to +1), the BUD32 ORF, and 217 bp downstream of the ORF in the vector pRS316 (URA3 CEN). The plasmid pYY132 containing the bud32D161A allele was constructed by site-directed mutagenesis of the parent plasmid pYY128.
Purification of tRNA
Yeast strains were grown in YPD or SC-Ura medium to O.D600=1.5 at 30°C, harvested, washed in cold distiled water and stored at −80°C. The conditional E. coli strain was grown at 37°C in LB supplemented with either 0.2% arabinose (YgjD+) or 0.2% Glucose (YgjD−) to OD600=0.3, harvested, washed in cold water and stored at −80°C. The yeast and bacterial cell pellets were thawed on ice and bulk tRNA was isolated using the mirVana™ miRNA isolation kit (Ambion) according to the manufacturer's instruction. The tRNA was eluted in water and stored at −80°C. RNA concentration was quantified using Nano Drop (Thermo Scientific). The RNA quality was checked by electrophoresis on a 15% denaturing polyacrylamide gel and visualized by ethidium bromide staining.
Primer extension assays
Primer extension experiments were carried out using 15–20 nucleotide primers (Operon) that were 5′-end-labeled with T4 polynucleotide kinase (NEB) and [γ-32P] ATP (6000 Ci/mmol, Perkin Elmer), followed by centrifugation through a Probe Quant G-50 micro column (GE Healthcare) to remove unincorporated ATP. In a 5-μl reaction, 1 pmol of each primer was annealed with 2 μg of total RNA (isolated as described above) in First-Strand buffer (Invitrogen), by heating to 95°C for 3 min followed by slow cooling to 37°C. The annealed RNA/primer was added to a 5-μl reaction containing 0.5 mM of each of the four dNTPs, 0.01 M DTT and 200 units of SuperScript™ II reverse transcriptase (Invitrogen) in First-Strand buffer. Extensions were allowed to continue at 42°C for 50 min, followed by heat inactivation at 70°C for 15 min. After addition of an equal volume (10 μl) of Gel Loading Buffer II (Ambion), the reactions were electrophoresed on a 15% denaturing polyacrylamide gel (SequaGel® sequencing system, National Diagnostics) and visualized using a PhosphorImager (Molecular Dynamics).
LC-MS/MS analysis
To quantify the t6A content of tRNAs, they were enzymatically hydrolysed to nucleosides as described (Crain, 1990). Approximately 30 μg of bulk tRNA was digested with five units of nuclease P1 (US Biological), 0.01 units of snake venom phosphodiesterase (Sigma) and 10 units of calf intestine alkaline phosphatase (Roche) in a total volume of 50 μl. The resulting nucleosides were lyophilized and resuspended in 10 μl of water. They were purified by passing them through a Millipore ZipTip C-18 column. The nucleosides were resolved in a Luna C18-RP (150 × 2 mm) analytical column using a solvent consisting of 5 mM of ammonium acetate pH 6.0 for 5 min followed by a gradient of 0–50% acetonitrile in the presence of 5 mM ammonium acetate for 10 min. They were analysed in a Thermo TSQ Quantum Access Triple Quadrupole Mass Spectrometer (Thermo-Fisher). The mass spectra were recorded in positive ion mode. Multiple/Selected reaction monitoring mode (MRM/SRM) was used with a collision energy of 22 eV. The mass transitions were m/z 268 → m/z 136 for adenosine, m/z 413.2 → m/z 280.9 for t6A and m/z 418.6 → m/z 286.0 for t6A containing heavy threonine (four C13 atoms and one N15 atom). Data acquisition and quantitative spectral analysis was conducted using the Xcalibur 2.0.7 software.
Preparation of yeast cell extracts
S. cerevisiae strains were grown in YPD at 30°C to an OD600=0.8–1, harvested and washed with cold water. The cell pellet was resuspended in cold lysis buffer (50 mM Tris (pH 8.0), 150 mM NaCl, 5 mM KCl, 1 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 2 mM benzamidine, 4 μg/ml pepstatin A, 4 μg/ml leupeptin and 10 μg/ml aprotinin) and slowly dropped into liquid nitrogen. The frozen pellets were ground in a coffee grinder (Krups model 203-70) with dry ice for 5 min. The powdered cell pellet/dry ice mixture was transferred to a cooled beaker and the dry ice allowed to sublime. After thawing on ice, the lysate was clarified at 5000 g at 4°C for 20 min and 16 000 g for 40 min at 4°C. The protein concentration of the lysate was determined by Bradford assay, and the lysates stored at −80°C.
Assay for in vitro t6A synthesis
The assay system consisted of 50 mM Tris–HCl (pH8.0), 20 mM MgC12, 25 mM KCl, 5 mM dithioerythritol, 10 mM ATP, 0.05 mM 14C threonine (20 Ci/mol), 25 mM NaHCO3, 10 μg t6A-deficient tRNA isolated from a sua5 mutant and 100 μg of cell extract in a final volume of 100 μl. After incubation for 20 min at 30°C, the reactions were treated with 1 ml of 10% trichloroacetic acid and salmon sperm DNA as a carrier to precipitate the macromolecules. The precipitates were collected on Whatman GF-C filters, washed with 5% trichloroacetic acid and then with 95% ethanol. The membranes were dried, placed in a vial with toluene-based scintillation fluid and counted in a Packard liquid scintillation counter. For assays with heavy threonine, the radioactive threonine was replaced by heavy threonine at the same concentration. After incubation at 30°C for 20 min, the tRNA in the reaction was purified using the mirVana miRNA isolation kit (Ambion) and processed for LC-MS/MS analysis as described above. Other methods are in the Supplementary data.
Supplementary Material
Acknowledgments
We thank V de Crécy-Lagard and E Phizicky for valuable information, M Hampsey, P Hieter, D Libri, V Lundblad and T Palmer for reagents, R Haltiwanger for advice, and R Rieger and T Koller of the Stony Brook University Proteomics Center for the mass spectroscopy. The Proteomics Center is supported by an NIH shared instrumentation grant, NCRR 1 S10 RR023680-1. The research was supported NIH grants R01 GM065319 to AWK and R01 GM055641 and P01 GM088297 to RS.
Author Contributions: MS, AWK and RS designed the experiments; MS, PM, YY and EP performed the experiments; MS, AWK, EVK and RS analysed the data and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abdullah KM, Lo RY, Mellors A (1991) Cloning, nucleotide sequence, and expression of the Pasteurella haemolytica A1 glycoprotease gene. J Bacteriol 173: 5597–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV (1999) Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J Mol Biol 287: 1023–1040 [DOI] [PubMed] [Google Scholar]
- Arigoni F, Talabot F, Peitsch M, Edgerton MD, Meldrum E, Allet E, Fish R, Jamotte T, Curchod ML, Loferer H (1998) A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol 16: 851–856 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston R, Moore D, Seidman J, Smith JA, Struhl K (1987) Current Protocols in Molecular Biology. New York: John Wiley & Sons [Google Scholar]
- Ben-Aroya S, Coombes C, Kwok T, O'Donnell KA, Boeke JD, Hieter P (2008) Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol Cell 30: 248–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crain PF (1990) Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol 193: 782–790 [DOI] [PubMed] [Google Scholar]
- Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, Guillard S, Partington M, Zubko MK, Krogan NJ, Emili A, Greenblatt JF, Harrington L, Lydall D, Durocher D (2006) A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 124: 1155–1168 [DOI] [PubMed] [Google Scholar]
- El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crécy-Lagard V (2009) The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res 37: 2894–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins BN, Keller EB (1974) The enzymatic synthesis of N-(purin-6-ylcarbamoyl)threonine, an anticodon-adjacent base in transfer ribonucleic acid. Biochemistry 13: 4622–4628 [DOI] [PubMed] [Google Scholar]
- Facchin S, Lopreiato R, Stocchetto S, Arrigoni G, Cesaro L, Marin O, Carignani G, Pinna LA (2002) Structure-function analysis of yeast piD261/Bud32, an atypical protein kinase essential for normal cell life. Biochem J 364: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV (2004) ‘Conserved hypothetical' proteins: prioritization of targets for experimental study. Nucleic Acids Res 32: 5452–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handford JI, Ize B, Buchanan G, Butland GP, Greenblatt J, Emili A, Palmer T (2009) Conserved network of proteins essential for bacterial viability. J Bacteriol 191: 4732–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker A, Leulliot N, Gadelle D, Graille M, Justome A, Dorlet P, Brochier C, Quevillon-Cheruel S, Le Cam E, van Tilbeurgh H, Forterre P (2007) An archaeal orthologue of the universal protein Kae1 is an iron metalloprotein which exhibits atypical DNA-binding properties and apurinic-endonuclease activity in vitro. Nucleic Acids Res 35: 6042–6051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker A, Lopreiato R, Graille M, Collinet B, Forterre P, Libri D, van Tilbeurgh H (2008) Structure of the archaeal Kae1/Bud32 fusion protein MJ1130: a model for the eukaryotic EKC/KEOPS subcomplex. EMBO J 27: 2340–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Mellors A (2004) Handbook of Proteolytic Enzymes. London: Elsevier [Google Scholar]
- Kisseleva-Romanova E, Lopreiato R, Baudin-Baillieu A, Rousselle JC, Ilan L, Hofmann K, Namane A, Mann C, Libri D (2006) Yeast homolog of a cancer-testis antigen defines a new transcription complex. EMBO J 25: 3576–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E (2010) New variants of known folds: do they bring new biology? Acta Crystallographica Section F 66: 1226–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner A, Soll D (1974) N-(purin-6-ylcarbamoyl)threonine: biosynthesis in vitro in transfer RNA by an enzyme purified from Escherichia coli. FEBS Lett 39: 301–306 [DOI] [PubMed] [Google Scholar]
- Leonard CJ, Aravind L, Koonin EV (1998) Novel families of putative protein kinases in bacteria and archaea: evolution of the ‘eukaryotic' protein kinase superfamily. Genome Res 8: 1038–1047 [DOI] [PubMed] [Google Scholar]
- Lin CA, Ellis SR, True HL (2010) The Sua5 protein is essential for normal translational regulation in yeast. Mol Cell Biol 30: 354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lopreiato R, Facchin S, Sartori G, Arrigoni G, Casonato S, Ruzzene M, Pinna LA, Carignani G (2004) Analysis of the interaction between piD261/Bud32, an evolutionarily conserved protein kinase of Saccharomyces cerevisiae, and the Grx4 glutaredoxin. Biochem J 377: 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao DY, Neculai D, Downey M, Orlicky S, Haffani YZ, Ceccarelli DF, Ho JS, Szilard RK, Zhang W, Ho CS, Wan L, Fares C, Rumpel S, Kurinov I, Arrowsmith CH, Durocher D, Sicheri F (2008) Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol Cell 32: 259–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng FL, Hu Y, Shen N, Tong XJ, Wang J, Ding J, Zhou JQ (2009) Sua5p a single-stranded telomeric DNA-binding protein facilitates telomere replication. EMBO J 28: 1466–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DK, Lundblad V (1997) Programmed translational frameshifting in a gene required for yeast telomere replication. Curr Biol 7: 969–976 [DOI] [PubMed] [Google Scholar]
- Murphy FVt, Ramakrishnan V, Malkiewicz A, Agris PF (2004) The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat Struct Mol Biol 11: 1186–1191 [DOI] [PubMed] [Google Scholar]
- Na JG, Pinto I, Hampsey M (1992) Isolation and characterization of SUA5, a novel gene required for normal growth in Saccharomyces cerevisiae. Genetics 131: 791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peggion C, Lopreiato R, Casanova E, Ruzzene M, Facchin S, Pinna LA, Carignani G, Sartori G (2008) Phosphorylation of the Saccharomyces cerevisiae Grx4p glutaredoxin by the Bud32p kinase unveils a novel signaling pathway involving Sch9p, a yeast member of the Akt/PKB subfamily. FEBS J 275: 5919–5933 [DOI] [PubMed] [Google Scholar]
- Stuart JW, Gdaniec Z, Guenther R, Marszalek M, Sochacka E, Malkiewicz A, Agris PF (2000) Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry 39: 13396–13404 [DOI] [PubMed] [Google Scholar]
- Weissenbach J, Grosjean H (1981) Effect of threonylcarbamoyl modification (t6A) in yeast tRNA Arg III on codon-anticodon and anticodon-anticodon interactions. A thermodynamic and kinetic evaluation. Eur J Biochem 116: 207–213 [DOI] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV (2001) Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res 11: 356–372 [DOI] [PubMed] [Google Scholar]
- Yarian C, Townsend H, Czestkowski W, Sochacka E, Malkiewicz AJ, Guenther R, Miskiewicz A, Agris PF (2002) Accurate translation of the genetic code depends on tRNA modified nucleosides. J Biol Chem 277: 16391–16395 [DOI] [PubMed] [Google Scholar]
- Yoshikawa A, Isono S, Sheback A, Isono K (1987) Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol Gen Genet 209: 481–488 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.