L1CAM regulates DNA damage checkpoint response of glioblastoma stem cells through NBS1
Radioresistance of glioblastomas has been correlated with increased DNA repair capacities of glioblastoma stem cells. This is now linked to a surface marker of such stem cells, whose radiation-induced intramembrane proteolysis induces expression of a key DNA repair gene.
Keywords: DNA damage checkpoint, glioblastoma stem cells, L1CAM, NBS1, radioresistance
Abstract
Glioblastomas (GBMs) are highly lethal brain tumours with current therapies limited to palliation due to therapeutic resistance. We previously demonstrated that GBM stem cells (GSCs) display a preferential activation of DNA damage checkpoint and are relatively resistant to radiation. However, the molecular mechanisms underlying the preferential checkpoint response in GSCs remain undefined. Here, we show that L1CAM (CD171) regulates DNA damage checkpoint responses and radiosensitivity of GSCs through nuclear translocation of L1CAM intracellular domain (L1-ICD). Targeting L1CAM by RNA interference attenuated DNA damage checkpoint activation and repair, and sensitized GSCs to radiation. L1CAM regulates expression of NBS1, a critical component of the MRE11–RAD50–NBS1 (MRN) complex that activates ataxia telangiectasia mutated (ATM) kinase and early checkpoint response. Ectopic expression of NBS1 in GSCs rescued the decreased checkpoint activation and radioresistance caused by L1CAM knockdown, demonstrating that L1CAM signals through NBS1 to regulate DNA damage checkpoint responses. Mechanistically, nuclear translocation of L1-ICD mediates NBS1 upregulation via c-Myc. These data demonstrate that L1CAM augments DNA damage checkpoint activation and radioresistance of GSCs through L1-ICD-mediated NBS1 upregulation and the enhanced MRN–ATM–Chk2 signalling.
Introduction
Glioblastoma (GBM) is the most common and fatal type of primary brain tumours. Despite recent therapeutic advances in other solid cancers, GBM treatment remains ineffective and most patients diagnosed with GBM die within 2 years (Furnari et al, 2007; Wen and Kesari, 2008; Stupp et al, 2009). Ionizing radiation (IR) has been the most effective non-surgical treatment modality for GBM patients, but tumour recurrence is essentially universal due to marked radioresistance. Therapeutic resistance is likely due to multiple factors, but we and others found that a highly tumourigenic subpopulation of cancer cells in GBM called GBM stem cells (GSCs) or stem cell-like glioma cells are highly resistant to radiation and chemotherapies (Eramo et al, 2006; Liu et al, 2006; Bao et al, 2006a). Like neural stem cells, GSCs express stem cell markers, display self-renewal capacity, and have the potential to differentiate into multiple lineages (neurons, astrocytes, and oligodendrocytes) (Hemmati et al, 2003; Galli et al, 2004; Singh et al, 2004; Vescovi et al, 2006; Bao et al, 2006a). However, GSCs exhibit significant distinctions from normal stem cells in frequency, proliferation, chromosomal abnormalities, and tumour formation (Vescovi et al, 2006; Bao et al, 2006a; Zhou et al, 2009; Cheng et al, 2010). The potent tumourigenic capacity of GSCs and increasing evidence of resistance to therapies supports critical roles for GSCs in tumour maintenance and recurrence, and suggests that targeting GSCs may overcome therapeutic resistance and improve patient outcome (Piccirillo et al, 2006; Bao et al, 2006a, 2008; Zhou et al, 2009; Cheng et al, 2010). In our previous study, we demonstrated that GSCs promote radioresistance through preferential activation of the DNA damage checkpoint response, and that an inhibitor blocking Chk1 and Chk2 checkpoint kinases reverses the radisoresistance of GSCs (Bao et al, 2006a). However, the molecular mechanisms associated with the preferential DNA damage checkpoint activation in GSCs in response to radiation remain poorly understood.
Radiation mainly causes cellular toxicity through induction of DNA double-strand breaks (DSBs) that activate DNA damage checkpoint signalling (Abraham, 2001; Shiloh, 2003; Lukas et al, 2004; Harper and Elledge, 2007). Activation of checkpoint pathways initiates cell-cycle arrest with attempted DNA repair or induces apoptosis when the extent of DNA damage exceeds the cellular capacity for repair (Abraham, 2001; Kastan and Bartek, 2004; Reinhardt and Yaffe, 2009). Thus, checkpoint pathways primarily have cytoprotective roles to promote cell survival. In the face of irreparable DNA damage, however, the checkpoints relay a pro-apoptotic signal to eliminate those highly damaged cells. The DNA damage checkpoint is a complex signal transduction pathway that includes the MRE11–RAD50–NBS1 (MRN) complex, ataxia telangiectasia mutated (ATM), the ATM and Rad3 related (ATR), Rad17, Chk2, Chk1, and other checkpoint proteins (Abraham, 2001; D'Amours and Jackson, 2002; Bartek and Lukas, 2007; Williams et al, 2007). The phophoinositide 3-kinases, ATM and ATR, are proximal components of the checkpoint signalling cascade that phosphorylate and activate downstream targets (Abraham, 2001; Bao et al, 2001; Shiloh, 2003). The MRN complex has critical roles in initiating DNA repair and early checkpoint activation by regulating ATM kinase activity after radiation (D'Amours and Jackson, 2002; Bartek and Lukas, 2007; Williams et al, 2007). A recent report showed that c-Myc is also required for the ATM-dependent checkpoint activation (Guerra et al, 2010). An earlier study demonstrated that c-Myc positively regulates NBS1 expression at transcriptional level (Chiang et al, 2003). These studies suggest that c-Myc–NBS1–ATM is a potential signalling link to regulate DNA damage checkpoint activation in response to radiation. Radiation-induced DSBs trigger the activation of ATM–Rad17–Chk2–CDC25A signalling cascade to induce cell-cycle arrest. The initiation of cell-cycle arrest permits cells to attempt to repair the damaged DNA, but prolonged presence of the DNA damage leads to an apoptotic cell death (Abraham, 2001; Bartek and Lukas, 2007). Thus, both DNA damage checkpoint activation and DNA repair capacity primarily protect cell survival. In response to DSBs, the activation of ATM, Chk2, and Rad17 are higher in GSCs than the matched non-stem tumour cells (Bao et al, 2006a). Understanding the molecular mechanisms underlying this GSC phenotype will be crucial for developing a therapeutic approach to overcome GSC radioresistance.
In the search for molecular regulators of GSCs, we previously identified L1CAM (CD171) as a differentially expressed protein in GSCs relative to bulk tumour cells and demonstrated that L1CAM supported GSC survival and tumour growth (Bao et al, 2008). L1CAM is a glycoprotein comprised of a cytoplasmic tail (intracellular domain), a transmembrane region and an extracellular domain that interacts with another L1CAM molecule, growth factor receptors, α5β1 and αvβ3 integrins, Neuropilin-1, and extracellular matrix proteins (Maness and Schachner, 2007; Raveh et al, 2009; Siesser and Maness, 2009). This surface protein regulates cell adhesion, survival, growth, migration, and invasion (Raveh et al, 2009; Siesser and Maness, 2009). Mutations in the L1CAM are associated with a number of genetic disorders, notably the X-linked recessive L1 syndrome that includes hydrocephalus and mental retardation (Weller and Gärtner, 2001). In contradistinction, a wide range of human cancers display increased L1CAM expression that informs tumour progression or metastasis of several types of cancer including GBM (Izumoto et al, 1996; Gavert et al, 2005, 2008; Suzuki et al, 2005; Sebens Müerköster et al, 2007; Stoeck et al, 2007, 2009; Raveh et al, 2009; Siesser and Maness, 2009). L1CAM has been shown to be expressed at the invasive front of colon cancers and is a target of β-catenin signalling, a key cancer stem cell pathway (Gavert et al, 2005, 2008; Raveh et al, 2009). An elegant study from the Altevogt's group demonstrated that the intracellular domain of L1CAM (L1-ICD; 28 kDa) is released from the membrane-bound L1CAM through specific cleavages mediated by ADAM10 (A Disintegrin and Metalloprotease 10) and Presenilin (γ-secretase) and then translocated into nuclei to regulate gene expression (Riedle et al, 2009). The proteolytic processing regulated by ADAM10 and γ-secretase has essential roles for the nuclear signalling of L1CAM (Riedle et al, 2009). This may explain why L1CAM overexpression is associated with multiple aspects of tumour progression. Furthermore, L1CAM mediates therapeutic resistance in ovarian and pancreatic cancer cells (Sebens Müerköster et al, 2007, 2009; Stoeck et al, 2007). We therefore attempted to examine whether L1CAM-mediated signalling may have a function in regulating checkpoint response and radioresistance of GSCs.
Results
DNA damage induces L1CAM expression in GSCs
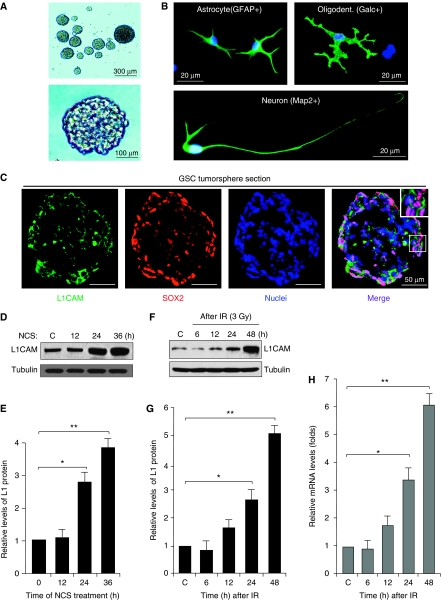
As L1CAM is preferentially expressed in GSCs (Bao et al, 2008) and L1CAM overexpression increases chemoresistance in several types of cancers (Sebens Müerköster et al, 2007, 2009; Stoeck et al, 2007), we speculated that elevated L1CAM expression may contribute to GSC radioresistance. To identify a potential link between L1CAM and radioresistance in GSCs, we initially examined L1CAM expression in GSCs after DNA damage. GSCs were isolated from primary GBM tumour specimens or xenografts as previously described (Bao et al, 2006a, 2006b, 2008; Li et al, 2009) and were validated for the enrichment of stem cell-like cancer cells by functional assays of self-renewal, multi-lineage differentiation, and tumour propagation. GSCs derived from GBM tumours formed tumorspheres (Figure 1A), and displayed differentiation potential into cells expressing makers for astrocytes (GFAP+), oligodentrocytes (Galc+), and neurons (Map2+) (Figure 1B). We further confirmed that L1CAM is co-expressed with SOX2 (a marker for GSCs) in GSC tumorspheres (Figure 1C). To examine L1CAM expression after DNA damage, we treated the isolated GSCs with the radiomimetic drug neocarzinostatin (NCS) or irradiation (IR, 3 Gy) to induce DNA damage over an early time course. Immunoblot analysis showed that L1CAM protein levels in GSCs were induced by NCS (Figure 1D and E) or IR (Figure 1F and G). Quantitative real-time PCR (RT–PCR) analysis indicated that L1CAM mRNA levels were also significantly increased after irradiation (Figure 1H). These data suggest that L1CAM expression is induced by DNA damage in GSCs, indicating a potential role of L1CAM in mediating the radioresistance of GSCs.
Figure 1.
L1CAM expression was induced by DNA damage in GSCs. (A) GSCs isolated from GBM surgical specimens or xenografts formed tumorspheres. Representative images of tumorspheres derived from a GBM specimen (G2038) are shown. (B) GSCs displayed potential to differentiate into cells expressing marker for astrocytes (GFAP+), oligodendrocytes (Galc+), and neurons (Map2+). Nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole) (blue). (C) L1CAM was expressed on the surface of GSCs expressing SOX2 (a stem cell transcription factor) in tumorspheres. The frozen sections of tumorspheres derived from the freshly isolated GSCs were stained with anti-L1CAM (green) and anti-SOX2 (red) antibodies. Nuclei were counterstained with DAPI (blue). The insert represents enlarged image of the part marked by a square. (D) L1CAM protein levels were induced by the radiomimetic drug NCS in GSCs. GSCs (G2038) were untreated or treated with 100 ng/ml NCS over a time course and then harvested for immunoblot analysis. (E) Relative L1CAM protein levels at different time points after NCS treatment from (D) were quantified. *P<0.005; **P<0.001. (F) L1CAM protein levels were induced by irradiation (IR) in GSCs. GSCs derived from a GBM tumour (CCF2170) were untreated (C) treated with IR (3 Gy) and cultured for different times (6, 12, 24, and 36 h), and then harvested for immunoblot analysis. (G) Relative L1CAM protein levels at different time points after IR treatment from (F) were quantified. *P<0.005; **P<0.001. (H) L1CAM mRNA levels were induced by IR treatment in GSCs. L1CAM mRNA expression levels in GSCs (CCF2045) at different time points after IR (3 Gy) were analysed by quantitative real-time PCR. *P<0.005; **P<0.001.
L1CAM regulates activation of checkpoint proteins in GSCs in response to DNA damage
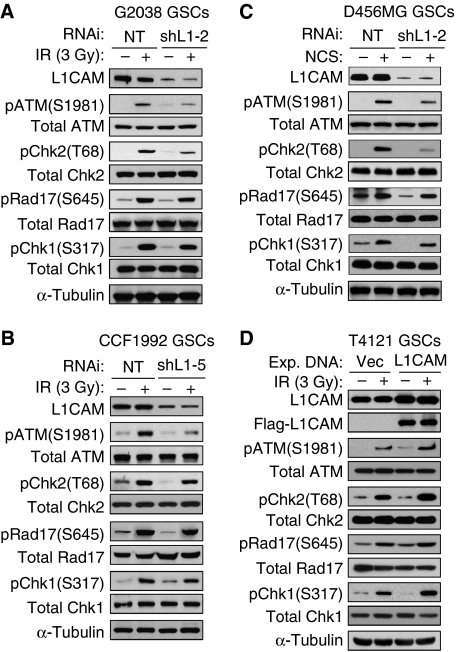
DNA damage checkpoint response has a critical role in determining cellular sensitivity to radiation. To interrogate the role of L1CAM in the preferential checkpoint activation in GSCs, we examined the effect of L1CAM loss of expression on the activating phosphorylation of several key checkpoint proteins including ATM kinase and downstream checkpoint proteins (Chk2, Rad17, and Chk1) in GSCs in response to DNA damage induced by IR or NCS. Isolated GSCs were transduced with L1CAM-targeting short hairpin RNA (shRNA) (shL1-2 or shL1-5) or non-targeting (NT) control shRNA through lentiviral infection, treated with NCS (100 ng/ml) or IR (3 Gy), and then analysed for activating phosphorylation of key checkpoint proteins. Targeting L1CAM expression (70–90% reduction) with the specific shRNA attenuated the activating phosphorylation of ATM (pS1981) and Chk2 (pT68) in response to DNA damage induced by IR or NCS treatment (Figure 2A–C). L1CAM knockdown also modestly reduced activating phosphorylation of Rad17 (pS645) and Chk1 (pS317) in response to NCS- or IR-induced DNA damage (Figure 2A–C). Of note, L1CAM knockdown did not alter the total protein levels of these checkpoint proteins in GSCs. As the DNA damage induced by IR or NCS mainly activates checkpoint response through ATM and Chk2 (Abraham, 2001; Bartek and Lukas, 2003; Reinhardt and Yaffe, 2009), the distinct reduction of ATM and Chk2 activating phosphorylation in response to IR in GSCs with loss of L1CAM expression (Figure 2A–C) indicated that the effect of L1CAM knockdown on checkpoint activation in GSCs was specific. In matched non-stem tumour cells that displayed much lower L1CAM expression (Bao et al, 2008), reduced L1CAM expression showed little or no effect on checkpoint activation (Supplementary Figure S1). These data demonstrate that reducing L1CAM expression indeed attenuated checkpoint activation in GSCs in response to DNA damage, suggesting that preferential expression of L1CAM in GSCs may contribute to the enhanced checkpoint activation in these cells. To further confirm these results, we examined the impact of increasing L1CAM expression in GSCs on the activation of DNA damage checkpoint, and found that ectopic expression of L1CAM increased the activating phosphorylations of ATM, Chk2, Rad17, and Chk1 in GSCs (Figure 2D). Interestingly, although L1CAM knockdown showed little effect on the checkpoint activation in non-stem tumour cells that generally express low levels of L1CAM, ectopic expression of L1CAM in non-stem tumour cells also increased the activating phosphorylations of ATM and Chk2 and cellular resistance to radiation (Supplementary Figure S2). Taken together, these data demonstrate that L1CAM promotes checkpoint activation in response to DNA damage, suggesting that the elevated expression of L1CAM in GSCs may be associated with the increased checkpoint activation and cytoprotection in GSCs.
Figure 2.
L1CAM enhanced DNA damage checkpoint activation in GSCs in response to irradiation (IR) or the radiomimetic drug NCS. (A, B) L1CAM knockdown attenuated activating phosphorylation of ATM and Chk2, and modestly reduced activating phosphorylation of Rad17 and Chk1 after DNA damage induced by IR in GSCs. GSCs derived from G2038 GBM (A) or CCF1992 GBM (B) surgical specimens were targeted with L1CAM shRNA (shL1-2 (A) or shL1-5 (B)) or non-targeting (NT) shRNA for 48 h through lentiviral infection, treated with IR (3 Gy) followed by a 3-h recovery, and then harvested for immunoblot analysis with specific antibodies against phosphorylated ATM(S1981), Chk2(T68), Rad17(S645), and Chk1(S317), and total checkpoint proteins. (C) Downregulation of L1CAM reduced checkpoint activation after NCS-induced DNA damage in GSCs. GSCs derived from D456MG GBM xenografts were targeted with shL1-2 or NT shRNA for 48 h and treated with NCS (100 ng/ml) for 3 h, and then harvested for immunoblot analysis similar to (A). (D) Increased L1CAM expression enhanced checkpoint activation after DNA damage induced by IR (3 Gy) in GSCs. T4121 GSCs were transfected with Flag-tagged L1CAM or vector control for 48 h through lentiviral infection, treated with IR (3 Gy) followed by a 3-h recovery, and then harvested for immunoblot analysis as described in (A).
L1CAM knockdown decreases DNA repair potential in GSCs
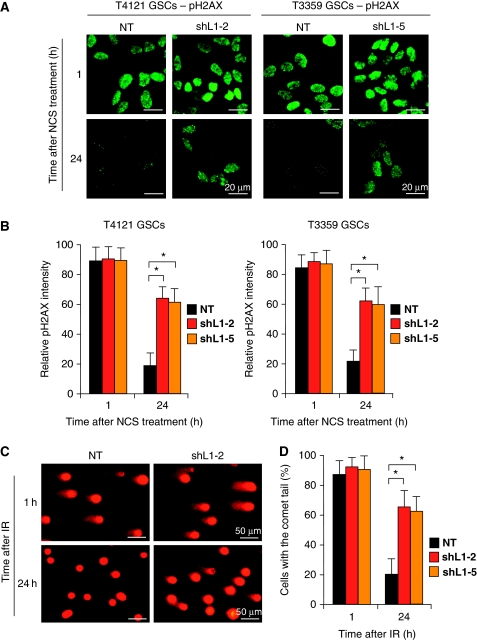
The activation of checkpoint pathways in response to DNA damage leads to cell-cycle arrest with attempt to repair damaged DNA. Based on the effects of L1CAM knockdown on checkpoint activation in GSCs upon DNA damage, we examined whether reduced L1CAM expression affects DNA repair potential and cell recovery after DNA damage in GSCs. To address this point, we assessed the recovery of GSCs targeted with L1CAM shRNA (shL1-2 or shL1-5) or NT shRNA in response to the NCS-induced DNA damage by assessing the resolution of phosphorylated histone 2AX (pH2AX) nuclear foci through immunofluorescence analysis, as pH2AX nuclear foci has been widely used as the indicator of the presence of damaged DNA (Celeste et al, 2003). GSCs isolated from GBM tumours (T4121 and T3359) were targeted with NT shRNA or L1CAM-targeting shRNA (shL1-2 or shL1-5) through lentiviral infection, and then treated without or with NCS (100 ng/ml) for 3 h. After removal of NCS from the culture medium, treated cells were allowed to recover over a time course before fixation and pH2AX staining to assess the resolution of nuclear foci after DNA damage. In the condition without IR or NSC treatment, L1CAM knockdown in GSCs did not affect pH2AX staining intensity relative to the control with NT shRNA (Supplementary Figure S3). At 1 h after the NCS treatment, almost all cells contained pH2AX nuclear foci with similar fractions in cells transduced with either NT shRNA or shL1CAM (Figure 3A and B), indicating that most cells suffered from the NCS-induced DNA damage. In contrast, at 24 h after recovery, the fraction of cells with the pH2AX nuclear foci was significantly higher in the GSCs targeted with shL1CAM than the control cells targeted with NT shRNA (Figure 3A and B). These data indicated that L1CAM knockdown delayed the resolution of the pH2AX nuclear foci in GSCs, suggesting that reduced L1CAM expression decreased DNA repair capacity in GSCs. To confirm this result, we also performed the alkaline single cell gel electrophoresis (comet) assay (Tice and Strauss, 1995; Bao et al, 2006a) that quantifies DNA damage by the frequency of comet tails. The results from the comet assay validated that reduced L1CAM expression decreased cellular ability to repair the damaged DNA induced by radiation (Figure 3C and D), suggesting that elevated L1CAM expression is associated with the preferential DNA repair potential in GSCs.
Figure 3.
L1CAM knockdown reduced DNA repair capacity after DNA damage in GSCs. (A) The resolution of phosphorylated histone 2AX (pH2AX) nuclear foci after DNA damage was delayed in GSCs after L1CAM knockdown. GSCs isolated from T4121 and T3359 GBMs were targeted with L1CAM shRNAs (shL1-2 or shL1-5) or the non-targeting (NT) shRNA and then treated with NCS (100 ng/ml) for 3 h. The treated cells were allowed to recover in the culture medium without NCS. Immunofluorescent staining of pH2AX nuclear foci was performed at 1 and 24 h after NCS removal. (B) Cells with pH2AX nuclear foci staining in (A) were quantified and statistically analysed. The relative intensity of pH2AX nuclear foci in the cells treated with L1CAM shRNA or NT control shRNA was analysed. The intensity of pH2AX nuclear foci was higher in cells expressing L1CAM shRNA than the control cells expressing NT shRNA at 24 h after NCS removal. *P<0.001. (C) Alkaline comet assay confirmed that L1CAM knockdown reduced DNA repair potential in GSCs. GSCs (CCF1683) were targeted with L1CAM shRNA or NT shRNA and then treated with IR (5 Gy). The comet assay was performed at 1 and 24 h after irradiation. (D) Data from the comet assay in (C) were quantified and statistically analysed. L1CAM knockdown significantly delayed the resolution of comet tails that served as indicators of DNA damage. *P<0.001.
Targeting L1CAM reduces radioresistance of GSCs
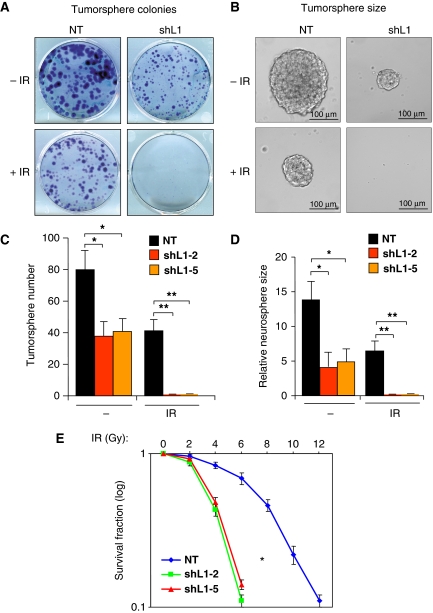
As L1CAM knockdown attenuated checkpoint activation and DNA repair capacity in GSCs, we next examined the effect of targeting L1CAM on GSC radioresistance. GSCs derived from GBM tumour specimens were targeted with shL1CAM (shL1-2 or shL1-5) or NT shRNA through lentiviral infection and treated without or with IR (5 Gy), allowed to recover and then form tumorspheres in neurobasal medium for 9 days. To stain, view, and count the formed tumorspheres after treatment, the tumorspheres in suspension were permitted to attach on plastic dish by culturing them for a short period (12 h) in Dulbecco's Modified Essential Media (DMEM) with 10% fetal bovine serum (FBS). As expected, treatment with 5 Gy of IR attenuated GSC tumorsphere formation but a significant number of GSCs survived (Figure 4A and C). In concordance with our previous report (Bao et al, 2008), L1CAM knockdown in GSCs reduced the efficiency and size (a potential surrogate of proliferation) of tumorspheres (Figure 4A–D). Furthermore, targeting L1CAM sensitized GSCs to radiation as the combination of shL1CAM and IR abolished GSC tumorsphere formation (Figure 4A–D). These data suggest that targeting L1CAM renders GSCs sensitive to the IR-induced cell death. This result was further validated in a dose response study with a range (0, 2, 4, 6, 8, 10, and 12 Gy) of IR treatment showing that reduced L1CAM expression with shRNA increased cellular sensitivity to radiation (Figure 4E). In addition, a cell-cycle profiling analysis showed that L1CAM knockdown reduced the IR-induced G2 arrest in GSCs and increased cell death (Supplementary Figure S4A), which could be due to the reduced ATM–Chk2 checkpoint activation caused by L1CAM knockdown (Figure 2A–C) and the increased p21-induced G1 arrest caused by L1CAM knockdown (Bao et al, 2008). Collectively, these data suggest that elevated expression of L1CAM likely contributes to the enhanced radioresistance of GSCs. L1CAM-mediated preferential checkpoint activation and DNA repair may enable GSCs more resistant to radiation, implicating that L1CAM is a potential target to overcome GSC radioresistance.
Figure 4.
Targeting L1CAM with shRNA increased radiosensitivity of GSCs. (A) Tumorsphere formation assay showed that L1CAM knockdown reduced tumorsphere growth of GSCs after irradiation (IR). GSCs (CCF1683) were targeted with L1CAM shRNA or NT shRNA through lentiviral infection for 48 h, and treated without or with IR (5 Gy), and then allowed to recover and grow for 9 days in the neurobasal medium. To view and count the tumorspheres formed by the survived cells, the spheres were allowed to attach on the dishes by culturing them for a short time (12 h) in DMEM with 10% FBS before they were fixed and stained for assessing the sphere number and size under different treatments. (B) Representative images of tumorspheres from (A) are displayed. (C) The number of formed tumorspheres from GSCs targeted with NT shRNA or L1CAM shRNA (shL1-2 or shL1-5) from (A) was quantified and analysed. *P<0.005; **P<0.001. (D) The tumorsphere size from the treatments in (A) was quantified and analysed. *P<0.002; **P<0.001. (E) Dose response survival curve of GSCs with L1CAM shRNA or NT shRNA in response to a range of IR treatment. GSCs (CW702) were targeted with L1CAM shRNA (shL1-2 or shL1-5) or NT shRNA through lentiviral infection for 48 h, and treated with different doses of IR as indicated. The survival fractions of GSCs were counted 4 days after IR treatment. L1CAM knockdown significantly increased radiosensitivity and decreased survival of GSCs. *P<0.002.
L1CAM functions through NBS1 upregulation to confer the preferential checkpoint response and radioresistance in GSCs
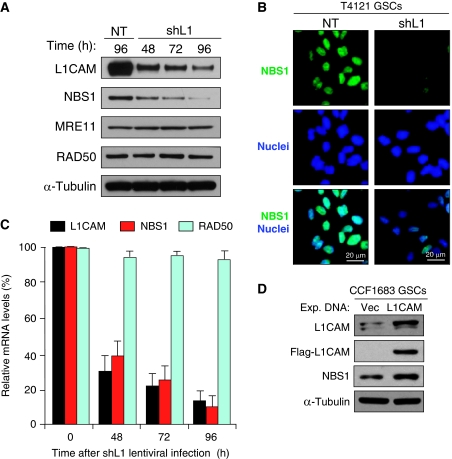
To elucidate the molecular mechanisms associated with the regulation of L1CAM on DNA damage checkpoint response, we examined the impact of L1CAM knockdown on the expression of several key checkpoint regulators involved in the early checkpoint response. These regulators include three core proteins in MRN (MRE11, RAD50, and NBS1) complex that has critical roles in mediating early checkpoint activation and initiating DNA repair process in response to radiation (D'Amours and Jackson, 2002; Lee and Paull, 2005; Williams et al, 2007). A functional MRN complex is required for ATM activation and the subsequent activation of its downstream checkpoint proteins such as Chk2 (Lee and Paull, 2005; Williams et al, 2007). We found that L1CAM knockdown reduced the expression of NBS1 but not the other two MRN components (MRE11 and RAD50) as demonstrated by immunoblot analysis (Figure 5A). Immunofluorescent staining with a specific antibody confirmed that nuclear NBS1 protein is reduced in GSCs targeted with shL1CAM (Figure 5B). RT–PCR analysis validated that L1CAM knockdown reduced NBS1 expression at mRNA level (Figure 5C). Moreover, forced expression of L1CAM upregulated NBS1 in GSCs and non-stem tumour cells (non-GSCs) (Figure 5D; Supplementary Figure S5). These data suggest that L1CAM upregulates the expression of a key DNA damage response component, NBS1, in GSCs.
Figure 5.
L1CAM regulated NBS1 expression in GSCs. (A) Immunoblot analysis showed that L1CAM knockdown reduced expression of NBS1 but not MRE11 and RAD50 of the MRN complex. GSCs (CCF1683) were targeted with the non-targeting (NT) control shRNA or L1CAM shRNA (shL1) for 48, 72, and 96 h through lentiviral infection, and then harvested for immunoblot analysis with specific antibodies against L1CAM, NBS1, MRE11, RAD50, and α-tubulin. (B) Immunofluorescent (IF) staining confirmed that L1CAM knockdown reduced NBS1 protein levels in nuclei of GSCs. The representative images of IF staining showed relative levels of NBS1 (green) in T4121 GSCs targeted with L1CAM shRNA (shL1) or NT shRNA for 48 h. Nuclei were counterstained with DAPI (blue). (C) Quantitative RT–PCR analysis confirmed that L1CAM knockdown reduced mRNA levels of NBS1 but not RAD50 in CCF1683 GSCs. (D) Ectopic expression of L1CAM upregulated NBS1 expression in GSCs. GSCs (CCF1683) were transfected with Flag-L1CAM (L1CAM) or vector control. L1CAM and NBS1 protein levels were analysed at 36 h after lentiviral vector-mediated transfection.
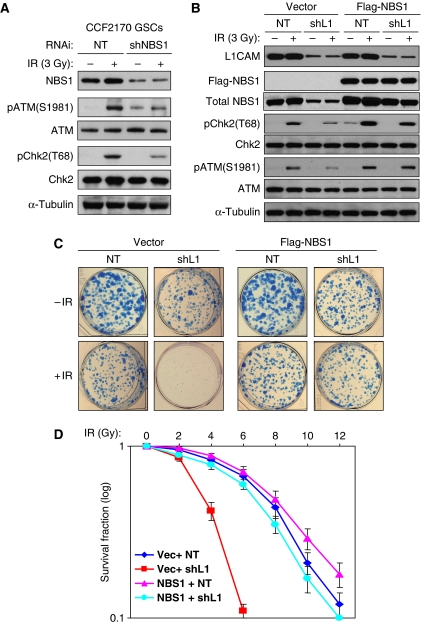
To address whether L1CAM functions through NBS1 to regulate checkpoint activation, we examined the effect of NBS1 knockdown on checkpoint activation in GSCs. Reduced NBS1 with a specific shRNA through lentiviral infection also attenuated checkpoint activation in GSCs in response to IR-induced DNA damage (Figure 6A), suggesting that NBS1 knockdown photocopied L1CAM knockdown on inhibition of checkpoint activation. To determine whether L1CAM confers the preferential checkpoint response and radioresistance in GSCs through NBS1 control, we attempted to rescue the effects of L1CAM knockdown on checkpoint activation and radioresistance by expressing NBS1 (Flag–NBS1). Ectopic expression of NBS1 not only rescued the reduced GSC checkpoint activation caused by L1CAM knockdown in response to radiation (Figure 6B) but also restored the preferential GSC survival after radiation as demonstrated by tumorsphere assay (Figure 6C) and a dose response study with a range (0, 2, 4, 6, 8, 10, and 12 Gy) of IR treatment (Figure 6D). Collectively, these data suggest that L1CAM functions through NBS1 to confer the preferential DNA damage checkpoint response and radioresistance in GSCs.
Figure 6.
Ectopic expression of NBS1 rescued the decreased checkpoint activation and radioresistance caused by L1CAM downregulation, and NBS1 knockdown photocopied L1CAM knockdown on inhibition of checkpoint activation. (A) NBS1 knockdown reduced checkpoint activation in GSCs in response to IR. GSCs (CCF2170) were targeted with NBS1 shRNA (shNBS1) or non-targeting (NT) shRNA for 48 h through lentiviral infection, treated with 3 Gy of IR followed by a 3-h recovery, and then harvested for immunoblot analysis with specific antibodies against phosphorylated ATM(S1981) and Chk2(T68), and total checkpoint proteins. (B) Ectopic expression of NBS1 (Flag–NBS1) restored the checkpoint activation that was reduced by L1CAM knockdown. GSCs (CW650) were transduced with Flag–NBS1 or vector control for 36 h, targeted with L1CAM shRNA (shL1) or NT shRNA for 48 h through lentiviral infection, then treated with IR (3 Gy) followed by 3 h recovery, and then harvested for immunoblot analysis with specific antibodies against L1CAM, Flag (Flag–NBS1), pATM(S1981), and pChk2(T68) phosphorylated checkpoint proteins, and the total checkpoint proteins. (C) Ectopic expression of NBS1 rescued the decreased radioresistance caused by L1CAM knockdown in GSCs. GSCs derived from CW650 GBM tumour were transduced with Flag–NBS1 or vector control for 36 h, targeted with L1CAM shRNA (shL1) or the NT control shRNA through lentiviral infection for 48 h, treated without or with irradiation (5 Gy), and then allowed to recover and grow for 9 days in neurobasal stem cell medium. To view and count the tumorspheres formed by the survived cells, tumorspheres were allowed to attach on dishes by culturing them in DMEM with 10% FBS for 12 h, and then fixed and stained for assessing the tumorsphere number and size under different treatments. (D) Dose response survival curve of GSCs without or with NBS1 ectopic expression and targeted with L1CAM shRNA (shL1) or NT shRNA in response to a range of IR treatment. GSCs (CW702) were transduced with Flag–NBS1 or vector control for 36 h, targeted with L1CAM shRNA (shL1-2) or NT shRNA through lentiviral infection for 48 h, and treated with different doses of IR as indicated. The survival fractions of GSCs were counted 4 days after IR treatment. Ectopic expression of NBS1 restored GSC radioresistance that was reduced by L1CAM downregulation.
Nuclear translocation of L1-ICD mediates NBS1 upregulation via c-Myc
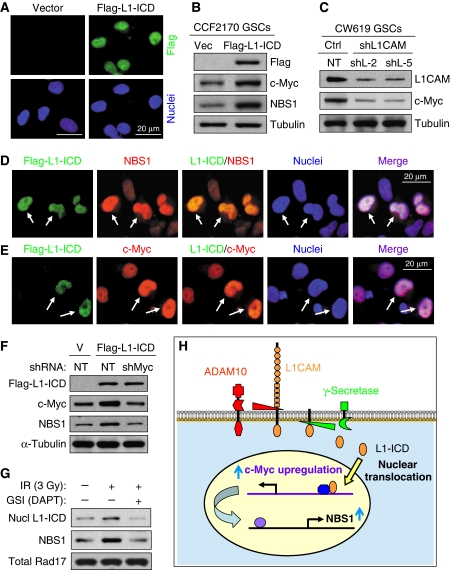
The L1CAM-mediated upregulation of NBS1 could be one of critical mechanisms that contribute to the preferential checkpoint response and the enhanced radioresistance in GSCs. Our results suggest that cellular signalling from cell surface may regulate checkpoint response in nuclei. To reveal the mechanistic link between L1CAM surface signalling and NBS1 expression and checkpoint response in nuclei, we investigated how L1CAM mediates a signalling to upregulate NBS1 expression. Although L1CAM is a membrane-bound surface protein that can interact with other membrane proteins such as growth factor receptors and integrins (Maness and Schachner, 2007; Raveh et al, 2009; Siesser and Maness, 2009), an elegant study demonstrated that L1-ICD (28 kDa) can be released from the membrane-bound L1CAM through specific cleavages mediated by ADAM10 (A Disintegrin And Metalloprotease 10) and Presenilin (γ-secretase) and that the nuclear translocation of L1-ICD is essential for the nuclear signalling of L1CAM to regulate gene expression (Riedle et al, 2009). Therefore, we examined whether the nuclear L1-ICD upregulates NBS1 expression. We confirmed that a Flag-tagged L1-ICD (Flag-L1-ICD) was translocated into nuclei in GSCs as demonstrated by immunofluorescent staining (Figure 7A). Furthermore, ectopic expression of Flag-L1-ICD indeed upregulates NBS1 expression in GSCs (Figure 7B). Immunofluorescent staining further confirmed that individual cells with nuclear Flag-L1-ICD also showed increased NBS1 (Figure 7D). These data suggest that the nuclear L1-ICD may directly or indirectly regulate NBS1 expression. As an earlier study demonstrated that c-Myc directly regulates NBS1 expression at transcriptional level (Chiang et al, 2003) and a recent study showed that c-Myc is required for the ATM-dependent checkpoint activation (Guerra et al, 2010), it is possible that L1-ICD may indirectly regulate NBS1 expression through c-Myc. This hypothesis was supported by our findings that L1CAM knockdown also reduced c-Myc levels (Figure 7C) and that L1-ICD expression upregulated c-Myc (Figure 7B and E). To determine whether L1-ICD upregulates NBS1 indirectly through c-Myc, we examined the effect of c-Myc knockdown on L1-ICD-induced NBS1 expression. We found that c-Myc knockdown attenuated the induction of NBS1 expression mediated by Flag-L1-ICD (Figure 7F), demonstrating that L1-ICD functions through c-Myc to upregulate NBS1 in GSCs. As radiation has been shown to activate γ-secretase (Presenilin) activities (Jin et al, 2008; Scharpfenecker et al, 2009), we confirmed that IR increased nuclear L1-ICD and NBS1 expression in GSCs, an effect was attenuated by treatment of GSCs with the γ-secretase inhibitor (GSI) (DAPT) (Figure 7G), suggesting that L1-ICD functions as a signal transducer to mediate L1CAM signalling from cell surface to nuclei to regulate NBS1 expression, and that the IR-induced nuclear translocation of L1-ICD depends on the activity of γ-secretase. Taken together, these data support that a signalling pathway mediated by the nuclear translocation of L1-ICD regulates NBS1 expression through c-Myc to enhance DNA damage checkpoint activation (Figure 7H). Thus, we identified the mechanistic link between L1CAM surface signalling and regulation of NBS1 expression and checkpoint response in nuclei.
Figure 7.
Nuclear translocation of L1CAM intracellular domain (L1-ICD) mediates NBS1 upregulation via c-Myc. (A) Immunofluorescent (IF) staining showed that a Flag-tagged L1-ICD (Flag-L1-ICD) was translocated into nuclei of GSCs. GSCs (CCF2170) were transduced with Flag-L1-ICD or vector control through lentiviral infection, and then immunostained with anti-Flag (green) and counterstained with DAPI for nuclei (blue). (B) Immunoblot analysis showed that ectopic expression of L1-ICD upregulated c-Myc and NBS1. GSCs (CCF2170) were transduced with Flag-L1-ICD or vector control through lentiviral infection for 48 h, and then harvested for immunoblot analysis. (C) L1CAM knockdown reduced c-Myc expression in GSCs. GSCs (CW619) were targeted with L1CAM shRNA (shL1-2 and shL1-5) or NT shRNA through lentiviral infection for 48 h, and then harvested for immunoblot analysis. (D, E) IF staining confirmed that individual cells expressing Flag-L1-ICD upregulated NBS1 (D) and c-Myc (E). GSCs (CW619) were transduced with Flag-L1-ICD expression for 48 h, and then immunostained with antibodies against Flag and NBS1 or c-Myc, and counterstained with DAPI (blue). Cells expressing Flag-L1-ICD (green) showed higher NBS1 (red) and c-Myc (red) in nuclei (indicated by arrows) than cells without Flag-L1-ICD. (F) c-Myc knockdown attenuated the L1-ICD-induced NBS1 upregulation. GSCs (CW619) were transduced with Flag-L1-ICD or vector control for 24 h, targeted with c-Myc shRNA (shMyc) or NT shRNA through lentiviral infection for 48 h, and then harvested for immunoblot analysis with antibodies against Flag, c-Myc, NBS1, and α-tubulin. (G) Radiation induced nuclear translocation of L1-ICD and NBS1 expression in GSCs, which was attenuated by the γ-secretase inhibitor (DAPT). GSCs (CW702) were untreated or treated with IR (3 Gy) in the absence or presence of DAPT (2 μm) and allowed to recovery for 48 h. Nuclear fractions were then isolated from these cells for immunoblot analysis with specific antibodies against the intracellular domain of L1CAM (L1-ICD), NBS1, and total Rad17 (control). (H) A schematic illustration shows that nuclear translocation of L1-ICD transduces L1CAM signalling from cell surface to nuclei to regulate NBS1 expression via c-Myc. The membrane-bound L1CAM can be cleaved by ADAM10 and γ-secretase/Presenilin to release the intracellular domain (L1-ICD) that is translocated into nuclei to upregulate c-Myc and NBS1 expression to regulate checkpoint activation in GSCs.
Discussion
GBMs are among the most aggressive and least successfully treated brain tumours. These tumours are highly resistant to current radiotherapy and chemotherapy (Furnari et al, 2007; Wen and Kesari, 2008; Stupp et al, 2009). New therapies on clinical trials have not translated into broad improvements in patient outcome, suggesting that new paradigms will be required. We and others have demonstrated that GSCs contribute to the therapeutic resistance (Eramo et al, 2006; Liu et al, 2006; Bao et al, 2006a; Bertrand et al, 2009; Frosina, 2009; Nakai et al, 2009). In addition, GSCs display enhanced invasive capacity and angiogenic potential (Bao et al, 2006b; Folkins et al, 2009; Wakimoto et al, 2009), indicating that targeting GSCs may significantly improve the treatment. We previously identified L1CAM as a cell surface molecule preferentially expressed on GSCs to maintain the cell survival and tumour growth (Bao et al, 2008). In this study, we identified a new function of L1CAM in promoting checkpoint activation and radioresistance of GSCs through regulating one of the key molecular regulators in cellular responses to DNA damage, NBS1. Therefore, L1CAM is a particularly attractive therapeutic candidate for GBM therapy as targeting L1CAM not only disrupts the maintenance of GBM propagating cells but also reduces GSC-mediated radioresistance. The cell surface location of L1CAM may present cues as to the role of the microenvironment in regulating a cancer stem cell phenotype (Gilbertson and Rich, 2007; Heddleston et al, 2009; Li et al, 2009).
L1CAM expression has been shown to be correlated with the likelihood of tumour progression in several types of solid cancers including GBM (Izumoto et al, 1996; Suzuki et al, 2005), ovarian cancer (Fogel et al, 2003; Stoeck et al, 2007; Wolterink et al, 2010), colon cancer (Gavert et al, 2005, 2007), malignant melanoma (Meier et al, 2006), and other tumours (Fogel et al, 2003; Sebens Müerköster et al, 2007; Gavert et al, 2008; Geismann et al, 2009). This surface protein has been demonstrated to be a molecular marker of poor prognosis in ovarian cancers and uterine carcinomas (Fogel et al, 2003). The mechanisms through which L1CAM acts to negatively impact patient outcome are not clear, but L1CAM mediates direct and indirect transmission of external signals regulating cell proliferation, differentiation, migration, and invasion (Raveh et al, 2009; Siesser and Maness, 2009). Ectopic expression of L1CAM increases cell motility and invasiveness in vitro as well as tumour growth and metastasis in nude mice (Gavert et al, 2005, 2007). In addition, L1CAM may have paracrine functions because the extracellular domain of L1CAM can be released from cell surface via proteolytic cleavage by plasmin, ADAM10, and ADAM17 (a disintegrin and metalloproteases) (Maretzky et al, 2005; Gavert et al, 2007). The soluble L1CAM can also promote cell migration, survival, growth, and angiogenesis through binding to integrins (Stoeck et al, 2007; Friedli et al, 2009; Raveh et al, 2009). Moreover, overexpression of both membrane-bound and soluble forms of L1CAM augments protection of ovarian and pancreatic carcinoma cells from apoptosis and contributes to chemoresistance (Sebens Müerköster et al, 2007, 2009; Stoeck et al, 2007). L1CAM knockdown or anti-L1CAM antibody has been shown to abolish chemoresistance and reduce cancer cell proliferation in vivo in xenograft models (Arlt et al, 2006; Bao et al, 2008; Gast et al, 2008; Weidle et al, 2009; Wolterink et al, 2010). Our results demonstrate that L1CAM signalling through the nuclear translocation of its intracellular domain (L1-ICD) may have an additional important role that was unappreciated through whole tumour analyses as L1CAM confers radioresistance in rare GSC population by enhancing DNA checkpoint activation and DNA repair. We are extending these studies to determine the contributions of L1CAM in other therapeutic resistance as well.
NBS1 is one of the three core components in MRN complex (MRE11, RAD50, and NBS1) that serves as an initial sensor of DNA DSBs (Lee and Paull, 2005; Williams et al, 2007). This critical complex is required for the activation of DNA damage checkpoint response after DSBs by activating ATM kinase and its downstream targets (Lee and Paull, 2005; Berkovich et al, 2007; Williams et al, 2007). NBS1 has been shown to localize to the DSBs in a pH2AX-dependent manner and facilitates recruitment of ATM to the damage site (Celeste et al, 2003; Falck et al, 2005). The MRN complex is also involved in the maintenance of telomere length (Chai et al, 2006; Wu et al, 2007). Furthermore, NBS1 has a crucial role in the initiation of DNA repair and is involved in the non-homologous end-joining pathway after DSBs (Berkovich et al, 2007; Deriano et al, 2009; Williams et al, 2009). NBS1 amplifies ATM activation by accumulating the MRN complex at break points and is a direct target of ATM kinase activity to stimulate the DNA repair process (Falck et al, 2005; Lee and Paull, 2005; Berkovich et al, 2007). Our studies demonstrate that L1CAM upregulates NBS1 expression through nuclear translocation of L1-ICD. We revealed that L1-ICD indirectly mediates NBS1 upregulation through c-Myc. This result is consistent with other studies showing that NBS1 expression is positively regulated by c-Myc at transcriptional level (Chiang et al, 2003) and that c-Myc is required for the ATM-dependent checkpoint activation (Guerra et al, 2010). Thus, differential expression of L1CAM in GSCs mediates MRN complex function through Myc–NBS1–ATM axis to enhance DNA damage checkpoint activation and DNA repair, which promotes radioresistance of GSCs (a working model shown in Supplementary Figure S6). The upregulation of NBS1 may also promote the maintenance of telomere length in GSCs that display longer telomere length than matched non-stem cancer cells (data not shown). Our results may explain why GSCs exhibit preferential DNA damage checkpoint responses and the increased radioresistance.
Augmenting the sensitivity of resistant cancers to conventional cytotoxic therapy has been the subject of great effort. The study of cancer stem cells has been theorized as a source of novel insights that may be translated directly into clinical approaches. We recently described the benefit of disrupting another key cancer stem cell pathway, Notch, in reversing GSC radioresistance. Treatment of GSCs with GSIs that block Notch activation enhanced cell death and impaired colony formation after radiation at clinically relevant doses (Wang et al, 2010). Although both Notch and L1CAM are cell–cell signalling molecules, Notch signalling does not alter DNA damage checkpoint activation in response to radiation (Wang et al, 2010), suggesting that L1CAM and Notch may regulate parallel pathways that could be useful to target simultaneously. Additional canonical cancer stem cell pathways such as Wnt/β-catenin signalling may also contribute to radioresistance (Woodward et al, 2007). In breast cancer, CSCs are relatively resistant to radiation potentially due to lower levels of reactive oxygen species (Diehn et al, 2009). It is unlikely that therapeutic resistance in any cancer is caused by a single process or pathway but likely results from several factors acting together.
In summary, we identified the L1CAM-mediated checkpoint activation through the NBS1–ATM axis as one of critical regulatory mechanisms underlying the preferential DNA damage checkpoint response and radioresistance of GSCs. We demonstrated that L1CAM, a cell surface molecule preferentially expressed in GSCs, enhanced checkpoint activation and DNA repair capacity of GSCs in response to radiation through nuclear translocation of L1-ICD that mediates c-Myc and NBS1 upregulation. Thus, anti-L1CAM therapy may synergize with radiotherapy and other current treatments to overcome the therapeutic resistance of GSCs. L1CAM represents a potential molecular target for developing novel therapeutics to improve the treatment outcome for GBM patients.
Materials and methods
Isolation and culture of glioma-derived cells
GSCs and non-stem tumour cells (non-GSCs) were isolated from GBM surgical specimens or xenografts and cultured as previously described (Bao et al, 2006a, 2008; Li et al, 2009). De-identified GBM specimens were collected from Cleveland Clinic Brain Tumor and Neuro-Oncology Center in accordance with an Institutional Review Board-approved protocol. GBM surgical specimens or xenografts maintained in athymic BALB/c nude mice were disaggregated using the Papain Dissociation System (Worthington Biochemical Corp.). Total tumour cells were recovered in stem cell medium (neurobasal-A medium with B27 supplement, 10 ng/ml epidermal growth factor and 10 ng/ml basic fibroblast growth factor) for at least 6 h to allow re-expression of surface markers and then sorted by fluorescence-activated cell sorting or magnetic cell sorting based on the presence of CD133 (Milenyi Biotech) or CD15 (SSEA-1, BD Bioscience). The GSC phenotype of these cells was confirmed by functional assays of self-renewal (serial neurosphere formation), stem cell marker expression, tumour propagation (in vivo limiting dilution assay), and differentiation potential as described in our previous studies (Bao et al, 2006a, 2008; Li et al, 2009).
L1CAM knockdown and lentivirus production
The lentiviral vector-mediated expression of shRNA for targeting human L1CAM was performed as described in our previous report (Bao et al, 2008). Two L1CAM shRNA (shL1-2 and shL-5, Sigma-Aldrich) clones targeting non-overlapping sequences that showed significant knockdown of L1CAM expression (70–90% reduction) and NT control shRNA (SHC002) were selected for the experiments. NBS1 shRNA (shNBS1) and c-Myc shRNA (shMyc) clones in lentiviral vector were also obtained from Sigma-Aldrich (Mission shRNA). Lentiviral particles expressing targeting or NT shRNAs were produced in HEK293FT cells with the pACK set of packing plasmids (System Biosciences) and the viruses were concentrated and titered as previously described (Bao et al, 2008; Li et al, 2009).
Immunoblot analysis
Immunoblot (western blot) analysis was performed as previously described (Bao et al, 2006a, 2008; Li et al, 2009). The anti-L1CAM antibody (Clone UJ127, mAb) was purchased from Lab Version or Genetex. The antibody against the intracellular (cytoplasmic) domain of L1CAM (L1-ICD) was obtained from Santa Cruz (SC-1508). Other antibodies against phospho-Chk1(S317), phospho-Chk2(T68), phospho-Rad17(S645), phospho-H2AX, phospho-ATM(S1981), total Chk1, Chk2, Rad17, MRE11, RAD50, and NBS1 (Cell Signaling Technology) and Flag and c-Myc mAbs (Sigma-Aldrich) were used for the immunoblotting.
Immunofluorescent staining
Immunofluorescent staining of GSCs or the differentiated cells was performed as previously described (Bao et al, 2006a, 2008; Li et al, 2009). Briefly, cells cultured in suspension or attached on cover glass or tumorsphere sections were fixed in 4% paraformaldehyde, incubated with primary antibodies (α-phospho-H2AX and α-NBS1 (Cell Signaling), α-L1CAM (Lab Version), α-L1-ICD (SC-1508, Santa Cruz), α-SOX2 (Millipore), α-Flag (M2, Sigma-Aldrich), α-c-Myc (Santa Cruz)) overnight at 4°C, and then incubated with the fluorescence-labelled secondary antibody for 1 h at room temperature. Nuclei were counterstained with DAPI. Tumorspheres were fixed in 4% formaldehyde for 15 min, and cryoprotected in 30% sucrose. Sections were post-fixed in methanol and processed as described above. Stained cells were viewed and analysed under a fluorescent microscope (Leica DMI3000B) or confocal microscope (Leica TCS SP5). To validate the differential potential of GSCs, cells were induced for differentiation in vitro and then immunostained with antibodies against the astrocyte marker GFAP (Covance), oligodendrocyte marker Galc (Millipore), and the neuronal markers Map2 and TUJ1 (Covance) by immunofluorescent staining as described (Bao et al, 2006a).
Tumorsphere formation assays
GSCs were transduced with L1CAM-targeting shRNA or NT control shRNA through lentiviral infection for 48 h, cultured or treated with IR (3 or 5 Gy), and then allowed to recover and grow for 9 days in 24-well plates. In order to stain, view, and analyse the number and size of tumorspheres formed by the surviving GSCs, the tumorspheres grown in the neurobasal medium were allowed to attach on plates by culturing them in DMEM with 10% FBS for 12 h, and then stained with the Quick-dip kit and analysed with Image J software.
Induction of DNA damage and comet assay
To induce DNA damage, GSCs or non-stem tumour cells were treated with 100 ng/ml NCS for 3 h or with IR (3 or 5 Gy), and then harvested for immunoblot analysis, fixed for immunofluorescent staining or cultured over a time course for tumorsphere formation assay. To examine the DNA repair capacity in GSCs by the comet assay, cells were subjected to the alkaline single cell gel electrophoresis assay to examine the resolution of DNA DSBs after IR or NCS treatment as previously described (Tice and Strauss, 1995; Bao et al, 2006a).
Quantitative RT–PCR analysis
Total RNA samples were isolated from GSCs after radiation treatment or L1CAM knockdown with an RNeasy kit (Qiagen), and then reverse transcribed into cDNA using the Superscript III Kit (Invitrogen). Quantitative RT–PCR was performed on an Applied Biosystems 7900HT cycler using SYBR-Green Mastermix (SA Biosciences) with the following primers: L1CAM (forward: 5′-TGC TCA TCC TCT GCT TCA TC-3′, and reverse: 5′-TCC TCG TTG TCA CTC TCC A-3′); NBS1 (forward: 5′-AGA CCA ACT CCA TCA GAA ACT AC-3′, and reverse: 5′-AAT GAG GGT GTA GCA GGT TG-3′); RAD50 (forward: 5′-CGA AGT ACC TAT CGT GGA CAA G-3′, and reverse: 5′-GAT CGT CCT CGC ATA TCC AAG-3′).
NBS1 rescue experiments
Flag–NBS1 was constructed by subcloning the human NBS1 open reading frame with a C-terminal Flag tag followed by a TGA stop codon into a lentiviral expression vector. To examine whether overexpression of NBS1 rescued the phenotype caused by L1CAM knockdown, GSCs were transduced with Flag–NBS1 or vector control through lentiviral infection for 36 h, targeted with L1CAM shRNA or NT shRNA for 48 h, treated with IR (3 or 5 Gy) followed by 3 h recovery, and then harvested for immunoblot analysis with specific antibodies as indicated to assess the rescue effect on checkpoint activation, or allowed to recover and grow for 9 days in neurobasal medium to form tumorspheres for assessing the rescue effect on radioresistance of GSCs.
Expression of L1-ICD
The cDNA fragment coding for the human L1-ICD (28 kDa) was amplified from the phL1A-pcDNA3 expression plasmid (Addgene plasmid 12307) with specific PCR primers (forward: 5′-ATC GAA TTC ACC ATG GAT TAC AAG GAT GAC GAC GAT AAG AAG CGC AGC AAG GGC GGC AAA-3′; and reverse: 5′-ATC GCG GCC GCC TAT TCT AGG GCC ACG GCA GG-3′), and then subcloned into pCDH-CMV-MCS-EF1-copGFP lentiviral vector (System Biosciences) or pLCMV-Neo lentivector (a kind gift of Dr Peter Chumakov) with Flag-tag coding sequences in frame and verified by sequencing. The expression of L1-ICD in GSCs was confirmed by immunoblot analysis and immunofluorescent staining.
Treatment of GSCs with GSI
The treatment of GSCs with the specific GSI was performed as previously described (Wang et al, 2010). To examine the effect of GSI on the radiation-induced nuclear translocation of L1-ICD, GSCs (CW702) were pre-treated with 2 μm of DAPT (Sigma-Aldrich) or DMSO for 4 h, then irradiated with radiation (3 Gy), and cultured for 48 h in the presence of DAPT or DMSO. The nuclear fractions from treated GSCs or control GSCs were isolated with a ProteoJET Cytoplasmic and Nuclear Protein Extraction Kit (Fermentas) and then analysed by immunoblotting with antibodies against the intracellular (cytoplasmic) domain of L1CAM (L1-ICD) (SC-1508, Santa Cruz), NBS1, and Rad17 (Cell Signaling). DAPT: N-[N-(3,5-difluorophenacetyl)-1-alanyl]-S-phenylglycine t-butyl ester.
Statistical analysis
Quantified data are presented as mean±s.d. Significance testing was performed by one-way analysis of variance or Student's t-test with JMP 8 software. Relative intensities were quantified via Adobe Photoshop 6.0.
Supplementary Material
Acknowledgments
We are grateful to The Brain Tumor and Neuro-Oncology Center at Cleveland Clinic and Dr Andrew Sloan for providing glioblastoma surgical specimens. We thank Sage O'Bryant and Cathy Shemo of the Flow Cytometry Core at Lerner Institute for their help. We also thank the members in Rich's laboratory for their help and scientific discussions. This work was supported by the Cleveland Clinic Foundation and a NIH R01 grant NS070315 to SB.
Author Contributions: LC and SB designed the experiments. LC, QW, ZH, and OG performed all the experiments. LC, SB, and JR analysed the data. SB and JR wrote the manuscript. QH and WS provided reagents and scientific inputs. SB coordinated the study and oversaw the research.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abraham RT (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15: 2177–2196 [DOI] [PubMed] [Google Scholar]
- Arlt MJ, Novak-Hofer I, Gast D, Gschwend V, Moldenhauer G, Grünberg J, Honer M, Schubiger PA, Altevogt P, Krüger A (2006) Efficient inhibition of intra-peritoneal tumor growth and dissemination of human ovarian carcinoma cells in nude mice by anti-L1-cell adhesion molecule monoclonal antibody treatment. Cancer Res 66: 936–943 [DOI] [PubMed] [Google Scholar]
- Bao S, Tibbetts RS, Brumbaugh KM, Fang Y, Richardson DA, Ali A, Chen SM, Abraham RT, Wang XF (2001) ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature 411: 969–974 [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, Li Z, Sathornsumetee S, Wang H, McLendon RE, Hjelmeland AB, Rich JN (2008) Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res 68: 6043–6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN (2006a) Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444: 756–760 [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN (2006b) Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res 66: 7843–7848 [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J (2003) Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3: 421–429 [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J (2007) DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol 19: 238–245 [DOI] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ Jr, Kastan MB (2007) Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 9: 683–690 [DOI] [PubMed] [Google Scholar]
- Bertrand J, Begaud-Grimaud G, Bessette B, Verdier M, Battu S, Jauberteau MO (2009) Cancer stem cells from human glioma cell line are resistant to Fas-induced apoptosis. Int J Oncol 34: 717–727 [DOI] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A (2003) Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5: 675–679 [DOI] [PubMed] [Google Scholar]
- Chai W, Sfeir AJ, Hoshiyama H, Shay JW, Wright WE (2006) The involvement of the Mre11/Rad50/Nbs1 complex in the generation of G-overhangs at human telomeres. EMBO Rep 7: 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Bao S, Rich JN (2010) Potential therapeutic applications of cancer stem cells in glioblastoma. Biochem Pharmacol 80: 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YC, Teng SC, Su YN, Hsieh FJ, Wu KJ (2003) c-Myc directly regulates the transcription of the NBS1 gene involved in DNA double-strand break repair. J Biol Chem 278: 19286–19291 [DOI] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP (2002) The Mre11 complex: at the crossroads of DNA repair and checkpoint signaling. Nat Rev Mol Cell Biol 3: 317–327 [DOI] [PubMed] [Google Scholar]
- Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB (2009) Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell 34: 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, Joshua B, Kaplan MJ, Wapnir I, Dirbas FM, Somlo G, Garberoglio C, Paz B, Shen J, Lau SK, Quake SR et al. (2009) Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 458: 780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramo A, Ricci-Vitiani L, Zeuner A, Pallini R, Lotti F, Sette G, Pilozzi E, Larocca LM, Peschle C, De Maria R (2006) Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ 13: 1238–1241 [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP (2005) Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434: 605–611 [DOI] [PubMed] [Google Scholar]
- Fogel M, Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Smirnov A, Edler L, Ben-Arie A, Huszar M, Altevogt P (2003) L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet 362: 869–875 [DOI] [PubMed] [Google Scholar]
- Folkins C, Shaked Y, Man S, Tang T, Lee CR, Zhu Z, Hoffman RM, Kerbel RS (2009) Glioma tumor stem-like cells promote tumor angiogenesis and vasculogenesis via vascular endothelial growth factor and stromal-derived factor 1. Cancer Res 69: 7243–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedli A, Fischer E, Novak-Hofer I, Cohrs S, Ballmer-Hofer K, Schubiger PA, Schibli R, Grünberg J (2009) The soluble form of the cancer-associated L1 cell adhesion molecule is a pro-angiogenic factor. Int J Biochem Cell Biol 41: 1572–1580 [DOI] [PubMed] [Google Scholar]
- Frosina G (2009) DNA repair and resistance of gliomas to chemotherapy and radiotherapy. Mol Cancer Res 7: 989–999 [DOI] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK (2007) Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 21: 2683–2710 [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A (2004) Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 64: 7011–7021 [DOI] [PubMed] [Google Scholar]
- Gast D, Riedle S, Issa Y, Pfeifer M, Beckhove P, Sanderson MP, Arlt M, Moldenhauer G, Fogel M, Krüger A, Altevogt P (2008) The cytoplasmic part of L1-CAM controls growth and gene expression in human tumors that is reversed by therapeutic antibodies. Oncogene 27: 1281–1289 [DOI] [PubMed] [Google Scholar]
- Gavert N, Ben-shmuel A, Raveh S, Ben-Ze'ev A (2008) L1-CAM in cancerous tissues. Expert Opin Biol Ther 8: 1749–1757 [DOI] [PubMed] [Google Scholar]
- Gavert N, Conacci-Sorrell M, Gast D, Schneider A, Altevogt P, Brabletz T, Ben-Ze'ev A (2005) L1, a novel target of beta-catenin signaling, transforms cells and is expressed at the invasive front of colon cancers. J Cell Biol 168: 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavert N, Sheffer M, Raveh S, Spaderna S, Shtutman M, Brabletz T, Barany F, Paty P, Notterman D, Domany E, Ben-Ze'ev A (2007) Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis. Cancer Res 67: 7703–7712 [DOI] [PubMed] [Google Scholar]
- Geismann C, Morscheck M, Koch D, Bergmann F, Ungefroren H, Arlt A, Tsao MS, Bachem MG, Altevogt P, Sipos B, Fölsch UR, Schäfer H, Müerköster SS (2009) Up-regulation of L1CAM in pancreatic duct cells is transforming growth factor beta1- and slug-dependent: role in malignant transformation of pancreatic cancer. Cancer Res 69: 4517–4526 [DOI] [PubMed] [Google Scholar]
- Gilbertson RJ, Rich JN (2007) Making a tumour's bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer 7: 733–736 [DOI] [PubMed] [Google Scholar]
- Guerra L, Albihn A, Tronnersjö S, Yan Q, Guidi R, Stenerlöw B, Sterzenbach T, Josenhans C, Fox JG, Schauer DB, Thelestam M, Larsson LG, Henriksson M, Frisan T (2010) Myc is required for activation of the ATM-dependent checkpoints in response to DNA damage. PLoS One 5: e8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN (2009) The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle 8: 3274–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI (2003) Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA 100: 15178–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumoto S, Ohnishi T, Arita N, Hiraga S, Taki T, Hayakawa T (1996) Gene expression of neural cell adhesion molecule L1 in malignant gliomas and biological significance of L1 in glioma invasion. Cancer Res 56: 1440–1444 [PubMed] [Google Scholar]
- Jin SM, Cho HJ, Jung ES, Shim MY, Mook-Jung I (2008) DNA damage-inducing agents elicit gamma-secretase activation mediated by oxidative stress. Cell Death Differ 15: 1375–1384 [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J (2004) Cell-cycle checkpoints and cancer. Nature 432: 316–323 [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT (2005) ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308: 551–554 [DOI] [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, Hjelmeland AB, Rich JN (2009) Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell 15: 501–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS (2006) Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer 5: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J (2004) Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair (Amst) 3: 997–1007 [DOI] [PubMed] [Google Scholar]
- Maness PF, Schachner M (2007) Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci 10: 19–26 [DOI] [PubMed] [Google Scholar]
- Maretzky T, Schulte M, Ludwig A, Rose-John S, Blobel C, Hartmann D, Altevogt P, Saftig P, Reiss K (2005) L1 is sequentially processed by two differently activated metalloproteases and presenilin/gamma-secretase and regulates neural cell adhesion, cell migration, and neurite outgrowth. Mol Cell Biol 25: 9040–9053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier F, Busch S, Gast D, Göppert A, Altevogt P, Maczey E, Riedle S, Garbe C, Schittek B (2006) The adhesion molecule L1 (CD171) promotes melanoma progression. Int J Cancer 119: 549–555 [DOI] [PubMed] [Google Scholar]
- Nakai E, Park K, Yawata T, Chihara T, Kumazawa A, Nakabayashi H, Shimizu K (2009) Enhanced MDR1 expression and chemoresistance of cancer stem cells derived from glioblastoma. Cancer Invest 27: 901–908 [DOI] [PubMed] [Google Scholar]
- Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, Vescovi AL (2006) Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature 444: 761–765 [DOI] [PubMed] [Google Scholar]
- Raveh S, Gavert N, Ben-Ze'ev A (2009) L1 cell adhesion molecule (L1CAM) in invasive tumors. Cancer Lett 282: 137–145 [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Yaffe MB (2009) Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Opin Cell Biol 21: 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedle S, Kiefel H, Gast D, Bondong S, Wolterink S, Gutwein P, Altevogt P (2009) Nuclear translocation and signalling of L1-CAM in human carcinoma cells requires ADAM10 and presenilin/gamma-secretase activity. Biochem J 420: 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharpfenecker M, Kruse JJ, Sprong D, Russell NS, Ten Dijke P, Stewart FA (2009) Ionizing radiation shifts the PAI-1/ID-1 balance and activates notch signaling in endothelial cells. Int J Radiat Oncol Biol Phys 73: 506–513 [DOI] [PubMed] [Google Scholar]
- Sebens Müerköster S, Kötteritzsch J, Geismann C, Gast D, Kruse ML, Altevogt P, Fölsch UR, Schäfer H (2009) alpha5-integrin is crucial for L1CAM-mediated chemoresistance in pancreatic adenocarcinoma. Int J Oncol 34: 243–253 [PubMed] [Google Scholar]
- Sebens Müerköster S, Werbing V, Sipos B, Debus MA, Witt M, Grossmann M, Leisner D, Kötteritzsch J, Kappes H, Klöppel G, Altevogt P, Fölsch UR, Schäfer H (2007) Drug-induced expression of the cellular adhesion molecule L1CAM confers anti-apoptotic protection and chemoresistance in pancreatic ductal adenocarcinoma cells. Oncogene 26: 2759–2768 [DOI] [PubMed] [Google Scholar]
- Siesser PF, Maness PF (2009) L1 cell adhesion molecules as regulators of tumor cell invasiveness. Cell Adh Migr 3: 275–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB (2004) Identification of human brain tumour initiating cells. Nature 432: 396–401 [DOI] [PubMed] [Google Scholar]
- Shiloh Y (2003) ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 3: 155–168 [DOI] [PubMed] [Google Scholar]
- Stoeck A, Gast D, Sanderson MP, Issa Y, Gutwein P, Altevogt P (2007) L1-CAM in a membrane-bound or soluble form augments protection from apoptosis in ovarian carcinoma cells. Gynecol Oncol 104: 461–469 [DOI] [PubMed] [Google Scholar]
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, et al. , European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10: 459–466 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Izumoto S, Fujimoto Y, Maruno M, Ito Y, Yoshimine T (2005) Clinicopathological study of cellular proliferation and invasion in gliomatosis cerebri: important role of neural cell adhesion molecule L1 in tumour invasion. J Clin Pathol 58: 166–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice RR, Strauss GH (1995) The single cell gel electrophoresis/comet assay: a potential tool for detecting radiation-induced DNA damage in humans. Stem Cells 13(Suppl 1): 207–214 [PubMed] [Google Scholar]
- Vescovi AL, Galli A, Reynolds BA (2006) Brain tumour stem cells. Nat Rev Cancer 6: 425–436 [DOI] [PubMed] [Google Scholar]
- Wakimoto H, Kesari S, Farrell CJ, Curry WT Jr, Zaupa C, Aghi M, Kuroda T, Stemmer-Rachamimov A, Shah K, Liu TC, Jeyaretna DS, Debasitis J, Pruszak J, Martuza RL, Rabkin SD (2009) Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res 69: 3472–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wakeman TP, Lathia JD, Hjelmeland AB, Wang XF, White RR, Rich JN, Sullenger BA (2010) Notch promotes radioresistance of glioma stem cells. Stem Cells 28: 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidle UH, Eggle D, Klostermann S (2009) L1-CAM as a target for treatment of cancer with monoclonal antibodies. Anticancer Res 29: 4919–4931 [PubMed] [Google Scholar]
- Weller S, Gärtner J (2001) Genetic and clinical aspects of X-linked hydrocephalus (L1 disease): mutations in the L1CAM gene. Hum Mutat 18: 1–12 [DOI] [PubMed] [Google Scholar]
- Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359: 492–507 [DOI] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, Tainer JA (2009) Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell 139: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA (2007) Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol 85: 509–520 [DOI] [PubMed] [Google Scholar]
- Wolterink S, Moldenhauer G, Fogel M, Kiefel H, Pfeifer M, Lüttgau S, Gouveia R, Costa J, Endell J, Moebius U, Altevogt P (2010) Therapeutic antibodies to human L1CAM: functional characterization and application in a mouse model for ovarian carcinoma. Cancer Res 70: 2504–2515 [DOI] [PubMed] [Google Scholar]
- Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM (2007) WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci USA 104: 618–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Xiao S, Zhu XD (2007) MRE11-RAD50-NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat Struct Mol Biol 14: 832–840 [DOI] [PubMed] [Google Scholar]
- Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB (2009) Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov 8: 806–823 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.