A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging
A protein complex including the vesicle coat protein clathrin is here shown to constitute a population of inter-microtubule bridges in the mitotic spindle, which crosslink kinetochore fibres to stabilise them.
Keywords: clathrin, crosslinking, kinetochore fibre, microtubule, mitotic spindle
Abstract
Kinetochore fibres (K-fibres) of the spindle apparatus move chromosomes during mitosis. These fibres are discrete bundles of parallel microtubules (MTs) that are crosslinked by inter-MT ‘bridges' that are thought to improve fibre stability during chromosomal movement. The identity of these bridges is unknown. Clathrin is a multimeric protein that has been shown to stabilise K-fibres during early mitosis by a mechanism independent of its role in membrane trafficking. In this study, we show that clathrin at the mitotic spindle is in a transforming acidic colied-coil protein 3 (TACC3)/colonic, hepatic tumour overexpressed gene (ch-TOG)/clathrin complex. The complex is anchored to the spindle by TACC3 and ch-TOG. Ultrastructural analysis of clathrin-depleted K-fibres revealed a selective loss of a population of short inter-MT bridges and a general loss of MTs. A similar loss of short inter-MT bridges was observed in TACC3-depleted K-fibres. Finally, immunogold labelling confirmed that inter-MT bridges in K-fibres contain clathrin. Our results suggest that the TACC3/ch-TOG/clathrin complex is an inter-MT bridge that stabilises K-fibres by physical crosslinking and by reducing rates of MT catastrophe.
Introduction
Chromosomes must be accurately segregated during mitosis for cells to remain viable (Holland and Cleveland, 2009). In higher organisms, the kinetochores of the chromosome are attached to the spindle via discrete bundles of parallel microtubules (MTs), termed kinetochore fibres (K-fibres), which extend from the spindle pole and terminate at their attachment at the kinetochore (McDonald et al, 1992; Compton, 2000; Rieder, 2005). The MTs of K-fibres are connected by inter-MT bridges that are thought to stabilise the fibre during chromosome movement (Hepler et al, 1970; Witt et al, 1981; Compton, 2000). Anti-parallel MTs at the spindle midzone are crosslinked via Ase1p/MAP65/PRC1 non-motor proteins (Peterman and Scholey, 2009; Bieling et al, 2010), yet the identity of inter-MT bridges in K-fibres is obscure and the stabilisation mechanism conferred by them has not been established (Compton, 2000; Manning and Compton, 2008).
A candidate for inter-MT bridges in K-fibres is clathrin: a triskelion consisting of three heavy (190 kDa) and three light (25–27 kDa) chains (Pearse, 1975; Kirchhausen and Harrison, 1981; Ungewickell and Branton, 1981). During interphase, clathrin is involved in coating membranes for vesicular transport (Brodsky et al, 2001), but during mitosis, a time when membrane trafficking is reduced (Warren, 1993; Schweitzer et al, 2005), a subset of clathrin localises to the spindle apparatus (Maro et al, 1985; Okamoto et al, 2000; Royle et al, 2005). Clathrin heavy chain (CHC) depletion by RNAi suggests that clathrin is important for stabilising K-fibres (Royle et al, 2005). The N-terminal domain (NTD) of CHC is important for clathrin to bind to the spindle (Royle et al, 2005; Royle and Lagnado, 2006), and replacement experiments indicate that the NTD and the trimerisation domain of CHC are essential for its mitotic function (Royle et al, 2005; Royle and Lagnado, 2006). This has led to the idea that clathrin could stabilise K-fibres by crosslinking MTs (Royle and Lagnado, 2006; Hood and Royle, 2009). However, direct experimental support for this notion is lacking and even the mechanism of clathrin's recruitment to the mitotic spindle is unclear.
Transforming acidic colied-coil protein 3 (TACC3) and colonic, hepatic tumour overexpressed gene (ch-TOG) are non-motor spindle proteins that promote MT outgrowth from the centrosome (Gergely et al, 2000a, 2000b, 2003; Barr and Gergely, 2008; Peset and Vernos, 2008). Both proteins are also found on spindle fibres, but their role here is uncertain (Gergely et al, 2003; Peset and Vernos, 2008). Depletion of TACC3 destabilises the mitotic spindle in a manner similar to depletion of clathrin (Gergely et al, 2003; Royle et al, 2005). Recently, it was reported that clathrin recruits TACC3, and by extension, ch-TOG to MTs (Fu et al, 2010; Hubner et al, 2010; Lin et al, 2010). In this model, the stabilisation of spindle fibres by clathrin is via the proposed anti-catastrophe activity of ch-TOG (Brouhard et al, 2008), and a role for clathrin as an inter-MT bridge for clathrin need not be invoked.
In this paper, we compared the two models for clathrin function in mitosis. We found little evidence to support a role for clathrin in the direct recruitment of TACC3 and ch-TOG to MTs. Instead, we now show direct evidence for clathrin acting as an inter-MT bridge in K-fibres. An alternative model is presented, in which clathrin is an inter-MT bridge that is recruited to MTs by TACC3 and ch-TOG. Clathrin crosslinks TACC3/ch-TOG subcomplexes on adjacent MTs, increasing their residence time on MTs and causing their accumulation on K-fibres. The TACC3/ch-TOG/clathrin inter-MT bridges, therefore, stabilise K-fibres by physical crosslinking and possibly by reducing the rate of MT catastrophe.
Results
A TACC3/ch-TOG/clathrin complex at the mitotic spindle
Recent work suggested that clathrin recruits TACC3 to mitotic spindles to promote stability (Fu et al, 2010; Hubner et al, 2010; Lin et al, 2010). However, clathrin is a highly abundant cytosolic protein (Goud et al, 1985), only a small subset of which is localised to the mitotic spindle (Tanenbaum et al, 2010), whereas TACC3 is strongly and consistently localised to spindles (Figure 1A). It is not clear how clathrin could recruit TACC3, specifically to the mitotic spindle when the majority of clathrin is in the cytoplasm or assembled in coated structures. This is further compounded by the observation that clathrin cannot bind MTs (Supplementary Figure 1), whereas maskin, a TACC3 homologue, can (O'Brien et al, 2005; Peset et al, 2005; Fu et al, 2010).
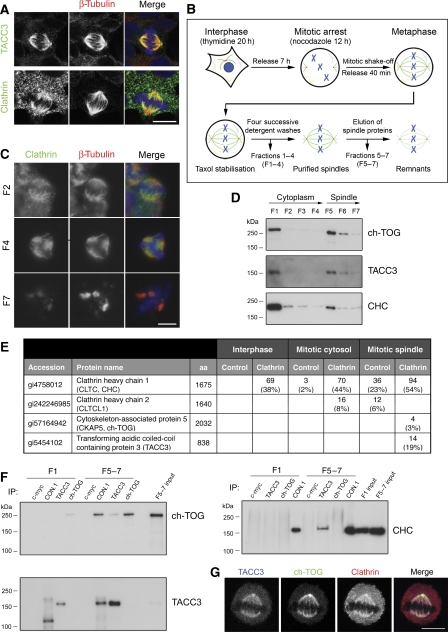
Figure 1.
Clathrin at the mitotic spindle is in a complex with TACC3 and ch-TOG. (A) Representative confocal micrographs to illustrate the different subcellular distributions of endogenous clathrin and TACC3 in mitotic HEK293 cells. (B) Schematic diagram of the experimental protocol to release proteins from mitotic spindles at metaphase. After each spin, the supernatant was collected (fractions 1–7, F1–7). (C) Fluorescence micrographs of pelleted material from the purification spotted onto coverslips, fixed and processed for immunofluorescence. Clathrin remains on spindles following detergent washes and was released from spindles using 0.3 M NaCl. The fraction number (F2, F4, F7) indicates the stage of the purification. (D) Western blots of fractions from the spindle purification to show the ‘release' of ch-TOG, TACC3 and clathrin from spindles. (E) Table to show the number of peptides and % coverage (in parentheses) for proteins detected by mass spectrometry in a single experiment. IPs were performed on interphase cell lysate, mitotic cytosol (fraction 1) and spindle fractions (combined fractions 5–7) using either CON.1, a monoclonal clathrin light chain antibody, or anti-myc/9E10 as a control. Immunoprecipitated material was separated by 12% SDS–PAGE and samples analysed by tandem mass spectrometry. (F) Reciprocal immunoprecipitation and western blotting of complex members from F1 or F5–7. In spindle fractions, TACC3 and ch-TOG were detected in clathrin IPs, clathrin and ch-TOG were detected in TACC3 IPs, but TACC3 and clathrin were not detected in ch-TOG IPs. This may be due to an excess of ch-TOG in the fractions that is not associated with TACC3/clathrin or that the ch-TOG antibody is not compatible with co-IP experiments. (G) Confocal micrographs to show the co-localisation of interaction partners at the metaphase spindle. HeLa cells co-expressing low levels of tdTomato–LCa and GFP–ch-TOG were immunostained with anti-TACC3. Scale bars, 10 μm.
To begin to address this paradox, we purified the native clathrin-containing complex from mitotic spindles (Figure 1B–D). A complex containing clathrin, TACC3 and ch-TOG was identified repeatedly using a variety of elution conditions. The complex was not detected in control immunoprecipitations (IPs) using anti-9E10/Myc, nor was it detected in clathrin IPs from mitotic or interphase cytosol, suggesting that this complex was specific to the mitotic spindle (Figure 1E). We largely confirmed the mass spectrometry results by reciprocal IP and western blotting (Figure 1F). During early mitosis, TACC3, ch-TOG and clathrin are all found on spindle MTs between the pole and chromosomes (Charrasse et al, 1998; Gergely et al, 2000a; Royle et al, 2005). Figure 1G shows a metaphase cell expressing GFP-ch-TOG and tdTomato-clathrin light chain a (LCa) and stained for TACC3. All three proteins co-localise on the spindle.
Order of recruitment of the TACC3/ch-TOG/clathrin complex components on spindle MTs
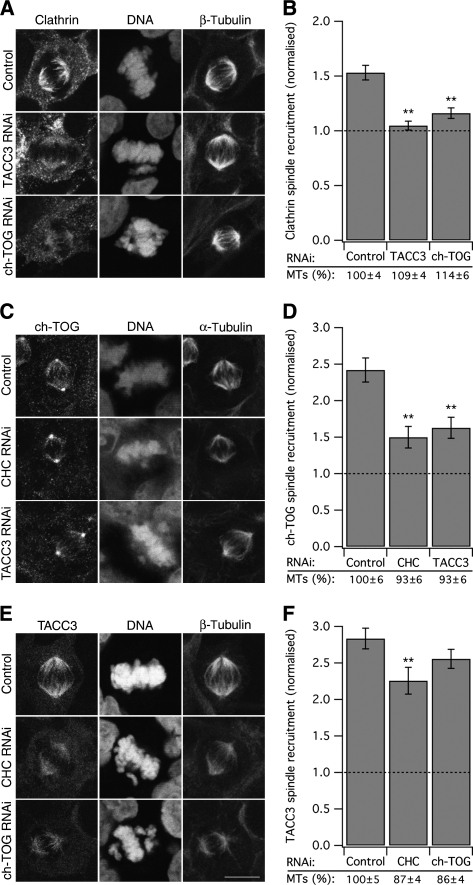
To understand how the TACC3/ch-TOG/clathrin complex is anchored to MTs, each member of the complex was depleted using RNAi and the spindle recruitment of the other two complex components was analysed by microscopy. In cells depleted of TACC3 or ch-TOG, clathrin recruitment to the spindle was dramatically reduced (Figure 2A and B). Cells depleted of TACC3 or clathrin exhibited reduced binding of ch-TOG to spindle MTs (Figure 2C and D; Gergely et al, 2003; Cassimeris et al, 2009). Finally, depletion of ch-TOG had very little effect on the spindle localisation of TACC3 (Figure 2E and F), as reported previously (Gergely et al, 2003). A small but significant reduction in TACC3 spindle localisation upon clathrin depletion was noted (Figure 2E and F). The spindle recruitment of clathrin was measured as the ratio of X22 immunofluorescence signal on the spindle versus that in the cytoplasm. Cells depleted of either TACC3 or ch-TOG had ratios that were reduced by 91 and 70%, respectively. These ratios were not significantly different to the ratio measured for a CHC that lacks the NTD and cannot bind the spindle (Royle and Lagnado, 2006), indicating that depletion of either TACC3 or ch-TOG completely blocks clathrin recruitment to the spindle (Figure 2B). In contrast, quantification of spindle recruitment of TACC3 showed only a 25 or 12% reduction following RNAi of clathrin or ch-TOG, respectively (Figure 2F). Similarly, ch-TOG recruitment was reduced, but not abolished by depletion of either CHC or TACC3 (Figure 2D). These results disagree with recent work that reported that depletion of clathrin mislocalised TACC3 completely, and that depletion of TACC3 had no effect on clathrin localisation (Hubner et al, 2010; Lin et al, 2010). The differences in TACC3 localisation are unlikely to be due to poor RNAi. We achieved good depletion of TACC3, ch-TOG and clathrin using shRNA in HEK293 cells at 3 days post-transfection (Supplementary Figure 2). Knockdown of clathrin in HeLa cells for 5 days gave an equivalent depletion, and following this manipulation we saw a slightly larger change in TACC3 spindle recruitment ratio, but not a complete block (2.52±0.09 versus 1.46±0.05, control versus CHC RNAi). In addition, inefficient RNAi in our hands would not explain why we observed mislocalisation of clathrin following depletion of TACC3 or ch-TOG and others did not.
Figure 2.
Spindle localisation of TACC3/ch-TOG/clathrin complex components following RNAi. Representative confocal micrographs to show the spindle recruitment of endogenous (A) clathrin (C) ch-TOG or (E) TACC3 following the individual depletion of the other two complex members. A fluorescent reporter confirmed the expression of shRNA (not shown). Bar charts to show spindle recruitment of endogenous (B) clathrin, (D) ch-TOG or (F) TACC3 following individual depletion of the other complex members. Spindle recruitment was measured as described in Materials and methods. Note that a value of 1 represents no detectable recruitment. The average MT intensity at the spindle normalised to control (%) is shown and did not account for the changes observed. Bars represent mean±s.e.m., ncell=45–116 from three independent experiments; **P<0.01. Scale bar, 10 μm.
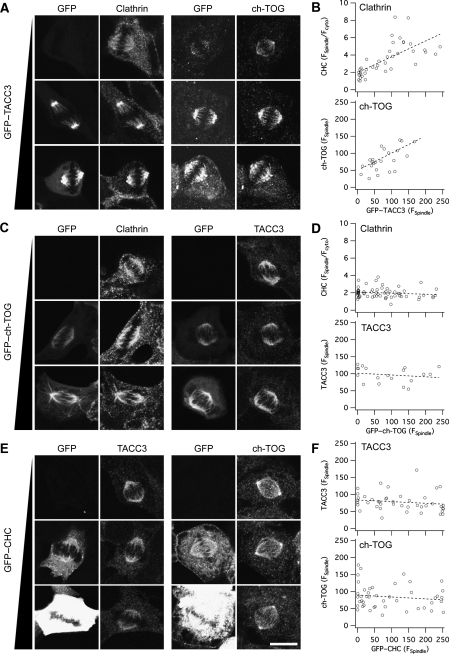
To further probe the order of recruitment of complex components to the mitotic spindle, we carried out similar experiments, but this time we individually overexpressed, rather than depleted, each component of the complex. Overexpression of GFP–TACC3 dramatically increased the amount of endogenous ch-TOG or clathrin that was recruited to the spindle (Figure 3A and B), indicating that TACC3 is the primary factor that recruits the other two complex components to the spindle. GFP–ch-TOG overexpression did not affect the levels of clathrin or TACC3 at the spindle (Figure 3C and D). Similarly, overexpression of GFP–CHC17 did not alter the levels of TACC3 or ch-TOG on the spindle (Figure 3E and F). Together, these results suggested an order of recruitment, where TACC3 binds directly to MTs, ch-TOG binds to TACC3 and clathrin binds to TACC3/ch-TOG subcomplexes, possibly as the most exterior component.
Figure 3.
Spindle localisation of TACC3/ch-TOG/clathrin complex components following overexpression. Representative confocal micrographs to show the spindle recruitment of the other two complex members following the individual overexpression of GFP-tagged TACC3 (A), ch-TOG (C) and clathrin (E) in HEK293 cells. Three conditions are shown: low or no expression (top), medium expression (middle) and high expression (bottom). In A, right panels, GFP–TACC3 was also detected with TACC3/A488. Spindle recruitment of endogenous proteins was visualised by immunostaining, and expressed protein was visualised by GFP fluorescence. (B, D, F) Plots of immunofluorescence as a function of GFP expression at the spindle are shown as indicated. A linear fit to the data is shown as a dotted line. Scale bar, 10 μm.
Recruitment of clathrin to the mitotic spindle by TACC3 is mediated via Aurora-A kinase
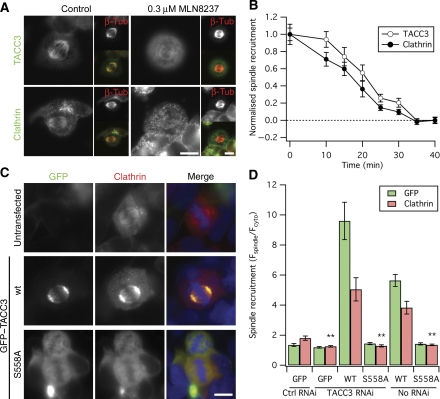
Recruitment of TACC3 to MTs is regulated by Aurora-A kinase-mediated phosphorylation (Kinoshita et al, 2005; Barr and Gergely, 2007; LeRoy et al, 2007). Incubation of cells with the Aurora-A kinase inhibitor MLN8237 (0.3 μM) induced a change in the localisation of endogenous TACC3 and clathrin from the mitotic spindle to the cytoplasm (Figure 4A and B). This effect was complete within 35 min, and both proteins were dispersed with similar kinetics (Figure 4B). To further understand this difference, we overexpressed a non-phosphorylatable mutant of TACC3, GFP–TACC3(S558A). In agreement with previous studies (LeRoy et al, 2007; Fu et al, 2010; Hubner et al, 2010; Lin et al, 2010), this mutant did not localise to the spindle. However, while expression of the wild-type GFP–TACC3 increased clathrin recruitment compared with untransfected cells, expression of GFP–TACC3(S558A) actually ‘removed' clathrin from spindles (Figure 4C and D). We found similar results in cells depleted of endogenous TACC3 (Figure 4D), which suggested that the reduction in clathrin recruitment was due to a removal of endogenous TACC3 from spindles by GFP–TACC3(S558A). In support of this idea, we found a reduction in TACC3 immunoreactivity at the spindle in cells expressing GFP–TACC3(S558A) (Supplementary Figure 3). Our data agree with previous work showing that the association of TACC3 with MTs is dependent upon phosphorylation by Aurora-A kinase (Barr and Gergely, 2007), but these results are incompatible with the suggestion that clathrin recruits TACC3 to spindles (Fu et al, 2010; Hubner et al, 2010; Lin et al, 2010). In summary, we found that TACC3 is the primary recruitment factor for the TACC3/ch-TOG/clathrin complex to bind MTs. These findings are in keeping with the hypothesis that clathrin acts as an inter-MT bridge in K-fibres.
Figure 4.
Recruitment of clathrin to the mitotic spindle is controlled by phosphorylation of TACC3 by Aurora-A kinase. (A) Representative micrographs of HEK293 cells incubated with 0.3 μM MLN8237 for 40 min. Cells were fixed and stained as indicated. (B) Time course of the reduction in spindle recruitment of endogenous clathrin and TACC3 by treatment of cells with 0.3 μM MLN8237. Data are mean±s.e.m., scaled to allow comparison of kinetics. The spindle recruitment for TACC3 and clathrin was 3.89±0.19 and 3.04±0.22 at 0 min and 1.32±0.08 and 1.08±0.07 at 40 min, respectively. (C) Representative fluorescence micrographs of mCherry-LCa (red) distribution in cells co-expressing GFP–TACC3 or GFP–TACC3(S558A). Triskelia are labelled by constitutive incorporation of mCherry-LCa (Hoffmann et al, 2010). (D) Bar chart to show recruitment of mCherry-LCa (red) in cells expressing GFP–TACC3 or GFP–TACC3(S558A). Note the decrease in clathrin from the spindle in cells expressing GFP–TACC3(S558A) compared to no co-expression. Bars, mean±s.e.m., **P<0.01. Scale bars, 10 μm.
Kinetochore fibres in clathrin-depleted cells lack short inter-MT bridges and have fewer MTs
With the order of recruitment of the complex established, we next tested directly whether clathrin is indeed an inter-MT bridge using electron microscopy. We reasoned that if clathrin is an inter-MT bridge, then we should be able to see a loss of bridges from K-fibres following clathrin depletion. A correlative light-electron microscopy approach was used to ensure that (i) all cells analysed had undergone RNAi, (ii) cells were close to metaphase and (iii) sections could be taken longitudinally or orthogonally relative to the spindle axis.
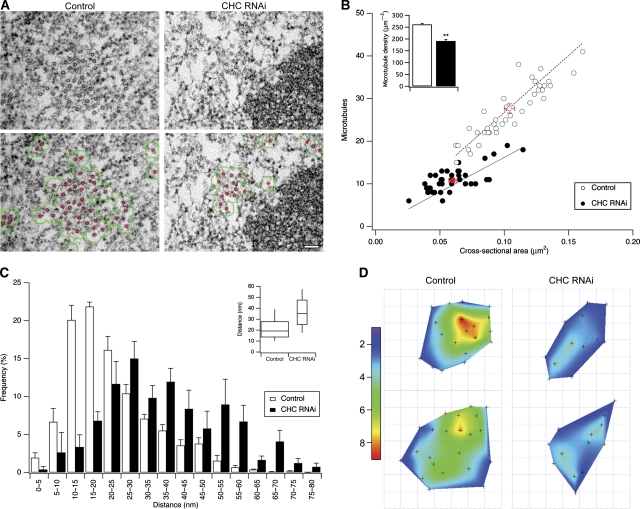
In clathrin-depleted cells, there were fewer MTs per K-fibre compared with control RNAi cells (27.7±1.1 versus 10.8±0.5, control versus CHC RNAi). The cross-sectional area of K-fibres in clathrin-depleted cells was also decreased compared with controls (Figure 5A and B). The effect of clathrin depletion was a loss of MTs and not simply a reduction in the thickness of K-fibres because clathrin-depleted K-fibres had a lower density of MTs than controls (Figure 5B). Loss of kinetochore MTs resulted in increased inter-MT spacing (Figure 5C), and a reduction in the number of neighbouring MTs within bridging distance (<80 nm) of one another (Figure 5D), which reinforced the conclusion that MT loss in clathrin-depleted K-fibres occurred throughout the fibre rather than selectively at superficial regions of the bundle.
Figure 5.
Clathrin-depleted K-fibres have fewer microtubules. (A) Typical electron micrographs of orthogonal sections from control or clathrin-depleted HeLa cells. Below, MTs are highlighted in red and the fibre outline is shown in green (see Materials and Methods). Scale bar, 100 nm. (B) Scatter plot showing the number of MTs per fibre versus cross-sectional area of the fibre. Markers show the value for individual fibres from control RNAi cells (open) or CHC RNAi cells (closed). Red markers show the mean±s.e.m. Dashed line is a fit constrained through the origin to indicate MT density. Inset: bar chart of MT density, ncell=4–6; **P<0.01. (C) Histogram to show the distance to the nearest neighbouring MT for each MT in the fibre. Bars show mean±s.e.m. ncell=4–6, nfibre=41–38, control versus CHC RNAi. Inset: Tukey plot for all nearest neighbour distances, nMT=1135–420. (D) ‘Heat maps' of a typical K-fibre from control or CHC RNAi cells. The co-ordinates of each MT in the fibre are plotted and the number of neighbouring MTs within 80 nm is indicated with a 2D surface of a Voronoi interpolation. The maximum observed length of inter-MT bridges was 80 nm and therefore this was the definition of neighbouring MTs within bridging distance of one another.
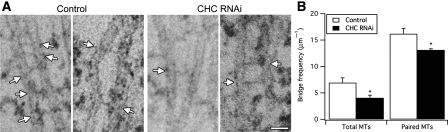
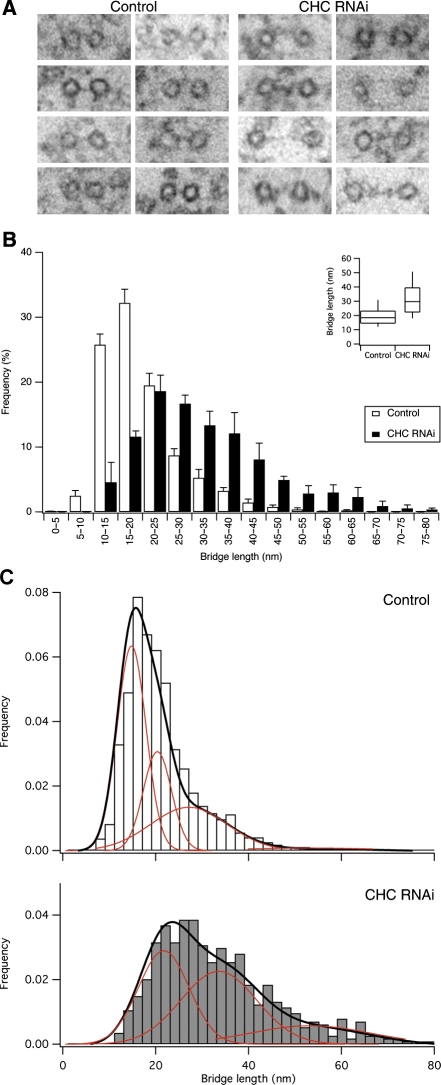
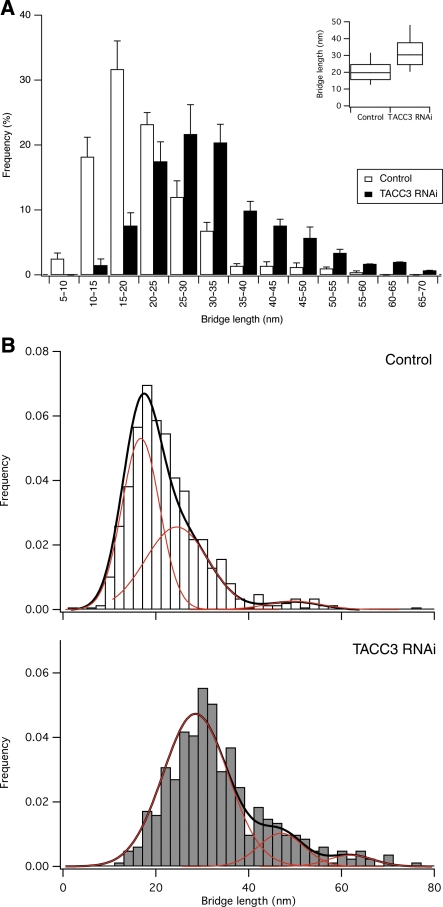
We next analysed the frequency of inter-MT bridges in control and clathrin-depleted cells. Inter-MT bridges were identified using established criteria (Hepler et al, 1970), that is, 2–5 nm-thick projections of electron-dense material that connect two MTs. We found that K-fibres in clathrin-depleted cells had fewer inter-MT bridges (Figure 6A and B). This reduction represented a loss of bridges from K-fibres and not a secondary effect of the change in MT packing, as the bridge frequency per unit length of MTs within bridging distance of one another was also significantly reduced (Figure 6B). Qualitative assessment of inter-MT bridges in orthogonal sections gave the impression that the bridges that remained in clathrin-depleted cells were longer than in controls (Figure 7A), so we returned to the longitudinal sections to quantify the inter-MT bridge lengths. Indeed, quantification of the lengths of inter-MT bridges showed that the median bridge length in controls was 18.4 nm, whereas in clathrin-depleted cells it was increased to 29.7 nm (Figure 7B). Had bridges become longer or was a specific length of bridge missing from clathrin-depleted cells? To address this, we examined the probability density functions of bridge lengths in clathrin-depleted cells versus controls. It was apparent that both distributions had multiple peaks and neither set could be fitted adequately by a single curve. We analysed the distributions by multi-peak analysis (see Materials and methods and Supplementary Figure 4) and found that while there was a small increase in longer bridges, the biggest change was that a prominent set of short bridges in control cells was missing upon clathrin depletion (Figure 7C). Multi-peak analysis of the data suggested four populations of bridges with mean lengths of ∼15, ∼21, ∼30 and ∼53 nm, respectively. In control cells, the ∼15 nm bridge population was most prominent (∼50% of total) and in clathrin-depleted cells this population was negligible. There were also small increases in the ∼30 and ∼53 nm populations. These changes in bridge population account for ∼40% decrease in bridges per unit length of MT and the apparent shift in bridge length. In summary, our ultrastructural analysis indicates that (i) there is a subset of inter-MT bridges in K-fibres whose presence is clathrin dependent and (ii) that K-fibres in clathrin-depleted cells have reduced numbers of MTs.
Figure 6.
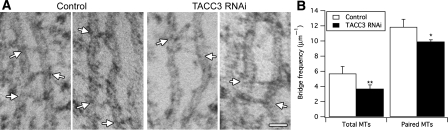
Clathrin-depleted K-fibres exhibit loss of inter-MT bridges. (A) Representative electron micrographs of K-fibres in longitudinal sections from control or clathrin-depleted cells. Arrows mark sites of inter-MT bridges. (B) Bar chart to show the frequency of bridges per μm of MT (total MTs) or per μm of paired MTs within 80 nm of each other (paired MTs). Bars show mean±s.e.m; *P<0.05. Scale bar, 40 nm.
Figure 7.
Clathrin-depleted K-fibres lack short inter-MT bridges. (A) Gallery of inter-MT bridges selected at random from electron micrographs of orthogonal sections to illustrate the apparent increase in bridge length in clathrin-depleted K-fibres. (B) Histogram to show the size distribution of inter-MT bridges from control or clathrin-depleted cells. Mean±s.e.m., ncell=3–4. Inset: Tukey plot for all bridge distances nbridge=1335–570. (C) Histograms of bridge size distribution of all bridges to illustrate putative bridge populations. Multi-peak analysis of CHC RNAi data (grey, below) indicated three populations with mean lengths of 21.8, 33.6 and 53.3 nm. Control RNAi cells (open, above) had an additional 14.8 nm bridge population, see Supplementary Figure 4. Individual populations, red; sum of populations, black.
Inter-MT bridges in K-fibres contain clathrin
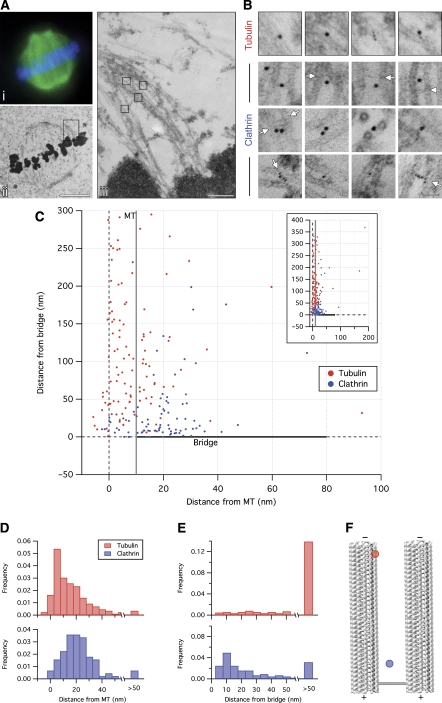
Clearly the presence of a subset of inter-MT bridges is clathrin dependent, but are the bridges actually composed of clathrin as our model predicts? To answer this question, we compared the distribution of clathrin and tubulin in relation to MTs and inter-MT bridges. Mitotic spindles were partially purified from HeLa cells expressing either GFP–α-tubulin or GFP–CHC and visualised by light and then electron microscopy following immunogold labelling (Figure 8A). As expected, tubulin labelling was tightly associated with MTs (Figure 8). The median distance from MT centres to tubulin-gold particles was 8.9 nm, whereas clathrin was more peripheral to MTs with a median distance of 18.8 nm (Figure 8C, D and F). However, clathrin was not equally associated with all MTs. The majority of clathrin labelling (88.8%) was found between paired MTs compared with 52.6% for tubulin. This suggests that, unlike tubulin, clathrin labelling was more often associated with paired MTs than single MTs. Strikingly, 53.9% of clathrin label was specific to inter-MT bridges, compared with only 8.8% of tubulin label (Figure 8E and F). Specificity is defined as gold <16 nm from an inter-MT bridge (Ottersen, 1989). The 37% of clathrin label that was between paired MTs, but was not associated with bridges, may reflect the labelling of bridges that are poorly stained/out-of-section or of partially assembled bridge structures. In summary, our results strongly suggest that inter-MT bridges in K-fibres contain clathrin.
Figure 8.
Inter-MT bridges in K-fibres contain clathrin. (A) A mitotic spindle purified from HeLa cells expressing GFP–α-tubulin, visualised by light (Ai) and electron (Aii and Aiii) microscopy following immunogold labelling (see Materials and methods). A view of the two K-fibres in the boxed region of Aii is shown in Aiii. Boxes in Aiii show the location of immunogold shown in B. Scale bar, 5 μm (ii) and 500 nm (iii). (B) Examples of immunogold labelling for tubulin or for clathrin. Spindles purified from cells expressing GFP–α-tubulin or GFP–CHC were labelled with anti-GFP and anti-tubulin or anti-GFP and anti-clathrin. Detection was with 5 and 10 nm gold-conjugated secondary antibodies. Arrows mark the site of bridges. A 2D plot (C) of gold particle locations relative to the MT centre (x axis) or the nearest bridge (y axis). Grey and black lines indicate the edge of the MT and the bridge, respectively. Inset: collapsed view of the data set. (D, E) Location of gold particles relative to MTs and bridges. Frequency histograms to show the distance from gold particles to the nearest MT centre (D) or to the nearest inter-MT bridge (E) (ntubulin=120, nclathrin=89). (F) Median distances from the data set are plotted on a scaled representation of two MTs and an idealised straight bridge.
Kinetochore fibres in TACC3-depleted cells lack short inter-MT bridges
If the TACC3/ch-TOG/clathrin complex is an inter-MT bridge then we would predict that depletion of either TACC3 or ch-TOG would also result in a loss of short inter-MT bridges, similar to depletion of clathrin. We tested this idea by analysing the ultrastructure of K-fibres from control, TACC3 or ch-TOG RNAi cells.
Depletion of ch-TOG results in a change in spindle morphology (Gergely et al, 2003) and at the ultrastructural level, the resultant derangement of K-fibres was in many cases so severe that they could not be analysed (Supplementary Figure 5). Briefly, the K-fibres did not lie in the plane of section and MTs were frequently in the wrong orientation (Supplementary Figure 5). We instead concentrated on TACC3-depleted cells. In these cells, K-fibres had fewer inter-MT bridges than controls (Figure 9). The loss of bridges was similar to the loss seen in clathrin-depleted cells with ∼35% fewer bridge per unit length of MT. Did this loss represent a loss in bridges or a change in MT spacing that results in fewer regions were bridging could occur? We again examined the frequency of inter-MT bridges per unit length of paired MT and found that there was a similar significant decrease in inter-MT bridges (Figure 9B). Finally, we wondered whether the same class of inter-MT bridge was missing following TACC3 depletion. We analysed the bridge lengths as described above and found that the median bridge length in TACC3-depleted K-fibres was 30.4 nm compared with 19.8 nm in controls (Figure 10A). The probability density functions of all bridge lengths from control or TACC3-depleted K-fibres again suggested multiple classes of inter-MT bridges (Supplementary Figure 4B). Multi-peak analysis of the data suggested three populations of bridges in controls with mean lengths of ∼17, ∼24 and ∼50 nm and three populations in TACC3-depleted cells with lengths of ∼29, ∼47 and 62 nm (Figure 10B). In control cells, the 16.7 nm bridge population was most prominent (∼54% of total) and in TACC3-depleted cells this population was negligible. These data suggest that TACC3 and clathrin mediate inter-MT bridging of short distance interactions between adjacent MTs in K-fibres.
Figure 9.
TACC3-depleted K-fibres exhibit loss of inter-MT bridges. (A) Representative electron micrographs of K-fibres in longitudinal sections from control or TACC3-depleted cells. Arrows mark sites of inter-MT bridges. (B) Bar chart to show the frequency of bridges per μm of MT (total MTs) or per μm of paired MTs within 80 nm of each other (paired MTs). Bars show mean±s.e.m.; **P<0.01, *P<0.05. Scale bar, 40 nm.
Figure 10.
TACC3-depleted K-fibres lack short inter-MT bridges. (A) Histogram to show the size distribution of inter-MT bridges from control or TACC3-depleted cells. Mean±s.e.m., ncell=6–7. Inset: Tukey plot for all bridge distances nbridge=732–406. (B) Histograms of bridge size distribution of all bridges to illustrate putative bridge populations. Multi-peak analysis of TACC3 RNAi data (grey, below) indicated three populations with mean lengths of 28.5, 47.0 and 61.9 nm. Control RNAi cells (open, above) had an additional 16.7 nm bridge population, see Supplementary Figure 4. Individual populations, red; sum of populations, black.
Discussion
In this study, we have identified a TACC3/ch-TOG/clathrin complex at the mitotic spindle. TACC3 anchors the complex at MTs and recruits ch-TOG and clathrin. Clathrin is a multimeric protein that could therefore crosslink TACC3/ch-TOG subcomplexes on adjacent MTs within a K-fibre. In agreement with this idea, we found that inter-MT bridges contain clathrin and that removal of clathrin or TACC3 resulted in a loss of bridges. These findings suggest that TACC3/ch-TOG/clathrin complexes are the molecular correlate of short inter-MT bridges in K-fibres.
The lengths of inter-MT bridges in K-fibres had a complex distribution that was best described as the sum of several populations of bridges of different length classes. In two independent series of experiments, we identified populations of bridges that were broadly similar (Supplementary Figure 4). These apparent populations are short (14–17 nm), medium (possibly two populations of 20 and 27 nm) and long (∼50 nm). It is the shortest class that is absent following depletion of TACC3 or clathrin, which suggests that the other bridges are comprised of other proteins. Our immunogold labelling results are consistent with the idea that the short form of inter-MT bridge density is composed of clathrin. Clathrin is a triskelion with a leg length of ∼53 nm (Jin and Nossal, 2000). How is it possible that this can form bridges that are <20 nm in length? Inter-MT bridges are measured from the inside edges of adjacent MTs, but clathrin could span the longer distance between the top or bottom edges of MTs. Orthogonal sections support this idea (Figure 7A). The model shown in Supplementary Figure 6 is the simplest configuration that fits the measured distances. We propose that two CHC legs associate with TACC3/ch-TOG subcomplexes on one MT with the other leg binding to a subcomplex on the adjacent MT.
In three recent studies, clathrin was identified as a TACC3-binding protein and it was suggested that clathrin recruits TACC3 to MTs and that spindle stability was conferred by TACC3/ch-TOG (Fu et al, 2010; Hubner et al, 2010; Lin et al, 2010). These results were surprising for several reasons. First, clathrin cannot bind MTs, whereas maskin, a TACC3 homologue, can bind to MTs with a Kd equivalent to other microtubule-associated proteins (MAPs; O'Brien et al, 2005; Peset et al, 2005), and also ch-TOG is also a known MT-binding protein (Charrasse et al, 1998). Second, the majority of clathrin in mitotic cells is not localised at the spindle, whereas TACC3 and ch-TOG are predominantly spindle associated. Third, if clathrin simply recruits TACC3 to the spindle then why are trimerisation-deficient mutants of CHC unable to function in mitosis (Royle and Lagnado, 2006)? In this study, we have tested the published model experimentally and found that it does not hold. Examples of results that are incompatible with this model are (i) overexpression of GFP–CHC did not cause more clathrin to accumulate at the spindle and it did not influence the spindle localisation of TACC3 or ch-TOG, (ii) overexpression of TACC3 increased the spindle localisation of ch-TOG and clathrin and (iii) blocking spindle recruitment of TACC3 by inhibition of aurora-A kinase or by mutating TACC3 inhibited the recruitment of clathrin to the spindle. Finally, the published model requires an additional factor for clathrin to bind to MTs. We purified the native clathrin-containing complexes from mitotic spindles and only identified clathrin, TACC3 and ch-TOG.
The model that we propose best explains our results and those of others. In this model, TACC3 recruits ch-TOG and clathrin to MTs (Supplementary Figure 6). Depletion of clathrin does not prevent the recruitment of TACC3 to spindles as previously suggested (Fu et al, 2010; Hubner et al, 2010; Lin et al, 2010); rather, it causes a reduction in TACC3 and ch-TOG at the spindle by preventing their accumulation. This would be achieved if the TACC3/ch-TOG/clathrin complex is more stable on MTs compared with TACC3/ch-TOG subcomplexes. This is perhaps to be expected if the recruitment of clathrin to MTs only requires the binding of one triskelion leg. Subsequent binding of TACC3 or TACC3/ch-TOG by the other legs would then ‘lock' more TACC3 and ch-TOG on MTs. In our model for TACC3/ch-TOG/clathrin binding to MTs, the effect observed previously of clathrin depletion on TACC3 is not to affect recruitment of TACC3 but rather its accumulation on the spindle.
The transcription factor B-Myb was reported to regulate the association of clathrin with the spindle (Yamauchi et al, 2008). It was not immediately obvious how B-Myb, which is not a spindle protein, could carry out this function. We found that the association of clathrin with the spindle is indirectly regulated by Aurora-A kinase, most likely via phosphorylation of TACC3 at S558 (Kinoshita et al, 2005; LeRoy et al, 2007). In addition, a recent study found that B-Myb is important for the transcription of several cell cycle-regulated genes, including Aurora-A kinase (Knight et al, 2009). In light of this work, we suggest that the effect of B-Myb on clathrin localisation may be via the regulation of expression of this kinase.
This study identifies TACC3/ch-TOG/clathrin as an inter-MT bridge in K-fibres, thus providing an ultrastructural explanation for the destabilisation of K-fibres observed following clathrin depletion (Royle et al, 2005). Loss of either MTs (Rubinstein et al, 2009) or bridges (Manning and Compton, 2008) is predicted to destabilise K-fibres. In clathrin-depleted K-fibres, we observed both. There were fewer short bridges and fewer MTs per K-fibre. This indicates that TACC3/ch-TOG/clathrin may stabilise K-fibres by (i) physically tethering adjacent MTs and (ii) by lowering the rate of MT catastrophe. It remains to be seen whether the latter property is common to all inter-MT bridges or is restricted to those that contain ch-TOG, a known anti-catastrophe factor (Brouhard et al, 2008).
Other proteins that crosslink spindle MTs have been identified (Manning and Compton, 2008; Peterman and Scholey, 2009). For example, non-motor proteins such as PRC1 (Zhu et al, 2006) and kinesin-5 motors (Sharp et al, 1999) can crosslink anti-parallel MTs in the spindle midzone. The TACC3/ch-TOG/clathrin complex, however, is the first inter-MT bridge to be defined in K-fibres. Interestingly, there are longer bridges that remain in clathrin-depleted or TACC3-depleted K-fibres. An alternative interpretation for our results is that flexible MAPs link MTs over longer distances (40–80 nm); these MAPs are compressed to make the shorter bridge type by clathrin binding to TACC3/ch-TOG on adjacent MTs. This is unlikely because an equivalent number of bridges would be predicted if this were the case, whereas we observed a net reduction in the number of bridges in clathrin-depleted or TACC3-depleted K-fibres. We predict, therefore, that there are other protein complexes in K-fibres that mediate the long form of bridging. Other proteins such as HURP (Wong and Fang, 2006) and the kinesin-related protein HSET/KIFC1 (Mountain et al, 1999; Zhu et al, 2006) are good candidates for inter-MT bridges in K-fibres. It is possible that these engage first to bring MTs in closer proximity, and the short TACC3/ch-TOG/clathrin bridges mediate the tighter, final bridges in a sequential stabilisation reaction. We anticipate the discovery of other bridge complexes that form longer inter-MT bridges in K-fibres.
Materials and methods
Cell biology
HeLa cells were maintained in DMEM containing 10% fetal bovine serum, 4 mM L-glutamine and 100 U/ml penicillin/streptomycin at 37°C and 5% CO2. HEK293 cell culture, transfection, RNAi and immunocytochemistry were as described previously (Royle and Lagnado, 2006; Hood and Royle, 2009). HeLa and HEK293 cells were transfected by lipofection or calcium phosphate-mediated transfection, respectively. Cells were fixed either with PTEMF (50 mM PIPES [1,4-Piperazinediethanesulfonic acid], pH 7.2, 10 mM EGTA, 1 mM MgCl2, 0.2% Triton X-100, 4% PFA) at room temperature, or methanol at −20°C for ch-TOG staining. The following antibodies were used: (1) mouse monoclonals: CHC (X22, CRL-2228 ATCC), cMyc (9E10, CRL-1729 ATCC), α-tubulin (TAT1, Cancer Research UK), TACC3 (ab56595, Abcam), clathrin light chain (CON.1, C1985, Sigma), hexa-His (HIS.H8, ab18184, Abcam) and α-tubulin (DM1A, ab7291, Abcam), (2) rabbit polyclonals: ch-TOG (34032, Autogen Bioclear), GFP (Abcam) and β-tubulin (ab6046, Abcam). Aurora-A kinase inhibitor MLN8237 was from Selleck chemicals.
Light microscopy
Confocal imaging was done using a Leica confocal microscope SP2 with a × 63 (1.4 NA) oil-immersion objective. GFP and Alexa568 and Alexa633 were excited using an Ar/Kr 488 nm and the 543 and 594 lines of a He/Ne laser, respectively. DAPI were excited using a multiphoton laser. Excitation and collection of emission were performed separately and sequentially. For quantitative experiments, identical laser power and acquisition settings were used. Images were captured at a depth of 8-bit. Epifluorescence images were taken using a Nikon-TE2000U microscope with standard filter sets for visualisation of DAPI, GFP and Alexa568.
Molecular biology
TACC3 and ch-TOG were amplified from IMAGE clones (6148176 and 40072423) by PCR and inserted into pEGFP-C1 using XmaI–XbaI (TACC3) and BspEI–BamHI (ch-TOG). To make constructs to co-express GFP and shRNA for TACC3 or ch-TOG, the appropriate shRNA oligos (based on siRNA sequences in Gergely et al, 2003) were cloned into pBrain–SpH vector using BglII–HindIII, and then an ApaLI–NheI fragment was subcloned into pEGFP-C1. For control shRNA co-expressed with GFP, pBrain–GFP–CHC1 was used. For CHC knockdown plus GFP expression, pBrain–GFP–CHC4 was used. To express CHC17ΔNTD on a background of clathrin depletion, pBrain–GFP–CHC(331–1639)–CHC4 was used; all plasmids were available from previous work (Royle and Lagnado, 2006; Hood and Royle, 2009). To make constructs to express knockdown-proof GFP–TACC3 and TACC3 shRNA, an NheI–MluI fragment from GFP–TACC3KDP was inserted into pBrain–GFP–shTACC3. The sequence targeted by shRNA (tgctccacagcaaactctc) was first mutated silently using PCR (gttacAggTagGtcAtctg). GFP–TACC3(S558A) was made by site-directed mutagenesis and a pBrain version made as described. MBP–CHC(1–1074)–His6 and MBP–CHC(1–579)*–His6 (exon 9 deleted) were made by PCR amplification of CHC coding sequences without stop codons from existing constructs, followed by insertion into pMal–Pre-His at BamHI–NotI. All constructs were verified by restriction digest and automated DNA sequencing (GATC, UK).
Purification of the spindle clathrin complex
The protocol for enrichment of spindle-localised clathrin complexes was adapted from methods described elsewhere (Sauer et al, 2005; Neef et al, 2007). HeLa cells were synchronised in S phase by addition of 2 mM thymidine for 20 h. After release (7 h), nocodazole was added to 40 ng/ml and incubated for 12 h to arrest cells in prometaphase. Mitotic cells were collected by shake-off, and nocodazole was washed out. Cells were replated in DMEM and incubated for ∼40 min to allow spindles to reform. Taxol was added to 5 μg/ml and cells incubated for 5 min at 37°C. Cells were collected by centrifugation at 290 × g, and washed once in PBS, 5 μg/ml taxol, 0.1 M PMSF. Cells were incubated in a series of buffers, followed by collection of cells each time by centrifugation at 1000 × g for 4 min at room temperature. Supernatant fractions were collected in each case. Incubations were (1) 5 min in PTEML (50 mM PIPES, pH 7.2, 10 mM EGTA, 1 mM MgCl2, 0.2% Triton X-100, 2 μg/ml latrunculin B, 1 × protease inhibitor cocktail (Roche), 100 nM okadaic acid, 20 mM β-glycerophosphate, 5 μg/ml taxol), (2) 3 min in PTEML, (3) 3 min in PTEM (50 mM PIPES, pH 7.2, 10 mM EGTA, 1 mM MgCl2, 0.2% Triton X-100, 1 × protease inhibitor cocktail, 100 nM okadaic acid, 20 mM β-glycerophosphate, 5 μg/ml taxol) and (4) 3 min in PTEM. Spindle proteins, including CHC, were then released from the spindles by three successive incubations in PTNM (50 mM PIPES, pH 7.2, 300 mM NaCl, 10 mM EGTA, 1 mM MgCl2, 0.2% Triton X-100, 1 × protease inhibitor cocktail, 100 nM okadaic acid, 20 mM β-glycerophosphate, 5 μg/ml taxol).
Immunoprecipitations were carried out using equal total protein concentrations of fraction 1 (cytoplasmic material), combined fractions 5–7 (spindle elutions) or an asynchronous whole-cell lysate (interphase), as described previously (Hood and Royle, 2009), all spun at 20 000 × g, 4°C, 15 min before use to clear debris. Lysate (200–500 μg total protein for mass spectrometry experiments, 700 μg for immunoblotting experiment) was precleared using 10 μl protein-G sepharose, at 4°C, 1 h, rotating. Antibody (1–2 μg) was incubated with cleared lysate at 4°C, 1.5 h, rotating, then 10 μl protein-G sepharose (Sigma) added and incubation continued for 1 h. Beads were washed 5 times in 1-ml wash buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Igepal, 1 × protease inhibitor cocktail), then beads resuspended in Laemmli buffer. For mass spectrometry analysis, IPs were separated by 12% SDS–PAGE and gels stained by Coomassie brilliant blue. Lanes were cut into 11 slices and subjected to in-gel digestion using trypsin. For immunoblotting, 20% of the IP'd material was initially run alongside the equivalent of 3.5% of this in whole-cell lysate. For further analysis, 40% of IP'd material for c-myc, TACC3 and ch-TOG IPs was run alongside 2.5% of the CON.1 IP, and 9.5% each cleared lysate input.
Mass spectrometry
Digests were reduced in volume to ∼10 μl using a speedvac. Peptides were separated using a nanoACQUITY UPLC system (Waters), coupled to an LTQ Orbitrap XL mass spectrometer (Thermo Scientific) with a Proxeon nano-electrospray source. Five μl of the digest was injected into a 5 cm × 180 μm BEH-C18 symmetry trapping column (Waters) before being resolved on a 25 cm × 75 μm BEH-C18 column (Waters), in a 99–37.5% acetonitrile gradient in 0.1% formic acid over 39 min, with a flow rate of 400 μl/min. Full scan MS spectra (m/z 300–2000) were generated at 30 000 resolution, and the top five most intense ions were fragmented and subjected to MS/MS in the linear quadrapole ion trap. Ions were fragmented using collision-induced dissociation (collision energy 35%, 30 ms). Raw files were converted into .mgf files using Proteome Discoverer (Thermo), for database searching carried out using a local MASCOT server. Searches were carried out against HumanIPI database.
Biochemistry
MBP–CHC–His and MBP–His recombinant proteins were purified using Ni-NTA agarose (Qiagen) according to manufacturer's instructions. Proteins were exchanged into PEM buffer (with 1 mM EGTA)+1 mM DTT+10 mM KCl using Sephadex G-25 columns (GE Healthcare). Interaction of recombinant CHC fragments with MTs in vitro was assessed using the MT Binding Protein Spin-down Assay kit (Cytoskeleton Inc.). In brief, MTs were assembled from purified tubulin (5 mg/ml) in PEM buffer (80 mM PIPES, pH 7.0, 1 mM MgCl2, 0.5 mM EGTA) plus 1 mM GTP for 20 min at 35°C. MTs were immediately stabilised by 10-fold dilution into PEM+20 μM taxol. For each assay, 10 μg test protein was incubated with 20 μl stabilised MTs for 30 min at 30°C, then layered onto a cushion of PEM+50% glycerol, and centrifuged at 100 000 × g, 40 min, 25°C, to pellet out MTs and associated proteins. Control proteins were 16 μg MAP fraction or 7.5 μg BSA (Cytoskeleton Inc.). Where cell extracts were used, the test protein and 30 μg cell extract (at ∼7 mg/ml) supplemented with 2.5 mM Mg.ATP were incubated for 30 min at 30°C, before the incubation with MTs. Cell extracts were made from nocodazole-arrested HeLa cells in EBS (80 mM β-glycerophosphate, pH 7.2, 20 mM EGTA, 15 mM MgCl2, 100 mM sucrose, 1 mM DTT, 1 mM PMSF)+protease inhibitor cocktail, 100 nM okadaic acid and 5 mM Na3VO4, as described previously (Hutchins et al, 2004). For SDS–PAGE and Coomassie staining/immunoblotting, 40% of supernatant or pellet were analysed. To determine RNAi efficiency, HEK293 transfected with pBrain plasmids were treated with 1.2 mg/ml G418 to improve transfection efficiency and lysed 3 days post-transfection. Cells were collected in Laemmli buffer, and 25 μg protein was analysed by SDS–PAGE and immunoblotting.
Electron microscopy
HeLa cells were transfected with pBrain–GFP–CHC1 (control RNAi), pBrain–GFP–CHC4 (clathrin RNAi), pBrain–GFP–shTACC3 (TACC3 RNAi) or pBrain–GFP–shch–TOG (ch-TOG RNAi) and analysed 5 days post-transfection for clathrin and 2–3 days post-transfection for TACC3 and ch-TOG. Cells were reseeded onto glass-bottomed gridded dishes (MatTek Corporation) at a density 10 000 cells per dish. GFP-positive cells at metaphase were identified by light and epifluorescence microscopy using a Nikon TE2000U microscope with a × 20 objective, and their position on the grid was documented. Cells were returned to the incubator for 5 min and then fixed with 3% gluteraldehyde, 0.5% PFA in 0.05 M PB. The cells were embedded in resin, 80 nm sections were post-stained with uranyl acetate and lead citrate. For longitudinal sectioning, cells of interest were reidentified in the resin and sectioned using a Leica UC6 microtome. For orthogonal sectioning, the cell was identified and the position of the spindle poles noted. The cells were remounted so that sections would progressively move through the spindle along the spindle axis. Images were taken at 100 kV on an FEI Tecnai G2 Spirit BioTWIN microscope. Multiple image alignments of 4 × 4 images were taken at × 60 000 or × 87 000 for longitudinal sections, respectively. For orthogonal sections, a tilt series was taken at × 87 000, which allowed the visualisation of ∼80% of the MTs in the section. K-fibres were identified in longitudinal sections as bundles of MTs that terminated at a kinetochore and in orthogonal sections by progressive sectioning in the vicinity of chromosomes.
For immunogold labelling, HeLa cells expressing GFP–α-tubulin or GFP–CHC17 were synchronised. Metaphase cells were collected by mitotic shake-off, pelleted and resuspended in PTEML for 5 min. Partially purified spindles were pelleted at 1000 × g. The resulting material was resuspended in a small amount of PTEML, mixed with equal amounts of fixative (2% glutaraldehyde and 2% PFA in 0.05 M PB), and spotted onto gridded dishes. Spindles were probed with rabbit anti-GFP (Abcam) and either anti-tubulin or mouse anti-clathrin (X22) antibodies in separate samples. Anti-rabbit 5 nm and anti-mouse 10 nm gold-conjugated secondary antibodies were used to detect anti-GFP and anti-tubulin or anti-clathrin, respectively. Spindles showing GFP fluorescence were selected by light microscopy and embedded in resin. Spindles were sectioned (150–200 nm) and post-stained with uranyl acetate and lead citrate. Sections were then imaged as described above.
Quantification and data analysis
To measure spindle recruitment, a single confocal image was taken using the β-tubulin stain to choose the focal plane. For analysis, an ROI of 12 × 12 pixels (1.95 μm2) was selected over the spindle (β-tubulin channel), and one in the cytoplasm (GFP channel). The antibody signals were measured, adjusted for background (secondary antibody only control), and the mean fluorescence of the spindle ROI was divided by the cytoplasmic ROI. These ratios were normalised to a control: X22 staining of pBrain–GFP–CHC17(331–1639)–CHC4 or anti-TACC3 or anti-ch-TOG staining of cells treated with 0.3 mM MLN8237 for 40 min. RNAi efficiency was assessed by measuring mean immunofluorescence over whole interphase cells for clathrin, whole mitotic cells for TACC3 and over the mitotic spindle, adjusted for nonspecific cytoplasmic signal, for ch-TOG. To analyse the effects of expression of GFP-tagged TACC3 wild type or S558A mutant on the localisation of endogenous TACC3, transfected HEK293 cells were stained with anti-TACC3/A568 and anti-β-tubulin/A633. For analysis, the mean pixel intensity of TACC3 and β-tubulin on one half-spindle was measured.
To quantify the effects of overexpression of GFP-tagged proteins on other endogenous proteins, GFP and antibody signal over a half-spindle were measured. A cytoplasmic region of 30 × 30 pixels (3.04 μm2) was also measured. All spindle and cytoplasm measurements were adjusted for background. For the experiments to examine the time course of Aurora-A inhibition, spindle recruitment was measured in a similar way from images taken on an epifluorescence microscope. The ratios were scaled so that the recruitment at time 0 was 1 and at 40 min was 0.
Longitudinal section EM multiple image alignments were analysed using AnalySIS (Soft Imaging System). Total MT length, and lengths of ‘paired MTs' (regions where two single parallel MTs were within 80 nm of each other) were measured. Inter-MT bridges observed in paired MT regions were counted and their individual lengths measured. Bridges were defined according to published criteria (Hepler et al, 1970): (i) thickness of 2–5 nm, (ii) connecting two MTs with attachments clearly observed, (iii) variety of morphology and tilts. K-fibres were identified in longitudinal sections according to their termination at the kinetochore. In orthogonal sections, some K-fibres were followed to the kinetochore by serial sectioning, but this was not feasible for all K-fibres. To ensure that we only analysed K-fibres, we restricted our analysis to sections to 1 μ of the chromosome. In this region, non-kMTs have very few interactions with kMTs (McDonald et al, 1992) and so should not interfere with our analysis. Also, reconstructions of interpolar MTs in metaphase cells show only bundles of 2–6 MTs (Mastronarde et al, 1993), we therefore quantified only bundles of >6 MTs. To measure K-fibre cross-sectional area, an 80 nm perimeter was computed around clusters of annotated MTs and measured. MTs within this boundary were counted as part of the K-fibre. To measure inter-MT distances (orthogonal section analysis), a map of MT centres was created against a matrix (ImageJ), and the distance between each point was calculated in Microsoft Excel, from here the distance for each MT and its nearest neighbouring MT was selected. These centre-to-centre measurements were corrected by subtracting 20 nm to give the edge-to-edge distance. This allows comparison of the nearest neighbour analysis with the bridge length data. For clustering analysis, the co-ordinates of every MT in a K-fibre were plotted against the number of neighbouring MTs within 80 nm. This 3D data set was interpolated using a Voronoi transformation in Igor Pro (Wavemetrics). K-fibres were identified in orthoganal sections; Tukey plots show the median, the box length denotes the interquartile range, the box width denotes the sample size and the whiskers show the 10th and 90th percentile. To analyse the putative inter-MT bridge populations, all bridge measurements were pooled for each condition and probability density functions with a 1 nm resolution were generated. Multi-peak fitting was done using Multipeak Fitting 2 in Igor Pro 6.12. Peaks were located using a noise level of 0–3, smoothing factor of 1–3 and a minimum fraction of 0.05. The multi-peak fits, as shown in full in Supplementary Figure 4, gave the lowest χ2-value of goodness of fit and were also a better fit than any single type of fit.
Immunogold labelling was quantified by recording four reference co-ordinates for each gold particle: (a) gold particle, (b) nearest MT centre, (c) nearest neighbouring MT centre and (d) nearest bridge (shortest route). The following distances were calculated: edge of gold particle to the nearest MT centre (from a to b minus the radius of gold particle), edge of gold particle to the nearest bridge (from a to d minus the radius of gold particle). Specific labelling was taken as gold label within 16 nm of a structure (Ottersen, 1989).
Normally distributed data were analysed by one-way ANOVA with Tukey's post hoc test. Data that were not normally distributed were compared by Kruskal–Wallis. For comparison of two data sets, Student's unpaired t-test was used. Calculations were performed with Minitab 15.1 (Minitab). Figures were assembled using Igor Pro (Wavemetrics), Chimera (UCSF) and Adobe Photoshop.
Supplementary Material
Acknowledgments
We thank Liam Cheeseman for early work on MLN8237 and TACC3(S558A). We are grateful to Maria Blixt for technical assistance and Francis Barr, Ulli Gruneberg and Anna Korniejewska for helpful suggestions. This work was supported by a Career Establishment Award from Cancer Research UK (C25425/A8722). DGB is the recipient of a Wellcome Trust Prize Studentship and IAP is a Royal Society University Research Fellow.
Author contributions: DGB performed the EM with input from IAP. FEH identified the complex and tested the order of recruitment. SJR contributed the Aurora-A experiments, wrote the paper and directed the work. All authors discussed the work, analysed the data and commented on the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barr AR, Gergely F (2007) Aurora-A: the maker and breaker of spindle poles. J Cell Sci 120: 2987–2996 [DOI] [PubMed] [Google Scholar]
- Barr AR, Gergely F (2008) MCAK-independent functions of ch-Tog/XMAP215 in microtubule plus-end dynamics. Mol Cell Biol 28: 7199–7211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey T (2010) A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell 142: 420–432 [DOI] [PubMed] [Google Scholar]
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE (2001) Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol 17: 517–568 [DOI] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA (2008) XMAP215 is a processive microtubule polymerase. Cell 132: 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L, Becker B, Carney B (2009) TOGp regulates microtubule assembly and density during mitosis and contributes to chromosome directional instability. Cell Motil Cytoskeleton 66: 535–545 [DOI] [PubMed] [Google Scholar]
- Charrasse S, Schroeder M, Gauthier-Rouviere C, Ango F, Cassimeris L, Gard DL, Larroque C (1998) The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J Cell Sci 111(Part 10): 1371–1383 [DOI] [PubMed] [Google Scholar]
- Compton DA (2000) Spindle assembly in animal cells. Annu Rev Biochem 69: 95–114 [DOI] [PubMed] [Google Scholar]
- Fu W, Tao W, Zheng P, Fu J, Bian M, Jiang Q, Clarke PR, Zhang C (2010) Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. J Cell Sci 123 (Part 21): 3645–3651 [DOI] [PubMed] [Google Scholar]
- Gergely F, Draviam VM, Raff JW (2003) The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev 17: 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F, Karlsson C, Still I, Cowell J, Kilmartin J, Raff JW (2000a) The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc Natl Acad Sci USA 97: 14352–14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F, Kidd D, Jeffers K, Wakefield JG, Raff JW (2000b) D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J 19: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goud B, Huet C, Louvard D (1985) Assembled and unassembled pools of clathrin: a quantitative study using an enzyme immunoassay. J Cell Biol 100: 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK, McIntosh JR, Cleland S (1970) Intermicrotubule bridges in mitotic spindle apparatus. J Cell Biol 45: 438–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Dannhauser PN, Groos S, Hinrichsen L, Curth U, Ungewickell EJ (2010) A comparison of GFP-tagged clathrin light chains with fluorochromated light chains in vivo and in vitro. Traffic 11: 1129–1140 [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW (2009) Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 10: 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood FE, Royle SJ (2009) Functional equivalence of the clathrin heavy chains CHC17 and CHC22 in endocytosis and mitosis. J Cell Sci 122: 2185–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M (2010) Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol 189: 739–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JR, Moore WJ, Hood FE, Wilson JS, Andrews PD, Swedlow JR, Clarke PR (2004) Phosphorylation regulates the dynamic interaction of RCC1 with chromosomes during mitosis. Curr Biol 14: 1099–1104 [DOI] [PubMed] [Google Scholar]
- Jin AJ, Nossal R (2000) Rigidity of triskelion arms and clathrin nets. Biophys J 78: 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Noetzel TL, Pelletier L, Mechtler K, Drechsel DN, Schwager A, Lee M, Raff JW, Hyman AA (2005) Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J Cell Biol 170: 1047–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T, Harrison SC (1981) Protein organization in clathrin trimers. Cell 23: 755–761 [DOI] [PubMed] [Google Scholar]
- Knight AS, Notaridou M, Watson RJ (2009) A Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene 28: 1737–1747 [DOI] [PubMed] [Google Scholar]
- LeRoy PJ, Hunter JJ, Hoar KM, Burke KE, Shinde V, Ruan J, Bowman D, Galvin K, Ecsedy JA (2007) Localization of human TACC3 to mitotic spindles is mediated by phosphorylation on Ser558 by Aurora A: a novel pharmacodynamic method for measuring Aurora A activity. Cancer Res 67: 5362–5370 [DOI] [PubMed] [Google Scholar]
- Lin CH, Hu CK, Shih HM (2010) Clathrin heavy chain mediates TACC3 targeting to mitotic spindles to ensure spindle stability. J Cell Biol 189: 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Compton DA (2008) Structural and regulatory roles of nonmotor spindle proteins. Curr Opin Cell Biol 20: 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maro B, Johnson MH, Pickering SJ, Louvard D (1985) Changes in the distribution of membranous organelles during mouse early development. J Embryol Exp Morphol 90: 287–309 [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR (1993) Interpolar spindle microtubules in PTK cells. J Cell Biol 123: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KL, O'Toole ET, Mastronarde DN, McIntosh JR (1992) Kinetochore microtubules in PTK cells. J Cell Biol 118: 369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton DA (1999) The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol 147: 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R, Gruneberg U, Kopajtich R, Li X, Nigg EA, Sillje H, Barr FA (2007) Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol 9: 436–444 [DOI] [PubMed] [Google Scholar]
- O'Brien LL, Albee AJ, Liu L, Tao W, Dobrzyn P, Lizarraga SB, Wiese C (2005) The Xenopus TACC homologue, maskin, functions in mitotic spindle assembly. Mol Biol Cell 16: 2836–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto CT, McKinney J, Jeng YY (2000) Clathrin in mitotic spindles. Am J Physiol Cell Physiol 279: C369–C374 [DOI] [PubMed] [Google Scholar]
- Ottersen OP (1989) Quantitative electron microscopic immunocytochemistry of neuroactive amino acids. Anat Embryol (Berl) 180: 1–15 [DOI] [PubMed] [Google Scholar]
- Pearse BM (1975) Coated vesicles from pig brain: purification and biochemical characterization. J Mol Biol 97: 93–98 [DOI] [PubMed] [Google Scholar]
- Peset I, Seiler J, Sardon T, Bejarano LA, Rybina S, Vernos I (2005) Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J Cell Biol 170: 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peset I, Vernos I (2008) The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol 18: 379–388 [DOI] [PubMed] [Google Scholar]
- Peterman EJ, Scholey JM (2009) Mitotic microtubule crosslinkers: insights from mechanistic studies. Curr Biol 19: R1089–R1094 [DOI] [PubMed] [Google Scholar]
- Rieder CL (2005) Kinetochore fiber formation in animal somatic cells: dueling mechanisms come to a draw. Chromosoma 114: 310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle SJ, Bright NA, Lagnado L (2005) Clathrin is required for the function of the mitotic spindle. Nature 434: 1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle SJ, Lagnado L (2006) Trimerisation is important for the function of clathrin at the mitotic spindle. J Cell Sci 119: 4071–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein B, Larripa K, Sommi P, Mogilner A (2009) The elasticity of motor-microtubule bundles and shape of the mitotic spindle. Phys Biol 6: 16005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer G, Korner R, Hanisch A, Ries A, Nigg EA, Sillje HH (2005) Proteome analysis of the human mitotic spindle. Mol Cell Proteomics 4: 35–43 [DOI] [PubMed] [Google Scholar]
- Schweitzer JK, Burke EE, Goodson HV, D'Souza-Schorey C (2005) Endocytosis resumes during late mitosis and is required for cytokinesis. J Biol Chem 280: 41628–41635 [DOI] [PubMed] [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM (1999) The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol 144: 125–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Vallenius T, Geers EF, Greene L, Makela TP, Medema RH (2010) Cyclin G-associated kinase promotes microtubule outgrowth from chromosomes during spindle assembly. Chromosoma 119: 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E, Branton D (1981) Assembly units of clathrin coats. Nature 289: 420–422 [DOI] [PubMed] [Google Scholar]
- Warren G (1993) Membrane partitioning during cell division. Annu Rev Biochem 62: 323–348 [DOI] [PubMed] [Google Scholar]
- Witt PL, Ris H, Borisy GG (1981) Structure of kinetochore fibers: microtubule continuity and inter-microtubule bridges. Chromosoma 83: 523–540 [DOI] [PubMed] [Google Scholar]
- Wong J, Fang G (2006) HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol 173: 879–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Ishidao T, Nomura T, Shinagawa T, Tanaka Y, Yonemura S, Ishii S (2008) A B-Myb complex containing clathrin and filamin is required for mitotic spindle function. EMBO J 27: 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Lau E, Schwarzenbacher R, Bossy-Wetzel E, Jiang W (2006) Spatiotemporal control of spindle midzone formation by PRC1 in human cells. Proc Natl Acad Sci USA 103: 6196–6201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.