Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers
This study investigates the structural and mechanistic basis for AMPAR subunit assembly and reveals that the N-terminal domain regulates the formation of diverse pools of AMPAR tetramers.
Keywords: AMPA receptor function, AMPA receptor assembly, N-terminal domain, crystallography
Abstract
The assembly of AMPA-type glutamate receptors (AMPARs) into distinct ion channel tetramers ultimately governs the nature of information transfer at excitatory synapses. How cells regulate the formation of diverse homo- and heteromeric AMPARs is unknown. Using a sensitive biophysical approach, we show that the extracellular, membrane-distal AMPAR N-terminal domains (NTDs) orchestrate selective routes of heteromeric assembly via a surprisingly wide spectrum of subunit-specific association affinities. Heteromerization is dominant, occurs at the level of the dimer, and results in a preferential incorporation of the functionally critical GluA2 subunit. Using a combination of structure-guided mutagenesis and electrophysiology, we further map evolutionarily variable hotspots in the NTD dimer interface, which modulate heteromerization capacity. This ‘flexibility' of the NTD not only explains why heteromers predominate but also how GluA2-lacking, Ca2+-permeable homomers could form, which are induced under specific physiological and pathological conditions. Our findings reveal that distinct NTD properties set the stage for the biogenesis of functionally diverse pools of homo- and heteromeric AMPAR tetramers.
Introduction
Ion channels largely assemble into hetero-oligomers. Subunit heteromerization greatly expands the functional repertoire and is mediated by specific assembly domains (Schwappach, 2008). Ionotropic glutamate receptors (iGluRs), the main mediators of excitatory neurotransmission, are either obligatory (NMDA subtype) or preferential (AMPA, kainate types) heteromers (Traynelis et al, 2010). The subunit combination dictates fundamental signalling parameters such as gating kinetics and ion conductance, which shape synaptic physiology (Erreger et al, 2004). AMPA-type iGluRs (AMPARs), in addition to initiating excitatory signalling, adjust synaptic strength via dynamic postsynaptic trafficking (Malinow and Malenka, 2002; Greger and Esteban, 2007; Shepherd and Huganir, 2007), which underlies various forms of experience-dependent synaptic plasticity (Kessels and Malinow, 2009). These processes are mediated by mobilization and recruitment of distinct pools of AMPAR heteromers.
AMPARs assemble from four subunits, GluA1–4, in various stoichiometries (Hollmann and Heinemann, 1994). Incorporation of GluA2 blocks Ca2+ flux through AMPARs; GluA2-containing receptors predominate throughout the brain (Isaac et al, 2007). The central role of AMPAR heteromer formation is well illustrated in the CA1 subfield of hippocampus. CA1 pyramidal neurons express comparable levels of GluA1 and GluA2 (but little GluA3 and no GluA4; Tsuzuki et al, 2001), which almost exclusively co-assemble (Lu et al, 2009). Pathological conditions can alter this balance, resulting in Ca2+-permeable, GluA2-lacking receptors and excitotoxicity (Kwak and Weiss, 2006; Liu and Zukin, 2007). GluA2-lacking receptors can also operate under specific physiological conditions (Cull-Candy et al, 2006), and have been detected in midbrain dopamine neurons, where enhanced signalling through these receptors is implicated in drug addiction (Carlezon and Nestler, 2002; Kauer and Malenka, 2007). How neurons control selective combinatorial assembly of subunits to generate this plethora of distinct compositional and functional phenotypes remains an open question. Furthermore, the composition of Ca2+-permeable (GluA2-lacking) AMPARs has not been established.
AMPAR assembly is mediated by three domains engaging in distinct subunit interactions in the endoplasmic reticulum (ER): the N-terminal domain (NTD), the ligand-binding domain (LBD) and the membrane-embedded ion channel (Madden, 2002; Greger et al, 2007). The first assembly step is initiated by the NTD, which spans ∼50% of polypeptide sequence and primes subunit dimerization via an extensive bipartite interface (Clayton et al, 2009; Jin et al, 2009; Kumar et al, 2009). The NTD is therefore expected to provide a major assembly platform during iGluR biogenesis (Hansen et al, 2010). The second assembly step, association of dimers into tetramers, is modulated by Q/R editing in the channel pore (Greger et al, 2003). Relative to the NTD scaffold, contacts mediated by the LBD are weak but are regulated by RNA recoding events (Greger et al, 2006; Penn et al, 2008; Penn and Greger, 2009). The interplay between these domains and their roles in the assembly of functionally distinct receptor heteromers remain elusive (Sukumaran et al, 2011a).
Here, we report that AMPAR heteromerization is driven by unexpectedly diverse, subunit-selective NTD interactions. A sensitive, fluorescence-based biophysical approach permitted direct quantification of homo- and heteromeric associations along the NTD axis. We find that GluA2 is dominantly incorporated into dimers at the level of the NTD and that assembly follows two distinct pathways: obligatory heterodimerization in case of GluA3 and preferential heterodimerization in GluA1 and GluA4. In the latter pathway, the non-constitutive, equilibrium-type assembly between homo- and heteromeric GluA1 NTDs potentially provides an explanation for the existence of GluA1 homomeric receptors, which have an enigmatic origin but have been detected under various conditions. Moreover, structural and functional mapping of the GluA1/2 NTD dimer interface identified evolutionarily variable hotspots mediating the balanced assembly function of the NTD, providing a mechanistic basis for the formation of Ca2+-permeable AMPARs. Therefore, a pivotal assembly function encoded in the NTD provides the organizing principle for the formation of diverse pools of AMPAR tetramers.
Results
Whereas tri-heteromeric NMDARs have been described (Hatton and Paoletti, 2005), an ongoing question for AMPARs is whether heteromers form initially at the level of the dimer or at the level of the tetramer via assembly of two different homodimers (Mansour et al, 2001; Gill and Madden, 2006). Solving this problem would hold the key to understanding the formation and organization of AMPAR heteromers. Because the NTD is believed to have a strategic role in subfamily-selective assembly (Leuschner and Hoch, 1999; Ayalon and Stern-Bach, 2001), we devised a sensitive, fluorescence-based ultracentrifugation assay to directly measure association of AMPAR NTD homo- and heterodimers.
AMPAR subunit NTDs feature vastly different assembly properties
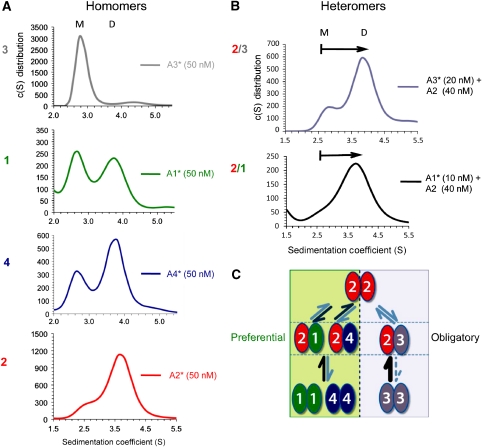
GluA1–4 AMPAR subunit NTDs, purified to homogeneity, were subjected to analytical ultracentrifugation with fluorescence detection (AU-FDS) (Rajagopalan et al, 2008). This technique facilitates measurements of monomer/dimer equilibria in the nanomolar range (MacGregor et al, 2004), and provides a means to quantify both homo- and heteromeric affinities. NTDs were fluorescently labelled at the N-terminus with 5,6-carboxyfluorescein (FAM) and subjected to velocity sedimentation. As illustrated in Figure 1A, this approach revealed a surprising spectrum of subunit-selective oligomerization properties. At a concentration of 50 nM labelled protein, both mono- and dimeric species were detected for the GluA1 NTD. GluA2 predominantly sedimented as a dimer, whereas the GluA4 NTD showed an intermediate phenotype. In stark contrast, GluA3 was exclusively monomeric (Figure 1A). This sedimentation behaviour was protein-concentration dependent; for example, a greater proportion of GluA2 monomers were detected when lowering the input from 50 nM to 2 nM (Supplementary Figure S1A). By titrating the protein concentration, we were able to derive Kd values of dimer dissociation from multiple runs. We find that AMPAR NTDs exhibit a wide range of dimeric affinities, with a ∼1000-fold disparity between the GluA2 and GluA3 NTDs (1.8 versus 1200 nM, respectively). GluA1 and GluA4 fell between these extremes, with an estimated Kd of ∼100 and 10 nM, respectively (Table I).
Figure 1.
GluA1–4 NTDs exhibit markedly different assembly behaviours. (A) Homomeric NTD assembly properties. Sedimentation coefficient (S) distributions were obtained for FAM-labelled GluA1–4 NTDs at 50 nM protein concentration. Labelled species are denoted with an asterisk (*). Continuous distribution c(S) models revealed a spectrum of homomeric affinities, from low (top) to high (bottom). Two species at ∼2.6S (monomer) and 3.7S (dimer) could be detected for all NTDs. GluA1* (green trace) showed incomplete homodimerization, as indicated by presence of the monomer peak at 2.6S. In contrast, GluA2* (red trace) and GluA4* (blue trace) were mostly dimeric, whereas GluA3* was mostly monomeric (grey trace). (B) Heteromeric NTD assembly properties. c(S) distributions are shown for FAM-labelled GluA3* (top) or GluA1* (bottom) at monomeric concentrations mixed with non-labelled GluA2 NTD. Adding non-labelled GluA2 was sufficient to shift the labelled monomeric species to a dimeric peak, indicating the presence of heterodimers. (C) Summary of the distinct assembly properties of GluA1–4 NTDs. In GluA1, GluA4 (left pathway), and GluA2 (top), heteromerization is favoured but must compete with possible homomerization, leading to preferential heterodimers; whereas in GluA3 (right pathway), homomers are disfavoured and therefore the NTD exhibits obligatory heteromerization. See also Supplementary Figures S1 and S2.
Table 1. Association behaviour of NTDs assayed by analytical ultracentrifugation.
| NTD protomers | Kd (nM)a | n measurements | s.e.m. |
|---|---|---|---|
| GluA1b | 98 | 8 | 36 |
| GluA2b | 1.8 | 12 | 0.2 |
| GluA3b | 1200 | 12 | 500 |
| GluA4b | 10.2 | 9 | 1.1 |
| GluA1b/GluA2 | 0.4 | 10 | 0.2 |
| GluA3b/GluA1 | 38 | 9 | 6.0 |
| GluA3b/GluA2 | 1.3 | 4 | 0.1 |
| GluA4b/GluA2 | 3.3 | 9 | 0.15 |
| GluA2-T78Ab | 37.1 | 7 | 3.7 |
| GluA2-N54Ab | 0.5 | 4 | 0.2 |
| aDissociation constant. | |||
| bFAM-labelled. | |||
Selective NTD affinities drive different assembly pathways
The vast, ∼1000-fold range of homomeric affinities suggests different overall assembly behaviours for GluA1–4. As AU-FDS allowed us to test whether AMPAR subunits can heteromerize at the level of the NTD, and thus at the level of dimers, we asked how the drastic differences in homomeric affinities affect heteromer formation. Heteromers were assayed by holding the labelled species (denoted with an asterisk (*)) constant at monomeric concentrations and adding unlabelled NTD (Supplementary Figure S2). The presence of heteromers was indicated by a shift of the labelled monomeric species to a dimeric peak (Figure 1B). Kds were calculated by titration of unlabelled assembly partner. We derived a tight GluA1*/2 heteromeric Kd of ∼0.5 nM. Similarly, titration of GluA2 onto GluA3* and GluA4* revealed low Kds of 1.3 and 3.3 nM, respectively (Table I). Therefore, AMPAR NTDs heteromerize with greater affinity, when compared with their respective homomeric Kds (Table I). This result provides a ready explanation for the predominance of AMPAR heteromers, which was recently assessed in hippocampal pyramidal neurons where GluA1/2 receptors prevail (Lu et al, 2009).
These data provide a roadmap for AMPAR heteromer assembly. First, they reveal two distinct AMPAR assembly pathways. Because of their relatively tight homomeric affinities, GluA1, GluA2 and GluA4 homomers are still likely to occur, but the existence of GluA3 homomers is unlikely in the presence of other subunits, especially GluA2 (Table I). Therefore, GluA1 and GluA4 preferentially heterodimerize with GluA2, whereas GluA2/3 dimers are obligatory (Figure 1C, bottom half). We note that GluA3 can form homomers in the absence of other assembly partners (e.g., Suzuki et al, 2008); hence, ‘obligatory' heteromerization pertains only to GluA3 expressed in neurons. Interestingly, this result also illustrates an apparent assembly dominance of GluA2. Preferential formation of GluA2-containing AMPARs is a well-established phenomenon; our results suggest that this can be determined at early stages of receptor biogenesis via the NTD. In addition to the edited Q/R site prolonging GluA2 ER dwell time (Greger et al, 2002; Greger and Esteban, 2007; Sukumaran et al, 2011a), this finding could explain why AMPARs harbouring this functionally critical subunit widely dominate throughout neuronal populations (Isaac et al, 2007). In the preferential assembly pathway, GluA2 appears to have greater affinity for GluA1 than for GluA4 (Figure 1C, top half). Therefore, the equilibrium between homo- and heterodimers of these subunits with GluA2 will differ, which ultimately could also have consequences for the arrangement and number of GluA2 subunits in a tetramer (Washburn et al, 1997).
Even though GluA2 has a dominant functional and assembly role, GluA2-lacking, Ca2+-permeable AMPARs have been detected and have a central physiological function (Cull-Candy et al, 2006; Kauer and Malenka, 2007). Their existence can be explained by the balanced assembly properties of the GluA1 (and GluA4) NTD (Figure 1A) and could be induced by increased GluA1 expression (Thiagarajan et al, 2005; Sutton et al, 2006; Aoto et al, 2008; see also Supplementary Figure S6). Hippocampal inter-neurons prominently express both GluA1 and 3 in the absence of GluA2 (Catania et al, 1998; Tsuzuki et al, 2001). In addition, a minor fraction of GluA1/3 co-precipitate from CA1/2 tissue, which increases in GluA2 knockout mice (Wenthold et al, 1996; Sans et al, 2003). The existence of pools of kinetically distinct, Ca2+-permeable AMPAR heteromers is therefore likely. Indeed, GluA1/3 NTDs produce a tight heteromeric Kd (∼38 nM; Table I) of greater affinity than their respective homomeric Kds. Similarly, GluA4/3 assemble in the low nanomolar range (data not shown). These data further suggest an obligatory heteromeric assembly mode for GluA3 in GluA2-lacking neurons and for the existence of functionally diverse Ca2+-permeable AMPAR populations. Importantly, the closely related GluK2 kainate receptor NTD does not shift GluA3* towards the dimer (data not shown); therefore, subfamily-specific assembly occurs at the level of the NTD. In summary, our results reveal that (1) MPAR heteromers form at the level of the dimer (which will affect the spatial arrangement of the channel tetramer), (2) subunit-selective assembly is driven by vastly distinct NTD associations, and (3) it results in preferential or obligatory AMPAR heteromers (Figure 1C).
Properties of the NTD dimer interface
The sedimentation data show that NTD heterodimers preferentially include GluA2 (Table I). This finding offers a solution for the long-standing question of how this key subunit is incorporated into the majority of AMPARs across neuronal populations; however, mechanistically, this poses an apparent paradox—how does the tight GluA2 homodimer permit selective heteromerization? Furthermore, our data also suggest that balancing between homo- and heteromerization via the NTD interface may explain the biogenesis of GluA1 homomers and possibly other populations of Ca2+-permeable AMPARs (Figure 1C). In vivo, neuronal populations tightly regulate synaptic recruitment of Ca2+-impermeable and Ca2+-permeable AMPARs; how these assembly modes are balanced is, therefore, critical for signalling dynamics (Toth and McBain, 1998; Cull-Candy et al, 2006; Liu and Zukin, 2007). To address these questions, we set out to pinpoint assembly determinants within the NTD interface.
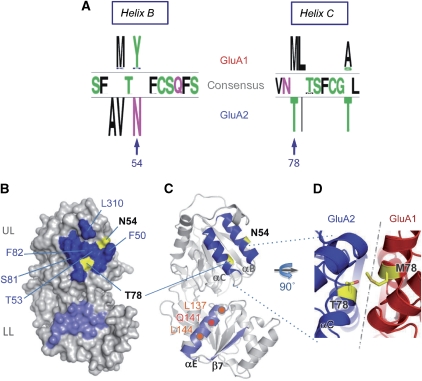
First, we mapped the evolutionary conservation within the GluA2 dimer interface, guided by our high-resolution GluA2 NTD structure (Supplementary Table I; Greger et al, 2009; Sukumaran et al, 2011b) and by position-specific comparative analysis of evolutionary conservation (Wuster et al, 2010). As conserved residues at interfaces are expected to maintain the affinity of subunit interactions (Landgraf et al, 2001), we first computed evolutionary conservation by generating an alignment of all currently available vertebrate AMPAR paralogs (>100 sequences). Overall, the NTD is the most divergent segment of the receptor, with a sequence identity score of 52–61% between the paralogs. This is in stark contrast to the conserved LBD/ion channel portion, which shows >80% sequence identity.
Within the NTD, the upper lobe (UL) interface is most highly conserved, indicating that contacts mediated by this segment are of functional importance. However, two UL interface positions featured a subunit-specific pattern of conservation: N54 at the upper edge and T78, which forms a polar contact in the core of the interface (Figure 2A and B). These residues are conserved between GluA2–4, whereas in GluA1, position 54 is occupied by a moderately conserved tyrosine and position 78 by a conserved methionine (Figure 2A and D). The fact that these two positions show systematic alteration between assembly partners and localize to the assembly interface indicates that they may be important for the specificity of interaction (Lichtarge et al, 1996). Therefore, we reasoned that these two ‘hotspots' might encode critical assembly determinants.
Figure 2.
Primary, secondary, and tertiary structure characteristics of the GluA2 NTD dimer interface. (A) Position-specific patterns of conservation in the GluA1 and GluA2 upper lobe (UL) interface. GluA1-specific, GluA2-specific, and consensus residues, generated from partitioning an alignment of 34 GluA1 and 33 GluA2 sequences, are indicated for positions in helices B and C, the major contributors of the UL interface. The top loop (not shown) is fully conserved between GluA1 and GluA2. N54 and T78 in GluA2 were identified as potential key determinants for assembly specificity, because they are located at the interface and are systematically mutated between GluA1 and GluA2. (B) The molecular surface of the GluA2 NTD is shown with UL and LL interfaces coloured dark and light blue, respectively, with the contribution of the variable interface residues shown in yellow. (C) Secondary structure contributions to the dimer interface. The NTD is oriented as in B, highlighting helices B and C in the UL-dimer and helix E and sheet 7 in the LL-dimer interfaces, respectively. Positions of N54 and T78 are in yellow, and important LL-interface contacts (L137, Q141, L144) on helix E are shown in orange. (D) Model of a potential heteromeric GluA1/GluA2 interface. A homology model of GluA1, associated with the crystal structure of GluA2 (3HSY) is shown, zoomed in on the T78 (A2)–M78 (A1) interaction in yellow.
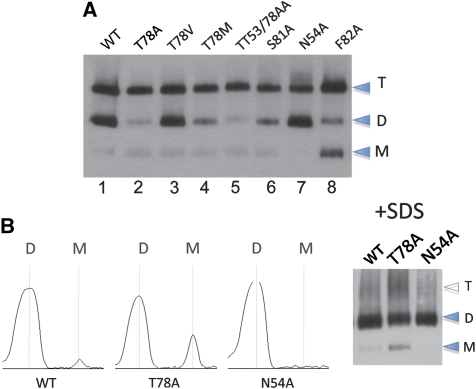
We first tested the impact of NTD dimer contacts on the stability of the whole receptor. Recent structural insights reveal that NTD associations are prominent contact points within the receptor and NTD dimers are major assembly interfaces in AMPAR dimers (Sobolevsky et al, 2009; Nakagawa, 2010; Shanks et al, 2010). Guided by the structure and our evolutionary analysis, we designed mutations in the assembly-critical GluA2 NTD interface. Blue native (BN) PAGE resolved full-length GluA2, expressed in HEK 293T cells, into stable mono-, di-, and tetrameric species (Figure 3A; Greger et al, 2003); this pattern was altered drastically by mutation. For example, F82A substantially weakened the tetrameric complex (Figure 3A, lane 8; see also Figure 2B). This is to be expected, as targeting the conserved hydrophobic core of the interface will destabilize the receptor non-specifically (Lichtarge et al, 1996). Similarly, targeting the variable hotspots markedly altered tetramer stability (Figure 3A, lanes 2–5, 7). Mild SDS treatment (1% SDS for 10 min at 30°C), which dissociates tetramers and thus facilitates a more direct assessment of dimer stability (Penn et al, 2008), uncovered bidirectional differences for the two hotspots, whereas for N54A, monomers were below detection limit and T78A displayed a five- to sixfold greater fraction of monomers relative to wild type (WT; Figure 3B). These opposing dimer stabilities suggest that mutation at N54 (stabilizing) and T78 (destabilizing) may affect assembly by mechanistically different routes, which in turn may underlie assembly specificity.
Figure 3.
Mutation in the NTD perturbs assembly of the full-length receptor. (A) Blue native PAGE (BN-PAGE) analysis of NTD interface mutants shows differential migration patterns. HEK293T cell suspensions of GluA2 flop (R/G unedited, Q/R edited) wild type (WT) and mutants (indicated on the top) were separated on 4–12% BN-PAGE and visualized by western blotting. Monomeric (M), dimeric (D), and tetrameric (T) assembly intermediates are denoted. Note the different phenotypes of N54A and T78A, as well as the intermediate phenotype of the N54A/T78A double mutant. (B) Mild SDS treatment before BN-PAGE dissociates tetramers and reveals that N54A (stabilizing) and T78A (destabilizing) have a bidirectional effect on tetramer and dimer assembly, relative to the wild type. The gel shown on right was scanned and bands computed as intensity peaks using ImageJ software (NIH).
Functional scanning of the GluA2 NTD dimer interface
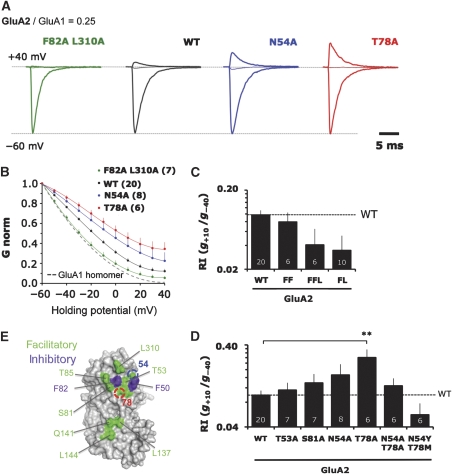
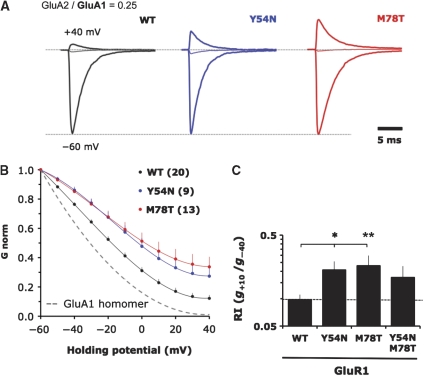
To extend our findings to heteromeric assembly, we employed a sensitive functional assay to characterize the determinants in the NTD dimer interface. We focused on GluA1/2, which are prominent heteromers throughout the brain and are the major AMPAR species in hippocampus (Lu et al, 2009). Guided by the structure, we introduced mutations into GluA2, which were co-expressed with GluA1 in HEK293T cells. The extent of GluA1/2 heteromerization was assayed by measuring current/voltage (I/V) relationships for an expression level of GluA2 giving optimal dynamic range (Supplementary Figure S3A). GluA1 homomers display inward rectification at positive holding potentials, reflecting blockade of the channel pore by intracellular polyamines. This block is alleviated by co-assembly with GluA2 edited at the Q/R site (Supplementary Figure S3B; Isaac et al, 2007). Thus, an increase of the rectification index (RI), computed as the ratio of slope conductance at +10 and −40 mV (g+10/g−40), indicates increased heteromer formation.
We first targeted the lower lobe (LL) of the NTD—residues in the LL interface are identical between GluA1 and GluA2. The inner face of helix E, including L137 and L144, forms a hydrophobic patch (Figure 2C). Breaking these hydrophobic contacts via the L137A/L144A double mutant elevated the heteromerization competence by 2.6-fold (Table II). In addition Q141A, which will abrogate a polar link with N158 and Y131, facilitated heteromerization (3.1-fold; Table II). Weakening the GluA2 LL interface, therefore, increased co-assembly with GluA1.
Table 2. Heteromerization of GluA1/GluA2 NTD mutants.
| GluA1 | GluA2 | Rectification index (RI) | Fold change in heteromer assembly competenceb | Reversal potential (Erev) | n | |||
|---|---|---|---|---|---|---|---|---|
| Average | LBa | UBa | Mean | s.e.m. | ||||
| WT | — | 0.027 | 0.025 | 0.029 | — | — | — | 4 |
| WT | WT | 0.097 | 0.085 | 0.111 | 1.0 | 6.1 | 0.5 | 20 |
| WT | F50A F82A | 0.079 | 0.060 | 0.104 | 0.8 | 5.7 | 0.8 | 6 |
| WT | F50A F82A L310A | 0.041 | 0.026 | 0.065 | 0.3 | 2.7 | 0.9 | 6 |
| WT | F82A L310A | 0.035 | 0.023 | 0.053 | 0.2 | 4.5 | 0.7 | 10 |
| WT | L310A | 0.173 | 0.114 | 0.262 | 2.2 | 4.9 | 1.0 | 5 |
| WT | T53A | 0.113 | 0.092 | 0.138 | 1.2 | 3.5 | 1.2 | 7 |
| WT | N54A | 0.172 | 0.127 | 0.233 | 2.2 | 5.4 | 0.4 | 8 |
| WT | T78A | 0.280 | 0.223 | 0.352 | 4.7 | 2.8 | 1.4 | 6 |
| WT | S81A | 0.138 | 0.107 | 0.177 | 1.6 | 5.6 | 0.5 | 7 |
| WT | T53A T78A | 0.137 | 0.114 | 0.166 | 1.6 | 3.7 | 0.9 | 10 |
| WT | N54A T78A | 0.127 | 0.103 | 0.157 | 1.4 | 5.0 | 1.2 | 6 |
| WT | N54Y T78M | 0.056 | 0.041 | 0.078 | 0.5 | 5.6 | 1.6 | 6 |
| M78T | WT | 0.230 | 0.178 | 0.299 | 3.4 | 5.9 | 0.8 | 13 |
| Y54N | WT | 0.207 | 0.166 | 0.258 | 2.9 | 5.0 | 0.1 | 9 |
| M78T Y54N | WT | 0.171 | 0.128 | 0.228 | 2.2 | 3.2 | 1.0 | 4 |
| WT | T78I | 0.041 | 0.026 | 0.063 | 0.3 | 5.4 | 0.7 | 5 |
| WT | T78L | 0.110 | 0.076 | 0.159 | 1.2 | 5.2 | 0.4 | 3 |
| WT | T78M | 0.109 | 0.089 | 0.134 | 1.2 | 5.3 | 0.1 | 7 |
| WT | T78V | 0.183 | 0.141 | 0.239 | 2.4 | 6.3 | 0.5 | 5 |
| M78T | T78A | 0.169 | 0.120 | 0.238 | 2.1 | 5.4 | 0.6 | 6 |
| M78T | T78M | 0.171 | 0.126 | 0.231 | 2.2 | 4.6 | 0.8 | 6 |
| M78T | T78V | 0.143 | 0.099 | 0.207 | 1.7 | 7.1 | 1.3 | 8 |
| M78T | T78A T53A | 0.120 | 0.086 | 0.166 | 1.3 | 5.8 | 0.9 | 9 |
| WT | L137A L144A | 0.191 | 0.145 | 0.253 | 2.6 | 5.4 | 0.6 | 6 |
| WT | Q141A | 0.216 | 0.147 | 0.318 | 3.1 | 6.2 | 1.2 | 5 |
| Abbreviations: LB, lower bound; UB, upper bound. | ||||||||
| aSee Materials and methods. | ||||||||
| bHeteromer assembly competence is defined as the equivalent wild-type GluA2/1 expression ratio required to give mutant rectification index. Determined from the titration curve in Figure S4C. | ||||||||
Surprisingly, mutation of most interfacing side chains in the UL, also further increased heteromerization with GluA1, seen by the larger outward currents at positive potentials (Figure 4D; Table II) and summarized by conductance–voltage (G–V) relationships (Figure 4B). It appears therefore that the NTD has not evolved to selectively facilitate heteromeric assembly, but rather to balance heteromerization together with homomerization. We observe a reduction in GluA1/2 heteromer formation only when targeting the hydrophobic core (F50, F82, L310) of the UL interface (Figure 4C and E), which is highlighted for the F82A/L310A (FL) double mutant (Figure 4A–C). These mutants were also destabilized on BN-PAGE (Figure 3A; and data not shown), likely reflecting the non-specific disruption of NTD-mediated associations, both for GluA2/2 homomers and GluA1/2 heteromers. Mutation of most other residues scattered throughout the UL interface facilitated heteromerization (Figure 4E; Table II).
Figure 4.
Mutation of the upper lobe (UL) interface results in bidirectional changes in heteromeric assembly. (A) Representative voltage clamp recordings of GluA2 NTD mutants in outside-out patches when co-transfected into HEK293T cells at a fixed, limiting ratio for heteromeric assembly with GluA1. Only currents for −60, 0, and +40 mV are shown to illustrate the differences in outward current for a series of key mutants. Currents are normalized to the absolute value at −60 mV. (B) Averaged conductance–voltage (G–V) plots for all patches of mutants described in A. Chord conductance (G) is normalized to the absolute value at −60 mV. Number of patches is shown in brackets. The dashed line denotes the G–V curve for homomeric GluA1 (n=4). Error bars represent s.e.m. and are only shown for the positive deviation. (C) Mutation of the conserved hydrophobic cluster in the UL interface in the GluA2 NTD generally disfavours heteromeric assembly with GluA1. I–V relationships were quantified by determining the slope conductance (g) at +10 and −40 mV and expressing these as a ratio, g+10/g−40, or rectification index (RI). The geometric mean of the RI is plotted on a logarithmically scaled axis, with increasing RI indicating greater heteromerization. Error bars show the back-transformed limits of the s.e.m. of the loge RI data. Number of patches is given at the base of each column. The dashed line denotes the RI value for WT GluA2. FF=F50A/F82A, FFL=F50A/F82A/L310A, FL=F82A/L310A. (D) Mutation of polar residues in the UL generally favour heteromeric assembly. Rectification indices of the indicated mutants are represented as in Figure 3D. Note that double mutation of N54 and T78 shows no additive effect and actually decreases heteromerization when mutated to GluA1 residues. The dashed line denotes the RI for GluA2 WT. **P<0.01 (one-way ANOVA with Dunnett multiple comparison: mutants versus wild type). (E) Molecular surface of the GluA2 dimerization interface is shown, with residues targeted for mutagenesis shown as either facilitating (green) or inhibiting (blue) heteromerization. See also Supplementary Figure S3.
Interestingly, evolutionarily variable positions N54 and T78 were most sensitive—mutation to alanine elevated heteromerization competence by approximately 2.2- and 4.7-fold (Figure 4A, B, D and Supplementary Figure 3C). A similar trend was also observed in GluA1 with the swap mutants Y54N and M78T (Figure 5A–C). Moreover, the fact that the N54A/T78A double mutant was not additive, but actually reversed the RI values closer to WT (Figure 4D), together with the fact that they show opposing homomeric phenotypes on BN-PAGE (Figure 3B), supports the hypothesis that these positions drive assembly by mechanistically different routes (see below). This observation could be extended to the GluA2 double swap mutant (N54Y/T78M), which heteromerized poorly (Figure 4D) and was also seen in the reciprocal experiment with GluA1 (Y54N/M78T; Figure 5C). We conclude that the capacity of the NTD to drive the assembly of heteromers is readily increased by mutation in both lobes, specifically at two non-conserved positions (54 and 78; Figure 2A and B). These two residues ‘buffer' the heteromerization competence of the NTD and may thereby facilitate the formation of AMPAR homomers by different mechanisms, which we tested next.
Figure 5.
Mutation of variable residues in the GluA1 upper lobe (UL) show increased heteromerization competence. (A) Voltage clamp recordings of GluA1 NTD mutants in outside-out patches when co-transfected into HEK293T cells with GluA2. Only currents for −60, 0, and +40 mV are shown. Currents are normalized to the absolute value at −60 mV. (B) Averaged conductance–voltage (G–V) plots for all patches of mutants described in A. Chord conductance (G) is normalized to the absolute value at −60 mV. Number of patches is shown in brackets. The dashed line denotes the G–V curve for homomeric GluA1 (n=4). Error bars represent s.e.m. and are only shown for the positive deviation. (C) Mutation of key polar interface residues in the GluA1 NTD UL favours heteromeric assembly. Rectification indices of the indicated mutants plotted as in Figure 3D. The dashed line denotes the RI value for WT GluA1. *P<0.05; **P<0.01 (one-way ANOVA with Dunnett multiple comparison: mutants versus Wild type).
Mechanism underlying balanced assembly via the NTD
To elucidate the mechanistic basis of the difference between N54 and T78, we conducted biophysical and crystallographic studies at the level of the NTD. In accord with the native gels, thermal stability measurements (Niesen et al, 2007) revealed that the melting temperature (Tm) was elevated for the more stable N54A homodimer (by ∼5°C, relative to WT), but was decreased for T78A (similarly by ∼5°C; Supplementary Figure S4A). In addition, an approximately 70-fold lower homodimer stability for T78A (Kd 37±3.7 nM), relative to N54A (Kd 0.5±0.2 nM) was measured by AU-FDS (Figure 6A; Supplementary Figure S4B; Table I). Ablating the polar link mediated by Thr78 (Supplementary Figure S5A, B), therefore, weakens GluA2 homodimers, which approached a Kd of GluA1 (Table I).
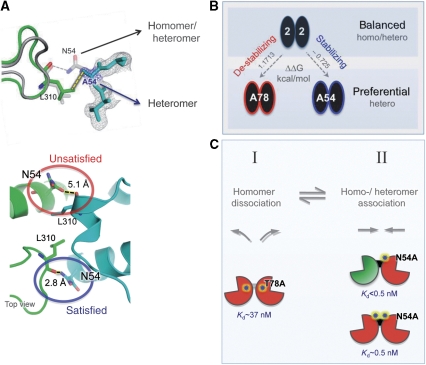
Figure 6.
Structural basis for balanced assembly. (A) Top: Crystal structure of the GluA2-N54A NTD shows subtle alterations in the dimeric packing. Structural alignment of the N54A (green) and the WT (grey) crystal structures are shown. As expected, disrupting the hydrogen bond between N54 and the L310 main-chain carbonyl resulted in a different top loop conformation, but the L310 side chain was not shifted from its interface location. 2Fo−Fc omit map of A54 contoured to 1.0σ is shown for the relevant residue (blue) and the neighbouring residues (grey) in N54A. Bottom: In the WT context (PDB 3HSY), N54 makes a cross-dimer polar contact only on one side of the dimer interface (circled in blue). The result is unsatisfied hydrogen bonding potential at the dimer interface (circled in red), a destabilizing component of the WT interface. (B) Model of the thermodynamic effect of NTD mutations and their effects on whole-channel assembly. Whereas WT GluA2 NTD (top) is capable of balancing homo- and heteromerization at the level of the NTD and the whole receptor, N54A and T78A (bottom), which have opposite effects on NTD dimer stability, preferentially heteromerize the whole channel. Free energy changes of dimer dissocation (ΔΔG) in kcal mol−1 are denoted. (C) Two interconnected steps dictate dimer assembly: homodimer dissociation (step I) and optimal homo- and heterodimer association (step II). Hotspots (stars) in the GluA2 NTD (red) upper lobe interface affect both these processes. Whereas the T78A mutation facilitates homomer dissociation, the N54A mutation stabilizes both homomerization and heteromerization (Kds of dimer dissociation measured by AU-FDS are shown below the relevant assemblies). As heterodimerization requires both homodimer dissociation and re-association of heterodimers, the functional outcome for both mutations in the context of the full receptor is, therefore, greater heteromerization. The GluA1 NTD is shown in green for reference.
The crystal structure of GluA2-T78A, solved to 3.2 Å (Supplementary Table I; PDB 3N6V), revealed that the mutation induces intra- and inter-helical rotations in the four-helical bundle of the UL (Supplementary Figure S5C, D), culminating in a rearranged interface with reduced contact area (approximately 1.4-fold reduced, solvent-accessible surface area). Therefore, alteration at position 78 in the core of the NTD interface facilitates heteromerization by destabilizing homodimeric contacts. Mutation of this core position likely weakens homodimers, increasing subunit availability for heteromeric assembly (Figure 6C, step I).
N54 locates to the edge of the interface and can engage the subunit partner in H-bonding via L310 (Figures 4E and 6A). This position shows greater variability than 78, and in addition to Met in GluA1, it features a His in GluA3 of ray-finned fish. As the N54A mutation is stabilizing (Table I; Supplementary Figure S4), the WT Asn at position 54 weakens GluA2 dimers, relative to the N54A mutant. Structural studies provide an explanation: the N54-L310 contact is asymmetric and is only seen on one side of the twofold dimer (in multiple X-ray data sets). The resulting unsatisfied H-bond of the ‘free' side (Figure 6A, bottom panel) will be energetically unfavourable (Hendsch et al, 1996) and may form the basis for relatively ‘destabilized' dimer contacts in the WT. The crystal structure of GluA2-N54A solved to 1.95 Å confirmed that the N54A mutation replaces this unsatisfied H-bond with a hydrophobic link (Figure 6A), resulting in a tighter, otherwise unaltered, homodimer (PDB 3O2J). This mutation likely stabilizes homo- and heterodimers (Table I; Figure 6C, step II). AU-FDS runs with the GluA1*/GluA2-N54A complex indeed revealed a Kd below the GluA1/2 WT heteromer (Kd<0.5 nM; unpublished observations). However, even though these measurements are reproducible, they approach the limit of resolution with AU-FDS. On the basis of these data, we suggest that the variable N54 interface ‘weakener' has evolved to reduce dimeric affinities in homodimers, facilitating formation of energetically favoured GluA1/2 heterodimers. The net effect is an NTD capable of undergoing both homomeric and heteromeric assembly as a function of subunit concentration in the ER (Figure 7 and Supplementary Figure S6).
Figure 7.
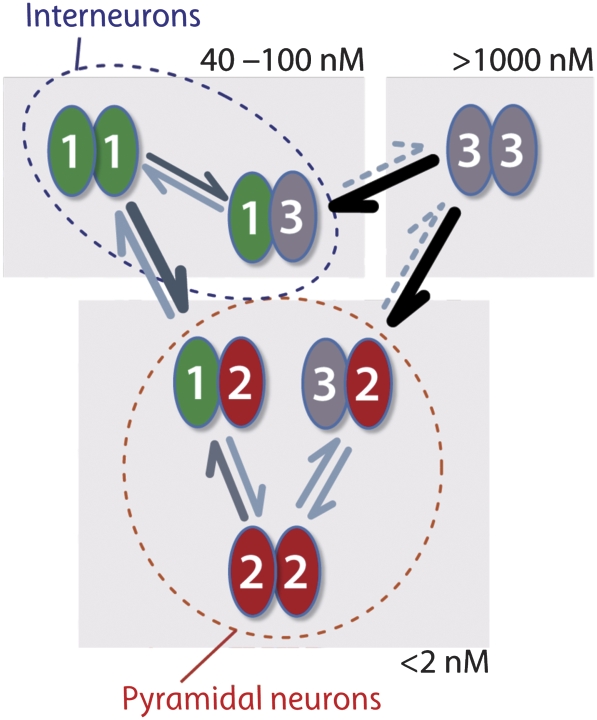
Differential affinities drive specific assembly. Summary of the experimentally derived association affinities. Note that the association affinities of NTDs span four orders of magnitude, from very tight Kds below 2 nM (bottom) for assembly driven by GluA2, to intermediate Kds in the range of 30–100 nM (top left) for assembly driven by GluA1 in the absence of GluA2, and to relatively loose, as in the case of Glua3 homomers (top right). Together with relative subunit concentrations, which differ in distinct neuronal populations (described in Supplementary Figure 6 and indicated by dashed outlines), these parameters will determine the assembly process.
Discussion
Herein, we provide organizing principles underlying the formation of distinct AMPAR hetero-tetramers. Together with single-cell gene expression analysis (Dixon et al, 2000), our results offer a roadmap for the actual assemblies expressed by a given neuron. The development of a high-resolution biophysical assay together with novel structural data reveal that (1) heterodimerization can be obligatory (GluA3) or preferential (GluA1, GluA2, and GluA4), (2) subunits (NTDs) heteromerize at the level of the dimer, (3) heterodimers form preferentially over homodimers, and (4) the NTD does not drive heteromeric assembly maximally and thereby facilitates a balance between homo- and heteromeric assembly modes.
Determinants of AMPAR subunit assembly
Like many (if not most) ion channels and receptors, iGluRs preferentially exist as hetero-oligomers. Although the cellular mechanisms underlying this phenomenon are unclear, the outcome has profound consequences for neuronal physiology and pathology (e.g. Zheng and Zagotta, 2004; Traynelis et al, 2010). Affinities between subunit interfaces and the relative concentration of subunit polypeptides in the ER are likely to be core assembly determinants (Figures 6B, 7 and Supplementary Figure S6), a basic principle applicable to other central cellular processes (Buchler and Cross, 2009).
In neurons, a relatively high local concentration of a given AMPAR subunit polypeptide will form during synthesis from a polyribosome. Each subunit is likely to dimerize at this point. Depending on determinants of ER exit kinetics (e.g., Q/R and R/G editing; Greger et al, 2002, 2006) homodimers will have time to diffuse away from the site of synthesis to where the subunit concentration is lower (Sukumaran et al, 2011a). Homodimers could thus dissociate, leaving them capable to assemble with other AMPAR subunits. In this regard, it can be rationalized that both homomer dissociation (Figure 6C, left panel) and heteromer association (Figure 6C, right panel) will influence heteromerization, as suggested empirically with the GluA2 NTD interface mutants N54A and T78A.
Quantification of subunit associations—the AMPAR subunit affinity network
iGluRs assemble as dimers of dimers into tetramers (Sun et al, 2002; Greger et al, 2003; Shanks et al, 2010). For AMPARs, it has been unclear whether heteromerization occurs at the level of the dimer or the tetramer. The outcome will have consequences for the ultimate organization of subunits within the receptor tetramer. In addition to the four core subunits, auxiliary factors such as TARPS, cornichons and CKAMP44 (reviewed in Guzman and Jonas, 2010) associate with the receptor tetramer and may impact the assembly process (Nakagawa, 2010). We developed a sensitive ultracentrifugation assay (AU-FDS), which facilitated a quantitative assessment of subunit interactions along the (∼60 Å) NTD axis for both homo- and heterodimeric combinations. In addition to revealing preferential heteromerization at the level of the dimer, we uncover a surprising spectrum of different affinities between the four subunits. These properties, together with subunit availability in the ER, will orchestrate the formation of distinct AMPAR heteromers. For example, in CA1 hippocampal neurons, expression levels of AMPAR subunits have been determined at the single cell level; equal proportions of GluA1 and GluA2 have been estimated, with a 10-fold lower concentration of GluA3 (GluA4 has not been detected; Tsuzuki et al, 2001). The single-cell profiling approach (Lambolez et al, 1992; Dixon et al, 2000), in combination with the affinity network measured by AU-FDS herein (Figure 7), provides a roadmap for the actual AMPAR assemblies in CA1, which can be extended to other neuronal populations. For example, the comparatively weak homodimeric affinity of the GluA3 NTD (Kd∼1200 nM) renders the existence of GluA3 homomers unlikely; GluA3/2 heterodimers are ∼1000-fold tighter and will therefore predominate. Similarly, the high affinity of GluA1/2 NTD heteromers is expected to outcompete formation of the respective homomers, which offers a mechanism for the observed abundance of functional GluA1/2 heteromers (Lu et al, 2009).
Altering the relative proportion of subunits after pathological insults (Kwak and Weiss, 2006; Liu and Zukin, 2007) or in response to physiological stimuli (Sutton et al, 2006; Aoto et al, 2008), will shift the balance towards homomers and in turn alter the signalling landscape of the neuron (Liu and Cull-Candy, 2000; Cull-Candy et al, 2006). Our data also suggest that in inter-neurons, in which Ca2+-permeable AMPARs prevail as a result of reduced GluA2 expression, GluA3 heteromers will form preferentially over GluA3 homomers. For example, preferred heteromeric assembly of GluA1/3 Ca2+-permeable AMPARs confers a kinetic profile to these receptors that is different from the GluA1 homomeric pool (Erreger et al, 2004) or GluA3 homomers. Similarly, GluA3/4 receptors will prevail over GluA3 homomers (data not shown), resulting in a more rapidly desensitizing receptor (Trussell, 1998).
The unique position of GluA3 at the low end of the affinity spectrum can be explained structurally—in contrast to the bipartite GluA2 dimer interface, which encompasses both lobes of the NTD clamshell, the LLs of GluA3 have separated as a result of like-charge repulsion (Sukumaran et al, 2011b), bearing an unexpected resemblance to the related mGluR agonist-binding domains (Kunishima et al, 2000).
Homo- versus heteromeric AMPARs—a balancing act of the NTD
In an effort to uncover assembly determinants within the GluA2 NTD, we were surprised to find that mutations scattered throughout the dimer interface mostly increased the fraction of heteromers. Specifically, two hotspots ‘disfavour' GluA1/2 heteromers, as they increase the fraction of heteromers when mutated. The NTD is thus not fully optimized to heterodimerize, which together with the intermediate homomeric affinity of the GluA1 interface (GluA2>GluA4>GluA1>GluA3) could provide the basis for GluA1 homomers.
Overall assembly will be dictated by two interconnected steps (Figure 6C)—availability for complex formation, which requires the dissociation of homodimers (step I), and optimal contacts within the newly formed complex (step II). The interface hotspots affect these two steps differently: N54 is ‘destabilizing' in the WT (relative to the N54A mutant), as a result of unsatisfied H-bonding potential at the edge of an interface (Hendsch et al, 1996; Pokkuluri et al, 2000). As WT GluA1/2 heterodimers only have one N54, heteromerization may partially remove the unsatisfied H-bonding potential. The Tyr at position 54 is unique to GluA1, and may also contribute hydrophobic character to the UL interface and may fully satisfy its H-bonding due to the increased ‘reach' of the Tyr side chain. Taken together, these properties may partially explain why GluA1/2 heterodimers are tighter than GluA2 homodimers, and in fact are the tightest WT assemblies we have measured (Table I). A full mechanistic understanding of the exact role of these determinants must await structure elucidation of the GluA1/2 NTD heterodimer.
Mutation of N54 to Ala introduced a more hydrophobic character to the GluA2 NTD interface and fully abolished the unsatisfied potential, resulting in stabilization of the dimer; thus, facilitating step II and also disfavouring step I (Figure 6C). The resulting impact of N54A on assembly depends on how the opposing effects on steps I and II are balanced; the overall assembly behaviour is readily probed with AU-FDS, and the GluA1/GluA2-N54A NTD heteromer was tighter than the respective homomers (Kd<0.5 nM; data not shown), and it indeed featured the tightest association measured in this study. This result indicates that, although the N54A mutation favours both homomer and heteromer formation, the effect on heteromer formation is stronger. Owing to its position at the interface edge, the N54A mutation did not result in rearrangements of dimeric contacts. Its interaction with a flexible loop via L310 may result in a modulatory role, as this loop is an integral part of the dimer interface and conformationally rearranges on mutation of N54 (Figure 6A).
Truncation of T78 to Ala destabilizes the homodimer, likely by severing water co-ordination across the interface, facilitating step I (Figure 6C). Mutation of this core position resulted in rearrangements of the NTD dimer. Destabilization is also apparent on BN-PAGE when introducing the GluA1-selective M78 (Figure 3A). Conservative mutation to Val will provide additional hydrophobic interactions across the interface and is expected to occupy the same space as the WT Thr. This mutant indeed stabilized dimers on BN-PAGE, similar to WT (Figure 3A). According to the scheme in Figure 6C, dissociation is likely to be energetically more costly for a GluA2-T78V dimer than for a T78A dimer (disfavouring step I). However, in a GluA1/GluA2-T78V heterodimer, Val is expected to interact favourably with M78 across the dimer interface (step II), ultimately explaining the increased heteromerization measured functionally (Table II).
In conclusion, we ascribe a novel role to the AMPAR NTD. Rather than simply being a rigid assembly module, this functionally and structurally diverse domain dictates the diverse subunit stoichiometries present in neuronal populations. In addition, as our data show that heterodimerization is overall favoured at the level of the NTD, we propose that, in principle, the assembly of tri-heteromeric AMPA receptors is possible (as tetramers formed from two different heterodimers), whereas preferential homodimerization would rule out this possibility. These findings, together with an emerging allosteric signalling capacity of the AMPAR NTD (Sukumaran et al, 2011b), uncover unexpectedly versatile functions of a previously poorly understood receptor domain.
Of note, recent kainate receptor NTD stuctures show that dimer packing in GluK3, which is know to form homotetramers, overall mirrors that of other nonNMDAR NTDs. However, the GluK5 dimer interface is rearranged and shows loosened contacts within the ULs (Kumar and Mayer, 2010). Interestingly, since GluK5 does not form homotetramers, the structure provides additional insight into the role of the NTD for selective assembly routes.
Furthermore, in a recent study Farina et al (in press) reveal that the NTD homodimer of the NMDA receptor NR1 subunit is a pivotal scaffolding element, and that this homodimer has to re-equilibrate into a heterodimer during NMDAR biogenesis. They also show that the NTD heteromerization equilibrium, and thus secretion competence of the NMDAR, can be influenced by mutations in the NR1 NTD dimer interface. Overall, these results provide a striking analogy to the results described in this study and reveals mechanistic parallels between the biogenesis of AMPA- and NMDARs.
Materials and methods
Protein preparation
GluA1 NTD (Ala1-Asn375) and GluA2 NTD (Val1-Thr375) with endogenous signal sequences and C-terminal His6-tags were cloned into the pFastBAC1 vector backbone and expressed from Sf9 (Invitrogen) and TriEx strain (Novagen) insect cells using standard baculovirus expression methods. GluA2, GluA3 (Gly1-Phe380) and GluA4 (Ala1-Asp380) NTDs were also subcloned into the pHLsec plasmid between a secretion signal sequence and a His6 tag, and expressed in transiently and stably transfected GntI− HEK293S cells, as described (Aricescu et al, 2006). The numbering of amino acids was according to the mature polypeptide, that is, without inclusion of the signal sequence. Point mutations (N54A, T78A) were introduced utilizing the QuikChange mutagenesis kit (Stratagene). NTD secreted into the culture medium was purified to monodispersity by metal affinity- and subsequent size-exclusion chromatography. Purified proteins were concentrated to 15–21 mg/ml for crystallization trials or analytical ultracentrifugation experiments.
X-ray crystallography
Crystallization screening was performed at 18°C using the sitting drop vapor-diffusion method by mixing of protein and reservoir solutions in the ratio 1:1. GluA2 NTD crystallized in 16–20% PEG3350 and 200–250 mM ammonium dihydrogen phosphate (pH 4.6). GluA2-T78A NTD crystallized in 100 mM Tris (pH 8.5), 100 mM MgCl2, and 18–20% PEG8000. GluA2-N54A NTD crystallized in 18–20% PEG3350, and 100 mM sodium citrate (pH 5.5). All crystals were cryoprotected in the mother liquor supplemented with 30% glycerol, 22–30% PEG400, or 35% PEG3350. Heavy atom derivatives for phasing were obtained by soaking native GluA2 NTD crystals in 200 mM ammonium sulphate (pH 4.6), 20% PEG3350, 1 mM p-chloromercuribenzoic acid (PCMBA; Hampton Research) overnight.
Diffraction data were collected from crystals at beamlines I03 at the Diamond Light Source (Oxford, UK), and ID14-4, ID23-1, and ID29 at the European Synchrotron Radiation Facility (ESRF; Grenoble, France; Supplementary Table I). Data were processed using the IMOSFLM (Leslie, 2006), SCALA (Evans, 2006), XDS, or HKL2000 packages (Kabsch, 1993; Otwinowski and Minor, 1997). The structure of the GluA2 NTD was solved by single anomalous dispersion on PCMBA-derivatized crystals. Initial phases and a partial model were obtained using the autoSHARP package (Vonrhein et al, 2007) and were used to solve the structure by molecular replacement using PHASER (McCoy et al, 2005). The structures of GluA2-N54A and GluA2-T78A were solved by molecular replacement using PHASER, with GluA2 NTD monomer (PDB: 3HSY) as a search model. The models were initially refined using REFMAC (Murshudov et al, 1997), and then alternately refined using PHENIX (Adams et al, 2002), and manually rebuilt in COOT (Emsley and Cowtan, 2004). Composite omit maps calculated with the CNS software package (Brunger and Rice, 1997) were used for model validation. Stereochemical properties of all models were assessed by MOLPROBITY (Davis et al, 2004) and PROCHECK (Laskowski et al, 1993).
FAM labelling and analytical ultracentrifugation
N-terminal labelling was performed by incubating protein with 30-fold excess of 5,6-carboxyfluorescein succinimidyl ester (FAM; Biotium) at room temperature in pH 7.0 for 30 min, and was quenched with excess primary amine (1 M Tris–HCl (pH 7.4)). Excess label was removed by dialysis, and labelled protein was repurified by size-exclusion chromatography. Labelling efficiency was estimated spectrophotometrically and was in the range of 0.9–1.3 fluorophore per protein molecule.
A ProteomeLab XL-I analytical ultracentrifuge (Beckman Coulter) equipped with a fluorescence detection system (AVIV Biomedical) was used for velocity sedimentation experiments, performed at 50 000 rpm at 10°C. A total of 400 scans were acquired in intervals of 2.5 min. Concentration gradients were fit with sedimentation coefficient distributions (c(s)) using SEDFIT (Schuck, 2000). Subsequent fitting of sedimentation profiles to normal distributions and Kd calculations were done with DataFitter software, developed in-house; c(s) distributions were deconvoluted into monomeric and dimeric fractions, and separately integrated to determine Kds. When c(s) distributions were concentration-dependent, Kds were calculated based on shifting peak positions. Determinations of heteromeric Kds was carried out by further deconvoluting contributions of labelled and unlabelled assembly species and numerically fitting data into binding models accounting for previously determined homomeric Kds of each assembly constituent (see also Supplementary Materials and methods).
Blue native PAGE
Analysis of AMPAR assembly by BN-PAGE was performed as described in detail previously (Greger et al, 2002; Penn et al, 2008). To dissociate AMPAR complexes for BN-PAGE, cell suspensions were treated with 1% SDS at 30°C for 10–15 min, cooled on ice, and subjected to BN-PAGE.
Cell culture and transfection
HEK293T cells were cultured using standard techniques. Cells were transfected in six-well plates using Effectene (Qiagen) with full-length GluA2-IRES-EGFP (flop; unedited at the R/G-site (position 743); edited at the Q/R site (position 586)) and pRK5-GluA1 flip (gift from Dr J Howe) to a total of 0.5 μg of plasmid.
Electrophysiology and analysis
We determined the optimal cDNA ratio for co-expression; a condition in which GluA2 was limiting was achieved with a GluA1/2 ratio of 4:1, resulting in an RI of ∼0.1 (Supplementary Figure 3). We also note that none of the mutants significantly affected protein levels detected by western blot, or traffic from the ER as assessed by endoglycosidase H sensitivity (Greger et al, 2002; data not shown). Voltage clamp recordings were performed on outside-out patches excised from HEK293T cells ∼48 h after transfection. AMPA receptor currents were evoked with fast piezo-driven L-Glu application (3 mM, 100 ms) via a theta pipette. Typical solution exchange rise times for the junction potential at the open tip were <0.2 ms (Penn et al, 2008). Extracellular solution contained (in mM): NaCl (145), KCl (3), CaCl2 (2), MgCl2 (1), HEPES (10), and glucose (10) (pH 7.4) with NaOH (∼310 mOsm). Patch pipettes had an open-tip resistance of 2–5 MΩ when filled with filtered intracellular solution (in mM): CsF (120), CsCl (10), EGTA (10), MgCl2 (2), Na2-ATP (2) QX314-Cl (1), Spermine (0.1), and HEPES (10) and adjusted to pH 7.2–7.3 with CsOH (∼280 mOsm). Current–voltage relationships of peak AMPAR currents (holding potentials −70 to +50 mV) were fit by polynomial regression, then the (real) root and first derivative of the fit were computed to obtain the reversal potential (Erev) and slope conductance (g), respectively, using custom scripts in GNU Octave. These parameters were used to calculate RI defined as the ratio of g at +10 mV over g at −40 mV from Erev (g+10/g−40). Rectification indices over the measured range showed log normal distributions and hence are summarized as the geometric mean. The s.e.m. limits of the log data were back transformed to give error bounds (Table II; also see Supplementary Materials and methods).
Accession numbers
Coordinates for WT GluA2 (3HSY), GluA2-T78A (3N6V) and GluA2-N54A (3O2J) were deposited in the Protein Data Bank.
Supplementary Material
Acknowledgments
We wish to thank David Komander and Masato Akutsu for support with structure determination, and Cyrus Chothia, Alexej Murzin and Ondrej Cais for stimulating discussions and helpful comments. We also wish to thank A Radu Aricescu for providing the pHLsec plasmid and GntI-HEK293S cells. MS, MR, ACP, MMB, DBV, and IHG were supported by the MRC. MS was also supported by the NIH and IHG by the Royal Society.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58(Part 11): 1948–1954 [DOI] [PubMed] [Google Scholar]
- Aoto J, Nam CI, Poon MM, Ting P, Chen L (2008) Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron 60: 308–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricescu AR, Lu W, Jones EY (2006) A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr 62(Part 10): 1243–1250 [DOI] [PubMed] [Google Scholar]
- Ayalon G, Stern-Bach Y (2001) Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron 31: 103–113 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Rice LM (1997) Crystallographic refinement by simulated annealing: methods and applications. Methods Enzymol 277: 243–269 [DOI] [PubMed] [Google Scholar]
- Buchler NE, Cross FR (2009) Protein sequestration generates a flexible ultrasensitive response in a genetic network. Mol Syst Biol 5: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlezon WA Jr, Nestler EJ (2002) Elevated levels of GluR1 in the midbrain: a trigger for sensitization to drugs of abuse? Trends Neurosci 25: 610–615 [DOI] [PubMed] [Google Scholar]
- Catania MV, Bellomo M, Giuffrida R, Stella AM, Albanese V (1998) AMPA receptor subunits are differentially expressed in parvalbumin- and calretinin-positive neurons of the rat hippocampus. Eur J Neurosci 10: 3479–3490 [DOI] [PubMed] [Google Scholar]
- Clayton A, Siebold C, Gilbert RJ, Sutton GC, Harlos K, McIlhinney RA, Jones EY, Aricescu AR (2009) Crystal structure of the GluR2 amino-terminal domain provides insights into the architecture and assembly of ionotropic glutamate receptors. J Mol Biol 392: 1125–1132 [DOI] [PubMed] [Google Scholar]
- Cull-Candy S, Kelly L, Farrant M (2006) Regulation of Ca2+-permeable AMPA receptors: synaptic plasticity and beyond. Curr Opin Neurobiol 16: 288–297 [DOI] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC (2004) MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32 (Web Server issue): W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AK, Richardson PJ, Pinnock RD, Lee K (2000) Gene-expression analysis at the single-cell level. Trends Pharmacol Sci 21: 65–70 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Part 12 Part 1): 2126–2132 [DOI] [PubMed] [Google Scholar]
- Erreger K, Chen PE, Wyllie DJ, Traynelis SF (2004) Glutamate receptor gating. Crit Rev Neurobiol 16: 187–224 [DOI] [PubMed] [Google Scholar]
- Evans P (2006) Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62(Part 1): 72–82 [DOI] [PubMed] [Google Scholar]
- Farina AN, Blain KY, Maruo T, Kwiatkowski W, Choe S, Nakagawa T Separation of domain contacts is required for heterotetrameric assembly of functional NMDA receptors. J Neurosci (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill A, Madden DR (2006) Glutamate Receptor Ion Channels: Structural Insights into Molecular Mechanisms. Cambridge, UK: RSC Publishing [Google Scholar]
- Greger IH, Akamine P, Khatri L, Ziff EB (2006) Developmentally regulated, combinatorial RNA processing modulates AMPA receptor biogenesis. Neuron 51: 85–97 [DOI] [PubMed] [Google Scholar]
- Greger IH, Esteban JA (2007) AMPA receptor biogenesis and trafficking. Curr Opin Neurobiol 17: 289–297 [DOI] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Kong X, Ziff EB (2003) AMPA receptor tetramerization is mediated by Q/R editing. Neuron 40: 763–774 [DOI] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Ziff EB (2002) RNA editing at Arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron 34: 759–772 [DOI] [PubMed] [Google Scholar]
- Greger IH, Rossmann M, Sukumaran M, Penn AC, Veprintsev DB (2009) Structure and function of the AMPA receptor N-terminal domain. Soc Neurosci Abstr 512: 4
- Greger IH, Ziff EB, Penn AC (2007) Molecular determinants of AMPA receptor subunit assembly. Trends Neurosci 30: 407–416 [DOI] [PubMed] [Google Scholar]
- Guzman SJ, Jonas P (2010) Beyond TARPs: the growing list of auxiliary AMPAR subunits. Neuron 66: 8–10 [DOI] [PubMed] [Google Scholar]
- Hansen KB, Furukawa H, Traynelis SF (2010) Control of assembly and function of glutamate receptors by the amino-terminal domain. Mol Pharmacol 78: 535–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton CJ, Paoletti P (2005) Modulation of triheteromeric NMDA receptors by N-terminal domain ligands. Neuron 46: 261–274 [DOI] [PubMed] [Google Scholar]
- Hendsch ZS, Jonsson T, Sauer RT, Tidor B (1996) Protein stabilization by removal of unsatisfied polar groups: computational approaches and experimental tests. Biochemistry 35: 7621–7625 [DOI] [PubMed] [Google Scholar]
- Hollmann M, Heinemann S (1994) Cloned glutamate receptors. Annu Rev Neurosci 17: 31–108 [DOI] [PubMed] [Google Scholar]
- Isaac JT, Ashby M, McBain CJ (2007) The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron 54: 859–871 [DOI] [PubMed] [Google Scholar]
- Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E (2009) Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J 28: 1812–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Kauer JA, Malenka RC (2007) Synaptic plasticity and addiction. Nat Rev Neurosci 8: 844–858 [DOI] [PubMed] [Google Scholar]
- Kessels HW, Malinow R (2009) Synaptic AMPA receptor plasticity and behavior. Neuron 61: 340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Mayer ML (2010) Crystal structures of the glutamate receptor ion channel GluK3 and GluK5 amino-terminal domains. J Mol Biol 404: 680–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Schuck P, Jin R, Mayer ML (2009) The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol 16: 631–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K (2000) Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 407: 971–977 [DOI] [PubMed] [Google Scholar]
- Kwak S, Weiss JH (2006) Calcium-permeable AMPA channels in neurodegenerative disease and ischemia. Curr Opin Neurobiol 16: 281–287 [DOI] [PubMed] [Google Scholar]
- Lambolez B, Audinat E, Bochet P, Crepel F, Rossier J (1992) AMPA receptor subunits expressed by single Purkinje cells. Neuron 9: 247–258 [DOI] [PubMed] [Google Scholar]
- Landgraf R, Xenarios I, Eisenberg D (2001) Three-dimensional cluster analysis identifies interfaces and functional residue clusters in proteins. J Mol Biol 307: 1487–1502 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthhur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Leslie AG (2006) The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr 62: 48–57 [DOI] [PubMed] [Google Scholar]
- Leuschner WD, Hoch W (1999) Subtype-specific assembly of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunits is mediated by their n-terminal domains. J Biol Chem 274: 16907–16916 [DOI] [PubMed] [Google Scholar]
- Lichtarge O, Bourne HR, Cohen FE (1996) An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol 257: 342–358 [DOI] [PubMed] [Google Scholar]
- Liu SJ, Zukin RS (2007) Ca2+-permeable AMPA receptors in synaptic plasticity and neuronal death. Trends Neurosci 30: 126–134 [DOI] [PubMed] [Google Scholar]
- Liu SQ, Cull-Candy SG (2000) Synaptic activity at calcium-permeable AMPA receptors induces a switch in receptor subtype. Nature 405: 454–458 [DOI] [PubMed] [Google Scholar]
- Lu W, Shi Y, Jackson AC, Bjorgan K, During MJ, Sprengel R, Seeburg PH, Nicoll RA (2009) Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron 62: 254–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor IK, Anderson AL, Laue TM (2004) Fluorescence detection for the XLI analytical ultracentrifuge. Biophys Chem 108: 165–185 [DOI] [PubMed] [Google Scholar]
- Madden DR (2002) The structure and function of glutamate receptor ion channels. Nat Rev Neurosci 3: 91–101 [DOI] [PubMed] [Google Scholar]
- Malinow R, Malenka RC (2002) AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci 25: 103–126 [DOI] [PubMed] [Google Scholar]
- Mansour M, Nagarajan N, Nehring RB, Clements JD, Rosenmund C (2001) Heteromeric AMPA receptors assemble with a preferred subunit stoichiometry and spatial arrangement. Neuron 32: 841–853 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61(Part 4): 458–464 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53(Part 3): 240–255 [DOI] [PubMed] [Google Scholar]
- Nakagawa T (2010) The biochemistry, ultrastructure, and subunit assembly mechanism of AMPA receptors. Mol Neurobiol 42: 161–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesen FH, Berglund H, Vedadi M (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2: 2212–2221 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallogr A 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Penn AC, Greger IH (2009) Sculpting AMPA receptor formation and function by alternative RNA processing. RNA Biol 6: 517–522 [DOI] [PubMed] [Google Scholar]
- Penn AC, Williams SR, Greger IH (2008) Gating motions underlie AMPA receptor secretion from the endoplasmic reticulum. EMBO J 27: 3056–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant K, Pelkey KA, Bortolotto ZA, Morita D, Terashima A, McBain CJ, Collingridge GL, Isaac JT (2006) Transient incorporation of native GluR2-lacking AMPA receptors during hippocampal long-term potentiation. Nat Neurosci 9: 602–604 [DOI] [PubMed] [Google Scholar]
- Pokkuluri PR, Cai X, Johnson G, Stevens FJ, Schiffer M (2000) Change in dimerization mode by removal of a single unsatisfied polar residue located at the interface. Protein Sci 9: 1852–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Jaulent AM, Wells M, Veprintsev DB, Fersht AR (2008) 14-3-3 activation of DNA binding of p53 by enhancing its association into tetramers. Nucleic Acids Res 36: 5983–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans N, Vissel B, Petralia RS, Wang YX, Chang K, Royle GA, Wang CY, O′Gorman S, Heinemann SF, Wenthold RJ (2003) Aberrant formation of glutamate receptor complexes in hippocampal neurons of mice lacking the GluR2 AMPA receptor subunit. J Neurosci 23: 9367–9373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 78: 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwappach B (2008) An overview of trafficking and assembly of neurotransmitter receptors and ion channels. Review. Mol Membr Biol 25: 270–278 [DOI] [PubMed] [Google Scholar]
- Shanks NF, Maruo T, Farina AN, Ellisman MH, Nakagawa T (2010) Contribution of the global subunit structure and stargazin on the maturation of AMPA receptors. J Neurosci 30: 2728–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Huganir RL (2007) The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol 23: 613–643 [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E (2009) X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462: 745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran M, Penn AC, Greger IH (2011a) AMPA receptor assembly—atomic determinants and built-in modulators. In Synaptic Plasticity—Dynamics, Development and Disease, Kreutz M, Sala C (eds) Wien, New York: Springer (in press) [DOI] [PubMed] [Google Scholar]
- Sukumaran M, Rossmann M, Shrivastava I, Dutta A, Bahar I, Greger IH (2011b) Dynamics and allosteric potential of the AMPA receptor N-terminal domain. EMBO J 30: 972–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E (2002) Mechanism of glutamate receptor desensitization. Nature 417: 245–253 [DOI] [PubMed] [Google Scholar]
- Sutton MA, Ito HT, Cressy P, Kempf C, Woo JC, Schuman EM (2006) Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell 125: 785–799 [DOI] [PubMed] [Google Scholar]
- Suzuki E, Kessler M, Arai AC (2008) The fast kinetics of AMPA GluR3 receptors is selectively modulated by the TARPs gamma 4 and gamma 8. Mol Cell Neurosci 38: 117–123 [DOI] [PubMed] [Google Scholar]
- Thiagarajan TC, Lindskog M, Tsien RW (2005) Adaptation to synaptic inactivity in hippocampal neurons. Neuron 47: 725–737 [DOI] [PubMed] [Google Scholar]
- Toth K, McBain CJ (1998) Afferent-specific innervation of two distinct AMPA receptor subtypes on single hippocampal interneurons. Nat Neurosci 1: 572–578 [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R, Sibley D (2010) Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62: 405–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trussell L (1998) Control of time course of glutamatergic synaptic currents. Prog Brain Res 116: 59–69 [DOI] [PubMed] [Google Scholar]
- Tsuzuki K, Lambolez B, Rossier J, Ozawa S (2001) Absolute quantification of AMPA receptor subunit mRNAs in single hippocampal neurons. J Neurochem 77: 1650–1659 [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G (2007) Automated structure solution with autoSHARP. Methods Mol Biol 364: 215–230 [DOI] [PubMed] [Google Scholar]
- Washburn MS, Numberger M, Zhang S, Dingledine R (1997) Differential dependence on GluR2 expression of three characteristic features of AMPA receptors. J Neurosci 17: 9393–9406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenthold RJ, Petralia RS, Blahos J II, Niedzielski AS (1996) Evidence for multiple AMPA receptor complexes in hippocampal CA1/CA2 neurons. J Neurosci 16: 1982–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuster A, Venkatakrishnan AJ, Schertler GF, Babu MM (2010) Spial: Analysis of sub-type specific features in multiple sequence alignments of proteins. Bioinformatics 26: 2906–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Zagotta WN (2004) Stoichiometry and assembly of olfactory cyclic nucleotide-gated channels. Neuron 42: 411–421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.