Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response
In the endoplasmic reticulum, unfolded proteins stimulate Ire1 autophosphorylation and RNase activity. The crystal structure of the dephosphorylated kinase/RNase domain of human Ire1 bound to ADP provides insight into the autophosphorylation reaction.
Keywords: autophosphorylation, dimerisation, inhibition, signal transduction, UPR
Abstract
Ire1 (Ern1) is an unusual transmembrane protein kinase essential for the endoplasmic reticulum (ER) unfolded protein response (UPR). Activation of Ire1 by association of its N-terminal ER luminal domains promotes autophosphorylation by its cytoplasmic kinase domain, leading to activation of the C-terminal ribonuclease domain, which splices Xbp1 mRNA generating an active Xbp1s transcriptional activator. We have determined the crystal structure of the cytoplasmic portion of dephosphorylated human Ire1α bound to ADP, revealing the ‘phosphoryl-transfer' competent dimeric face-to-face complex, which precedes and is distinct from the back-to-back RNase ‘active' conformation described for yeast Ire1. We show that the Xbp1-specific ribonuclease activity depends on autophosphorylation, and that ATP-competitive inhibitors staurosporin and sunitinib, which inhibit autophosphorylation in vitro, also inhibit Xbp1 splicing in vivo. Furthermore, we demonstrate that activated Ire1α is a competent protein kinase, able to phosphorylate a heterologous peptide substrate. These studies identify human Ire1α as a target for development of ATP-competitive inhibitors that will modulate the UPR in human cells, which has particular relevance for myeloma and other secretory malignancies.
Introduction
The endoplasmic reticulum (ER) is a major compartment within the eukaryotic cell responsible for folding of secretory proteins. Disruption of the balance between secretory protein synthesis and the folding capacity of the ER activates a signalling network called the unfolded protein response (UPR). This attempts to minimise ER overload by transcriptional upregulation of ER chaperones, attenuation of translation, and activation of ER-associated degradation (ERAD) pathways (Ron and Walter, 2007; Todd et al, 2008; Zhang and Kaufman, 2008). If the effort to rectify the imbalance is not successful, the UPR switches from being pro-survival to eliciting an apoptotic response (Groenendyk and Michalak, 2005).
Ire1 (a.k.a Ern1) is one of the three transmembrane ‘sensor' proteins that propagate the signal from the lumen of the ER to the cytosol. Ire1 consists of an N-terminal luminal domain, a single-pass transmembrane spanning segment, and a cytosolic region subdivided into a Ser/Thr protein kinase domain and a C-terminal endoribonuclease (RNase) domain with homology to RNaseL (Tirasophon et al, 1998). The ER luminal domain of Ire1 is maintained in its inactive monomeric state by binding to the ER Hsp70 chaperone Bip. Accumulation of misfolded protein in the ER is thought to cause release of Bip (Bertolotti et al, 2000), allowing the luminal domain to dimerise (Credle et al, 2005; Zhou et al, 2006). This brings associated cytoplasmic domains into close proximity on the other side of the ER membrane, facilitating transautophosphorylation of the kinase domain, which results in activation of the RNase domain (Ron and Walter, 2007; Zhang and Kaufman, 2008). In budding yeast, Ire1 RNase activity is specific for Hac1 mRNA and excises a 252-nucleotide segment in a spliceosome-independent manner (Sidrauski and Walter, 1997; Gonzalez et al, 1999). Human Ire1 excises 26 nucleotides from Xbp1 mRNA. The spliced transcript Xbp1s encodes a potent transcriptional activator protein that upregulates expression of UPR target genes (Yoshida et al, 2001), including ER chaperones and components of the ERAD pathway (Friedlander et al, 2000).
The UPR is activated in many human tumours and is a mechanism tumour cells exploit to survive hypoxia (Koumenis, 2006). The Ire1-Xbp1 branch of signalling is particularly important in multiple myeloma; a cancer resulting from malignant transformation of plasma cells. The survival rate for this cancer is low and thus far there is no direct curative therapy. Alongside its role in the UPR, Xbp1 controls plasma cell differentiation, and when misregulated, causes uncontrollable proliferation in myeloma cells (Davies et al, 2003; Iwakoshi et al, 2003; Carrasco et al, 2007; Davenport et al, 2007; Todd et al, 2008). As Xbp1s mRNA levels are directly dependent upon splicing by Ire1, preventing splicing of Xbp1 mRNA by pharmacological inhibition of the kinase/RNase activity of Ire1 would appear to be a very good strategy for selectively targeting multiple myeloma. However, the lack of a clear mechanistic understanding of Ire1 function, and in particular the coupling of the kinase and RNase activities, has clouded the issue. Early work in the field indicated that Ire1 acts like a typical receptor protein kinase, undergoing autophosphorylation in trans, following dimerisation of its ER luminal signalling domain (Shamu and Walter, 1996; Welihinda and Kaufman, 1996). Furthermore, a kinase-dead Ire1 was unable to splice substrate RNA (Shamu et al, 1994; Tirasophon et al, 1998), demonstrating that the Ire1 phosphoryl-transfer reaction was essential for RNase activation. However, the remarkable finding that binding of an ATP-competitive inhibitor to an active site mutant of yeast Ire1 resulted in RNase activation rather than inhibition has suggested that it is a conformational change in the kinase domain induced by nucleotide binding, rather than phosphorylation, that provides the trigger to activate the RNase (Papa et al, 2003). This phenomenon has also been reported for wild-type yeast Ire1 with the FDA-approved anti-cancer drug sunitinib (Korennykh et al, 2009).
Recently, there have been several structures of the C-terminal kinase-RNase domains of yeast Ire1. The first of these structures (Lee et al, 2008) revealed a symmetric Ire1 dimer arranged in a back-to-back orientation and reported to be in an RNase ‘active' conformation, but with the kinase active sites facing outwards in a manner that would preclude transphosphorylation. The biological significance of this arrangement is supported by mutations in the back-to-back interface that impair RNase activity. A second structure of yeast Ire1 has been described (Korennykh et al, 2009), in which the same back-to-back dimer is arranged in a rod-shaped helical assembly, giving rise to an oligomeric RNase ‘active' complex. More recently, a yeast Ire1–quercetin–ADP complex (Wiseman et al, 2010) showed essentially the same back-to-back orientation that has been seen with the previous structures. All of the yeast structures have been crystallised from similar constructs that have an internal ∼24 amino-acid deletion within the kinase domain. Furthermore, all the structures are derived from phosphorylated yeast Ire1 protein and all describe the RNase ‘active' conformation of yeast Ire1.
We have now determined the crystal structure of the cytoplasmic kinase-RNase region of human Ire1α at 2.7 Å resolution, in a dephosphorylated state, and in a complex with Mg2+-ADP, revealing a face-to-face dimer of the kinase domains, reminiscent of dimerisation-activated kinases such as Chk2, LOK, SLK, and DAPK3 (Oliver et al, 2006, 2007; Pike et al, 2008), in which the kinase active site and activation segment of the Ire1 protomers are in a suitable orientation and proximity for transphosphorylation. We believe that this orientation is an early state ‘phosphoryl-transfer' competent conformation of Ire1, which precedes and is distinct from the RNase ‘active' back-to-back conformation, that has been visualised in the yeast structures. Furthermore, our biochemical analysis of human Ire1α shows direct dependence of RNase activity on autophosphorylation, and that inhibition of autophosphorylation by ATP-competitors blocks Ire1α RNase activity in vivo. Our combined structural and biochemical data show that human Ire1 behaves mechanistically like a typical receptor protein kinase and that inhibition of its kinase activity is likely to provide an important new therapeutic approach to multiple myeloma.
Results
Crystal structure of human Ire1α
A cytoplasmic region of human Ire1α (residues 547–977) was expressed in insect cells, chromatographically purified, dephosphorylated and crystallised by vapour diffusion in a buffer containing Mg2+-ADP. Diffraction data to 2.7 Å were collected on station I03 at the Diamond Light Source and the structure was phased by molecular replacement using the structure of yeast Ire1. The crystals contain four molecules of Ire1α in the asymmetric unit (see Materials and methods).
The human Ire1α cytoplasmic region has a very similar overall structure to those previously determined for yeast Ire1 (Lee et al, 2008; Korennykh et al, 2009; Wiseman et al, 2010) (Figure 1A), consisting of a bilobal protein kinase fold with the ATP-binding site in the cleft between the N-terminal β-sheet lobe and the α-helical C-terminal lobe. A distinct feature of Ire1 is an addition helical domain, fused to and structurally continuous with the C-terminal lobe of the kinase, which provides the sequence-specific ribonuclease activity by which Ire1 is able to initiate splicing of Xbp1 mRNA (Sidrauski and Walter, 1997; Tirasophon et al, 2000).
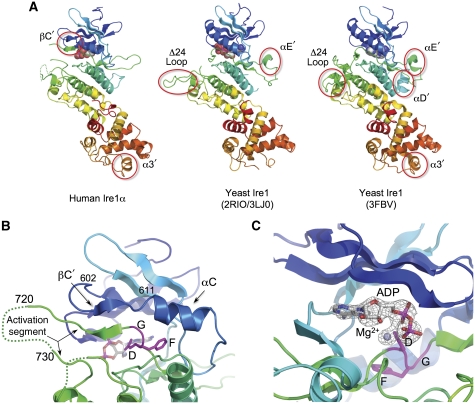
Figure 1.
Structure of human Ire1α. (A) Secondary structure cartoons of the crystal structures of cytoplasmic regions of human Ire1α (left) and yeast Ire1 (middle and right), rainbow coloured blue → red, N → C-terminus. Non-canonical secondary structural elements in the yeast structures (αD′, αE′), which mediate protein–protein interactions in the different yeast Ire1 crystal lattices, are not conserved in human Ire1α. Both yeast crystal structures have an engineered loop (Δ24 loop) connecting α-helices EF and F, from which 24 residues were deleted to obtain crystals. The equivalent loop in human (and other metazoan) Ire1α is much shorter, and was not modified. A small helix in the C-terminal RNase domain (α3′), which has been suggested to contribute to the catalytic function of this domain is fully ordered in human Ire1α. Human Ire1α contains an additional β-strand (βC′) formed by the unwinding of the N-terminal end of the kinase domain C-helix, which makes a β-sheet interaction with the N-terminal end of the activation segment. (B) Close-up of the β-sheet interaction between the βC′-strand and the N-terminal part of the activation segment immediately following the conserved DFG motif at residues 711–713. This β-sheet interaction directs the subsequent residues of the activation segment away from the body of the kinase, towards the kinase active site of a second Ire1α molecule (see below). To our knowledge, this type of interaction, involving the unwinding of the C-helix, has not previously been described in protein kinases. The tip of the activation segment between residues 720 and 730 is poorly ordered. (C) Close-up of Mg2+-ADP bound to dephosphorylated human Ire1α. The DFG motif is in the ‘in' conformation associated with an activated kinase, and the side chain of Asp711 in the motif provides a direct ligand interaction with the Mg2+ ion. Electron density mesh is from a Fo–Fc difference Fourier map calculated with Mg-ADP omitted, and contoured at 3.0 σ.
The human Ire1α structure is visible from residue 561 to 966, which is 11 residues upstream of the biological C-terminus. The central ∼10 residues (720–729) of the kinase activation segment, containing a putative phosphorylation site at Ser724, conserved in yeast Ire1, are disordered. The activation segment is similarly disordered in the high-resolution yeast structure (2RIO) and Ire1–quercetin complex structure (3LJ0), but is ordered in the low-resolution oligomeric yeast structure (3FBV), where it projects sideways and forms an interface with another monomer. The conformation and orientation of the yeast activation segments are markedly different to human Ire1α (see below). Residues 746–748 in the human Ire1α kinase domain, at the tip of the loop connecting helix αF and the short αEF helix carrying the APE motif, are also disordered. In yeast Ire1, this loop is substantially longer, and was artificially ‘short circuited' by excision of 24 residues in the published structures of the yeast enzyme, which contributes to crystal lattice interactions. The connection between helices 3 and 4 in the RNase domain, which was disordered in the higher-resolution yeast Ire1 structure, is fully ordered in human Ire1α where it has a helical conformation. Two helical segments (αD′ and αE′) observed in the yeast Ire1 structures (Figure 1A) do not occur in human Ire1, where the equivalent segments are short loops.
The most significant differences between the human Ire1 and yeast Ire1 monomer structures occur in the N-lobe and cleft region of the kinase domain. The kinase N-lobe overall is rotated ∼6° in human Ire1 with respect to the C-lobe, compared with the juxtaposition of these lobes in the yeast structures. Accompanying this is a substantial unravelling of the N-terminal end of the C-helix in the Ire1 kinase N-lobe, which adopts a fully ordered extended conformation for residues 602–611. The rest of this segment, which maintains a helical conformation, is displaced from its position in yeast Ire1 by ∼6 Å, separating the conserved Lys-Glu ion pair, required for activity (Huse and Kuriyan, 2002). The DFG motif, however, has an ‘in' conformation, consistent with a catalytically competent state of the protein, with the carboxylate side chain of Asp711 interacting with the Mg2+ associated with the bound ADP (Figure 1B and C).
In both the human and the two yeast Ire1 structures (3LJ0, 2RIO), as in many protein kinases in their unphosphorylated states, the activation segment (Johnson et al, 1996) connecting the protein kinase DFG and APE motifs is disordered. However, in the lower-resolution phosphorylated yeast Ire1, an ordered activation segment projects out and away from the active site and forms an interface with another monomer in a ‘side-to-side' orientation. The phosphorylated residues are intimately involved in this interaction, with the invariant pSer841 interacting with Lys678 from the opposing monomer, and pSer840 associating with Asp763 and Glu764 in a negatively charged pocket formed by the αD–αE loop. This arrangement is not seen in the human structure, since the orientation of the molecules relative to each other is different to that seen in all the yeast structures (see below).
In the higher-resolution yeast Ire1 structure, the ordered N-terminal part of the activation segment, extending from the DFG motif, is directed away from the bound ADP, in the same direction as the Δ24-αEF–αF loop. The C-terminal part leading into the APE motif and the Δ24–αEF–αF loop heads in the opposite direction, so that the ends of the disordered 15 residues of the activation segment, that connects them, are separated by >30 Å. By contrast, in human Ire1α, the N-terminal end of the activation segment immediately following the DFG motif projects directly out from the body of the kinase towards the active site of the opposing monomer, making an anti-parallel β-sheet with the residues from the N-terminal end of the C-helix, in their unwound extended conformation. The C-terminal segment upstream of the APE motif heads in the same direction, so that the visible tips of the ordered parts of the human activation segment are only ∼9 Å apart, and could be readily bridged by the intervening residues making a reverse loop (Figure 1B).
Ire1 dimerisation and autophosphorylation
In the current models for the activation mechanism of Ire1, the protein is believed to be held in a monomeric and inactive state by the ER Hsp70 homologue Bip, which binds to the ER luminal domain of Ire1 (Bertolotti et al, 2000; Okamura et al, 2000; Kimata et al, 2003). Accumulation of unfolded protein in the ER promotes release of Bip, permitting dimerisation of the luminal domains (Credle et al, 2005; Zhou et al, 2006). This in turn drives oligomeric association and transphosphorylation of the cytoplasmic kinase domain (Shamu and Walter, 1996; Welihinda and Kaufman, 1996), which is an essential prerequisite for activation of the splicing activity of the cytoplasmic ribonuclease domain (Tirasophon et al, 2000).
The requirement for formation of dimers (and possibly higher oligomers) in the activation of Ire1 has dominated the mechanistic interpretation of the previously described crystal structures of yeast Ire1 (Lee et al, 2008; Korennykh et al, 2009). In both studies (which utilised extremely similar constructs incorporating the engineered Δ24–αEF–αF loop), an intimate dimer could be identified in the crystal lattice, in which two Ire1 molecules formed a back-to-back interaction, with the nucleotide-binding site facing outwards. In both cases, this dimeric arrangement is further expanded by helical symmetry to generate the full crystal lattice—in the high-resolution structure (Lee et al, 2008), the helix has a six-fold periodicity and is consequently subsumed in the space group description of the lattice, while in the lower-resolution structure (Korennykh et al, 2009), the helix has a much longer periodicity and 14 copies of the back-to-back dimer constitute the crystallographic asymmetric unit. The arrangement of molecules in the human Ire1α crystals is quite different to that of the yeast Ire1 structures. Only two protein–protein interfaces of any substance are observed. The largest (burying >1700 Å2 of molecular surface) is an approximately two-fold symmetric parallel interaction, involving residues from the kinase domain only, and generates a ‘face-to-face' dimer arrangement in which the open mouths of the interlobe clefts, containing bound Mg2+-ADP, face each other across the dimer (Figure 2A). The next largest interface (burying ∼1600 Å2) is an anti-parallel arrangement, involving the last part of the kinase C-lobe and the ribonuclease domain. This arrangement would be incompatible with the membrane tethering of Ire1 and is likely to be a crystal lattice contact only.
Figure 2.
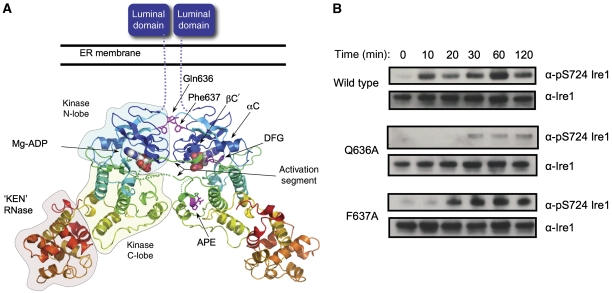
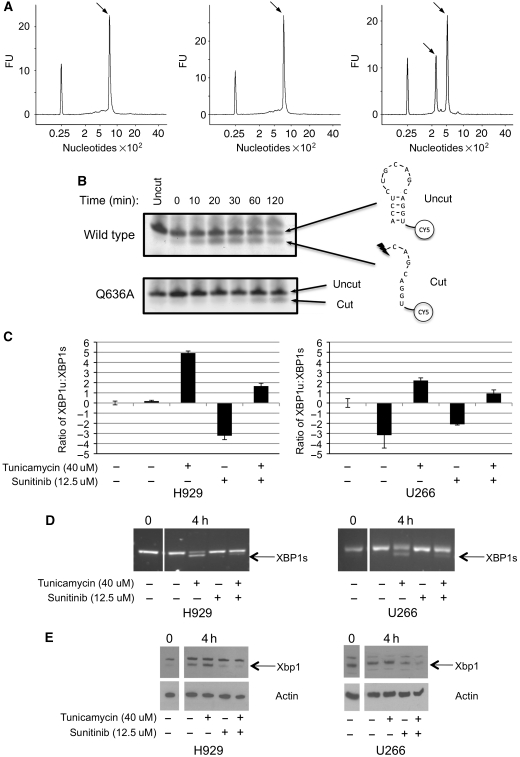
Face-to-face kinase domain dimerisation of human Ire1α. (A) Human Ire1α forms a dimer in which the kinase active sites of the two monomers face each other. The activation segment of one monomer is directed towards the other, so that the target substrate residue, Ser724 would come into close proximity of the Mg2+-ATP bound in the opposite active site, and be phosphorylated by it. This arrangement of Ire1α molecules provides a straightforward mechanistic model for how dimerisation of Ire1 N-terminal domains in the lumen of the ER would facilitate association and transphosphorylation of their associated kinase domains on the cytoplasmic side of the membrane. (B) Autophosphorylation of dephosphorylated dimer interface mutants Q636A and F637A Ire1 as compared with wild type. Both wild-type and mutant proteins were incubated with 5 mM MgCl and 5 mM ATP at 37°C and samples were run at specific time points. Protein samples were visualised by western blot with generic Ire1α or the phospho-specific pS724-Ire1α.
No interaction resembling the back-to-back dimer seen in the yeast structures can be identified in the human Ire1α crystal structure. This maybe in part due to features such as the αE′ helix, which has a major role in forming the yeast back-to-back dimer but is absent from human Ire1α (and all metazoan homologues), and only some of the residues involved in the back-to-back interaction of the yeast ribonuclease domain are conserved in the human enzyme. The additional side-to-side interface observed in the other yeast structure (3FBV), and invoked in a formation of a biologically important high-order Ire1 assembly, also depends upon interactions of a secondary structural element (helix αD′) that is absent from human (and other metazoan) Ire1. Whether the back-to-back orientation exists in the human Ire1 mechanism remains to be seen. However, what is certain is that the face-to-face orientation is distinct from the back-to-back arrangement visualised in the yeast structures. As the human protein was extensively dephosphorylated prior to crystallisation, this face-to-face arrangement most likely represents a functional state that precedes the back-to-back ‘RNase active' arrangement seen in the phosphorylated yeast Ire1 structures.
The face-to-face interaction observed in the human Ire1α crystals provides a straightforward mechanistic explanation for how the cytoplasmic kinase domains of two Ire1 molecules, brought into close proximity by the association of their luminal domains on the opposite side of the ER membrane, might achieve transphosphorylation. The conformation of the activation segment, bolstered and supported by the partial unwinding of the C-helix, directs the conserved phosphorylation target Ser724 at the tip of the activation segment from one molecule, towards the active site of the opposite Ire1α molecule in the face-to-face dimer (Figure 2A). This type of reciprocal face-to-face transphosphorylation has been demonstrated structurally and biochemically for a number of kinases from different branches of the kinome (e.g. Chk2, LOK, SLK, DAPK3, etc), which like Ire1 are activated by dimerisation (Oliver et al, 2006, 2007; Pike et al, 2008). The secondary structural elements involved in the human Ire1α face-to-face interaction (the β1–β2 hairpin, β3–αC loop, and αG helix) are present in all Ire1 homologues, and many of the interfacial residues are invariant or only conservatively varied in metazoa and yeast, suggesting that, this face-to-face interaction would be available to all Ire1 homologues.
To test whether this interface is indeed biologically significant in the autophosphorylation process, we made mutations in key interfacial residues, Q636A and F637A, and determined their ability to autophosphorylate as compared with wild type (Figure 2B). Ire1 autophosphorylation was measured over several time points and using an antibody developed to measure phosphorylation of Ser724 on the activation segment of human Ire1α (see below). Both wild-type and mutant protein were extensively dephosphorylated prior to incubation with ATP-Mg2.
Mutation of residues implicated in the face-to-face interaction observed in the Ire1α crystals, significantly diminishes the ability of Ire1 to autophosphorylate compared with wild type, indicating the importance of this interface. In particular, the mutant Q636A, which disrupts the hydrogen bond with Asp634 and its Gln637 counterpart in the other monomer, shows significant retardation of autophosphorylation especially over the initial 30 min compared with wild type. The mutation of phenylalanine to a lesser hydrophobic alanine has less of an impact, but is still significantly slower to autophosphorylate than wild type.
Ire1α nucleotide binding
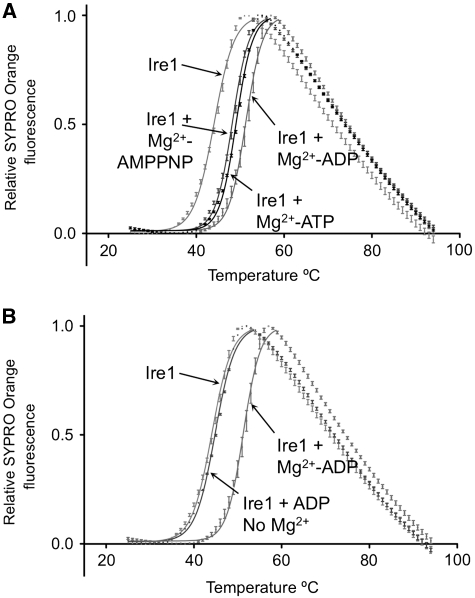
Previous structural and biochemical studies have suggested a mechanisms of Ire1 activation, whereby ADP binding to the phosphorylated protein promotes a conformational change that is essential for activation of the ribonuclease activity (Sidrauski and Walter, 1997; Lee et al, 2008). This notion is supported by studies, showing that a mutant form of yeast Ire1 can be directly activated by a mutant-specific ATP-competitive inhibitor (Papa et al, 2003), and most radically, that wild-type yeast Ire1 can be directly activated by the clinically licensed kinase inhibitor sunitinib (Korennykh et al, 2009). A key feature of this ADP-activation mechanism is the highly unusual observation, unique to Ire1, that only the phosphorylated state of Ire1 is competent to bind ADP (Lee et al, 2008). Nonetheless, the human Ire1α crystals grown from protein that has been very substantially dephosphorylated by λ-phosphatase (see Materials and methods) clearly contain Mg2+-ADP at full occupancy (Figure 1C). To test this further, we analysed the binding of ADP to human Ire1α using a thermal-shift assay (Cummings et al, 2006), and observed clear Mg2+-dependent ADP binding to dephosphorylated protein (Figure 3A and B), fully consistent with its presence in the crystals. Given the disparity between our observations and those previously reported, we re-examined the yeast study, and noted that the unphosphorylated yeast Ire1, used to demonstrate non-binding of ADP, was a kinase-dead mutant, inactivated by an alanine mutation of Asp797 in the catalytically essential HRD motif (Lee et al, 2008). As this residue is in close proximity to a bound Mg2+ and its associated solvent molecules, it is likely that the observed loss of affinity in the Asp797Ala mutant results from disruption of the structure and electrostatic complementarity of the Mg2+-binding site, with consequent loss of affinity for ADP. The enzymatically dephosphorylated human Ire1α with a native-binding site, used here, is fully able to bind Mg2+-ADP in its unphosphorylated state like all other known protein kinases.
Figure 3.
Nucleotide binding. (A) Thermal-shift analysis of adenine nucleotide binding to dephosphorylated Ire1α. ATP and AMPPNP stabilise Ire1 by +4.7°C and +4.1°C, respectively, while ADP shows the tightest binding, stabilising Ire1α by +7.2°C. Reaction conditions are given in Materials and methods section. (B) ADP binding to Ire1α is totally dependent on Mg2+ and in its absence ADP has minimal effect on Ire1α stability.
Ire1α phosphorylation activity and inhibition
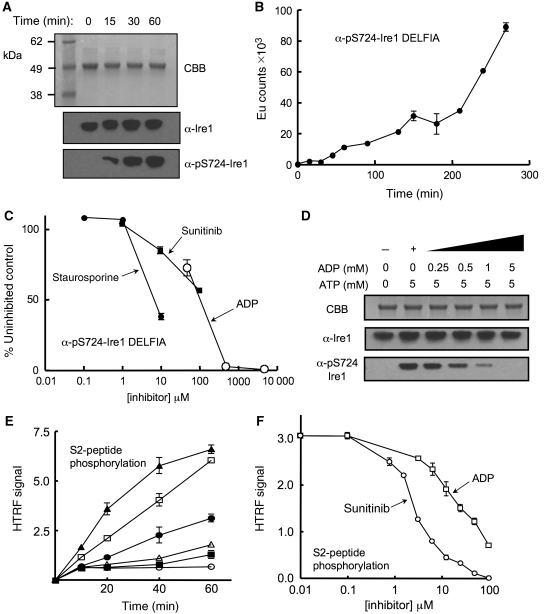
The involvement of Ire1α in human cancers in general, and multiple myeloma in particular, identifies it as an important potential target for therapeutic intervention (Lee et al, 2003). However, the uncertainties regarding the activation mechanism, and the observation of paradoxical activation rather than inhibition of the yeast enzyme by ATP-competitive inhibitors, have placed a question mark over its suitability as a target. To try and clarify these issues, we developed a phospho-specific antibody to a peptide derived from residues 718–729 in the activation segment of human Ire1α, incorporating a phospho-serine at 724, and used this to measure autophosphorylation by Ire1α (see Materials and methods). We were able to measure robust autophosphorylation on Ser724 with the previously dephosphorylated human Ire1α when incubated with Mg2+-ATP (Figure 4A), which showed non-linear kinetic behaviour consistent with a transautophosphorylation reaction (Figure 4B). This autophosphorylation was substantially inhibited when the broad-specificity protein kinase inhibitor staurosporin was included in the reaction (Figure 4C). Furthermore, we found that ADP was also able to efficiently inhibit autophosphorylation of Ire1α (Figure 4C and D), consistent with it being able to bind to the nucleotide-binding site in the unphosphorylated enzyme contrary to previous suggestions (see above). Most significantly, we found that Ire1α autophosphorylation was also inhibited by sunitinib (Figure 4C).
Figure 4.
Autophosphorylation and inhibition. (A) Autophosphorylation of dephosphorylated Ire1α monitored by α-pS724-Ire1α phospho-specific antibody. Ire1α was incubated with 5 mM ATP and 5 mM MgCl2 at 37°C for the times indicated. Protein was visualised by SDS–PAGE with Coomassie Brilliant Blue (CBB) and by western blot with a generic Ire1α antibody or the phospho-specific α-pS724-Ire1α antibody. (B) Quantitative DELFIA assay measuring autophosphorylation of dephosphorylated Ire1α monitored by α-pS724-Ire1α phospho-specific antibody. Reaction used 700 nM Ire1α at 37°C in the presence of 0.5 mM ATP and 20 mM Mg2+. The non-linear kinetics are consistent with a transphosphorylation reaction. (C) Inhibition of Ire1α autophosphorylation by the broad-specificity kinase inhibitor staurosporine, the licensed anti-cancer drug sunitinib, and ADP. Autophosphorylation reaction was followed by DELFIA assay as in (D) after 2.5 h. (D) Inhibition of Ire1α autophosphorylation by ADP. Autophosphorylation of previously dephosphorylated Ire1α, indicated by α-pS724-Ire1α western blot, is progressively inhibited in the presence of increasing concentrations of ADP, and essentially blocked at equimolar ADP to ATP, consistent with the higher affinity of ADP for the nucleotide-binding site in the unphosphorylated kinase indicated by the thermal-shift analysis (A). Reactions were carried out in the presence of 5 mM MgCl2. (E) Hetero-phosphorylation of the HTRF biotinylated peptide S2 (200 nM) by phosphorylated Ire1α (○=no enzyme; ▪=50 nM; ▵=100 nM; •=200 nM; □=400 nM; and ▴=800 nM) in the presence of 30 μM Mg-ATP. (F) Concentration-dependent inhibition of Ire1α hetero-phosphorylation activity. Curves are shown for sunitinib (IC50=3.7 μM±1.2) and ADP (IC50=38 μM±17). Staurosporine (data not plotted) had an IC50=13 nM±8.4. Reactions used 200 nM Ire1α, 200 nM peptide S2, and 30 μM ATP. IC50 values are averages of three experiments.
To date, no other biological substrate for Ire1α phosphorylation other than itself, has been identified. However, using a commercial homogeneous time-resolved fluorescence assay system (Cisbio, France), we identified a peptide, S2, that was efficiently phosphorylated by Ire1α (Figure 4E). While autophosphorylation may be the only significant activity of the Ire1α kinase in vivo, that it is nonetheless able to phosphorylate a heterologous substrate in vitro, shows that phosphorylated Ire1 retains the conformation required for the phosphoryl-transfer reaction, including a presumably ordered activation segment (Johnson et al, 1996), and is able to bind Mg2+-ATP and a peptide substrate productively. As with the autophosphorylation activity, hetero-phosphorylation of the S2 peptide was inhibited by staurosporin, ADP, and sunitinib (Figure 4F).
Dependence of Ire1α RNase activity on phosphorylation
The truly unique feature of Ire1 is its possession of both protein kinase and ribonuclease activities. Furthermore, there is clear evidence that the ribonuclease activity is dependent on the kinase activity, so that kinase-dead Ire1 mutants, and mutants in which the activation-segment autophosphorylation site are mutated, are defective in ribonuclease activity and UPR signalling (Shamu and Walter, 1996; Welihinda and Kaufman, 1996; Tirasophon et al, 2000). To analyse this dependence with the human Ire1α protein used here, we developed a semi-quantitative assay for in vitro cleavage of unspliced human Xbp1 mRNA, using an Agilent BioAnalyzer (Agilent Technologies, Cheshire, UK). We found that phosphorylated Ire1α, showing strong immunoreactivity to the pS724 antibody, was fully able to cleave the Xbp1 mRNA, whereas Ire1α that was incubated with λ-phosphatase and was devoid of S724 phosphorylation could not (Figure 5A).
Figure 5.
Xbp1 mRNA cleavage by human Ire1α. (A) Elution profiles from Agilent Bioanalyser of (left) untreated Xbp1 mRNA, (middle) Xbp1 mRNA incubated with dephosphorylated Ire1α, and (right) Xbp1 mRNA incubated with phosphorylated Ire1α. Reaction conditions are given in Materials and methods section. The peak eluting at a position corresponding to 800 nucleotides in the left and middle panels is the unspliced Xbp1 mRNA, whereas the pair of peaks eluting at 300 and 500 nucleotides, respectively, in the right panel, corresponds to the two cleaved products of Xbp1 mRNA. The peak at 25 nucleotides in all three panels is a calibration marker. (B) In vitro cleavage activity of Ire1α, wild-type, and Q636A mutant, on a fluorescently tagged oligonucleotide encapsulating the sequence and secondary structure of a known Ire1α cleavage site in human Xbp1 mRNA. Cleavage generates a fluorescent species with higher gel mobility. The Q636A mutation, which causes a defect in autophosphorylation, results in a similar decrease in RNase activity, confirming the dependence of RNase activation on autophosphorylation. (C) Sunitinib inhibits Xbp1 mRNA splicing in vivo. Myeloma cell lines (U266+H929) were treated with tunicamycin to induce ER stress, in the presence or absence of sunitinib, and levels of spliced (Xbp1s) and unspliced (Xbp1u) mRNAs in the treated cells were determined by quantitative real-time PCR using LUX primers (see Materials and methods), and plotted as relative Xbp1u:Xbp1s ratio. In both cell lines, the addition of the Ire1α kinase inhibitor sunitinib significantly inhibits the splicing of Xbp1 mRNA. (D) Similar results were obtained by RT–PCR amplification. (E) Western blot of protein extracts from cells treated as in (C), showing inhibition of Xbp1 protein production in ER-stressed cells treated with sunitinib. Actin is shown as a loading control.
The dimerisation interface mutant Q363A showed a markedly decreased ability to autophosphorylate (Figure 2B). As activation-segment phosphorylation correlated with the ability to cleave Xbp1 mRNA (see above), we determined the effect of the Q363A mutation on the RNase activity of Ire1α using an RNA hairpin-nicking assay. Dephosphorylated Ire1α-Q363A showed substantially lower RNase activity than the wild-type protein, when incubated with an Xbp1 mRNA-derived hairpin oligonucleotide in the presence of Mg-ATP (Figure 5B). The diminished RNase activity of the autophosphorylation-defective Q636A mutant is fully consistent with a direct dependence of RNase activity on the ability of Ire1α to autophosphorylate, and argues very strongly that the observed dimer crystal structure is a key step in the activation mechanism of Ire1α.
As sunitinib was able to effectively inhibit autophosphorylation of Ire1α, on which the development of Ire1α RNase activity depends (see above), we sought to determine whether sunitinib could affect splicing of Xbp1 mRNA in vivo. We treated H929 and U266 myeloma cells with tunicamycin, which generates ER stress (Davenport et al, 2007), and is an effective activator of the UPR, and observed robust increases in the levels of Xbp1s mRNA that could be detected by real-time quantitative polymerase chain reaction (PCR) and by gel electrophoresis of reverse transcribed PCR products 4 h after treatment (Figure 5C and D). When sunitinib was added, Xbp1s mRNA levels were substantially reduced. Xbp1 protein levels, visualised by western blot, reflected changes in the cellular Xbp1s mRNA levels, with sunitinib causing a marked decrease in detectable Xbp1 protein (Figure 5E). Thus, inhibition of Ire1α autophosphorylation by sunitinib has the consequence of inhibiting development of the autophosphoryation-dependent RNase activity of Ire1α, and thereby blocks Xbp1 mRNA splicing and consequent Xbp1 protein expression in myeloma cells.
Discussion
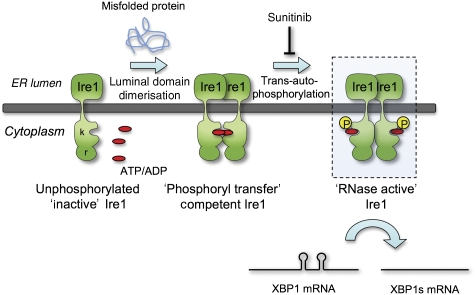
The crystal structure of human Ire1α described here indicates a face-to-face association of the cytoplasmic kinase domains, such that the activation segment of one monomer can reciprocally access and be phosphorylated by the active site of the other. This orientation is distinct from the ‘RNase active' back-to-back orientation that has been seen in all the yeast structures thus far, and represents a ‘phosphoryl-transfer' competent state. The difference in this conformation is due to our protein being extensively dephosphorylated prior to crystallisation and therefore most likely represents an early stage of the Ire1 mechanism of action, in which the initial dimerisation of the luminal domains allow the face-to-face orientation to occur in order to transphosphorylate.
Furthermore, we show that the dephosphorylated protein is able to bind nucleotide (both ADP and ATP) in the presence of Mg2+ and rather than Ire1α being allosterically locked in an active state by ADP binding post-phosphorylation, as suggested, Ire1α, is fully able to bind ADP in its inactive unphosphorylated state, and indeed its autophosphorylation, on which its RNase activity depends, is competitively inhibited by ADP. Based on our observations and previous studies, we propose a model (Figure 6) whereby in the absence of any unfolded protein, Bip associates with the luminal domains of Ire1. Upon accumulation of misfolded protein Bip dissociates, which in turn allows dimerisation of the luminal domains to occur. This juxtaposes the kinase domains in a face-to-face orientation and upon binding of nucleotide, strengthens this interaction such that the activation segment of one monomer can access the active site of the opposite monomer in trans with the projecting activation segment supported by formation of an anti-parallel β-sheet with the residues from the N-terminal end of the C-helix, in their unwound extended conformation. The effect of the interfacial mutations on both kinase and RNase activity strongly suggests that the crystal structure we have obtained represents a biologically authentic state that is the necessary prelude to Ire1 transphosphorylation and its consequent ability to cleave Xbp1 mRNA.
Figure 6.
Autophosphorylation-dependent activation of Ire1α RNase activity. Model of Ire1 dimer-induced mechanism of action, highlighting the role of the face-to-face ‘phosphoryl-transfer' competent state defined here that facilitates transphosphorylation and phosphorylation-dependent downstream activation of the RNase activity. Whether the putatively RNase active back-to-back interaction observed in yeast Ire1 structures also occurs in the mammalian system is yet to be determined—the main structural components, not being conserved—and the mechanism by which autophosphorylation mediates RNase activation is yet to be directly demonstrated. Nonetheless, that inhibition of autophosphorylation by an ATP-competitive inhibitor such as sunitinib blocks consequent RNase activity provides a means by which therapeutic manipulation of the Ire1 arm of the UPRs might be achieved.
From the structures observed for phosphorylated yeast Ire1, the protein must rearrange on autophosphorylation from the face-to-face orientation to the back-to-back orientation that is suggested to be the state of the protein required for RNase activity. Whether this occurs in the human protein remains to be seen—the residues involved in the yeast dimers not being conserved. Indeed, we still await an Ire1–RNA complex structure from either yeast or human that will give us direct experimental insights into the true RNase ‘active' conformation. A corollary of the back-to-back arrangement for the activated Ire1 is that the kinase active sites and activation segments would be easily accessible to phosphatases, providing a facile mechanism for dephosphorylation and deactivation that returns Ire1 to the ‘ground' state. Both the back-to-back and face-to-face interfaces contain residues that have now been shown to be functionally significant by mutational studies at least in the species tested, so that both arrangements may be required for the complete Ire1 mechanistic cycle.
Our structure provides a clear mechanism for how transphosphorylation occurs on the activation segment and has many precedents in other dimeric protein kinases. What remains to be defined is a satisfactory mechanistic explanation as to how transphosphorylation of the activation segment is biochemically coupled to activation of the C-terminal RNase domain and specifically how autophosphorylation switches Ire1 from a face-to-face to back-to-back orientation, if that is indeed the RNase active configuration.
Finally, our biochemical and mutational analysis of human Ire1α shows direct dependence of RNase activity on autophosphorylation, and that inhibition of autophosphorylation by ATP-competitors blocks Ire1α RNase activity in vivo, including sunitinib that has the conventional and predictable effect of inhibiting autophosphorylation and consequent RNase activation, unlike in the yeast system. The demonstration that an ATP-competitive inhibitor of Ire1α autophosphorylation can inhibit Xbp1 mRNA splicing in myeloma cells opens the way for the development of selective and potent new drugs for the treatment of this disease.
Materials and methods
Cloning, expression, and purification of dephosphorylated Ire1α
The coding sequence for human Ire1α was PCR amplified from a human liver cDNA library, with primers designed to encompass the intact kinase and endoribonuclease domains (residues 547–977). The PCR product was cloned into a modified version of the pFastBac vector that contains a 6xHis tag followed by a rhinovirus 3C protease site. Recombinant baculovirus was prepared using the Bac-to-Bac (Invitrogen, Paisley, UK) method and were transfected into Sf9 insect cells. Three rounds of amplification were undertaken, and at each stage, the baculovirus titer was measured by plaque assay. Protein was expressed after 3 days of growth at 27°C. The cell pellet was lysed by sonication in 50 mM Hepes pH 7.5, 300 mM NaCl, 10% glycerol, supplemented with protease inhibitors and centrifuged to remove cell debris and insoluble material at 40 000 g for 60 min. The supernatant was then passed through a batch/gravity column containing 15 ml of talon resin (Clontech) and protein was eluted off with 300 mM imidazole in a total volume of 50 ml. Ire1 was dephosphorylated and the his-tag was removed by incubating with λ-phosphatase (300 μl at 1.6 mg/ml) in the presence of 2 mM MnCl2 and Rhinovirus 3C protease (PreScission protease, Amersham Biosciences, 300 μl at 2 mg/ml) for 30 min at room temperature for phosphatase activity and then overnight at 4°C. At this stage, both 1 mM EDTA and 5 mM DDT was added and kept in all subsequent buffers. Dephosphorylation was confirmed by western blot using a pSer724 phospho-specific antibody. Human Ire1 was further purified from contaminants by ion exchange using a Mono-Q column and by size exclusion chromatography on a Superdex200 column (GE Healthcare).
Cloning, expression, and purification of dephosphorylated interface mutants of Ire1α
Interface mutants, Q636A, F637A, of the intact kinase and endoribonuclease domains (residues 547–977) in a modified version of pFastBac vector, were cloned using a QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) following the manufacturer's recommended protocol. Mutant protein was generated using the baculovirus expression system and purified in the same way as wild-type protein (as mentioned above), with the omission of the final purification step of size exclusion chromatography, due to the relatively low expression level of the mutant proteins.
Immunoblotting
Protein (5–10 μg) was electrophoresed on 4–12% precast Bis-Tris gels (Invitrogen) and transferred to nitrocellulose. Blocking solution, (40 ml PBS, 0.1% Tween-20, 2 g of Marvel Dried Milk Powder) was used to clarify the membrane and 5 μl of primary antibody was added to 10 ml of blocking solution in a 1/2000 dilution, and this was mixed for 40 min. The membrane was washed three times with PBS buffer (PBS+0.1% Tween-20), followed by addition of the horseradish peroxidase-conjugated secondary antibody at 1/5000 dilution and mixed for 1 h. This was washed a further three times, and bands were visualised using ECL chemiluminescent reagents (GE Healthcare) and developed on Biomax film (Kodak).
Crystallisation
Protein was prepared for crystallisation by concentrating to 9 mg/ml and incubated with 1 mM ADP, 2 mM MgCl2 on ice for 30 min. Initial crystallisation experiments were conducted using a Phenix robot (Art Robbins Instruments) using sparse matrix screens. Crystals were obtained in two conditions, 2 M (NH4)2SO2, 2.2 M NaSCN (condition 1) and 25% PEG 3350, 0.2 M (NH4)2SO2, Hepes pH 7.5 (condition 2). Condition 1 gave crystals that were large and readily reproducible, but diffracted poorly, whereas condition 2 gave small crystals that were small and hard to reproduce, but did diffract. Several rounds of microseeding were used to improve crystals grown from condition 2 that were suitable for data collection. Crystals were cryoprotected with 30% glycerol and flash cooled in liquid nitrogen.
Data collection and processing
Data to 2.7 Å of the human Ire1α–ADP–MgCl2 complex were collected on station I03 at Diamond light source from a single crystal at 100 K. The crystals belonged to the P21 space group (cell dimensions a=109 Å, b=83 Å, c=116 Å, β=94°), with four molecules in the asymmetric unit. Images were integrated using MOSFLM (Leslie, 1995), and reduced/scaled using programs from the CCP4 suite (CCP4, 1994). Human Ire1α crystal structure was solved by molecular replacement using the program PHASER (McCoy, 2007), with the yeast Ire1 crystal structure used as the search model (PDB:2rio). The generated maps were of good quality especially around the active site and in particular for ADP-Mg2+ ligand. Phenix.autobuild (Adams et al, 2004) was used for initial model building and then iterative rounds of manual building in Coot (Emsley and Cowtan, 2004) and simulated annealing refinement in phenix yielded a structure with good agreement with observed data and stereochemistry (96% of residues in favoured and allowed regions in the Ramachandran plot) (see Table I). Coordinates and structure factors have been deposited in the Protein Databank with accession code: 3P23. Buried surface areas were calculated using the PISA server http://www.ebi.ac.uk/msd-srv/prot_int/pistart
Table 1. Crystallographic data collection and refinement statistics.
| Data collection | |
| Space group | P21 |
| Cell dimensions, a, b, c, (Å) | 109.8, 83.2, 116.0, β=94° |
| Resolution (Å) | 58.8–2.7 |
| Rsym (%) | 9 (32) |
| I/σI | 7.9 (3.2) |
| Completeness (%) | 98.5 (98.5) |
| Redundancy | 3.2 |
| Refinement | |
| Reflections | 56 524 |
| Rwork/Rfree | 0.24/0.29 |
| RMSD | |
| Bond length | 0.003 |
| Bond angle | 0.914 |
| No. of atoms | |
| Protein | 12 500 |
| Ligand | 1554 |
| Water | 504 |
| Overall B factor | 39 |
Quantitative measurement of autophosphorylation and peptide phosphorylation by Ire1α
DELFIA assay buffer (4002-0010), europium-labelled anti-rabbit IgG (AD0105), and enhancement solution (4001-0010) were obtained from Perkin-Elmer Life Sciences. Rabbit phospho-specific Ire1α antibody was raised to a KLH conjugate of the peptide CLAVGRHS(p)FSRRSG and affinity purified using the non-phosphorylated peptide (Open Biosystems, Huntsville, AL). The HTRF® KinEASE kit (Cisbio Bioassays, Bagnols/Ceze Cedex, France) was used to detect the peptide phosphorylation activity of the Ire1α proteins. All other chemicals were from Sigma-Aldrich Co. Ltd.
For the autophosphorylation of Ire1α, non-phosphorylated Ire1α, ATP, inhibitor or 3% DMSO/buffer were incubated at 37°C for 1 h (40 mM Tris, 20 mM MgCl2, 1 mM DTT pH 7.4) and the reaction stopped by addition of 20 mM EDTA. The DELFIA assay measured phosphorylated Ire1α in 96-well plates (Thermo Immulon2-HB) using a coated-antigen format. The phosphorylation mix (50 μl) was transferred into the microtitre plates and incubated at 4°C overnight. After washing and blocking (5% dried milk/PBS), phosphorylated Ire1α was detected using specific primary antibody (165 ng/ml) and europium-labelled anti-rabbit IgG (0.25 μg/ml). Fluorescence (615 nm) was measured using an Envision multilabel counter (Perkin-Elmer Life Sciences) using a time-resolved measurement mode.
Peptide phosphorylation was measured using the STK Substrate 2-biotin (Biotinyl-Ahx-RRRLSFAEPG-CONH2) substrate from the HTRF KinEASE kit. This peptide gave the optimum activity with a Km of 200 nM. Assays were carried out in low volume, black 384-well plates (3676 from Corning Life Sciences, MA), with a 10 μl assay volume containing 30 μM ATP (Km=26 μM), 600 nM STK Substrate 2-biotin and 100 nM of phosphorylated or non-phosphorylated Ire1. Following incubation at 37°C for 1 h, the reaction was stopped with buffered EDTA, which contained the detection reagents, streptavidin-XL665 and the STK-antibody labelled with Eu3+-cryptate. The resulting TR-FRET signal, calculated as the fluorescence ratio at 665/620 nm, was read on an Envision and was proportional to the level of phosphorylation of the peptide.
Thermal shift ligand binding assays
To monitor the binding of ligands to Ire1, the thermal-shift assay was performed. The method is based on the observation that ligands change protein thermal stability upon binding to the protein. This results in a change of the midpoint temperature for the thermal protein unfolding transition, Tm.
The protein unfolding process is monitored using the environmentally sensitive dye Sypro Orange. Its quantum yield increases upon binding to hydrophobic surfaces exposed during protein unfolding and so does the fluorescence signal.
The thermal-shift assay was conducted in the DNA Engine Opticon 2 (Bio-Rad), originally designed for RT–PCR. Solutions of 4 μM Ire1, 4.5 X Sypro Orange, 50 mM HEPES pH 7.5, and 200 mM NaCl final concentration and a volume of 50 μl were added to the wells of a 96-well PCR plate. The final concentration of ADP, AMPPNP, and ATP was 2 mM for each, with 0 or 5 mM MgCl, for Sunitinib it was 1 mM with 5 mM MgCl.
Data were analysed with the Excel-based worksheet DSF analysis. To obtain Tm, a Boltzmann model was used to fit the fluorescence imaging data using the curve-fitting software Prism (GraphPad):
where I is the fluorescence intensity at temperature T, A and B are pretransitional and posttransitional fluorescence intensities, respectively, and C is a slope factor.
Data points after the fluorescence intensity maximum were excluded from fitting. In the absence of ligands (control, with 2% DMSO for nucleotides and 10% DMSO for Sunitinib), Tm=T0, and the ligand-dependent changes in midpoint temperature, ΔTm=Tm−T0, could be calculated for each ligand.
In vitro full-length XBP1 RNAse cleavage assay
Full-length Xbp1 mRNA was synthesised from cDNA from the myeloma cell line U266, which only expresses the unspliced form of Xbp1 (Xbp1u) using the following PCR primers: XBP1-F NDE1 site: GGAATTCCATATGATGGTGGTGGTGGCAGCCGCG, XBP1-R HindIII site: CGGACGAAAGCTTTTAGTTCATTAATGGCTTCCAGCTTGG. The Xbp1 fragment was inserted into TOPO-TA vector (TOPO-TA kit, Invitrogen) according to the manufacturer's instructions. Transformation was performed using XL-blue competent cells (Stratagene). Following restriction with HindIII, the construct was reverse transcribed using the MEGAscript® T7 Kit (Ambion, Austin, TX), and purified using the GeneChip® IVT cRNA Cleanup Kit (Affymetrix, High Wycombe, UK) to produce a full-length Xbp1u mRNA. In all, 150 ng Ire1α was incubated with 500 ng Xbp1u mRNA at 37°C for 90 min, purified (GeneChip IVT cRNA Cleanup Kit) and analysed using the Agilent Bioanalyser system (Agilent Technologies) according to the manufacturer's instructions.
In vitro hairpin-RNA cleavage assay
Single-stranded RNA mimicking the stem loop structure of Ire1's Xbp1 RNA substrate was synthesised with a Cy5 tag at the 5′ end: Cy5ACCUCUGCAGCAGGU (Sigma Genosys). The substrate RNA was then incubated with wild-type and mutant protein at a concentration of 10 μM in the presence of MgCl2 and ATP at 2 mM and 1 mM, respectively. The reaction was kept at 37°C and at specific time intervals, samples were taken and run on a 15% TBE-Urea polyacrylamide gel. Gels were analysed by excitation at 635 nm wavelength using a Fulji FLA5000 laser scanner.
In vivo XBP1 mRNA splicing
Myeloma cell lines were grown in RPMI1640 GlutaMAX™ (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum and 100 μg/ml streptomycin. Cultures were kept at 37°C in a humidified atmosphere of 95% air/5% carbon dioxide. Cells were treated with 100 μM tunicamycin to induce stress ±12.5 μM Sunitinib for 2 and 4 h. Following RNA extraction and cDNA synthesis, samples were subjected to PCR using 100 pmol/μl LUX primers (Invitrogen) and the ratio of Xbp1s:Xbp1u was determined. Primers were as follows: XBP1-s F (FAM-tag): CACCTCTGAGTCCGCAGCAGG[FAM]G, XBP1-u R (JOE-tag): CAGAGGCTCAGACTACGTCACCTC[JOE]G, XBP1 common R (unlabelled): CCAGAATGCCCAACAGGATA. Thermal-cycling conditions were 10 min at 95°C, 40 cycles at 95°C for 15 s followed by 1 min at 60°C. Samples were run and analysed on the 7500 Fast time real-time PCR machine (Applied Biosystems).
To determine relative expression levels of XBP1/XBP1s within a sample, PCR was performed in a 50-μl platinum Taq reaction containing 4 pmol of primers (forward 5′-CCTTGTAGTTGAGAACCAGG-3′ and reverse 5′ GGGGCTTGGTATATATGTGG-3′; Invitrogen), 1 unit of platinum Taq DNA polymerase, and 200 μM dNTP. The temperature profile was at 94°C for 2 min, followed by 30 cycles of 94°C for 15 s, 60°C for 1 min, and 72°C for 30 s. Products were run on 2% agarose gels containing ethidium bromide.
In addition, protein was obtained at similar time points following drug treatment and subjected to western blotting using standard conditions. Antibodies were Xbp1 (Biolegend) with an anti-rabbit secondary (GE Healthcare Ltd).
Supplementary Material
Acknowledgments
We are grateful to Valerie Good, Kirsten Wood, Daniel Itzhak, Lauren Aronson, Mark Roe, and Mathieu Rappas for assistance and advice. We are grateful to the Diamond Light Source PLC for access to synchrotron radiation. LHP acknowledges support from The Wellcome Trust (Programme Grant 080041/Z/06/Z/) and Cancer Research UK (Infrastructure Support Grant C302/A7803). FED acknowledges support from Kay Kendall Leukaemia Fund, Department of Health and Myeloma UK. WA, IC, MR, MC, and AH acknowledge support from Cancer Research UK (Programme Grants C309/A8274 and C309/A8365). MA acknowledges support from Cancer Research UK Fellowship (C33269/A11161). We acknowledge NHS funding to the NIHR Biomedical Research Centre.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC (2004) Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat 11: 53–55 [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D (2000) Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2: 326–332 [DOI] [PubMed] [Google Scholar]
- Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE, Zheng M, Mani M, Henderson J, Pinkus GS, Munshi N, Horner J, Ivanova EV, Protopopov A, Anderson KC, Tonon G, DePinho RA (2007) The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell 11: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) Programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P (2005) On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA 102: 18773–18784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings MD, Farnum MA, Nelen MI (2006) Universal screening methods and applications of ThermoFluor. J Biomol Screen 11: 854–863 [DOI] [PubMed] [Google Scholar]
- Davenport EL, Moore HE, Dunlop AS, Sharp SY, Workman P, Morgan GJ, Davies FE (2007) Heat shock protein inhibition is associated with activation of the unfolded protein response pathway in myeloma plasma cells. Blood 110: 2641–2649 [DOI] [PubMed] [Google Scholar]
- Davies FE, Dring AM, Li C, Rawstron AC, Shammas MA, O'Connor SM, Fenton JA, Hideshima T, Chauhan D, Tai IT, Robinson E, Auclair D, Rees K, Gonzalez D, Ashcroft AJ, Dasgupta R, Mitsiades C, Mitsiades N, Chen LB, Wong WH et al. (2003) Insights into the multistep transformation of MGUS to myeloma using microarray expression analysis. Blood 102: 4504–4511 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T (2000) A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol 2: 379–384 [DOI] [PubMed] [Google Scholar]
- Gonzalez TN, Sidrauski C, Dorfler S, Walter P (1999) Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J 18: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenendyk J, Michalak M (2005) Endoplasmic reticulum quality control and apoptosis. Acta Biochim Pol 52: 381–395 [PubMed] [Google Scholar]
- Huse M, Kuriyan J (2002) The conformational plasticity of protein kinases. Cell 109: 275–282 [DOI] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Glimcher LH (2003) The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol Rev 194: 29–38 [DOI] [PubMed] [Google Scholar]
- Johnson LN, Noble ME, Owen DJ (1996) Active and inactive protein kinases: structural basis for regulation. Cell 85: 149–158 [DOI] [PubMed] [Google Scholar]
- Kimata Y, Kimata YI, Shimizu Y, Abe H, Farcasanu IC, Takeuchi M, Rose MD, Kohno K (2003) Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol Biol Cell 14: 2559–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P (2009) The unfolded protein response signals through high-order assembly of Ire1. Nature 457: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumenis C (2006) ER stress, hypoxia tolerance and tumor progression. Curr Mol Med 6: 55–69 [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH (2003) Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci USA 100: 9946–9951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Dey M, Neculai D, Cao C, Dever TE, Sicheri F (2008) Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell 132: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW (1995) MOSFLM Users Guide. Cambridge, UK: MRC Laboratory of Molecular Biology [Google Scholar]
- McCoy AJ (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63: 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K (2000) Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem Biophys Res Commun 279: 445–450 [DOI] [PubMed] [Google Scholar]
- Oliver AW, Knapp S, Pearl LH (2007) Activation segment exchange: a common mechanism of kinase autophosphorylation? Trends Biochem Sci 32: 351–356 [DOI] [PubMed] [Google Scholar]
- Oliver AW, Paul A, Boxall KJ, Barrie SE, Aherne GW, Garrett MD, Mittnacht S, Pearl LH (2006) Trans-activation of the DNA-damage signalling protein kinase Chk2 by T-loop exchange. EMBO J 25: 3179–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa FR, Zhang C, Shokat K, Walter P (2003) Bypassing a kinase activity with an ATP-competitive drug. Science 302: 1533–1537 [DOI] [PubMed] [Google Scholar]
- Pike AC, Rellos P, Niesen FH, Turnbull A, Oliver AW, Parker SA, Turk BE, Pearl LH, Knapp S (2008) Activation segment dimerization: a mechanism for kinase autophosphorylation of non-consensus sites. EMBO J 27: 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Shamu CE, Cox JS, Walter P (1994) The unfolded-protein-response pathway in yeast. Trends Cell Biol 4: 56–60 [DOI] [PubMed] [Google Scholar]
- Shamu CE, Walter P (1996) Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J 15: 3028–3039 [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Walter P (1997) The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90: 1031–1039 [DOI] [PubMed] [Google Scholar]
- Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ (2000) The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev 14: 2725–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ (1998) A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev 12: 1812–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd DJ, Lee AH, Glimcher LH (2008) The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol 8: 663–674 [DOI] [PubMed] [Google Scholar]
- Welihinda AA, Kaufman RJ (1996) The unfolded protein response pathway in Saccharomyces cerevisiae. Oligomerization and trans-phosphorylation of Ire1p (Ern1p) are required for kinase activation. J Biol Chem 271: 18181–18187 [DOI] [PubMed] [Google Scholar]
- Wiseman RL, Zhang Y, Lee KP, Harding HP, Haynes CM, Price J, Sicheri F, Ron D (2010) Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell 38: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107: 881–891 [DOI] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ (2008) From endoplasmic-reticulum stress to the inflammatory response. Nature 454: 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu CY, Back SH, Clark RL, Peisach D, Xu Z, Kaufman RJ (2006) The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci USA 103: 14343–14348 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.