Re-dicing the pancreatic β-cell: do microRNAs define cellular identity?
A role for Dicer and miRNAs in regulating pancreatic β-cell differentiation and insulin production is discussed here in the broader context of miRNA functions in cell fate specification and cellular stress responses.
EMBO J 30 5, 835–845 (2011); published online February012011
Understanding the mechanisms which contribute to successful growth and function of the pancreatic β-cell is essential to our ability to combat the global epidemic of diabetes. To date, there is limited in vivo data addressing the role of microRNAs or components of the RNAi machinery in the pancreatic β-cell. In this issue of The EMBO Journal, Melkman-Zehavi et al provide further evidence of how small RNAs contribute to the normal growth and function of this cell type, characterizing the metabolic consequences of Dicer depletion. Their study highlights the role of miR-24, miR-26, and miR-148 in promoting insulin mRNA levels and begins to suggest potential for many other microRNAs to contribute to an already complex story.
The ribonuclease III (RNaseIII) enzyme Dicer is widely recognized for its role in the processing of double-stranded RNA (Bartel, 2009). As the initial publications by Greg Hannon and colleagues reporting its identification and role in development, subsequent conditional knockout studies have continuously re-emphasized that Dicer is essential for the processing of mature microRNAs and for cellular survival. Furthermore, as total loss of Dicer in mice results in embryonic lethality, and also impairs the assembly of centromeric heterochromatin, it is clear that this gene impacts a broad range of cellular processes (Siomi and Siomi, 2009). What remains to be determined is how its functions are interwoven, and how the microRNAs processed by Dicer coordinately facilitate the needs of the cell. To date, the compound loss of both miR-106b∼25 and miR-17∼92 is the only microRNA mutant to result in lethality (Ventura et al, 2008). Also, murine loss of function models for microRNAs, such as miR-21 (Patrick et al, 2010), have been reported with phenotypes apparent only under stress. Taken together, these observations make it difficult to weigh the relative contribution of each Dicer function to the lethal phenotype. Does the global decrease in microRNAs that are processed by Dicer outweigh the effects on chromatin assembly or other functions yet to be ascribed to this gene?
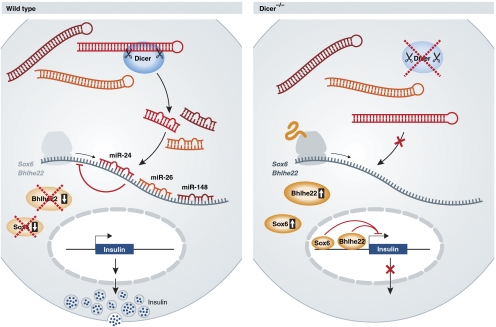
Nonetheless, given previous studies using Dicer conditional mutants, the observations reported here by Melkman-Zehavi et al (2011) show a somewhat surprising outcome. Strikingly, Dicer-deficient pancreatic β-cells lose their expression of insulin with only a slight but insignificant effect on pancreatic β-cell mass. A subtle decrease in cell number due to apoptosis would not be surprising as genetic ablation of Dicer is frequently associated with cell death. Evidence is provided that miR-24, miR-26, and miR-148 contribute to the direct regulation of two target genes known to repress expression of insulin, Sox6 and Bhlhe22 (Peyton et al, 1996; Iguchi et al, 2005) (Figure 1). Both genes are well-established regulators of developmental processes, but the relevance of their regulation by these microRNAs with respect to the growth of β-cells remains to be explored. Still, the question remains: is the decrease in insulin expression due solely to alterations in the expression of specific microRNAs or also to changes on the heterochromatin? In addition, if pancreatic β-cells in this model have lost expression of Dicer, and in turn insulin, can they still be considered β-cells? The expression of insulin is generally seen as the sole attribute to distinguish pancreatic β-cells from all other cell types. If this population of mutant cells has lost expression of this hormone, have they in fact transformed into another cell type? Immunofluorescence has been employed to confirm expression of established pancreatic markers such as Pdx1, MafA, Nkx6.1, and Pax6 to suggest that these Dicer-deficient cells maintain a part of their β-cell identity. Of note, electron microscopy could have proved useful to determine dramatic changes in their morphological composition such as the complete absence of insulin-containing secretory granules or changes to β-cell size. With the establishment of a link between insulin levels and the expression of Dicer, this raises the point that the small RNA profile can be used to identify specific cell types or their origins. Can the microRNAs addressed in this study, miR-24, miR-26, and miR-148, and their expression levels along with miR-375 and miR-7, be used to establish a β-cell-specific expression ‘signature'? As the expression of microRNAs is known to differ between tissues, it is possible the small RNA profile may one day be used for classification.
Figure 1.
Loss of Dicer decreases the expression of insulin in pancreatic β-cells. Sox6 and Bhlhe22 are targeted by miR-24, miR-26, and miR-148 to facilitate insulin production. Dicer deficiency decreases these microRNAs and allows for these transcriptional repressors to inhibit the expression of insulin mRNA.
Of note, the data presented here are in contrast to a report implementing a constitutively active RIP2-Cre line to deplete Dicer, where the authors reported no abnormalities with respect to pancreatic β-cell maintenance, islet development, or on circulating glucose levels (Lynn et al, 2007). Curiously, data on this mouse line were only discussed at ages E18.5 and 8 months. It may be that the absence of a reported phenotype in this Dicer knockout results from proliferation of Cre-negative cells that can form a sufficient β-cell mass to maintain normoglycemia.
In light of the numerous reports illustrating the effect of depleting Dicer and a global reduction of its associated microRNAs, the data presented here further demonstrate the influence of the microRNA pathway on the integrity and perhaps the identity of the β-cell. It comes as no surprise that fundamental processes such as growth of pancreatic β-cells and synthesis of insulin mRNA are dependent on the expression of numerous key genes, including miRNAs. One intriguing hypothesis that appears to be gaining momentum in the microRNA field has recently been illustrated by Sharp and colleague (Leung and Sharp, 2010). Within this review the authors point out the emergence of in vivo data suggesting microRNAs act to mediate the cellular response to stress. Previous observations with the miR-375 knockout mouse illustrate an effect on pancreatic β-cell mass that becomes more pronounced when crossed with the leptin-deficient obob mouse, a model of severe hyperglycaemia and insulin resistance (Poy et al, 2009). Furthermore, a recent study systematically quantified microRNA expression by RT–PCR from pancreatic islets, liver, and adipose from diabetic mice revealing significant changes in the expression of several microRNAs (Zhao et al, 2009). Taken together, these observations support the idea that many microRNAs are responsive to stress and are necessary for cells to adjust to changes in the environment. What remains of great interest now are the signals or mechanistic pathways leading from hyperglycaemia, hyperinsulinemia, or obesity to changes in microRNA expression.
Future studies should also address the coordinate regulation of gene expression by small RNAs as dozens of microRNAs are expressed at high levels in all cells. These types of experiments will help to understand how microRNAs cooperate to maintain the essential functions of specific cell types such as β-cells. The work of Melkman-Zehavi et al illustrates that the maintenance of glucose homoeostasis and insulin production is dependent on the expression of Dicer and its associated microRNAs. New models, which highlight specifically the individual contribution of miR-24, miR-26, and miR-148 are sure to provide insight into the bigger picture that constitutes the landscape of gene regulation in the pancreatic β-cell.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi H, Ikeda Y, Okamura M, Tanaka T, Urashima Y, Ohguchi H, Takayasu S, Kojima N, Iwasaki S, Ohashi R, Jiang S, Hasegawa G, Ioka RX, Magoori K, Sumi K, Maejima T, Uchida A, Naito M, Osborne TF, Yanagisawa M et al. (2005) SOX6 attenuates glucose-stimulated insulin secretion by repressing PDX1 transcriptional activity and is down-regulated in hyperinsulinemic obese mice. J Biol Chem 280: 37669–37680 [DOI] [PubMed] [Google Scholar]
- Leung AK, Sharp PA (2010) MicroRNA functions in stress responses. Mol Cell 40: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS (2007) MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes 56: 2938–2945 [DOI] [PubMed] [Google Scholar]
- Melkman-Zehavi T, Oren R, Kredo-Russo S, Shapira T, Mandelbaum AD, Rivkin N, Nir T, Lennox KA, Behlke MA, Dor Y, Hornstein E (2011) miRNAs control insulin content is pancreatic β-cells via downregulation of transcriptional repressors. EMBO J 30: 835–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN (2010) Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest 120: 3912–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyton M, Stellrecht CM, Naya FJ, Huang HP, Samora PJ, Tsai MJ (1996) BETA3, a novel helix-loop-helix protein, can act as a negative regulator of BETA2 and MyoD-responsive genes. Mol Cell Biol 16: 626–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, Zavolan M, Stoffel M (2009) miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci USA 106: 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC (2009) On the road to reading the RNA-interference code. Nature 457: 396–404 [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T (2008) Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao E, Keller MP, Rabaglia ME, Oler AT, Stapleton DS, Schueler KL, Neto EC, Moon JY, Wang P, Wang IM, Lum PY, Ivanovska I, Cleary M, Greenawalt D, Tsang J, Choi YJ, Kleinhanz R, Shang J, Zhou YP, Howard AD et al. (2009) Obesity and genetics regulate microRNAs in islets, liver, and adipose of diabetic mice. Mamm Genome 20: 476–485 [DOI] [PMC free article] [PubMed] [Google Scholar]