The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana
Mediator is a large multi-subunit complex that is required for the transcription of genes by RNA polymerase II. Here, Mediator also has a role in the transcription of miRNAs in plants and in generating large noncoding scaffold transcripts by RNA polymerase V during siRNA-mediated transcriptional gene silencing.
Keywords: Mediator, microRNA, noncoding RNA, Pol II, siRNA
Abstract
Mediator is a conserved multi-subunit complex known to promote the transcription of protein-coding genes by RNA polymerase II (Pol II) in eukaryotes. It has been increasingly realized that Pol II transcribes a large number of intergenic loci to generate noncoding RNAs, but the role of Mediator in Pol II-mediated noncoding RNA production has been largely unexplored. The role of Mediator in noncoding RNA production in plants is particularly intriguing given that plants have evolved from Pol II two additional polymerases, Pol IV and Pol V, to specialize in noncoding RNA production and transcriptional gene silencing at heterochromatic loci. Here, we show that Mediator is required for microRNA (miRNA) biogenesis by recruiting Pol II to promoters of miRNA genes. We also show that several well-characterized heterochromatic loci are de-repressed in Mediator mutants and that Mediator promotes Pol II-mediated production of long noncoding scaffold RNAs, which serve to recruit Pol V to these loci. This study expands the function of Mediator to include Pol II-mediated intergenic transcription and implicates a role of Mediator in genome stability.
Introduction
Mediator is a multi-subunit complex first identified in yeast as required for activator-dependent stimulation of RNA polymerase II (Pol II) transcription (Kelleher et al, 1990; Flanagan et al, 1991). It promotes transcription initiation by bridging transcription activators that bind to upstream promoter or enhancer elements and the Pol II general transcriptional machinery located at the basal promoter. Structural studies show that the 21-subunit yeast Mediator forms three submodules, the head, middle, and tail, with the head submodule serving as the Pol II-interacting interface and the tail submodule interacting with sequence-specific transcription factors (reviewed in Chadick and Asturias, 2005). While some studies support a strict activator-dependent role of Mediator in transcription (Fan et al, 2006; Fan and Struhl, 2009), others favour an additional role of Mediator in basal transcription as a general transcription factor (Thompson and Young, 1995; Holstege et al, 1998; Ansari et al, 2009). Mediator is highly conserved in a wide range of eukaryotes including mammals and plants, although the mammalian and Arabidopsis Mediator contains more subunits than the yeast Mediator (Malik and Roeder, 2005; Backstrom et al, 2007). Almost all studies on yeast and mammalian Mediator have been focused on the role of Mediator in the expression of protein-coding genes. A recent study shows that the mouse Mediator promotes the expression of small nuclear RNAs (snRNAs) (Krebs et al, 2010). But the role of Mediator in noncoding RNA transcription remains largely unexplored.
The Arabidopsis Mediator was biochemically characterized only recently and found to contain 21 subunits with yeast or metazoan counterparts and six plant-specific subunits (Backstrom et al, 2007). A few Arabidopsis genes encoding Mediator subunits have been genetically characterized. Loss-of-function mutants in MED21 are embryo lethal (Dhawan et al, 2009), indicating that Mediator is essential. On the other hand, mutants in STRUWWELPETER/MED14 and PHYTOCHROME AND FLOWERING TIME1/MED25 are viable but defective in specific developmental processes or responses to environmental stimuli (Autran et al, 2002; Cerdan and Chory, 2003; Kidd et al, 2009). This led to the hypothesis that Mediator integrates various signalling pathways at the molecular level (Backstrom et al, 2007). Although these studies suggest that the plant Mediator is important in gene expression, the scope of Mediator's function in transcription remains unknown.
Transcriptome studies in fungi, plants, and animals have revealed pervasive transcription of the genome to generate a multitude of noncoding RNAs. microRNAs (miRNAs) constitute a class of well-studied noncoding RNAs that regulates gene expression at posttranscriptional levels (reviewed in Chen, 2009; Kim et al, 2009). Small interfering RNAs (siRNAs) constitute another class of endogenous small RNAs that maintains genome stability by triggering heterochromatin formation at repeats and transposons in plants and Schizosaccharomyces pombe (Reinhart and Bartel, 2002; Volpe et al, 2002; Xie et al, 2004). In these organisms, long noncoding RNAs are also produced at repeats and transposons and serve to recruit siRNAs to these loci to result in heterochromatin formation (Volpe et al, 2002; Wierzbicki et al, 2008; Zheng et al, 2009). The role of Mediator in the production of these small and long noncoding RNAs has not been evaluated. The fact that plants have evolved from Pol II two new polymerases, Pol IV and Pol V, to produce siRNAs and long noncoding RNAs, respectively, to silence repeats and transposons (reviewed in Chen, 2009), raises the question of whether Mediator acts in transcriptional gene silencing (TGS) and whether Mediator promotes the activities of Pol IV or Pol V.
Here, we evaluate the effects of three Mediator mutations in the expression of protein coding as well as noncoding RNA genes in Arabidopsis. We show that Mediator is likely a general transcription factor that promotes Pol II transcription of a large number of protein-coding genes. We also show that Mediator promotes the transcription of miRNA genes (MIR) by recruiting Pol II to their promoters. In addition, we reveal a previously unsuspected role of Mediator in noncoding RNA production at, and TGS of, loci regulated by endogenous siRNAs. These findings broaden our knowledge of the role of Mediator in gene expression and genome defense.
Results
Arabidopsis mutants in three Mediator subunit genes display pleiotropic developmental defects
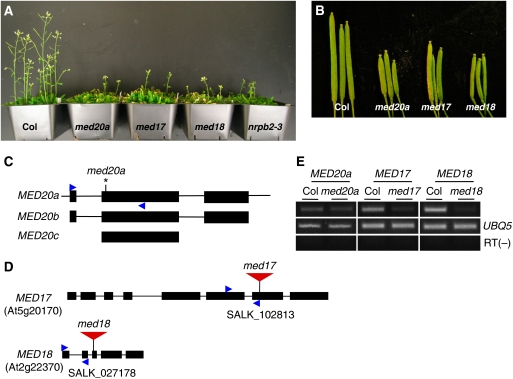
A mutant with pleiotropic developmental phenotypes was isolated in a forward genetic screen in the Columbia (wild type) background. The mutant was smaller than wild type in stature and late flowering (Figure 1A). The leaves had shorter petioles and curled downward (Supplementary Figure S1A), the phyllotaxy of flowers was abnormal (Supplementary Figure S1B), and fertility was reduced (Figure 1B; Supplementary Figure S1D). At a frequency of ∼20%, the mutant developed three cotyledons and three first true leaves instead of a pair of each in wild type (Supplementary Figure S1A).
Figure 1.
Isolation and characterization of mutants in Mediator genes. (A) Four-week-old plants of wild type (Col), med20a, med17, med18, and nrpb2-3. (B) Siliques from Col, med20a, med17, and med18 plants. (C) Schematic diagrams of MED20 paralogs MED20a, MED20b, and MED20c. The asterisk indicates the mutation causing a premature stop codon in the med20a mutant. Black rectangles represent exons and lines represent introns. MED20a and MED20b are composed of three exons showing 85% identity at the nucleotide level. MED20c only encodes the conserved second exon compared with MED20a and MED20b. (D) Schematic diagrams of T-DNA insertion mutants of MED17 and MED18. Large triangles represent T-DNA insertions. Black rectangles represent exons. (E) RT–PCR analysis of MED20a, MED17, and MED18 expression in med20a, med17, and med18 mutants, respectively. The images in the top row represent the indicated Mediator genes. UBIQUITIN5 (UBQ5) was used as an internal loading control. The ‘−RT' reactions were performed with the UBQ5 primers. The PCR primers used are indicated by small arrowheads in (C, D).
The mutation was mapped to a 120-kb region covered by the BACs T3B23 and T1B3 on chromosome 2. Sequencing candidate genes in this region revealed a C-to-T mutation in At2g28230, which encodes a subunit of the head submodule in Mediator (Figure 1C). The mutation is at the 58th nucleotide of the coding region in the second exon and results in a premature stop codon (Figure 1C). A genomic construct covering 2.5 kb of the promoter and the entire coding region of At2g28230 was introduced into the mutant—the morphological phenotypes of the mutant were rescued in all 39 independent transgenic lines (Supplementary Figure S1C and data not shown). Therefore, the med20a mutation was responsible for the morphological defects. A homology-based search identified two more paralogs in Arabidopsis: At2g28020 and At4g09070. Therefore, At2g28230, At4g09070, and At2g28020 were named MED20a, MED20b, and MED20c, respectively (Figure 1C). MED20a and MED20b have 85% identity at the nucleotide level. MED20c is shorter than MED20a and MED20b; there is 93% identity at the nucleotide level in exon 2 among the three genes. Transcriptome studies using Affymetrix ATH1 microarrays found MED20a and MED20c to be expressed throughout the plant but probes for MED20b were not present in the microarray (Winter et al, 2007). Given the high degree of sequence similarity among the genes, it is unlikely that the microarray studies were able to differentiate the paralogs.
To determine whether the developmental phenotypes of the med20a mutant reflect the function of the head submodule of Mediator, we obtained two T-DNA mutant lines of genes encoding two other head submodule subunits, MED17 and MED18, from the SALK collection and analysed their phenotypes (Figure 1D). Transcript levels of MED17 and MED18 were significantly reduced in med17 and med18, respectively, indicating that the T-DNA insertions caused at least a partial loss of function of MED17 and MED18 genes (Figure 1E). The med17 and med18 mutants exhibited similar developmental defects as med20a, although the phenotypes of med17 were weaker overall as compared with those of the other two mutants (Figure 1A and B; Supplementary Figure S1A, B, and D). These results indicate that Mediator is broadly involved in developmental processes in Arabidopsis.
The plant Mediator is likely a general transcription factor
Mediator is a conserved multi-protein complex in animals, fungi, and plants. Even for the yeast Mediator, which has been studied much more extensively than the plant Mediator, there is still controversy as to whether Mediator promotes basal transcription as a general transcriptional factor for Pol II (Thompson and Young, 1995; Holstege et al, 1998; Fan et al, 2006; Ansari et al, 2009; Fan and Struhl, 2009). A few studies on mutants in several Arabidopsis Mediator genes implicate a role of Mediator in integrating various environmental signals (Cerdan and Chory, 2003; Backstrom et al, 2007; Kidd et al, 2009), but it is unknown whether the plant Mediator serves as a general transcription factor or as a bridge between sequence-specific transcription factors and Pol II.
To investigate the molecular function of the plant Mediator, we compared the phenotypic as well as transcriptomic effects of the med20a mutation with those of nrpb2-3, a weak allele in the second largest subunit of Pol II that we had previously isolated (Zheng et al, 2009). med20a and nrpb2-3 exhibited similar morphological phenotypes—both mutants were small in stature, had reduced fertility, and exhibited altered phyllotaxy and delayed leaf emergence indicative of abnormal shoot apical meristem activity (Figure 1A, Supplementary Figure S1A, and data not shown). Affymetrix ATH1 microarray-based transcript profiling using inflorescence tissues from wild type and nrpb2-3 revealed that 448 genes were downregulated and 95 genes were upregulated by two-fold in nrpb2-3 (Zheng et al, 2009). To examine the overlap between Pol II- and Mediator-dependent genes, we conducted similar transcript profiling between wild-type and med20a inflorescences. We found that a total of 754 genes were downregulated and 140 genes were upregulated in med20a by two-fold in three biological replicates (Supplementary Tables 1 and 2). Genes upregulated in the two mutants were unlikely direct targets of Pol II or Mediator and thus were not studied further. The number of genes downregulated in med20a was almost twice that in nrpb2-3, which was not surprising given that nrpb2-3 is a weak allele. Intriguingly, 84% (377 genes) of the downregulated genes (448 genes) in nrpb2-3 were also downregulated in med20a (Supplementary Figure S2), suggesting a large degree of overlap between Pol II and Mediator in gene expression and supporting the role of Mediator as a general transcription factor. The small number of genes (754) downregulated in med20a is likely due to the presence of two other MED20 paralogs in the genome.
Pol II and Mediator are required for miRNA accumulation in Arabidopsis
Mediator is well known for its role in Pol II-mediated transcription of protein-coding genes and has been recently shown to promote the expression of snRNAs (Krebs et al, 2010), but its role in noncoding RNA transcription remains largely unexplored. Although the structures of MIR genes, such as the promoters and introns, and the presence of polyA tails in primary precursors of miRNAs (pri-miRNAs) suggest that MIR genes are transcribed by Pol II in plants (Xie et al, 2005), direct evidence for Pol II transcription of MIR genes is still lacking. We explored the role of Pol II and Mediator in miRNA biogenesis using the nrpb2-3 and med mutants.
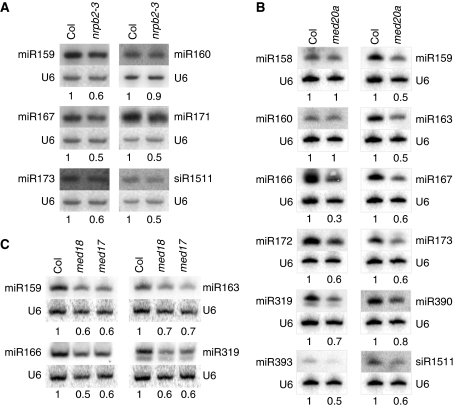
We first examined the accumulation of various mature miRNAs in nrpb2-3 and med20a mutants by northern blotting. siR1511, a trans-acting siRNA (ta-siRNA), whose biogenesis requires a miRNA (Peragine et al, 2004; Vazquez et al, 2004; Allen et al, 2005; Yoshikawa et al, 2005), was also examined. Nine out of 12 tested miRNAs and siR1511 were present at reduced levels in nrpb2-3, with the exceptions being miR160, miR163, and miR166 (Figure 2A; Supplementary Figure S3A). In the med20a mutant, 8 out of 11 examined miRNAs (with miR158, miR160, and perhaps miR390 being the exceptions) and siR1511 were at lower levels (Figure 2B). The reduced accumulation of miRNAs in med20a was rescued by the introduction into med20a a MED20a transgene (Supplementary Figure S3B). We also examined the levels of four miRNAs in med17 and med18. Consistently, all four miRNAs were at reduced levels in med17 and med18 (Figure 2C). Therefore, both Pol II and Mediator are required for miRNA biogenesis in Arabidopsis.
Figure 2.
The accumulation of miRNAs and a ta-siRNA in nrpb2-3 and Mediator mutants as determined by northern blotting. (A) The accumulation of five miRNAs and a ta-siRNA (siR1511) in wild type (Col) and nrpb2-3. (B) The accumulation of 11 miRNAs and one ta-siRNA (siR1511) in Col and med20a. (C) The accumulation of four miRNAs in Col, med17, and med18. Total RNAs were extracted from inflorescences. The levels of each small RNA were normalized to those of U6 and compared with Col. The numbers below the gel images indicate the relative abundance of the small RNAs. Two to three biological replicates were performed for all small RNAs except for miR390 and miR159 and yielded similar results.
Mediator regulates MIR gene expression at the transcriptional level
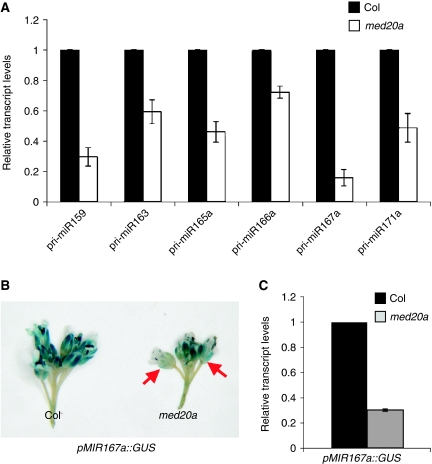
The reduced miRNA accumulation in Mediator mutants may be due to reduced transcription of MIR genes, compromised posttranscriptional processing of the precursors, or decreased stability of the mature miRNAs. Given the known role of Mediator in promoting transcription, it is likely that the reduced miRNA accumulation in Mediator mutants is attributable to reduced transcription of MIR genes. To test this hypothesis, we first examined the levels of six pri-miRNAs by real-time RT–PCR. Indeed, the levels of all examined pri-miRNAs were decreased in the med20a mutant to 20–70% of the wild-type levels (Figure 3A; Supplementary Figure S4). We further examined the levels of individual pri-miRNAs from each member of the miR166 family (Supplementary Figure S5). Four of the six MIR166 loci were expressed at lower levels in med20a as compared with wild type, suggesting that different MIR family members have different degrees of dependence on MED20a for their expression.
Figure 3.
med20a affects the transcription of MIR genes. (A) The accumulation of six pri-miRNAs was determined by real-time RT–PCR in Col and the med20a mutant. Total RNAs were extracted from inflorescences. The pri-miRNA levels were normalized to those of UBQ5 and compared with Col. Standard deviations were calculated from three technical replicates. Three biological replicates yielded similar results. (B) GUS expression driven by the MIR167a promoter was monitored in isogenic Col and med20a transgenic lines through GUS staining. GUS expression in old flowers was reduced in med20a compared with Col (arrows). (C) The accumulation of GUS mRNA from the lines in (B) was determined by real-time RT–PCR. The GUS mRNA levels were normalized to those of UBQ5 and compared with Col. Standard deviations were calculated from three technical replicates. Two biological replicates yielded nearly identical results.
To directly demonstrate that the med20a mutation affected the activity of an MIR promoter, we crossed a GUS transgene under the control of the MIR167a promoter (pMIR167a∷GUS) (Wu et al, 2006) into med20a. In the F2 population, 3 to 1 segregation of the transgene was observed, which indicated that the transgene was at a single locus. Wild-type and med20a plants containing the transgene were obtained from the F2 population of the cross and their progeny were screened to identify ones that were homozygous for the transgene. GUS staining performed on these single insertion, isogenic med20a and wild-type plants revealed decreased GUS expression in med20a compared with wild type, especially in old flowers (Figure 3B). Since GUS staining was not a quantitative measure of reporter gene expression, we performed real-time RT–PCR to determine the levels of GUS mRNA in inflorescences. Indeed, GUS mRNA levels were lower in med20a than in wild type (Figure 3C), indicating that Mediator acts through the promoter of MIR167a.
The plant Mediator has been implicated as an integrator in response to environmental cues in Arabidopsis (Cerdan and Chory, 2003; Backstrom et al, 2007; Kidd et al, 2009). Meanwhile, the expression of some MIR genes is known to be regulated by abiotic stresses. For example, the expression of miR398b is transcriptionally downregulated by oxidative stress (Sunkar et al, 2006), whereas the expression of MIR167a and MIR171a is transcriptionally upregulated by cold stress and that of MIR159a is upregulated by salt stress (Liu et al, 2008). To determine whether Mediator is required for the stress-induced changes in MIR gene expression, we monitored levels of pri-miRNAs of these stress-inducible MIR genes in response to stress treatments in wild type, med20a, and med18. Our semi-quantitative RT–PCR reproduced the induced expression of MIR159a, MIR167a, and MIR171a and the repressed expression of MIR398b by various stresses (Supplementary Figure S6). The stress-induced changes in the expression of these MIR genes occurred also in med20a and med18 mutants (Supplementary Figure S6). Given that med20a is likely a null allele and med18 is a strong allele in that MED18 RNA levels were strongly reduced in this mutant (Figure 1E), these results suggest that MED20a and MED18 are dispensable for stress-induced changes in MIR gene expression.
Mediator promotes Pol II recruitment to MIR genes
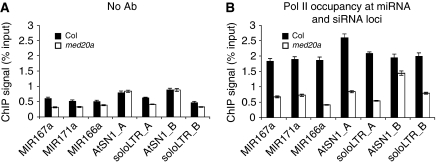
The reduced pri-miRNA and mature-miRNA levels in the med20a mutant are unlikely attributable to reduced expression of miRNA biogenesis genes because none of the known genes in miRNA biogenesis were affected in med20a as determined by microarray analyses (see Supplementary Table 3 for a list of the genes). This prompted us to examine whether Mediator has a direct role in the transcription of MIR genes. We examined Pol II occupancy at promoters of several MIR genes by chromatin immunoprecipitation (ChIP) using an antibody against the second largest subunit of Pol II (RPB2). We monitored three MIR genes, MIR167a, MIR171a, and MIR166a, of which pri-miRNAs were at reduced levels in the med20a mutant (Figure 3A). Pol II was enriched at the regions encompassing the transcription start sites at these three loci, as compared with the ‘no antibody' ChIP that served as a negative control (Figure 4A and B). Pol II occupancy at these loci was significantly reduced in the med20a mutant (Figure 4B). These results indicate that Mediator promotes Pol II recruitment to MIR genes.
Figure 4.
Pol II occupancy at miRNA and siRNA loci. Pol II occupancy at several miRNA and siRNA loci was determined by ChIP using anti-RPB2 antibodies in Col and med20a. DNA present in the immunoprecipitates was quantified by real-time PCR relative to total input DNA. (A) ChIP performed with no antibodies as negative controls. (B) ChIP with anti-RPB2 antibodies. The results were reproduced in two biological replicates. Standard deviations were calculated from three technical repeats. AtSN1_A and soloLTR_A, regions A of these two loci as depicted in Figure 5B. AtSN1_B and soloLTR_B, regions B of these two loci as in Figure 5B.
Mediator is required for TGS and Pol II-mediated long noncoding RNA production at repeats and transposons
Plants have evolved from Pol II two polymerases, Pol IV and Pol V, to specialize in endogenous small RNA-based genome defense against repeats and transposons (Herr et al, 2005; Kanno et al, 2005; Onodera et al, 2005; Pontier et al, 2005; Huang et al, 2009; Ream et al, 2009). These repeated elements generate 24 nt endogenous siRNAs in a Pol IV-dependent manner (Herr et al, 2005; Onodera et al, 2005; Pontier et al, 2005) and the siRNAs are thought to be recruited back to these elements by nascent transcripts, also known as scaffold transcripts, generated by Pol V from these elements (Herr et al, 2005; Kanno et al, 2005; Wierzbicki et al, 2008, 2009). At some loci, such as solo LTR and siR02, Pol II is responsible for the production of noncoding scaffold transcripts and TGS of the loci (Zheng et al, 2009). The presence of two additional polymerases derived from Pol II and the recent finding that Pol II generates long noncoding RNAs at heterochromatic loci raised the question of whether Mediator acts with any of the three polymerases in TGS.
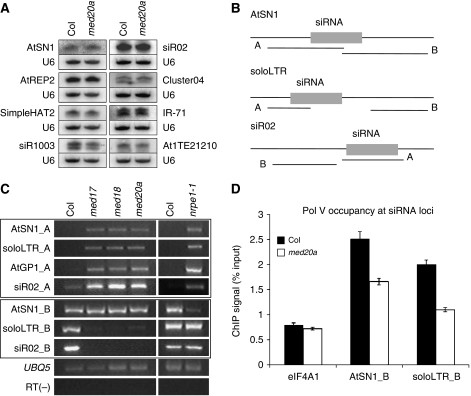
We first determined whether Mediator is required for the production of 24 nt siRNAs, a process that requires Pol IV. We examined the levels of 24 nt siRNAs from eight loci known to undergo siRNA production and siRNA-mediated TGS (Herr et al, 2005; Onodera et al, 2005; Pontier et al, 2005; Huettel et al, 2006) in wild type and med20a by northern blotting. The levels of the siRNAs were not affected in med20a (Figure 5A). These results do not support a role of Mediator in promoting Pol IV-mediated production of endogenous siRNAs, although such a role cannot be excluded given that the med20a mutation is unlikely to completely eliminate Mediator activity.
Figure 5.
Mediator in TGS of repeats and transposons. (A) The accumulation of endogenous siRNAs from heterochromatic loci. Small RNA northern blotting was performed with total RNAs extracted from inflorescences from Col and med20a. Small RNA accumulation was not affected in the med20a mutant. The U6 blots served as loading controls for the overlying small RNA blots. (B) Diagrams of AtSN1, soloLTR, and siR02 genomic regions. These regions are based on analysis of transcription units by Wierzbicki et al (2008) and Zheng et al (2009). The ‘A' regions are where the siRNAs are derived, while the ‘B' regions are where scaffold transcripts are produced. (C) RT–PCR analysis of noncoding transcripts from regions A and B at siRNA loci in Col and Mediator mutants. UBQ5 was used as an internal control. The RT (−) control PCR was performed with UBQ5 primers. nrpe1-11 is a loss-of-function allele in the largest subunit of Pol V. The results shown were reproduced in three biological replicates. (D) Pol V occupancy at regions B of AtSN1 and soloLTR was determined by ChIP using anti-FLAG antibodies in pNRPE∷NRPE1-FLAG and pNRPE∷NRPE1-FLAG med20a. DNA that co-purified with Pol V was measured by real-time PCR against total input DNA. eIF4A1, a gene not bound by Pol V, served as a negative control. Standard deviations were calculated from three technical repeats. The results were reproduced in two other biological replicates shown in Supplementary Figure S8.
Next, we examined the expression of these loci as a measure of their silencing status by real-time RT–PCR. It is known that these loci are de-repressed in Pol IV and Pol V mutants (Herr et al, 2005; Onodera et al, 2005; Pontier et al, 2005; Huettel et al, 2006). At all tested loci, transcript levels from regions A, to which siRNAs map (Figure 5B), were higher in med17, med18, and med20a mutants as compared with wild type, although the degree of de-repression was not as large as in nrpe1-11, a mutant in the largest subunit of Pol V (Pontier et al, 2005) (Figure 5C). Note that among these loci, AtSN1 and AtGP1 are known as type I loci that are silenced in a Pol V-dependent manner whereas solo LTR and siR02 are known as type II loci that are silenced in a Pol II- and Pol V-dependent manner (Zheng et al, 2009). Pol V generates scaffold transcripts at type I loci, whereas Pol II generates these RNAs at type II loci (Wierzbicki et al, 2008; Zheng et al, 2009). Since siRNA levels were not affected in med20a (Figure 5A), we suspected that the de-repression of these loci in the Mediator mutants was due to reduced levels of Pol II- or Pol V-dependent scaffold RNAs from these loci. We examined the levels of scaffold RNAs in wild type, med17, med18, med20a, and nrpe1-11 (a control for type I loci) corresponding to regions B (Figure 5B) at these loci. At type II loci (soloLTR and siR02), the levels of Pol II-dependent scaffold transcripts were obviously reduced in all med mutants (Figure 5C). However, at the type I locus AtSN1, the scaffold transcripts were reduced in abundance in nrpe1-11 but not affected in any of the Mediator mutants (Figure 5C). The levels of Pol V-dependent scaffold transcripts at two other loci, IGN6 (Wierzbicki et al, 2008) and MEA-ISR (Law et al, 2010), were also not affected in the Mediator mutants (Supplementary Figure S7).
To determine whether Mediator has a direct role in promoting Pol II-based noncoding transcription at the type II locus soloLTR, we performed ChIP to examine Pol II occupancy at this locus in wild type and med20a. At both region A and region B, from which siRNAs and scaffold transcripts are produced, respectively (Figure 5B), Pol II was enriched at soloLTR in comparison to the ‘no antibody' control (Figure 4A and B), consistent with previous observations (Zheng et al, 2009). Pol II occupancy at both regions A and B at this locus was reduced in med20a, supporting a role of Mediator in recruiting Pol II to this locus. This, together with the reduced levels of Pol II-dependent scaffold transcripts at soloLTR and siR02 (Figure 5C), indicates that Mediator promotes Pol II-mediated noncoding RNA production at loci undergoing siRNA-mediated TGS.
Intriguingly, although analysis of the weak nrpb2-3 mutant did not reveal a role of Pol II in TGS at type I loci (Zheng et al, 2009), Pol II was enriched at the type I locus AtSN1 (Figure 4A and B), which was also observed previously (Zheng et al, 2009). Moreover, Pol II occupancy at both regions A and B at AtSN1 was reduced in med20a (Figure 4B). In addition, although Pol V-dependent scaffold transcripts at AtSN1 were not affected in the med20a mutant (Figure 5C), ChIP detected reduced occupancy of Pol V at AtSN1 region B (Figure 5D; Supplementary Figure S8), where Pol V-dependent scaffold transcripts are derived (Wierzbicki et al, 2008). At soloLTR, which requires Pol V for TGS and to which Pol V recruitment to region B relies on Pol II-dependent scaffold transcripts (Zheng et al, 2009), Pol V occupancy was reduced in med20a (Figure 5D; Supplementary Figure S8), consistent with the reduced scaffold RNAs in this mutant (Figure 5C). There are two possibilities regarding the relationship between Mediator and Pol V. In one, Mediator promotes Pol V-mediated noncoding transcription at type I loci, but the weak nature of the med20a, med17, and med18 mutants (due to the presence of paralogs or partial loss of function) precluded the assignment of such a role to Mediator. The second possibility is that Pol II also acts in the TGS of type I loci, and the de-repression of these loci in med mutants reflects the role of Mediator acting with Pol II.
Discussion
In this study, we provided evidence supporting that MED20a, a subunit of the head submodule of the plant Mediator, is broadly required for Pol II transcription of protein-coding genes. Transcript profiling with med20a and nrpb2-3 mutants revealed a high degree of overlap in genes that are decreased in expression in the two mutants. Although only 750 genes were affected in the med20a mutant, it is likely that these genes represent only a portion of those requiring MED20 since two other MED20a paralogs may compensate for the absence of MED20a. It is likely that the plant Mediator, or at least the head submodule, functions as a general transcription factor to promote basal Pol II transcription.
Mediator also promotes Pol II transcription of MIR genes. Plant MIR genes contain canonical basal promoter elements and produce polyadenylated pri-miRNAs, suggesting that MIR genes are transcribed by Pol II. In this study, we provided direct evidence for a role of Pol II in MIR gene expression by showing that miRNAs are reduced in abundance in the nrpb2-3 mutant and that Pol II is present at the transcription start sites of MIR genes. The reduced accumulation of mature miRNAs in mutants in three head submodule subunits of Mediator suggests a general role of Mediator in MIR gene expression. The role of Mediator in promoting Pol II transcription of MIR genes is supported by the reduced levels of pri-miRNAs, compromised MIR167a promoter activity, and decreased Pol II occupancy at MIR transcription start sites in med20a.
This work also revealed a previously unknown role of Mediator in silencing repeats and transposons. Well-characterized elements that undergo siRNA-mediated TGS were found to be de-repressed in all three med mutants. The decreased Pol II occupancy and reduced levels of noncoding scaffold RNAs transcribed by Pol II support a role of Mediator in Pol II-mediated noncoding RNA production in genome defense. While a previous study with the weak nrpb2-3 mutant established a role of Pol II in TGS only at type II loci, the analysis of three Mediator mutants in this study shows that Mediator is involved in TGS at both type I and type II loci. How Mediator acts in TGS of type I loci remains to be determined. One possibility is that Mediator acts with Pol II, which also acts at type I loci but such a function of Pol II has not been established since it is not compromised by the weak nrpb2-3 mutation. Another possibility is that Mediator acts with Pol V. This is consistent with the observation that Pol V occupancy is reduced in med20a (Figure 5D). Intriguingly, the plant-specific Mediator subunit MED36 (Backstrom et al, 2007) was found in a proteomic study to co-purify with the largest subunit of Pol V (Huang et al, 2009).
Materials and methods
Plant materials
All mutants are in the Col background. Arabidopsis plants were grown at 23°C under continuous light. T-DNA insertion lines of MED17 and MED18 were obtained from the Salk T-DNA insertion collection. nrpb2-3 was isolated from our laboratory (Zheng et al, 2009). nrpe1-11 (formerly known as nrpd1b-1) (Pontier et al, 2005) and the pMIR167a∷GUS transgenic line (Wu et al, 2006) were gifts from Dr Thierry Lagrange and Dr Jason W Reed, respectively. For med20a genotyping, genomic DNA was amplified with the primers At2g28230-F (CATACCTCAATTTCGATTGGG) and At2g28230-R4 (GAAGAATCAGCTTCCAAGAC) and PCR products were digested with AluI. The PCR products from wild type could be digested by AluI, whereas those from med20a could not.
For NaCl and cold stress treatments, seeds were surface sterilized and sown on MS-agar plates. Seeds were stratified at 4°C for 2 days and transferred to 22°C for 10 days. For salt stress treatments, seedlings were transferred to filter paper saturated with either water (control) or 300 mM NaCl and incubated for 6 h. For cold stress treatments, seedlings on plates were incubated either at 22°C (control) or at 4°C for 6 h. For copper treatment, surface-sterilized seeds were sown on MS-agar plates with or without 10 μM Cu2+ and 10-day-old seedlings were harvested.
Map-based cloning of MED20a
med20a (Col) was crossed to Ler. In the F2 population, plants showing med20a phenotypes were identified and used as the mapping population. Rough mapping using 27 med20a plants showed that MED20a is linked to the marker nga168 on chromosome 2. For fine mapping, we designed new SSLP or CAPS markers in this region according to polymorphisms between Ler and Col according to Monsanto Arabidopsis polymorphism database (http://www.arabidopsis.org/Cereon). Using these markers and ∼500 plants of med20a phenotypes, we mapped MED20a to a 120-kb region covered by the BACs T3B23 and T1B3. Among the 23 genes in this region, sequencing analysis revealed a point mutation in MED20a.
Plasmid construction
MED20a genomic region was amplified with primers At2g28230-pro2 (CACCCTTGGATTGTACTGCTGGT) and At2g28230-R2 (GCCTCTCACAGC-TTGAACG) and cloned into the pEG301 gateway vector (Earley et al, 2006) to generate pMED20a∷MED20a-HA. The plasmid was used to transform med20a plants by agroinfiltration.
RT–PCR
Total RNA from seedlings or inflorescences was extracted with TRI reagent (Molecular Research Center) and reverse transcribed using SuperScriptII reverse transcriptase (Invitrogen) and an oligo-d(T) primer according to the manufacturer's instructions. For pri-miRNA detection, quantitative PCR was carried out in triplicate on a Bio-Rad IQcycler apparatus with the Quantitech SYBR green kit (Bio-Rad). To detect Pol II- or Pol V-dependent noncoding transcripts, RT–PCR was done according to Zheng et al (2009). Primers used are listed in Supplementary Table 4.
Small RNA northern blot analysis
RNA isolation and hybridization for miRNA and endogenous siRNA detection were performed as described (Park et al, 2002; Pall et al, 2007). In all, 10 μg total RNA and 5 μg small RNA-enriched RNA from inflorescences were used in northern blotting to detect miRNAs and endogenous siRNAs, respectively. 5′-End-labelled 32P antisense DNA or LNA oligonucleotides were used to detect miRNAs and endogenous siRNAs. Oligonucleotide probes used are listed in Supplementary Table 5.
Histochemical staining
GUS staining was performed as described (Jefferson et al, 1987; Rodrigues-Pousada et al, 1993). Inflorescences were fixed in 90% cold acetone for 15–20 min and rinsed with the rinse solution (50 mM NaPO4 pH 7.2, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6). Then, they were vacuum infiltrated in the staining solution (50 mM NaPO4 pH 7.2, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 2 mM X-Gluc)) and incubated at 37°C.
ATH1 Affymetrix microarray analysis
GeneChip arrays were hybridized according to the manufacturer's instructions (Affymetrix). Data analysis was done according to Horan et al (2008). Normalization of raw intensities across all probe sets was performed in R using RMA algorithms. To calculate P-values for increases or decreases in expression, the Wilcoxon signed-rank test was applied to each pair of chips after normalization using R & Bio Conductor (developed by Dr Thomas Girke). A P-value of ⩽0.05 in combination with a two-fold difference was used to define changes in gene expression.
Chromatin immunoprecipitation
ChIP was performed according to Zheng et al (2009). The results shown were reproduced in three biological replicates. Commercial anti-RPB2 (catalog no. ab10338, Abcam) and anti-FLAG (F7425, Sigma) antibodies were used. Quantitative real-time PCR was performed on bound and input DNAs. The primer sets used for the PCR are listed in Supplementary Table 4.
Supplementary Material
Acknowledgments
We thank Drs Thierry Lagrange and Jason W Reed for providing nrpe1-11 and pMIR167a∷GUS seeds, respectively; Dr Stefan Björklund for sharing antibodies against MED6 and MED32, although they were not used in this study; and Thanh Theresa Dinh, Lijuan Ji, and Yuanyuan Zhao for critical reading of the manuscript. This work was supported by grants from Chinese National Science Foundation (30970265) to BM and National Science Foundation (MCB-1021465) and National Institutes of Health (GM61146) to XC.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Ansari SA, He Q, Morse RH (2009) Mediator complex association with constitutively transcribed genes in yeast. Proc Natl Acad Sci USA 106: 16734–16739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, Inze D, Traas J (2002) Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J 21: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backstrom S, Elfving N, Nilsson R, Wingsle G, Bjorklund S (2007) Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Cerdan PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Chadick JZ, Asturias FJ (2005) Structure of eukaryotic Mediator complexes. Trends Biochem Sci 30: 264–271 [DOI] [PubMed] [Google Scholar]
- Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Mittelsten Scheid O, Mengiste T (2009) HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fan X, Chou DM, Struhl K (2006) Activator-specific recruitment of Mediator in vivo. Nat Struct Mol Biol 13: 117–120 [DOI] [PubMed] [Google Scholar]
- Fan X, Struhl K (2009) Where does mediator bind in vivo? PLoS One 4: e5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan PM, Kelleher RJ III, Sayre MH, Tschochner H, Kornberg RD (1991) A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350: 436–438 [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120 [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Horan K, Jang C, Bailey-Serres J, Mittler R, Shelton C, Harper JF, Zhu JK, Cushman JC, Gollery M, Girke T (2008) Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol 147: 41–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Jones AM, Searle I, Patel K, Vogler H, Hubner NC, Baulcombe DC (2009) An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol 16: 91–93 [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJ, Matzke M (2006) Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J 25: 2828–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37: 761–765 [DOI] [PubMed] [Google Scholar]
- Kelleher RJ III, Flanagan PM, Kornberg RD (1990) A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61: 1209–1215 [DOI] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Krebs AR, Demmers J, Karmodiya K, Chang NC, Chang AC, Tora L (2010) ATAC and Mediator coactivators form a stable complex and regulate a set of non-coding RNA genes. EMBO Rep 11: 541–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE (2010) A protein complex required for polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Curr Biol 20: 951–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14: 836–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG (2005) Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci 30: 256–263 [DOI] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A (2007) Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res 35: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T (2005) Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev 19: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu JK, Hagen G, Guilfoyle TJ, Pasa-Tolic L, Pikaard CS (2009) Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell 33: 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP (2002) Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Pousada RA, De Rycke R, Dedonder A, Van Caeneghem W, Engler G, Van Montagu M, Van Der Straeten D (1993) The Arabidopsis 1-aminocyclopropane-1-carboxylate synthase gene 1 is expressed during early development. Plant Cell 5: 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CM, Young RA (1995) General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA 92: 4587–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS (2009) RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 41: 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An ‘Electronic Fluorescent Pictograph' browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218 [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X (2009) Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev 23: 2850–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.