Abstract
Alteration of epidermal growth factor receptor (EGFR) is involved in various human cancers and has been intensively investigated. A plethora of evidence demonstrates that posttranslational modifications of EGFR play a pivotal role in controlling its function and metabolism. Here, we show that EGFR can be acetylated by CREB binding protein (CBP) acetyltransferase. Interestingly, EGFR acetylation affects its tyrosine phosphorylation, which may contribute to cancer cell resistance to histone deacetylase inhibitors (HDACIs). Since there is an increasing interest in using HDACIs to treat various cancers in the clinic, our current study provides insights and rationale for selecting effective therapeutic regimen. Consistent with the previous reports, we also show that HDACI combined with EGFR inhibitors achieves better therapeutic outcomes and provides a molecular rationale for the enhanced effect of combination therapy. Our results unveil a critical role of EGFR acetylation that regulates EGFR function, which may have an important clinical implication.
Introduction
EGFR, an essential mediator for various growth factors, plays a pivotal role in regulating multiple signaling pathways, cell proliferation, cell cycle, and cell migration [1; 2]. Posttranslational modifications of EGFR such as phosphorylation, ubiquitination, and neddylation confer EGFR a multipotent player and arbitrate the fate of EGFR in mediating signal transduction, shuttling to different subcellular locations, or committing to degradation in cellular processes [3; 4; 5]. Upon ligand binding, such as epidermal growth factor (EGF), EGFR forms a dimer and activates several downstream signal pathways to promote cell growth [6; 7; 8]. At the same time, EGFR itself needs to be tightly regulated through a variety of posttranslational modifications, and then subjected to recycle, degradation, or nuclear localization [4; 5]. However, little is known about whether EGFR is dynamically regulated prior to ligand stimulation. As EGFR is a critical surface molecule responsible for pathological abnormities of cellular function as well as many diseases and cancers, dissecting the early stage regulation of EGFR would likely provide important information for tackling EGFR-associated life-threatening diseases.
Most recently, a growing number of non-histone protein acetylation has been reported to play critical roles in cellular processes alongside the phosphorylation and ubiquitination of affected proteins [9; 10], suggesting that regulation of protein acetylation may be useful for therapeutic settings [9; 11; 12; 13; 14; 15]. While histone deacetylase inhibitors (HDACIs) have shown promising signs for treating various cancers, the detailed mechanism by which HDACIs act on and the subset of cancers that benefit the most from HDACI regimen are not completely understood. These issues need to be further addressed in order to effectively and safely treat patients in clinical settings.
Since EGFR is a common target for anticancer therapy, a combination of HDACI and EGFR inhibitor or other receptor tyrosine kinase inhibitors (TKI) was proposed for cancer therapy. The initial results were encouraging and a synergistic effect was reported [16; 17]. However, the molecular mechanism that contributes to the synergistic effect is not completely understood. Interestingly, a report showed that trichostatin A (TSA), an HDACI, induces EGFR phosphorylation in a dose- and time-dependent manner in ovarian cancer cells [18]. More recently, EGFR is shown to acetylated at lysine 1155, 1158, and 1164 sites, and the acetylation affected its endocytosis in endothelial cells [19], and HDAC6 was reported to regulate EGFR turnover [20; 21; 22]. Collectively, these observations raised an interesting question of whether EGFR acetylation is related to phosphorylation which could therefore contribute to synergistic effect by combination treatment of TKI and HDACI.
Here, we report that acetylation of EGFR is linked to enhanced-EGFR function. Specifically, we observed that suberoylanilide hydroxamic acid (SAHA) has an adverse effect in the treatment of a subset of EGFR-expressing cancers such as breast cancer. Our study further suggests that elevated EGFR acetylation by SAHA may contribute to enhanced EGFR phosphorylation. Since SAHA has been used as an anti-cancer drug and accounts for over 50% of the existing clinical trials that are associated with HDACI, our observations provide an insight of potential adverse effect of SAHA derived from EGFR acetylation in cancer treatment. For high EGFR-expressing cancers, it may be critical to include TKI while using SAHA to treat these cancers. Taken together, our finding unveils a critical role of EGFR acetylation, which may have an important clinical implication.
Materials and Methods
Cell lines and antibodies
HEK293, MCF7, A431, MDA-MB-453, and MDA-MB-468 cell lines were obtained from ATCC and cultured according to ATCC’s instructions. Antibodies were purchased from companies as follows: polyclonal anti-acetyl-lysine (Upstate, Billerica, MA; CalBiochem, Gibbstown, NJ; Immunechem, Burnaby British Columbia, Canada) anti-phosho-Erk, anti-Erk, anti-phospho-Akt, anti-Akt, anti-phospho-Stat3, and anti-Stat3 (Cell Signaling, Danvers, MA); anti-EFGR (Santa Cruz Biotechnology, Santa Cruz, CA); monoclonal anti-EGFR (NeoMarkers, Fremont, CA); anti-p300, anti-CBP, anti-PCAF and CBP siRNA, and p300 siRNA (Santa Cruz); anti-phospho-tyrosine (4G10, Upstate); TrueBlot HRP-labeled anti-rabbit IgG (eBioscience, San Diego, CA).
Immunoprecipitation and immunoblot
For detecting EGFR acetylation, the cells were lysed in RIPA buffer containing 3 mM PMSF, 3 mM Na3VO4, 3 mM NaF, 5 mM sodium butyrate, and 20 µM TSA. TSA and sodium butyrate are essential for inhibiting HDACs from deacetylating EGFR. For immunoprecipitation, a total of 500 µg of cell lysate was used and diluted in 500µl of RIPA buffer with the corresponding antibodies. After addition of 2 µg EGFR (mouse monoclonal, NeoMarkers) antibody, the lysate was incubated with gentle rotation for 2 h at 4 °C, and then 80µl of protein-G beads added to the mixture and incubated for another 2 h. The protein complexes were resolved on 8% SDS-polyacrylamide gel and probed with antibodies as indicated. For immunoblot, a total of 20 µg of cell lysate was used.
Nano-HPLC-MS/MS spectrophotometry
A431 cells were treated with 2 µM TSA and 5 mM sodium butyrate for 24 h. Cells were lysed by RIPA buffer plus 20 mM Nicotinamide. 5 mg of cell lysate extracted from pretreated A431 cells were immunoprecipitated by using monoclonal anti-EGFR antibody (NeoMarkers) and resolved on 8% SDS-polyacrylamide gel. The detailed procedures were essentially the same as described above for immunoprecipitation. The separated gel bands were excised and subjected to trypsin for gel digestion, followed by nano-HPLC-MS/MS system analysis [23].
Site-directed mutagenesis
Ultra-Blue site-directed mutagenesis kit was purchased from Stratagene (La Jolla, CA), and mutations were created according to the manufacturer’s instructions. The 3KR-EGFR mutant was confirmed by DNA sequencing (MD Anderson Sequencing Core Facility).
Transfection and siRNA knockdown
Transfection of wild-type EGFR (wt-EGFR) or 3KR-EGFR or cotransfection with p300 or CBP, or PCAF was performed using liposomes. To compare the acetylation of 3KR-EGFR with wt-EGFR by CBP, HEK293 cells were cotransfected with either 2 µg wt-EGFR or 2 µg 3KR-EGFR with 7 µg CBP. After 48 h incubation, the cell lysates were subjected to immunoprecipitation analysis for EGFR acetylation. siRNA of EGFR, p300 or CBP (Santa Cruz) were transfected by electroporation as described in manufacturer’s instructions (Lonza, Walkersville, MD).
Immunofluorescence assay
A431 cells were serum-starved and treated with EGF prior to collection. The cells were fixed with methanol and incubated with anti-CBP antibody for 30 min at room temperature. A FITC labeled anti-IgG antibody was used to probed CBP protein.
Cell viability assay
1 × 104 cells were seeded in a 96-well plate. The cells were cultured for 24 h and then treated with SAHA or TKIs (Erlotinib, Gefitinib, or Lapatinib) or different combinations as indicated for 72 h. After incubation, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added to final 0.5 mg/ml and continued culturing for 2–4 h. Then 100 µl of β-isopropanol was added into each well to dissolve precipitated substrate. The absorbance was measured by a microplate reader (Bio-Rad, Hercules, CA).
In vivo mammary fat pad tumor cell injection
5 × 106 MDA-MB-468 cells were inoculated into the mammary fat pad of each mouse. The treatment was initiated when tumor size reached 100 mm3. SAHA (20mg/kg) and /or erlotinib (15mg/kg) were fed orally everyday until the control group reached maximal tumor size that was allowed by institutional guideline. The tumor size was measured twice per week. All animal handling procedures were performed in accordance with the institutional policies and regulations.
Statistical analysis
All data are described as mean ± standard derivation. Statistical analyses were performed by student’s t-Test and ANOWA analysis. P<0.05 is considered as statistical significance.
Results
CBP acetylates EGFR at K684, K836, and K843
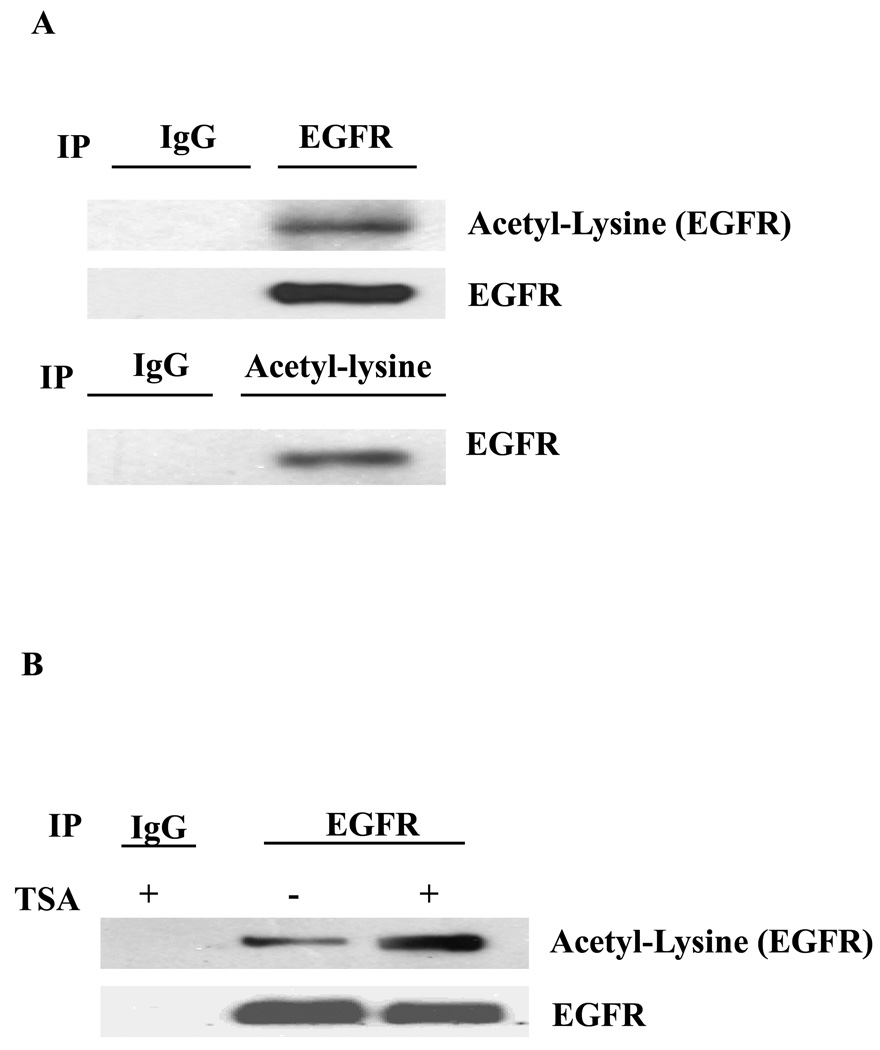
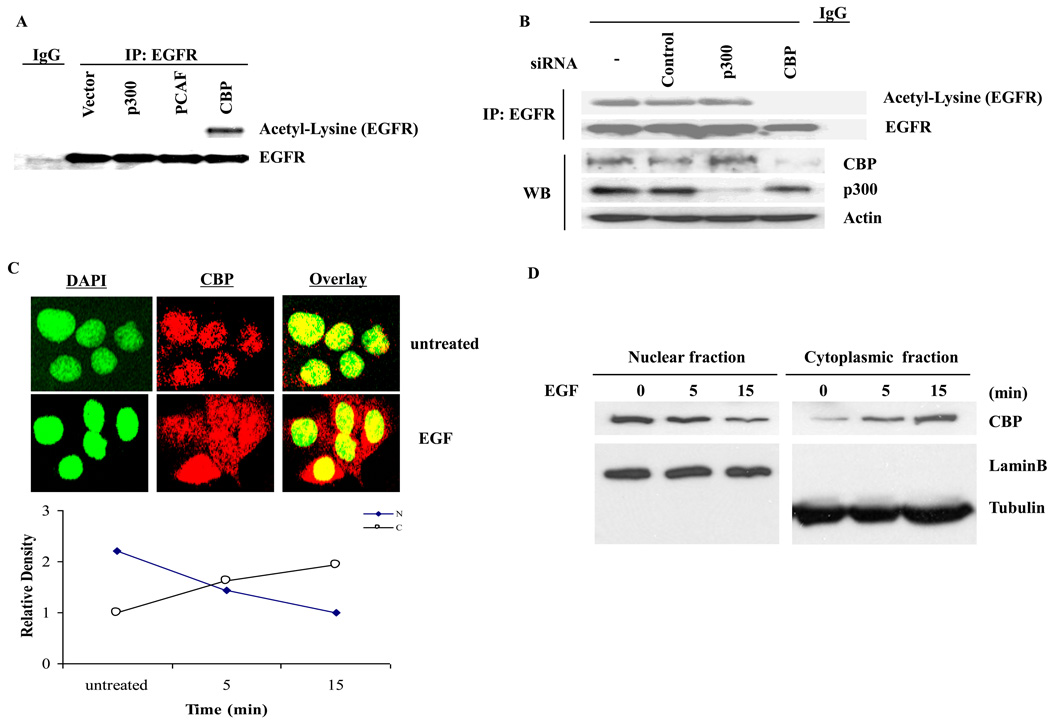
To determine if EGFR is acetylated, we performed a reciprocal immunoprecipitation and immunoblot analysis of endogenous EGFR extracted from A431 cells and demonstrated EGFR lysine acetylation by an antibody against acetyl-lysine (Fig. 1A). This acetylation was enhanced in the presence of TSA (Fig. 1B). In addition, acetylation of EGFR can be detected by using two other polyclonal antibodies against acetyl-lysine (data not shown). Next, to determine the specific acetylation sites, tandem mass spectrometry (MS/MS) analysis was performed [23], and three sites, K684, K836, and K843, located within the intracellular domain of EGFR were identified (Supplementary Figs. 1A, B, and C). We also found that EGFR acetylation was dramatically induced by co-transfection of wt-EGFR and CBP acetyltransferase but not others acetyltransferases such as p300 and PCAF (Fig. 2A). Acetylation of EGFR by CBP was blocked by siRNA against CBP but not p300 (Fig. 2B). Although CBP is mainly a nuclear protein, it can translocate to the cytoplasm in response to interferon stimulation to acetylate INFαR2 [24]. Similarly, EGF also triggers CBP shuttling from the nucleus to the cytoplasm, which may be responsible for EGFR acetylation in response to ligand stimulation (Fig. 2C and D).
Fig. 1.
Acetylation of EGFR at K684, K836, and K843. A total of 500 µg cell lysate was immunoprecipitated and immunoblotted with antibodies as indicated. EGFR acetylation was detected by using polyclonal anti-acetyl-lysine antibodies. (A) A431 cells were cultured in DMEM containing 10% FBS and used to analyze endogenous EGFR acetylation. (B) A431 cells were serum-starved then treated with 20 µM TSA for 5 h prior to collecting the cells for Western blot analysis.
Fig. 2.
CBP acetylase is responsible for acetylating EGFR. (A) Wild-type EGFR (wt-EGFR) was co-transfected with CBP, p300 or PCAF into HEK293 cells for 48 h in DMEM with 10%FBS, respectively. Then the cells were lysed and immunoprecipitation was performed. (B) siRNA for silencing p300 or CBP was transfected into A431 cells by electroporation for 72 h, and then the cells were lysed for immunoprecipitation and immunoblot analysis. (C) A431 cells were serum-starved and probed with anti-CBP antibody for immunostaining. (D) The nuclear fraction and cytoplasm fraction of proteins from A431 cells were immunoblotted with indicated antibodies.
SAHA enhances EGFR acetylation that is associated with cancer cell resistance to HDACI
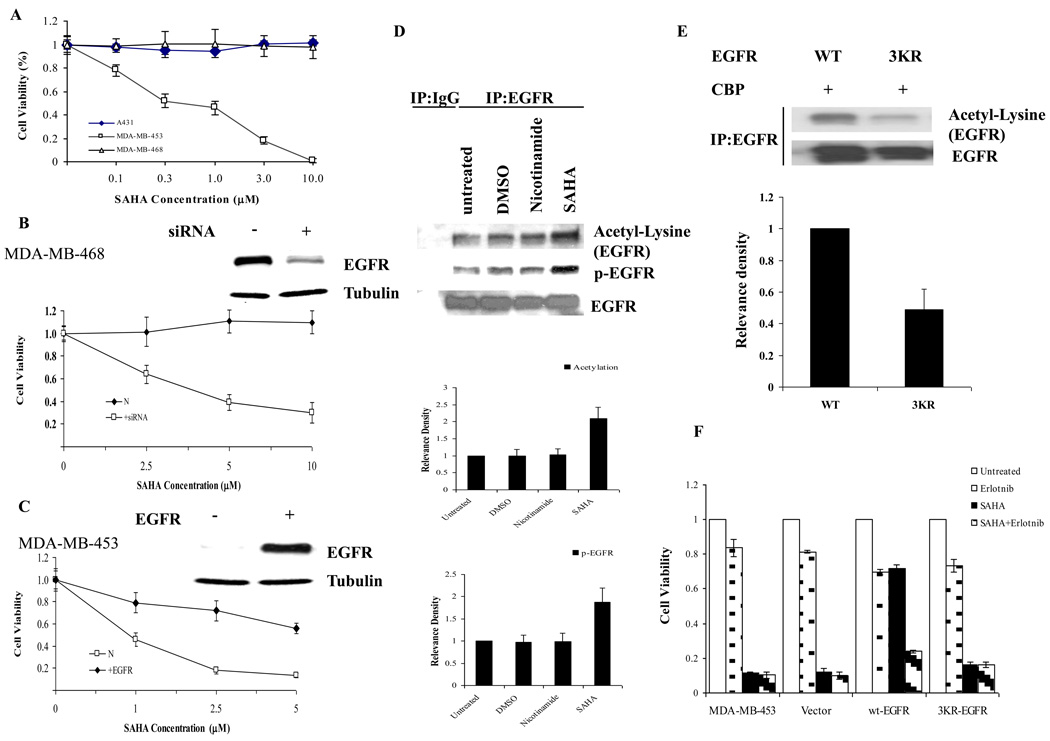
To determine the biological affects of EGFR acetylation, we tested several cancer cells lines in the presence of increasing concentrations of SAHA, a potent class I and II HDAC inhibitor. Presumably, since EGFR is acetylated by CBP, the addition of a deacetylase inhibitor would enhance its acetylation. We first determined the cell viability of A431, MDA-MB-468, and MDA-MB-453 cancer cell lines by MTT assay. As shown in Fig. 3A, both A431 and MDA-MD-468 cell lines were resistant to SAHA treatment even at a concentration of 10 µM. In contrast, cell viability of MDA-MB-453 breast carcinoma cells decreased with a concomitant increase in SAHA concentration. Interestingly, both cell lines (A431 and MDA-MB-468) that were resistant to SAHA are known to express high levels of EGFR whereas the sensitive cell line (MDA-MB-453) has low levels of EGFR expression (data not shown) [25]. When we knocked down EGFR by siRNA transfection, MDA-MB-468 cells became sensitive to SAHA treatment (Fig. 3B) while ectopically expressed wt-EGFR in MDA-MB-453 cells rendered them resistant to SAHA treatment (Fig. 3C).
Fig. 3.
Acetylation of EGFR is associated with cancer cell resistance to HDACI. (A) A431, MDA-MB-468, and MDA-MB-453 cells were treated with various concentrations of SAHA as indicated for 72h. (B) Knockdown of EGFR in MDA-MB-468 cells rendered cells sensitive to SAHA. (C) Ectopic expression of EGFR in MDA-MB-453 cells rendered cells resistant to SAHA. (D) A431 cells were serum-starved then treated with SAHA (5 µM) or Nicotinamide (4mM) for 5 h prior to collecting cells. (E) For comparison of EGFR acetylation between wt-EGFR and or 3KR-EGFR mutant, 7 µg CBP was cotransfected with either 2 µg wt-EGFR or 2 µg 3KR-EGFR in HEK293 cells. After 48h expression, the cell lysates were subject to immunoprecipitation analysis for EGFR acetylation. The bar charts represent an average of four independent experiments. (F) The wt-EGFR or 3KR-EGFR mutant was stably transfected into MDA-MB-453 cells, and cells were treated with either 2.5 µM SAHA or 10 µM erlotinib for 72 h as indicated. All data are calculated with mean ± SD and statistically analyzed by ANOWA and student’s t-Test. P<0.05 is considered as statistical significance.
Nest, we asked whether the resistance to SAHA treatment is associated with increased EGFR acetylation. Indeed, A431 cells treated with SAHA but not nicotinamide, a class III HDAC inhibitor, demonstrated an increase in acetylation as shown by Western blot analysis using the acetyl-lysine antibody (Fig. 3D). In addition, we found that EGFR phosphorylation also increased in the presence of SAHA. We quantitated the intensity of the bands and determined that SAHA increased both acetylation and phosphorylation of EGFR by about 2-fold (Figure 3D, right). We constructed an EGFR triple mutant (3KR-EGFR) containing arginine substitutions at K684, K836, and K843 acetylation sites that we identified from MS/MS analysis above and showed that the 3KR-EGFR mutant was more resistant to acetylation by CBP as compared with wt-EGFR (Figure 3E). In addition, we demonstrated that expression of wt-EGFR rendered the SAHA-sensitive MDA-MB-453 cells resistant to SAHA treatment which was likely caused by the acetylation-enhanced tyrosine phosphorylation of EGFR (Figure 3D), as combination treatment of SAHA and erlotinib, an EGFR tyrosine kinase inhibitor, could sensitize the EGFR-expressing cells (Figure 3F). Interestingly, The 3KR-EGFR mutant lost its ability to cause resistance to SAHA treatment (Figure 3F). Together, the increase in EGFR acetylation and phosphorylation under SAHA treatment suggests that EGFR phosphorylation might contribute to SAHA resistance, and therefore, also implying that an intrinsic link between elevated EGFR acetylation and cancer cell resistance to HDACI exists.
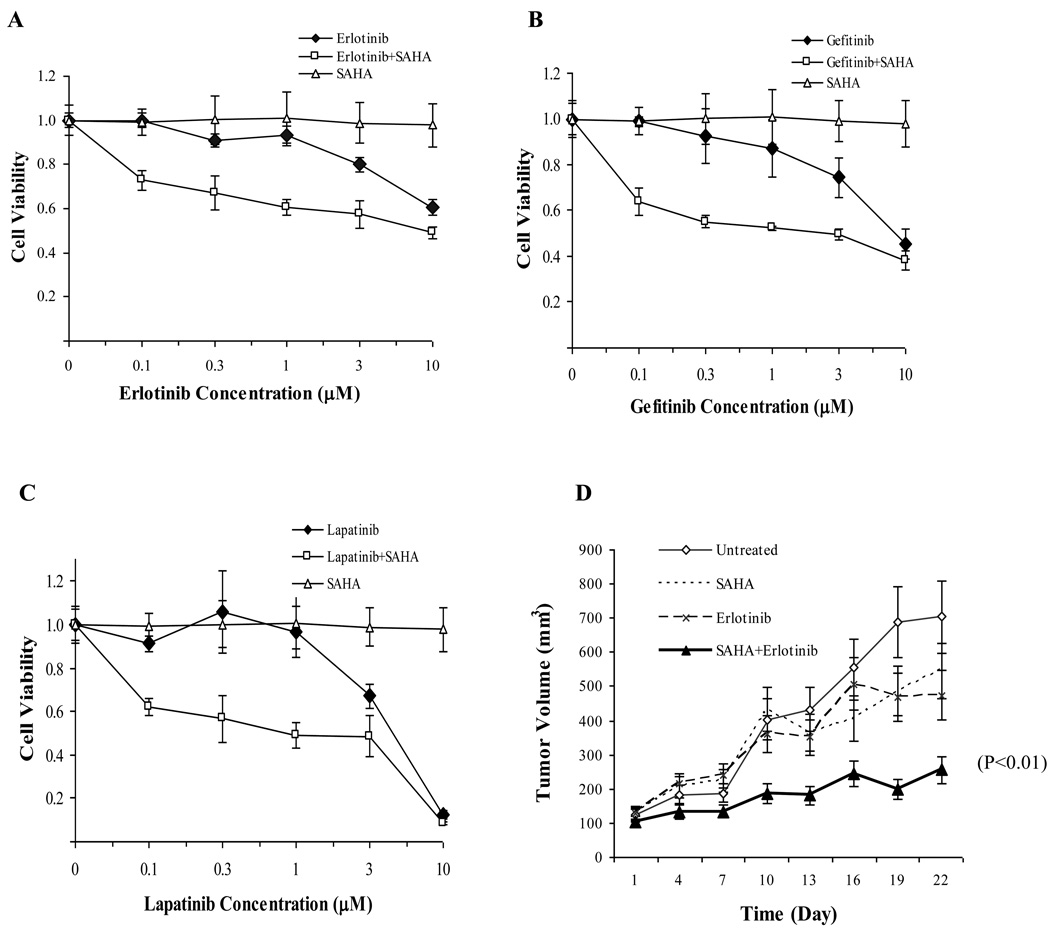
SAHA in combination with EGFR tyrosine inhibitors enhances therapeutic efficacy
EGFR inhibitors have been widely used for treating various cancer types, including lung and breast cancer. Since elevated EGFR phosphorylation by acetylation may be associated cancer cell resistance to HDACI, combing HDACI with EGFR tyrosine inhibitors may offset the adverse effect. In MDA-MB-468 breast cancer cell line, which is resistant to SAHA, we treated cells with SAHA in combination with erlotinib, gefitinib, or laptinib and showed that the combined treatment effectively inhibited cell growth in vitro compared with the individual agents (Fig. 4, A, B, and C, respectively). In an orthotopic breast cancer mouse model, MDA-MB-468 cells were injected into mammary fat pad of nude mice, when tumor size reached 100 mm3, mice were administered SAHA (concentration), erlotinib (concentration) or a combination of both (Fig. 4D). The results showed that the combination of SAHA and TKI had the most inhibition of tumor growth compared with either agent alone.
Fig. 4.
Combination of SAHA and EGFR inhibitors offsets cancer resistance to SAHA. MTT assays were performed by plating 1 × 104 cells into 96-well plates and cultured for 72 h in the presence or absence of indicated TKIs and/or SAHA. All treatments were set up at least as triplets and represent three independent experiments. MDA-MB-468 cells were treated with 2.5 µM SAHA in combination with either erlotinib (A), gefitinib (B), or lapatinib (C) with indicated concentrations for 72 h. (D) MDA-MB-468 cells were injected into nude mice in the mammary fat pad. The drugs were fed orally everyday for 3 weeks. The tumor volume was measured twice per week. All data are calculated with mean ± SD and statistically analyzed by ANOWA and student’s t-Test. P<0.05 is considered as statistical significance.
Discussion
In this study, we indentified three acetylation sites in EGFR by MS/MS and found that CBP is able to acetylate EGFR. In addition, in response to EGF, CBP can shuttle from the nucleus into cytoplasm. It should be mentioned that MS/MS analysis in our study did not detect the lysine acetylation at 1155, 1158, and 1164 sites that were shown to be acetylated in endothelial cells recently [19]. The EGFR acetylation in the endothelial cells was involved in its endocytosis in that study. It is not yet clear the relationship between these two sets of acetylation sites that were identified in the epithelial and endothelial cells. It would also be of interest to determine whether they are caused by different acetyltransferases and/or due to different cell type. However, in either case, EGFR acetylation seems to play an important role in EGFR function.
HDACIs are an attractive group of anticancer agents and have been increasingly used in the clinic as anticancer drugs. Pre-clinical and clinical studies have demonstrated varied efficacies in cancer treatment. To date, EGFR has been considered as a common target for anti-cancer therapy and several FDA approved drugs including small molecule inhibitors and antibodies targeting EGFR have been used for anticancer therapy. However, single HDACI or EGFR inhibitor regimens only achieved minimal sufficient efficacy in cancer cell growth inhibition. Frequently, tumors develop resistance to both drugs within a short period of time. The combination of HDACI and TKIs has been proposed to tackle this pitfall [16; 17]. Initial data suggest that the combination regimen achieved significantly better outcomes or even synergistic effect in some cases compared with single drug treatment, and different working mechanisms have been proposed [16; 17; 18; 26]. However, several issues still remain elusive. For example, it is not known whether HDACI directly interacts with receptor tyrosine kinases and which function of HDACI plays a dominant role in this synergistic anticancer effect or if their interaction is through transcriptional and translational regulation or local protein complex interruption.
In our study, we demonstrated that the HDACI such as TSA and SAHA, which are potent class I and II HDACIs, enhanced EGFR acetylation and phosphorylation (Figs. 1B and 3D). The universal inhibition of HDAC family proteins resulted in an increase of acetylation of many cellular proteins including EGFR. Recently, a study showed that TSA enhanced EGFR phosphorylation level in ovarian cancer cells in a dose- and time-dependent manner [18]. The observation of HDACI-enhanced EGFR activity raised an interesting and clinically important question whether HDACI treatment alone may promote tumorigenesis or malignancy through EGFR activation.
The recent discovery of non-histone proteins as acetylation substrates [9; 23; 24; 27; 28] provides plausible evidence that protein acetylation may be critical for cellular processes and signal transduction [23; 24; 29]. This study not only opens up a new avenue to examine possible phosphorylation regulation of cell surface receptors by acetylation but also has an important clinical implication, predicting that EGFR-expressing cancer cells may be resistant to single HDACI treatment due to acetylation-enhanced EGFR tyrosine phosphorylation.
Supplementary Material
Mass spectrometry analysis of EGFR acetylation. (A to C) A431 cells pretreated with 2 µM TSA and 5 mM sodium butyrate for 24 h were lysed and immunoprecipitated by an EGFR antibody and then subjected to nano-HPLC-MS/MS analysis.
Acknowledgements
We thank Drs. Stephanie A. Miller and Jennifer L. Hsu for proofreading the manuscript. We are grateful to Su Zhang and Drs. Zhenbo Han, Hirohito Yamaguchi, Ming-Chuan Hsu, and Chi-Hong Chao for technical assistance. We also thank Drs. E.Y. Chin (Brown University) and T.P. Yao (Duke University) for generously supplying the expression plasmids for p300, CBP, and PCAF. This work was supported in part by NIH CA1109311 and CA099031 (to M.-C.H.), and CA126832 (to Y.Z.); the Breast Cancer SPORE CA116199 and CCSG CA16672; National Breast Cancer Foundation, Inc. (to M.-C.H.); NSC 96/97/98-3111-B-039 (to M.-C.H., L.-Y. L. and S.-P.Y) and 99-2632-B-039-001-MY3 (to M.-C. H and L.-Y.L.); NHRI-EX98-9603BC; DOH97-TD-I-111-TM003, DOH98-TD-I-111-TM002 (to L.-Y.L.), DOH99-TD-C-111-005, and DOH97-TD-G-111-041; the MD Anderson Cancer Center-China Medical University and Hospital Sister Institution Fund; and Kadoorie Charitable Foundations (to M.-C.H.). H.S. was a recipient of the NIH-NRSA Award (1UL1RR024148). In memoriam, Tiong-Loi Ang for his courageous fight against cancer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 3.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 4.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 5.Oved S, Mosesson Y, Zwang Y, Santonico E, Shtiegman K, Marmor MD, Kochupurakkal BS, Katz M, Lavi S, Cesareni G, Yarden Y. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J Biol Chem. 2006;281:21640–21651. doi: 10.1074/jbc.M513034200. [DOI] [PubMed] [Google Scholar]
- 6.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Poppleton HM, Wiepz GJ, Bertics PJ, Patel TB. Modulation of the protein tyrosine kinase activity and autophosphorylation of the epidermal growth factor receptor by its juxtamembrane region. Arch Biochem Biophys. 1999;363:227–236. doi: 10.1006/abbi.1998.1095. [DOI] [PubMed] [Google Scholar]
- 9.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 10.Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol. 2006;7:391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- 11.Marks PA. Discovery and development of SAHA as an anticancer agent. Oncogene. 2007;26:1351–1356. doi: 10.1038/sj.onc.1210204. [DOI] [PubMed] [Google Scholar]
- 12.Pulukuri SM, Gorantla B, Rao JS. Inhibition of histone deacetylase activity promotes invasion of human cancer cells through activation of urokinase plasminogen activator (uPA) J Biol Chem. 2007 doi: 10.1074/jbc.M705867200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Munshi A, Kurland JF, Nishikawa T, Tanaka T, Hobbs ML, Tucker SL, Ismail S, Stevens C, Meyn RE. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin Cancer Res. 2005;11:4912–4922. doi: 10.1158/1078-0432.CCR-04-2088. [DOI] [PubMed] [Google Scholar]
- 14.Glaser KB. HDAC inhibitors: Clinical update and mechanism-based potential. Biochem Pharmacol. 2007;74:659–671. doi: 10.1016/j.bcp.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Kelly WK, Marks PA. Drug insight: Histone deacetylase inhibitors--development of the new targeted anticancer agent suberoylanilide hydroxamic acid. Nat Clin Pract Oncol. 2005;2:150–157. doi: 10.1038/ncponc0106. [DOI] [PubMed] [Google Scholar]
- 16.Chinnaiyan P, Allen GW, Harari PM. Radiation and new molecular agents, part II: targeting HDAC, HSP90, IGF-1R, PI3K, and Ras. Semin Radiat Oncol. 2006;16:59–64. doi: 10.1016/j.semradonc.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Edwards A, Li J, Atadja P, Bhalla K, Haura EB. Effect of the histone deacetylase inhibitor LBH589 against epidermal growth factor receptor-dependent human lung cancer cells. Mol Cancer Ther. 2007;6:2515–2524. doi: 10.1158/1535-7163.MCT-06-0761. [DOI] [PubMed] [Google Scholar]
- 18.Zhou C, Qiu L, Sun Y, Healey S, Wanebo H, Kouttab N, Di W, Yan B, Wan Y. Inhibition of EGFR/PI3K/AKT cell survival pathway promotes TSA's effect on cell death and migration in human ovarian cancer cells. Int J Oncol. 2006;29:269–278. [PubMed] [Google Scholar]
- 19.Goh LK, Huang F, Kim W, Gygi S, Sorkin A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J Cell Biol. 2010;189:871–883. doi: 10.1083/jcb.201001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao YS, Hubbert CC, Yao TP. The microtubule-associated histone deacetylase 6 (HDAC6) regulates epidermal growth factor receptor (EGFR) endocytic trafficking and degradation. J Biol Chem. 2010;285:11219–11226. doi: 10.1074/jbc.M109.042754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deribe YL, Wild P, Chandrashaker A, Curak J, Schmidt MH, Kalaidzidis Y, Milutinovic N, Kratchmarova I, Buerkle L, Fetchko MJ, Schmidt P, Kittanakom S, Brown KR, Jurisica I, Blagoev B, Zerial M, Stagljar I, Dikic I. Regulation of epidermal growth factor receptor trafficking by lysine deacetylase HDAC6. Sci Signal. 2009;2:ra84. doi: 10.1126/scisignal.2000576. [DOI] [PubMed] [Google Scholar]
- 22.Kamemura K, Ito A, Shimazu T, Matsuyama A, Maeda S, Yao TP, Horinouchi S, Khochbin S, Yoshida M. Effects of downregulated HDAC6 expression on the proliferation of lung cancer cells. Biochem Biophys Res Commun. 2008;374:84–89. doi: 10.1016/j.bbrc.2008.06.092. [DOI] [PubMed] [Google Scholar]
- 23.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 24.Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, Ramratnam B, Chin YE. Acetylation-Dependent Signal Transduction for Type I Interferon Receptor. Cell. 2007;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 25.Yamasaki F, Zhang D, Bartholomeusz C, Sudo T, Hortobagyi GN, Kurisu K, Ueno NT. Sensitivity of breast cancer cells to erlotinib depends on cyclin-dependent kinase 2 activity. Mol Cancer Ther. 2007;6:2168–2177. doi: 10.1158/1535-7163.MCT-06-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, Brandstetter K, Wittner B, Ramaswamy S, Classon M, Settleman J. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munshi N, Agalioti T, Lomvardas S, Merika M, Chen G, Thanos D. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science. 2001;293:1133–1136. doi: 10.1126/science.293.5532.1133. [DOI] [PubMed] [Google Scholar]
- 28.Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, Frye R, Ploegh H, Kessler BM, Sinclair DA. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 29.Cohen T, Yao TP. AcK-knowledge reversible acetylation. Sci STKE. 2004;245:pe42. doi: 10.1126/stke.2452004pe42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mass spectrometry analysis of EGFR acetylation. (A to C) A431 cells pretreated with 2 µM TSA and 5 mM sodium butyrate for 24 h were lysed and immunoprecipitated by an EGFR antibody and then subjected to nano-HPLC-MS/MS analysis.