Abstract
We conducted a two-stage genome-wide association study of renal cell carcinoma (RCC) in 3,772 cases and 8,505 controls of European background from 11 studies, and followed up 6 SNPs in three replication studies of 2,198 cases and 4,918 controls. Two loci on the regions of 2p21 and 11q13.3 were associated with RCC susceptibility below genome-wide significance. Two correlated variants (r2 = 0.99 in controls), rs11894252 (P = 1.8×10−8) and rs7579899 (P = 2.3×10−9), map to EPAS1 on 2p21, which encodes hypoxia-inducible- factor-2 alpha, a transcription factor previously implicated in RCC. The second locus, rs7105934, at 11q13, contains no characterized genes (P = 7.8×10−14). In addition, we observed a promising association on 12q24.31 for rs4765623 which maps to the scavenger receptor class B, member 1 (SCARB1) gene (P = 2.6×10−8). Our study reports novel genomic regions associated with RCC risk that may lead to new etiological insights.
Kidney cancer accounts for approximately 2% of new cancer diagnoses worldwide1 and is the deadliest urologic malignancy with an estimated 5-year survival rate between 50% and 60%2. Approximately 80–90% of kidney cancers develop in the renal parenchyma, and are known as renal cell carcinoma (RCC). Epidemiological studies have conclusively identified three risk factors, all modifiable: hypertension, obesity and smoking2, 3. Furthermore, there is evidence that genetic factors influence susceptibility to RCC; for instance, the life-time risk increases approximately twofold for those with a first-degree relative with RCC4–7. The tumor is also commonly observed in pedigrees with von Hippel-Lindau (VHL) syndrome as well as other genetic disorders, such as hereditary papillary renal cell carcinoma, Birt-Hogg-Dubé syndrome, and hereditary leiomyomatosis and renal cell cancer (HLRCC)2, 8. However, familial RCC cases represent less than 5% of RCC overall9. To date, candidate gene studies have not yielded genetic variants that conclusively replicate. In search of common genetic variants with moderate effect sizes, we have therefore conducted a genome-wide association study (GWAS) of RCC.
We report the findings of a two-stage GWAS of RCC, based on two parallel scans followed by replication of six notable SNPs in three studies. The two scans were coordinated by (i), the International Agency for Research on Cancer (IARC) and the Centre National de Génotypage (CNG), based on 2,639 RCC cases and 5,392 controls of European background drawn from 7 studies conducted in Europe with the Illumina Infinium HumanHap 300 and 610 Bead Chips; and (ii), the U.S. National Cancer Institute (NCI) scan, based on 1,453 RCC cases and 3,531 controls of European background from 4 studies with the Illumina Infinium HumanHap 500 and 610 chips (Supplementary Table 1, Online Methods and Supplementary note). All subjects from the IARC/CNG study were genotyped at the CNG with the exception of 305 cases and 323 controls from Russia that were genotyped at the Center “Bioengineering” and at the “Kurchatov Institute” in Moscow. All subjects from the NCI study were scanned at the NCI Core Genotyping Facility. In addition, 1,438 controls from the Wellcome Trust Case-Control Consortium were genotyped at the Sanger Institute, UK10. All RCC cases were defined on the basis of the International Classification of Diseases for Oncology, Second Edition (ICD-O-2), and included all cancers that were coded as C64.
Comparable quality control metrics were applied to the two scanned data sets and following sample and SNP exclusions, genotype data for up to 577,547 SNPs were available for 2,461 cases and 5,081 controls in the IARC/CNG scan, while data for 585,576 SNPs were available for 1,311 cases and 3,424 controls in the NCI scan (Online Methods). Primary analyses were conducted using unconditional logistic regression models for genotype trend effects (1 degree of freedom) and adjusted for sex, country, eigenvectors, and study for the USA (Online Methods). In order to compute summary findings across both scans, a meta-analysis was performed using a fixed effects model with inverse variance weighting followed by a pooled analysis with individual level data. Quantile-quantile plots of the combined results showed little evidence for inflation of the test statistics compared to the expected distribution (λ = 1.018, overall, Supplementary Fig. 1). Genomic control was subsequently applied, and all reported p-values and confidence intervals were corrected for the observed inflation. A Manhattan plot summarizing the combined results of 586,069 SNPs is shown in Supplementary Figure 2.
Based on the meta-analysis using SNPs genotyped in both centers, six SNPs associated with RCC at a significance level approaching or surpassing genome-wide statistical significance (P < 5×10−7 in two-tailed tests)10 were selected for replication in three additional case-control series from Europe and the US (2,198 RCC cases, 4,918 controls) (Supplementary Table 1). Performing genomic control showed that hidden population substructures or differential genotype calling between cases and controls did not substantively influence these results (Online methods). Three SNPs on 2p21 (rs11894252, rs7579899 and rs6758592) were selected as well as single SNPs on 3q26.31 (rs9839909), 11q13.2 (rs7105934), and 12q24.31 (rs4765623). For the replication study, rs11894252 could not be optimized; thus a highly correlated SNP, rs1867785 (r2 = 1.0 in HapMap CEU11), was genotyped (Online Methods). For the other five SNPs, there was a high concordance between genotype calls on the Illumina bead chip and optimized TaqMan assays in both centers (100% for IARC/CNG and 98.9–100% for NCI)12. Because rs9839909 (3q26.31) and rs7105934 (11q13.2) were not included on the Illumina HumanHap 300 bead chip, subjects genotyped with this chip in the GWAS (908 cases and 2,415 controls) were also genotyped by TaqMan and included in the replication phase. In a meta-analysis of the pooled GWAS and replication results, SNPs in three of the four regions achieved genome-wide significance and mapped to 2p21, 11q13.3 and 12q24.31 (Table 1 and Fig. 1). Imputing SNPs in the implicated regions 2p21, 11q13.3 and 12q24.31, using the 1000 Genomes data13 as scaffold did not reveal additional SNPs with stronger, independent associations to those genotyped directly (Supplementary Table 2).
Table 1.
Summary results for 6 SNPs selected for replication in renal cell carcinoma genome-wide association study
| IARC+NCIa 3,772/8,505d |
Replicationb 2,198/4,918d |
All combinedc 5,970/13,423d |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus (Gene region) | SNP ID (Minor allele frequency) |
OR* | 95% CI* | P value* | OR* | 95% CI* | P value* | OR* | 95% CI* | P value* |
| 2p21 (EPAS1) | rs11894252 (0.40) | 1.18 | (1.12–1.26) | 1.9×10−8 | 1.08 | (1.00–1.16) | 0.06 | 1.14 | (1.09–1.20) | 1.8×10−8 |
| 2p21 (EPAS1) | rs7579899 (0.40) | 1.18 | (1.11–1.25) | 5.9×10−8 | 1.11 | (1.03–1.20) | 0.008 | 1.15 | (1.10–1.21) | 2.3×10−9 |
| 2p21 (EPAS1) | rs6758592 (0.47) | 1.13 | (1.07–1.20) | 2.5×10−5 | 1.05 | (0.97–1.14) | 0.20 | 1.10 | (1.05–1.15) | 4.0×10−5 |
| 3q26 (PP13439) | rs9839909 (0.34) | 0.82 | (0.76–0.89) | 4.3×10−6 | 0.96 | (0.90–1.03) | 0.30 | 0.90 | (0.86–0.95) | 4.0×10−5 |
| 11q13 (chr 11) | rs7105934 (0.07) | 0.65 | (0.55–0.76) | 1.7×10−7 | 0.71 | (0.62–0.81) | 6.8×10−7 | 0.69 | (0.62–0.76) | 7.8×10−14 |
| 12q24 (SCARB1) | rs4765623 (0.34) | 1.18 | (1.11–1.25) | 6.4×10−8 | 1.07 | (0.99–1.16) | 0.09 | 1.15 | (1.09–1.20) | 2.6×10−8 |
Odds ratios (OR) were estimated using the per rare allele log-additive model and unconditional logistic regression (Online methods).
All scanned samples from IARC/CNG and NCI, combined by meta-analysis (Online methods)
Samples include subjects from three replication studies: the MD Anderson Renal Cell Cancer Study, the Dutch Renal Cell Cancer Study, and the IARC Replication Study (Supplementary note).
Column shows combined results of the pooled GWAS data, and the three replication studies by meta-analysis.
Number of cases / controls
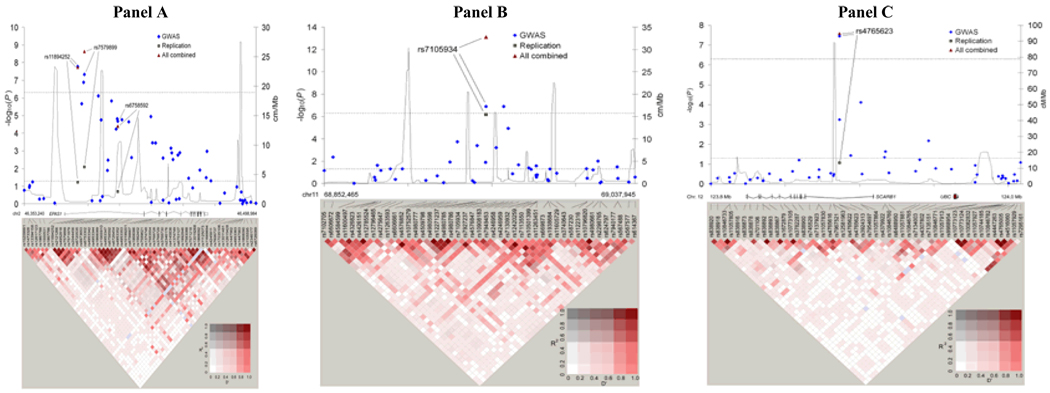
Figure 1. Association results, recombination and linkage disequilibrium plots for three regions achieving genome-wide significance in RCC GWAS.
Results of pooled IARC/CNG and NCI GWAS data (GWAS), and for SNPs selected for replication in replication studies combined by meta-analysis (replication), and of all studies combined by meta-analysis (all combined). P-values for log-additive association results (negative base 10 logarithm) are shown with recombination rates (cm/Mb) based on HapMap phase II data, and pairwise r2 and superimposed D' values are displayed at the bottom for all SNPs included in the GWAS analysis. Coordinates refer to genome build 36.1. Panel A depicts the region of 2p21 including the EPAS1 gene region (46,353,240 – 46,498,984). Panel B depicts the region of 11q13 (68,852,465 – 69,037,945). Panel C depicts the region of 12q24.31 including the SCARB1 gene region (123,800,267 – 124,008,657).
In the combined analysis14, two SNPs on 2p21 achieved genome-wide significance, rs7579899 (P = 2.3×10−9; per allele odds ratio (OR) = 1.15, 95% confidence interval (CI): 1.10–1.21) and rs11894252 (P = 1.8×10−8; OR = 1.14, 95% CI: 1.09–1.20). Further, rs7579899 was significant in the independent replication analysis (P = 0.008; OR = 1.11, 95% CI: 1.03–1.20) whereas rs1867785, a highly correlated surrogate for rs11894252, suggested a comparable effect that did not achieve independent significance (P = 0.06; OR = 1.08, 95% CI: 1.00–1.16) (Table 1). When stratified by either SNP marker, the signal of the second was extinguished (data not shown). Together with the high correlation between the two markers (r2 = 0.99 in controls), these results point towards a single common susceptibility locus. An additional SNP rs4952818 achieved genome-wide significance in the combined scan (P = 1×10−7, Figure 1), but its association was accounted for by rs11894252 and rs7579899 (Padjusted = 0.45 and Padjusted = 0.36, respectively) and was therefore not selected for replication. The third SNP selected for replication, rs6758592, was minimally correlated with the previous two (r2 = 0.12 and 0.11 with rs11894252 and rs7579899, respectively), and only showed an association in the NCI data (PNCI = 1.8×10−7, PIARC = 0.16, Pheterogeneity = 0.0004, Supplementary Table 3) not accounted for by rs11894252 and rs7579899 (Padjusted = 1x10−5 for both). While rs6758592 did not replicate, the combined analysis yielded P= 4.0×10−5, suggesting that in the NCI scan data there could be evidence for a more complex genomic architecture underlying the association of this locus with RCC.
Our finding on 2p21 is notable because the candidate gene, EPAS1, has already been implicated in RCC15–19. The two SNPs, rs11894252 and rs7579899, are distributed across a 4.2 kb region of intron 1 in the EPAS1 gene, which encodes the hypoxia-inducible factor 2α (HIF-2α), a key gene in the VHL-HIF pathway. The VHL complex targets HIF subunits for ubiquitin-mediated degradation20. Accumulation of HIF-2α leads to up-regulation of vascular endothelial growth factor (VEGF) and epidermal growth factor receptor (EGFR). The inactivation of VHL in renal carcinoma cell lines leads to unchecked HIF-2α mediated expression of HIF-responsive tumorigenic factors, most notably VEGF16, 17. Further, tumor formation in VHL-deficient renal carcinoma cells has been found to be suppressed by inhibition of HIF-2α18,19. The findings from our GWAS provide further evidence that EPAS1 is a key gene in RCC development, but additional studies are needed to identify the functionally relevant common variants associated with increased risk.
A variant, rs7105934, on 11q13 was associated with RCC in the combined analysis (P = 7.8×10−14, OR = 0.69, 95% CI: 0.62–0.76). The SNP was independently replicated with a comparable risk estimate to the initial GWAS results (P = 6.8×10−7; OR = 0.71, 95% CI: 0.62–0.81). Overall, the magnitude of the association with this relatively uncommon SNP (minor allele frequency = 0.08 in controls) is comparatively large compared to risk markers previously identified in the GWAS of other cancers21. This SNP maps to a 350 kb region of 11q13 containing no characterized genes; flanking genes are Homo sapiens myeloma over-expressed (in a subset of t(11;14) positive multiple myelomas) (MYEOV) and cyclin D1 (CCND1), situated approximately 140 kb centromeric and 220kb telomeric, respectively, from rs7105934. In the control samples, there is little evidence for linkage disequilibrium with markers in these genes (r2 < 0.01 in scanned controls). Similarly, we did not observe LD with a complex susceptibility locus for prostate cancer also identified within 11q1322, 23, nor with a SNP marker, rs614367, 89kb telomeric to rs7105934 recently associated with breast cancer risk24.
A third locus, marked by rs4765623 on 12q24, also achieved genome-wide significance overall (P = 2.6×10−8; OR = 1.15, 95% CI: 1.09–1.20), although it did not independently replicate using a two-tailed significance test (P = 0.09; OR = 1.07, 95% CI: 0.99–1.16). The SNP maps to intron 1 of the scavenger receptor class B, member 1 (SCARB1) gene, a cell surface receptor that binds to high-density lipoprotein cholesterol (HDL-C) and mediates HDL-C uptake25–27. Its role in cancer biology is not as well established, and the signal was stronger in the European studies (scan and replication studies) than in the US studies (Supplementary Table 3 and Fig. 2). While this SNP marks a promising association, further confirmatory work is required to establish its association with RCC risk.
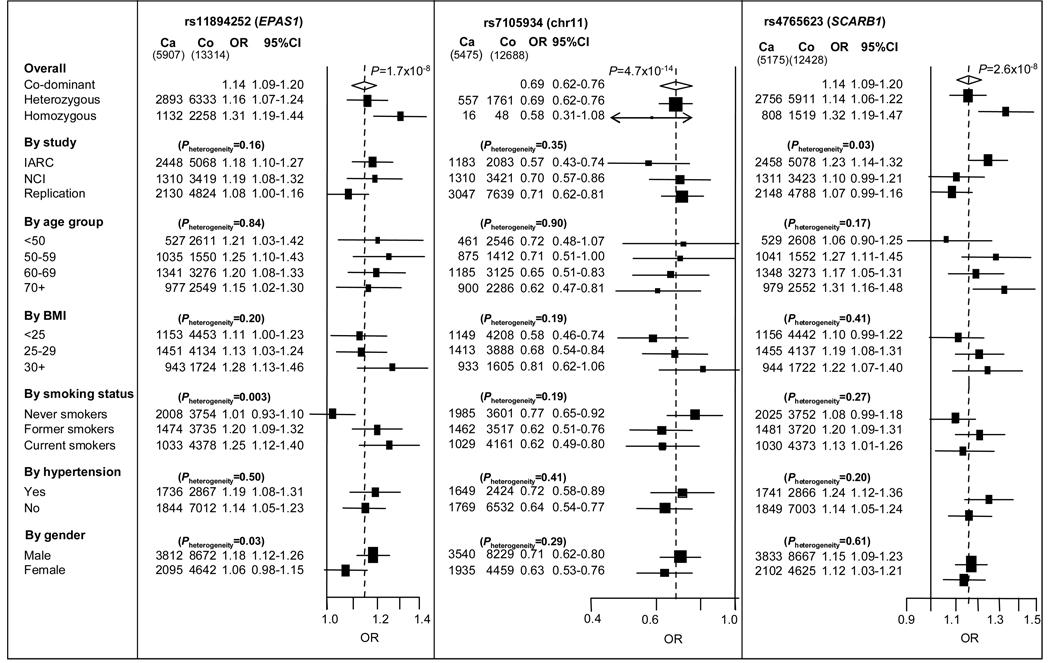
For each of the three regions associated with RCC risk, we conducted further pooled analyses stratified by study, age, gender and established modifiable risk factors: body mass index, smoking status and history of diagnosed hypertension. The associations with rs11894252 and rs7579899 were notable in former and current smokers but not in never-smokers, suggesting an interaction with smoking (P heterogeneity = 0.003) (Fig. 2). This observation raises the possibility that the effect of EPAS1 could be dependent on tobacco smoking, but further studies are needed to explore this promising finding. The associations with the two 2p21 (EPAS1) SNPs were stronger among men than women, possibly a result of the different risks by smoking status. The stratified analyses suggested no other evidence of interaction.
Figure 2. Forest plots for three SNPs achieving genome-wide significance in RCC GWAS.
Forest plots show stratified odds ratios (OR) for SNPs selected for replication and achieving genome-wide significance. The two highly correlated SNPs located at 2p21, rs7579899 and rs11894252, gave very similar results in stratified analysis, and only the results of one of the SNPs (rs11894252) are shown in the figure. Apart from the odds ratios for heterozygous and homozygous, odds ratios and 95% confidence intervals were estimated by the per rare allele log-additive trend model. All models were adjusted for sex, study and country. The overall log-additive OR is shown by the broken vertical line. P-values indicate heterogeneity for OR within each group.
This study was well powered to detect common alleles with large effect sizes (greater than 90% power to detect a per-allele OR of 1.5 for a variant of allele frequency of 20% at an alpha of 5×10−7), but the statistical power was limited for detecting effects of weaker size or those due to uncommon SNPs. Additional studies are needed to identify susceptibility markers of weaker effects or lower allele frequency.
Our study has identified novel regions of the genome associated with risk of RCC. Two regions on 2p21 and 12q24 map to candidate genes EPAS1 and SCARB1, respectively, while one maps to a region of 11q13 with no characterized genes. Further fine-mapping of these regions is required prior to investigating the optimal variants for studies into the biological underpinnings of the observed associations. Moreover, these loci should be pursued in follow-up studies in distinct populations, such as African Americans who have an increased risk of RCC2, 3. Similarly, it will be important to evaluate these regions in studies that address clinical endpoints such as response to therapy and survival. The discovery of additional susceptibility loci should lead to further advances in understanding the etiology of RCC as well its risk prediction and early detection.
Supplementary Material
Acknowledgments
The authors thank all of the participants who took part in this research, and the funders and support staff who made this study possible. Funding for the genome-wide genotyping was provided by the Institut National du Cancer (INCa), France, for those studies coordinated by IARC/CNG, and by the intramural research program of the National Cancer Institute (NCI), National Institute of Health (NIH), USA, for those studies coordinated by the NCI. Additional acknowledgments can be found in the supplementary note.
Footnotes
Author contributions
M.P.P., M.J., J.R.T., G.S., L.E.M., V.G., W.H.C., J.D.M., N.R., S.J.C., and P. Brennan contributed to the design and execution of the overall study. M.P.P., M.J., J.R.T., G.S., L.E.M., L.A.K., X.W., V.G., K.B.J., J.D.M., N.R., S.J.C., and P Brennan contributed to the statistical analysis. M.P.P., M.J., S.J.C. and P. Brennan wrote the first draft of the manuscript. D. Zeleniak, E.P., L.A.K., X.W., K.B.J., S.H.V., S.L.M., Y.Y., A.M.M., E.S.B., N.N.C., M.F., D.L., I.G., S.H., H. Blanche, A.H., G.T., Z.W., M.Y., K.G.S., S.J.C., and M.L. supervised or conducted the genotyping. The remaining authors conducted the epidemiologic studies and contributed samples to the GWAS and/or replication. All authors contributed to the writing of the manuscript.
References
- 1.Ferlay J, Bray F, Pisani P, Parkin DM. IARC CancerBase No. 5. version 2.0. Lyon: IARCPress; 2004. GLOBOCAN 2002: Cancer Incidence, Mortality and Prevalence Worldwide. [Google Scholar]
- 2.Scelo G, Brennan P. The epidemiology of bladder and kidney cancer. Nat. Clin Pract. Urol. 2007;4:205–217. doi: 10.1038/ncpuro0760. [DOI] [PubMed] [Google Scholar]
- 3.Chow WH, Dong LM, Devesa SS. Epidemiology and risk factors for kidney cancer. Nature Reviews Urology. 2010;7:1–13. doi: 10.1038/nrurol.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLaughlin JK, et al. A population--based case--control study of renal cell carcinoma. J. Natl. Cancer Inst. 1984;72:275–284. [PubMed] [Google Scholar]
- 5.Schlehofer B, et al. International renal-cell-cancer study. VI. the role of medical and family history. Int. J. Cancer. 1996;66:723–726. doi: 10.1002/(SICI)1097-0215(19960611)66:6<723::AID-IJC2>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 6.Gago-Dominguez M, Yuan JM, Castelao JE, Ross RK, Yu MC. Family history and risk of renal cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 2001;10:1001–1004. [PubMed] [Google Scholar]
- 7.Hung RJ, et al. Family history and the risk of kidney cancer: a multicenter case-control study in Central Europe. Cancer Epidemiol. Biomarkers Prev. 2007;16:1287–1290. doi: 10.1158/1055-9965.EPI-06-0963. [DOI] [PubMed] [Google Scholar]
- 8.Linehan WM, et al. Hereditary kidney cancer: unique opportunity for disease-based therapy. Cancer. 2009;115:2252–2261. doi: 10.1002/cncr.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peto J, Houlston RS. Genetics and the common cancers. Eur. J. Cancer. 2001;37 Suppl 8:S88–S96. doi: 10.1016/s0959-8049(01)00255-6. [DOI] [PubMed] [Google Scholar]
- 10.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Packer BR, et al. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34:D617–D621. doi: 10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Via M, Gignoux C, Burchard EG. The 1000 Genomes Project: new opportunities for research and social challenges. Genome Med. 2010;2:3. doi: 10.1186/gm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 15.Higgins JP, et al. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am. J. Pathol. 2003;162:925–932. doi: 10.1016/S0002-9440(10)63887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia G, et al. Regulation of vascular endothelial growth factor transcription by endothelial PAS domain protein 1 (EPAS1) and possible involvement of EPAS1 in the angiogenesis of renal cell carcinoma. Cancer. 2001;91:1429–1436. doi: 10.1002/1097-0142(20010415)91:8<1429::aid-cncr1149>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 17.Sowter HM, Raval RR, Moore JW, Ratcliffe PJ, Harris AL. Predominant role of hypoxia-inducible transcription factor (Hif)-1alpha versus Hif-2alpha in regulation of the transcriptional response to hypoxia. Cancer Res. 2003;63:6130–6134. [PubMed] [Google Scholar]
- 18.Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS. Biol. 2003;1:E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmer M, Doucette D, Siddiqui N, Iliopoulos O. Inhibition of hypoxia-inducible factor is sufficient for growth suppression of VHL−/− tumors. Mol. Cancer Res. 2004;2:89–95. [PubMed] [Google Scholar]
- 20.Gunaratnam L, Bonventre JV. HIF in kidney disease and development. J. Am. Soc. Nephrol. 2009;20:1877–1887. doi: 10.1681/ASN.2008070804. [DOI] [PubMed] [Google Scholar]
- 21.Chanock S. High marks for GWAS. Nat. Genet. 2009;41:765–766. doi: 10.1038/ng0709-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 23.Eeles RA, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 24.Turnbull C, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozarsky KF, et al. Overexpression of the HDL receptor SR-BI alters plasma HDL and bile cholesterol levels. Nature. 1997;387:414–417. doi: 10.1038/387414a0. [DOI] [PubMed] [Google Scholar]
- 26.Rigotti A, et al. A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc. Natl. Acad. Sci. U. S. A. 1997;94:12610–12615. doi: 10.1073/pnas.94.23.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueda Y, et al. Lower plasma levels and accelerated clearance of high density lipoprotein (HDL) and non-HDL cholesterol in scavenger receptor class B type I transgenic mice. J. Biol. Chem. 1999;274:7165–7171. doi: 10.1074/jbc.274.11.7165. [DOI] [PubMed] [Google Scholar]
- 28.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 29.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landi MT, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amundadottir L, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen GM, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu K, et al. Population substructure and control selection in genome-wide association studies. PLoS. One. 2008;3:e2551. doi: 10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 36.de Bakker PI, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC. Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.