Summary
Many bacteria have historically been classified as obligate anaerobes. They have been construed as wholly intolerant of oxygen, a feature that was originally ascribed to their lack of superoxide dismutases and catalases. Clostridial species were regarded as classic examples. We now know this view is quite wrong: enzymes that scavenge superoxide, hydrogen peroxide, and even oxygen itself abound in anaerobes. In the current issue, Hillmann et al. demonstrate that full expression of these proteins can allow Clostridium acetobutylicum to survive and even grow in oxygenated culture medium. Evidently oxidative defenses in anaerobes can be robust. In all likelihood they are critical for the movement of bacteria thru aerobic environments to new anaerobic habitats.
In 1971 Joe McCord and Irwin Fridovich were casting about for ways of testing whether the superoxide dismutase activity of a novel metalloprotein did indeed constitute its biological function. They hit upon the idea of surveying both aerobic and obligately anaerobic microbes, reasoning that since superoxide can only be derived from molecular oxygen, bona fide superoxide dismutases would be found only in organisms that dwell in oxygen-containing habitats. And in fact their data supported this conclusion: superoxide dismutase activity was scant or nill in obligate anaerobes, but it was ubiquitous among aerobes. And, gratifyingly, the correlation extended to catalase: it, too, seemed to be abundant only among aerobes (McCord et al., 1971).
These results supported the proposed role of superoxide dismutase. But the correlation induced the microbiology community to go a step further: Perhaps, in fact, obligate anaerobes cannot grow in oxygen precisely because they lack enzymes to defend themselves against superoxide and hydrogen peroxide. According to this theory, life evolved and diversified in an anaerobic world. Two billion years later, oxygenic photosynthesis led to the accumulation of so much oxygen that extant life forms were forced into one of two choices: either to acquire enzymes to protect themselves, or else to sequester themselves in anaerobic microenvironments. The obligate anaerobes made the latter choice.
In subsequent decades many details of superoxidc and hydrogen peroxide toxicity have been filled in (reviewed in Imlay, 2008). These partially reduced oxygen species are generated by adventitious reactions inside aerobic cells, when molecular oxygen steals one or two electrons from reduced flavoenzymes (Messner and Imlay, 2002). If not scavenged, both of these reactive oxygen species (ROS) can destroy the exposed [4Fe-4S] clusters of a family of dehydratases, thereby disrupting any metabolic pathways that include such enzymes (Gardner and Fridovich, 1991; Flint et al., 1993; Jang and Imlay, 2007). Target enzymes of this class belong to the TCA cycle and to amino-acid biosynthetic pathways, for example; therefore, excessive levels of intracellular oxidants will bring growth to a halt. Most ominously, H2O2 can react with unincorporated intracellular iron—including that which is leeched from damaged iron-sulfur clusters—in the Fenton reaction (Imlay et al., 1988):
The product is the hydroxyl radical, an oxidant powerful enough to directly damage DNA and thereby kill cells. The overarching lesson is that any organism that seeks to live in the presence of oxygen must synthesize high titers of scavenging enzymes to avoid toxicity by endogenous oxidants. Indeed, mutants of E. coli and Saccharomyces that lack these enzymes can no longer thrive in aerobic habitats (Carlioz and Touati, 1986; van Loon et al., 1986; Chang et al., 1991). Obligate anaerobes, it seemed, were like these mutants: devoid of scavenging enzymes, they were incapable of tolerating exposure to oxygen.
Problems with the traditional model
Even as this tidy narrative was incorporated into the biochemistry textbooks, its premise was a little hard to swallow. No habitat is free from the occasional incursion of oxygen, and it seemed odd that obligate anaerobes would not pay the price of synthesizing just a couple more enzymes if doing so would allow them to survive and even grow during periods of aeration. And, in time, further work revealed that superoxide and hydrogen peroxide are not the sole effectors of obligate anaerobiosis. Anaerobic biochemistry must sometimes operate upon highly reduced metabolites, and the optimal solutions can involve enzymes that employ glycyl radicals or low-potential metal centers in solvent-exposed active sites. These moieties can be directly oxidized by molecular oxygen itself; therefore, central pathways can be poisoned in aerobic habitats, irrespective of intracellular levels of superoxide and hydrogen peroxide (Sawers and Watson, 1998; Meinecke et al., 1989; Pan and Imlay, 2001).
Furthermore, as more genomic sequences have been collected, it has become apparent that the original McCord/Fridovich survey was, by chance, somewhat misleading: Quite a few microbes that are classified as obligate anaerobes are capable of synthesizing superoxide dismutases and catalases—in some cases, they do so in abundant amounts. And within the last decade workers have realized that those anaerobes that lack these scavengers often employ superoxide reductases (Jenney, Jr. et al., 1999; Lombard et al., 2000) and peroxidases (Poole, 2005) in their place. So at this point it is more reasonable to wonder whether there is any organism at all that lacks enzymatic defenses against partially reduced oxygen species.
This reappraisal comes to a head in the study reported by Hillmann et al. in this issue (“PerR acts as a switch for oxygen tolerance in the strict anaerobe Clostridium acetobutylicum”). Clostridia are often regarded as quintessential anaerobes: neither they nor key enzymes inside them are able to function in aerobic lab conditions, and rigorously anaerobic conditions are used in their cultivation. However, by scanning the genome of Clostridium acetobutylicum, Hillman and co-workers in the lab of Hubert Bahl discovered a homologue of PerR, which is the master repressor of peroxide stress responses in gram-positive bacteria. PerR was originally identified in Bacillus subtilis by John Helmann and colleagues at Cornell (Lee and Helmann, 2006). Helmann’s group determined that H2O2 directly oxidizes a mononuclear iron atom within PerR, creating a hydroxyl-radical-like species that directly oxidizes an iron-coordinating histidine residue. The subsequent dissociation of iron ramifies to a global change in protein structure, and the oxidized PerR protein loses the capacity to bind DNA. PerR normally represses the synthesis of proteins that suppress Fenton chemistry, including catalase, peroxidase, and the iron-sequestering protein MrgA; therefore, the oxidative inactivation of PerR exploits the Fenton reaction as a trip wire to prepare the cell for potentially stressful levels of H2O2.
The presence of a PerR homologue in C. acetobutylicum suggested to Bahl and his co-workers that this obligate anaerobe might be more capable of withstanding oxidative stress than had been appreciated. In the present study the elimination of perR through mutation provided surprisingly emphatic support for this notion: Whereas wild-type C. acetobutylicum rapidly lost viability upon aeration, the perR null mutant was able to maintain full viability for hours—and, in fact, it was able to slowly grow.
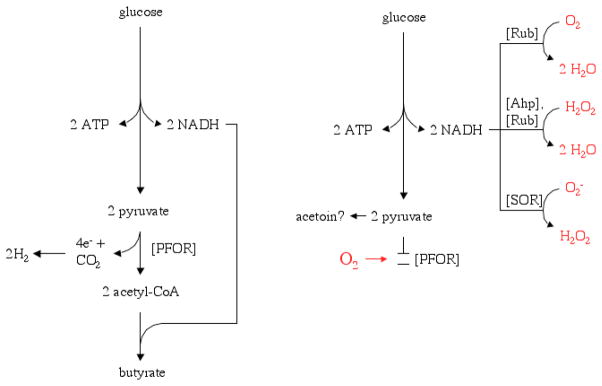
How does the mutant do it? As a starting point Hillman et al. found that two different kinds of trouble incapacitated wild-type cells when they were aerated. First, cell viability declined by four orders of magnitude within the first hour of exposure. This irreversible injury was evidently due to Fenton-mediated DNA damage, since the cells were almost completely protected when the experimenters first added dipyridyl, a cell-permeable agent that chelates intracellular iron and keeps it from reacting with H2O2. The H2O2 itself was presumably formed by the oxidation of redox enzymes. Overlaid upon that kind of injury was additional damage that blocked metabolism and growth. Although its basis was not explored in detail, the effect likely resulted when molecular oxygen inactivated pyruvate:ferredoxin oxidoreductase (PFOR), the key pyruvate-dissimilating enzyme in this anaerobe (Fig. 1).
Figure 1.
Possible re-routing of electron flow during oxidative stress. Anaerobic C. acetobutylicum ferments glucose to butyrate and hydrogen (left). The introduction of oxygen (right) probably inactivates pyruvate:ferredoxin oxidoreductase (PFOR). Redox balance may be restored by the reduction of oxygen by the Hsp21 ruberythrin (Rub); the excretion of pyruvate or acetoin is speculative. The NADH pool also provides reducing equivalents for the scavenging of H2O2 and superoxide by peroxidases (Ahp, Rub) and superoxide reductase (SOR). The intermediary electron donors to rubrerythrin and SOR are not known.
In contrast, the perR mutant completely escaped the Fenton-mediated lethality—it even survived when high doses of H2O2 were directly added to the culture. And it continued to grow for some hours after oxygenation, suggesting that central metabolism was substantially functional. On its face these data indicate that this fastidious anaerobe has a heretofore unappreciated capacity to cope with oxygen.
Which PerR-controlled genes confer oxygen tolerance?
Hillman et al. found that the perR mutant synthesizes defensive enzymes that act at several stages of oxidative stress. First among these was an oxidase that speeds the clearance of oxygen. The perR mutant exhibited a rate of oxygen consumption that, amazingly, matched that of aerobic E. coli: several million molecules of oxygen per cell per second. They suspect that the direct catalyst is the rubrerythrin Hsp21, an iron-containing protein that was synthesized in such quantities in the perR mutant that the cells turned red. Rubrerythrins have been associated in vitro with both peroxidase and oxygen-scavenging activities (Lumppio et al., 2001; Gomes et al., 2001), and presumably one or both of these activities is crucial to the oxygen tolerance of the mutant. This soluble enzyme presumably does not conserve energy, and so its role must be expressly to restore anaerobiosis whenever oxygenated waters invade the cell habitat. Its di-iron site is the likely location of oxygen reduction; if so, electrons are probably received at a distal mononuclear iron site from an as-yet unidentified electron donor, such as rubredoxin. The ultimate electron source is probably the NAD(P)H pool—and so for the system to work, the cell must be capable of continuously generating NADH when oxygen is present. On its face this requirement sounds daunting, since molecular oxygen is broadly regarded as being incompatible with Clostridial metabolism. However, if the primary site of poisoning is pyruvate:ferredoxin oxidoreductase, then glycolytic NADH formation could continue (Fig. 1). Indeed, if C. acetobutylicum excretes pyruvate or acetoin as a terminal product, then the scavenging of oxygen will serve a second purpose of reoxidizing the dinucleotide pool and thereby support continued glycolytic flux and ATP formation. The Bahl group noted that the perR mutant retained motility during oxygen exposure, confirming that its energy charge remained high despite the presumptive inactivity of PFOR. Growth did not continue indefinitely, however—perhaps because of an inability to produce acetyl-CoA, a necessary precursor for membrane synthesis.
Oxygen clearance could not have been the sole mechanism of oxygen tolerance in the Hillman experiments, however, since rigorous aeration did not restore sensitivity to the perR mutant. The authors speculate that its abundant synthesis of rubrerythrin may have had the collateral effect of depleting unincorporated iron from the cytosol—and may have thereby protected the DNA from Fenton chemistry. Interestingly, peroxide-stressed aerobes accomplish the same end through the synthesis of MrgA and Dps (Chen and Helmann, 1995; Grant et al., 1998), ferritin-like iron-storage proteins that C. acetobutylicans apparently lacks.
Finally, C. acetobutylicum, like many anaerobes, uses multiple, dissociated proteins to deliver electrons to its terminal peroxidases and superoxide reductases. For a long time these systems escaped discovery because, unlike single-protein disproportionation systems such as catalases and superoxide dismutases, they cannot function in dilute extracts and so cannot easily be detected. The advantages of superoxide and peroxide reduction systems have been debated and may include the capacity to drive ROS concentrations to especially low levels. Alternatively, evolution may simply have favored reduction rather than disproportionation, because in low-oxygen environments resting enzymes will tend to be reduced by promiscuous low-potential electron carriers such as flavodoxins, rubredoxins, and ferredoxins (Imlay, 2002). In any case, the scarcity of catalases and superoxide dismutases clearly misled the community to conclude that anaerobes passively expire when oxygen seeps into their habitat.
Earlier studies by the Niimura and Bahl groups had indicated that these ROS scavenging systems are induced when C. acetobutylicum is exposed to low doses of oxygen (May et al., 2004; Kawasaki et al., 2005; Hillman et al., 2006; Riebe et al., 2007). The present report indicates that PerR controls all of these responses, and the induction of these enzymes must certainly be a key element in perR survival. This regulatory scheme is exceptional: in aerobes the enzymes that react with oxygen, superoxide, and hydrogen peroxide are typically controlled by discrete regulators—such as Fnr, SoxR, and OxyR, in E. coli--that are specific for their distinct substrates. In contrast, C. acetobutylicum seems to employ a single oxygen species--hydrogen peroxide--as a general sign of aeration. The rationale may be that hydrogen peroxide is so efficiently formed when oxygen invades low-potential environments that it comprises the earliest signal of impending stress.
Is adaptation effective in wild-type cells?
It is striking that deletion of the perR repressor gene was necessary to confer oxygen resistance in these experiments. Implicitly, when wild-type cells were aerated, they did not synthesize adequate levels of protective enzymes. Why not? Perhaps the full-bore oxygenation of erstwhile anaerobic cultures generates an ROS stress that is so severe that metabolism is immediately paralyzed and adaptation is impossible. A similar effect has been noted for aerobic bacteria: Only sub-toxic doses of H2O2 effectively induce the protective response. In nature, perhaps, any entry of oxygen into Clostridial environments occurs gradually, so that robust induction is possible. If so, then the adaptability of anaerobes to oxygen may have been overlooked because experimental protocols failed to mimic the dynamics of aeration in natural environments. Further investigation is called for, but the Hillman study indicates that when it comes to oxidative stress, anaerobes may not be so different from their aerobic cousins after all.
Acknowledgments
Research in the author’s lab is supported by GM49640 from the National Institutes of Health.
References
- Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: Is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EC, Crawford BF, Hong Z, Bilinski T, Kosman DJ. Genetic and biochemical characterization of Cu, Zn superoxide dismutase mutants in Saccharomyces cerevisiae. J Biol Chem. 1991;266:4417–4424. [PubMed] [Google Scholar]
- Chen L, Helmann JD. Bacillus subtilis MrgA is a Dps (PexB) homologue: evidence for metalloregulation of an oxidative-stress gene. Mol Microbiol. 1995;18:295–300. doi: 10.1111/j.1365-2958.1995.mmi_18020295.x. [DOI] [PubMed] [Google Scholar]
- Jenney FE, Jr, Verhagen MF, Cui X, Adams MW. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science. 1999;286:306–309. doi: 10.1126/science.286.5438.306. [DOI] [PubMed] [Google Scholar]
- Flint DH, Tuminello JF, Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- Gardner PR, Fridovich I. Superoxide sensitivity of the Escherichia coli aconitase. J Biol Chem. 1991;266:19328–19333. [PubMed] [Google Scholar]
- Gomes CM, Le Gall J, Xavier AV, Teixeira M. Could a diiron-containing four-helix-bundle protein have been a primitive oxygen reductase? Chembiochem. 2001;2:583–587. doi: 10.1002/1439-7633(20010803)2:7/8<583::AID-CBIC583>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- Hillmann F, Fisher RJ, Bahl H. The rubrerythrin-like protein Hsp21 is a general stress protein. Arch Microbiol. 2006;185:270–276. doi: 10.1007/s00203-006-0091-y. [DOI] [PubMed] [Google Scholar]
- Imlay JA. What biological purpose is served by superoxide reductase? J Biol Inorg Chem. 2002;7:659–663. doi: 10.1007/s00775-002-0361-3. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Ann Rev Biochem. 2008 doi: 10.1146/annurev.biochem.77.061606.161055. epub ahead of print Jan 2 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S, Watamura Y, Ono M, Watanabe T, Takeda K, Niimura Y. Adaptive response to oxygen stress in obligatory anaerobes Clostridium acetobutylicum and Clostridium aminovalericum. Appl Environ Microbiol. 2005;71:8442–8450. doi: 10.1128/AEM.71.12.8442-8450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalyzed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- Lombard M, Fontecave M, Touati D, Niviere V. Reaction of the desulfoferrodoxin from Desulfoarculus baarsii with superoxide anion. Evidence for a superoxide reductase activity. J Biol Chem. 2000;275:115–121. doi: 10.1074/jbc.275.1.115. [DOI] [PubMed] [Google Scholar]
- Lumppio HL, Shenvi NV, Summars AO, Voordouw G, Kurtz DM., Jr Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J Bacteriol. 2001;183:101–108. doi: 10.1128/JB.183.1.101-108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May A, Hillmann F, Riebe O, Fischer RJ, Bahl H. A rubrerythrin-like oxidative stress protein of Clostridium acetobutylicum is encoded by a duplicated gene and identical to the heat shock protein Hsp21. FEMS Microbiol Lett. 2004;238:249–254. doi: 10.1016/j.femsle.2004.07.042. [DOI] [PubMed] [Google Scholar]
- McCord JM, Keele BB, Jr, Fridovich I. An enzyme-based theory of obligate anaerobiosis: the physiological function of superoxide dismutase. Proc Natl Acad Sci USA. 1971;68:1024–1027. doi: 10.1073/pnas.68.5.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinecke B, Bertram J, Gottschalk G. Purification and characterization of the pyruvate-ferredoxinoxidoreductase from Clostridium acetobutylicum. Arch Microbiol. 1989;152:244–250. doi: 10.1007/BF00409658. [DOI] [PubMed] [Google Scholar]
- Messner KR, Imlay JA. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J Biol Chem. 2002;277:42563–42571. doi: 10.1074/jbc.M204958200. [DOI] [PubMed] [Google Scholar]
- Pan N, Imlay JA. How does oxygen inhibit central metabolism in the obligate anaerobe Bacteroides thetaiotaomicron? Mol Microbiol. 2001;39:1562–1571. doi: 10.1046/j.1365-2958.2001.02343.x. [DOI] [PubMed] [Google Scholar]
- Poole LB. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys. 2005;433:240–254. doi: 10.1016/j.abb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Riebe O, Fischer RJ, Bahl H. Desulfoferrodoxin from Clostridium acetobutylicum functions as a superoxide reductase. FEBS Lett. 2007;581:5605–5610. doi: 10.1016/j.febslet.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Sawers G, Watson G. A glycyl radical solution: oxygen-dependent interconversion of pyruvate formate-lyase. Mol Microbiol. 1998;29:945–954. doi: 10.1046/j.1365-2958.1998.00941.x. [DOI] [PubMed] [Google Scholar]
- van Loon APGM, Pesold-Hurt B, Schatz G. A yeast mutant lacking mitochondrial manganese-superoxide dismutase is hypersensitive to oxygen. Proc Natl Acad Sci USA. 1986;83:3820–3824. doi: 10.1073/pnas.83.11.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]