Abstract
Paracrine Hedgehog (Hh) signaling regulates growth and patterning in many Drosophila organs. We mapped chromatin binding sites for Cubitus interruptus (Ci), the transcription factor that mediates outputs of Hh signal transduction, and we analyzed transcription profiles of control and mutant embryos to identify genes that are regulated by Hh. Putative targets that we identified included several Hh pathway components, mostly previously identified targets, and many targets that are novel. Every Hh target we analyzed that is not a pathway component appeared to be regulated by Hh in a tissue-specific manner; analysis of expression patterns of pathway components and target genes provided evidence of autocrine Hh signaling in the optic primordium of the embryo. We present evidence that tissue specificity of Hh targets depends on transcription factors that are Hh-independent, suggesting that `pre-patterns' of transcription factors partner with Ci to make Hh-dependent gene expression position specific.
Keywords: Drosophila, Hedgehog, Cubitus interruptus
INTRODUCTION
Hedgehog (Hh) is a secreted signaling protein that regulates the growth and patterning of many organs. First identified because of its roles in Drosophila embryonic and imaginal disc development, it is now understood to be essential to most organs in Drosophila and higher vertebrates. Despite its systemic importance, Hh signaling is local and is dependent upon Hh that is expressed and secreted by discrete sets of cells in each tissue that it regulates. In each context, organ-specific programs of gene expression, morphogenesis and cell-cycle regulation depend upon Hh regulation. The mechanism that endows Hh signaling with tissue specificity has not been fully elucidated.
Hh signaling has been most thoroughly analyzed in the Drosophila wing imaginal disc, where Hh is expressed in posterior compartment cells. Signaling to adjacent anterior cells begins when Hh engages the participation of two membrane proteins in target cells: Patched (Ptc) and Smoothened (Smo). Hh activates Smo (Stone et al., 1996; Taipale et al., 2002) and initiates the transformation of a complex of proteins that includes an inactive form of the transcription factor Ci (Jia et al., 2003; Lum et al., 2003; Ruel et al., 2003). Ci mediates most, and perhaps all, of the output of the Hh pathway (Alexandre et al., 1996; Méthot and Basler, 2001). In the absence of Hh, cleavage of Ci generates a truncated peptide that functions as a transcriptional repressor (CiRep) (Aza-Blanc et al., 1997). The presence of Hh reduces CiRep and enhances production of a transcriptional activator form (CiAct) (Aza-Blanc et al., 1997; Méthot and Basler, 1999).
Pleiotropic phenotypes result from loss or inactivation of Hh pathway components, suggesting that the pathway is similarly constituted in the affected tissues (reviewed by McMahon et al., 2003). Ci is an essential core pathway component (Alexandre et al., 1996; Forbes et al., 1993) and is stabilized by Hh signal transduction (Aza-Blanc et al., 1997; Ruel et al., 2003). Expression of ptc (Forbes et al., 1993; Ingham, 1991; Tabata and Kornberg, 1994) and roadkill (rdx) (Kent et al., 2006; Zhang et al., 2006) are also hallmarks of Hh action. Many of the pathway components are evolutionarily conserved in higher vertebrates and most of these homologs have been found to play similar roles in the corresponding Hh pathways (reviewed by Varjosalo and Taipale, 2008). The deep homology extends to the three Gli transcription factors (Orenic et al., 1990), homologs of Ci that share a consensus binding sequence (TGGGTGGTC) (Hallikas et al., 2006; Kinzler and Vogelstein, 1990; Pavletich and Pabo, 1993).
As with all signaling systems that employ a common mechanism of signal transduction to regulate a variety of target tissues, the question arises of how tissue-specific responses are induced. For the Hh pathway, the question of specificity also applies to CiAct and CiRep, which regulate expression of targets in the on and off states, respectively. Analysis of the imaginal disc enhancer of decapentaplegic (dpp) revealed that both CiAct and CiRep can bind to, and regulate, the same binding site sequence (Müller and Basler, 2000). However, we do not yet know whether such shared access is a feature of all Ci binding sites and, if it is, how the distinct activator and repressor functions are realized in the context of their common sites. Possible mechanisms include differential affinity, input from linked cis-regulatory elements or participation of co-factors (Müller and Basler, 2000). Although support for the latter might come from the observation that full activation of the dpp heldout enhancer requires both Hh signaling and the wing `selector' protein Vestigial (Hepker et al., 1999), the mechanisms that determine tissue-specific activation of other Hh targets remain unidentified. Recent studies that used chromatin binding to identify target sequences for mouse Gli1 and Gli3 proteins in neural tissues and whole limb buds, respectively (Vokes et al., 2007; Vokes et al., 2008), identified many novel Gli-responsive cis-regulatory elements. Characterization of these elements led to the conclusion that the Gli1 activator and the Gli3 repressor recognize and regulate common sequences.
The number of known Hh targets in Drosophila is small. The work described here was undertaken to obtain a better understanding of targets and to investigate the mechanism of Ci action and specificity. We employed expression array assays to identify genes that have Hh-dependent expression in embryos, and chromatin-binding assays to identify genes linked to genomic regions that are recognized by CiAct and CiRep. Putative targets were selected that bind both Ci forms and are Hh-dependent. Characterization of several novel targets highlighted two key aspects of Hh signaling. We found evidence for autocrine signaling and also found that, with the exception of several genes that encode core components of the Hh signal transduction pathway, all targets are expressed in restricted domains within Hh-responsive tissues. For Ecdysone-inducible gene L2 (ImpL2), a novel Hh-regulated gene that is expressed in the tracheal primordium, we show that expression is dependent upon the transcription factor Trachealess (Trh), expression of which is not dependent upon Hh, and that ImpL2 expression is restricted to only a portion of the cells that express Trh. Expression of this Hh target is, therefore, specified by the combined activities of Hh-dependent Ci and Hh-independent Trh.
MATERIALS AND METHODS
Fly stocks and crosses
Homozygous null mutant embryos lacking ptc or hh function were generated from the following alleles: hh13c, hhAC, ptcB98 and ptc13C. ci-null embryos were generated from a cross of ciRES (Méthot and Basler, 1999) in a ci-null background (ci94/ci94). smo germline clones were generated from a cross of y w hs-FLP/+; smoQ FRT40A/CyO females to +/Y; OvoD1, FRT40A/CyO males (Chou and Perrimon, 1996). Embryos and first instar progeny were heat shocked daily at 37°C for 1 hour prior to eclosion. UAS-Cim1-m4 (CiAct) was obtained from S. Smolik (Oregon Health and Science University, Portland, OR, USA) and the Dam alone transgenic line was obtained from S. Parkhurst (Fred Hutchinson Cancer Research Center, Seattle, WA, USA).
In situ hybridization and immunostaining
Digoxygenin-labeled anti-sense RNA probes were hybridized to whole-mount embryos (O'Neill and Bier, 1994). Antibodies used for immunostaining were: rat anti-2A1 (CiFl; R. Holmgren, Northwestern University, Evanston, IL, USA; 1:2000), mouse anti-Patched (I. Guerrero, Universidad Autonoma de Madrid, Madrid, Spain; 1:200), mouse anti-Fasciclin 2 (Y. N. Jan, University of California, San Francisco, CA, USA; 1:1000) and rabbit anti-β-galactosidase (Y. N. Jan; 1:5000). The signal was visualized with Alexa 488-conjugated secondary antibodies (Molecular Probes) or the ABC Vectastain Kit (Vector Laboratories) as described previously (Aza-Blanc et al., 1997).
DamID constructs
N-terminally fused DamID constructs were generated by PCR amplification of BglII-XbaI fragments of Ci76 (Aza-Blanc et al., 1997) and Cim1-m4 (S. Smolik) using the forward primer 5′-TAAGATCTTATGGACGCCTACGCGTTACCTAC-3′ and reverse primers 5′-TAATCTAGAGTCTGCCACGTCCACGTCATCGT-3′ for Ci76 and 5′-TAATCTAGACTGCATCATTTGAAGGTATCTATTTTCC-3′ for Cim1-m4. PCR products were digested with BglII and XbaI and ligated to pNDamMyc (B. van Steensel, Netherlands Cancer Institute, Amsterdam, The Netherlands). DamCi fusion cassettes were subcloned into pUAST.
DamID analysis
DamID probes were prepared following the protocol described at http://research.nki.nl/vansteensellab/damid.htm. Genomic DNA was isolated from 2- to 6-hour-old embryos containing DamCi or Dam transgenes. DNA (2.5 μg) was ethanol-precipitated, digested with DpnI and ligated to annealed adaptor oligos for 2 hours at 16°C (5′-CTAATACGACTCACTATAGGGCAGCGTGGTCGCGGCCGAGGA-3′ and 5′-TCCTCGGCCG-3′). After heat inactivation, the ligation mixture was digested with DpnII, which cuts unmethylated DNA. PCR amplification of Dam-methylated DNA was carried out using the PCR Advantage Mix (Clontech) and the primer 5′-GGTCGCGGCCGAGGATC-3′. Labeling and hybridizations were performed by NimbleGen Systems (Reykjavík, Iceland). The amplified methylated fragments were labeled with Cy3 or Cy5 in a dye-swap configuration to eliminate bias and were hybridized to NimbleGen Systems' whole genome tiling arrays consisting of 375,000 60mer probes spaced ∼300 bp apart (Choksi et al., 2006). Three replicates of control (Dam alone) and experimental (DamCiAct and DamCiRep) samples were analyzed. Raw intensity data were analyzed as described below.
All analyses were performed in R v2.7.1 (R Development Core Team, http://www.R-project.org/) unless otherwise stated. At each tiling array feature, log ratios between the experimental and control samples were calculated and normalized using `limma' (Smyth, 2005), applying `loess' and `Aquantile' for normalization within arrays and between arrays, respectively (Wormald et al., 2006). To predict binding regions, each feature was tested for significant positive intensity using a one-sided t-test. Because sample size was small (n=3), information was pooled from all features to obtain more stable variance estimates for the t-statistic using the limma package. To identify genomic regions of high signal intensity, P-values for consecutive features were `averaged' using meta-analysis based on combining P-values (for details, see Kechris et al., 2010). Because of the large number of significance tests, P-values were corrected for false discovery rate (FDR) control (Benjamini et al., 2001) with window size set at four. Binding regions were created by scanning features as they are ordered along each chromosome. If a feature had an adjusted P-value below the set FDR cutoff (0.02 for DamCiRep; 0.001 for DamCiAct), a new binding region was formed. The next feature passing the cutoff in the linear chromosome was then evaluated. This procedure was continued in a step-wise fashion.
Overlap of DamID protected regions
The overlap binding regions were evaluated using two metrics: (1) overlap of set of all proximal transcripts and (2) base pair overlap between binding regions. To compare these values with chance occurrence, binding regions for CiRep were generated randomly fifty times based on length distribution and number of binding regions. Each random set of binding regions was compared with the CiAct binding region, and the two overlap metrics were calculated. To obtain gene and base pair overlap, enrichment scores of original values for overlap metrics were divided by values based on an average of 50 randomizations. This analysis was also repeated by performing the binding region randomizations on each of the 21 transcription factors analyzed in a previous study (MacArthur et al., 2009) and comparing those with the CiAct binding regions. We found extensive overlap to the same regions identified by MacArthur and colleagues. Given the extensive overlap within the MacArthur data set for the many different transcription factors that they and others have profiled, these binding sites appear to be detected by several contrasting techniques and are of uncertain significance.
Incidence and frequency of Ci motifs
The sequences of the DamID binding regions were extracted from FlyBase v4.0, and occurrence of TGGGTGGTC and reverse complement were counted (Perl script v5.10.0). The rate of this motif, as well as the occurrence of `relaxed consensus' sequences, in binding regions and genome were calculated by dividing counts of motif by the total length of binding regions or genome, respectively. Enrichment was the rate of motif occurrences in the peak set divided by the rate in the genome. P-values for enrichment assumed a Poisson distribution. The strict consensus (TGGGTGGTC) occurs 2.5 and 2.4 times more frequently in CiAct and CiRep binding sites, respectively, than would be expected by chance (P=4×10–24 and 7.2×10–8, respectively). The relaxed degenerate Ci motif (YGSGDGGNC) occurs 1.2 and 1.3 times more frequently in CiAct and CiRep binding sites (P=1.4×10–9 and 4.7×10–11, respectively). The clusters of the degenerate binding motif YGSGDGGNC in the vicinity of Hh targets ptc, rdx, hh, wg, en, drm, odd and ImpL2 (see Table 2 in the supplementary material) are depicted in Figs 1, 3 and 6 with the presence of clusters of degenerate Ci recognition motifs shown as red boxes. The motif is based on identified Ci recognition motifs (Müller and Basler, 2000; Sasaki et al., 1997; Von Ohlen et al., 1997) as well as binding affinities of Ci for random nucleotides at each position of the Gli consensus TGGGTGGTC (Hallikas and Taipale, 2006). Sequence coordinates refer to Drosophila genome R5.29. The clusters were identified using Flyenhancer software (Markstein et al., 2002), which scans the genome for clusters of motifs. We set the following parameters: at least two matches to the motif pattern (YGSGDGGNC) within 600 bp, or at least one of the ten known Ci recognition sites within 1000 bp.
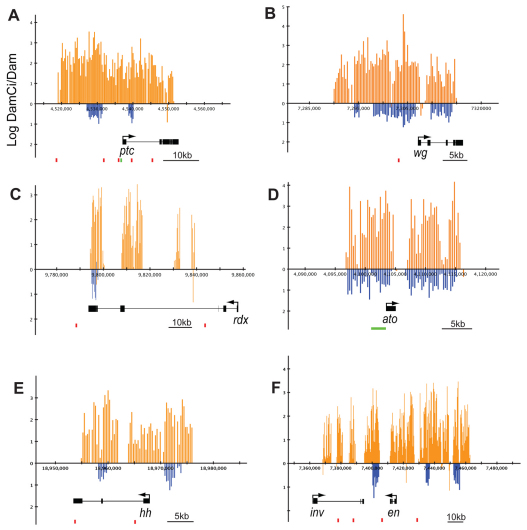
Fig. 1.
Ci binds at known targets of Hedgehog signaling in Drosophila. (A-F) Averaged log2-transformed ratios of DamCiAct/Dam alone (orange vertical bars) and DamCiRep/Dam alone (blue vertical bars) at statistically relevant regions of the genome (x-axes) for patched (ptc; A), wingless (wg; B), roadkill (rdx; C), atonal (ato; D), hedgehog (hh; E) and engrailed/invected (en/inv; F) genes, all of which had a significant DamID signal. Transcription units are indicated below. Red boxes indicate clusters of Ci binding motifs with at least two motifs matching the consensus YGSGDGGNC within a 600 bp window (for coordinates and sequence of the motifs see Table S1 in the supplementary material). Green boxes (A,D) indicate known enhancers (Forbes et al., 1993; Sun et al., 1998).
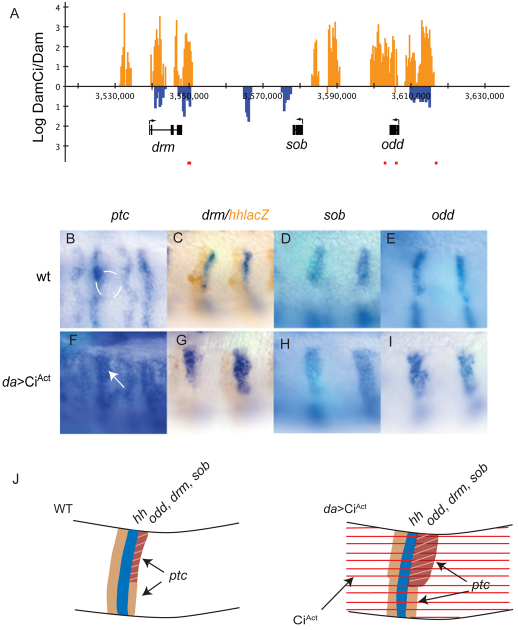
Fig. 3.
Genes in the odd skipped cluster are targets of Hh signaling in the dorsal ectoderm of Drosophila. (A) DamCiAct (orange vertical bars) and DamCiRep (blue vertical bars) signal at drumstick (drm), sister of odd and bowl (sob) and odd skipped (odd). Red boxes indicate Ci motif clusters matching the motif YGSGDGGNC (see Table S1 in the supplementary material). (B-E) Wild-type expression patterns of ptc (B), drm (C), sob (D) and odd (E). Expression of ptc coincides with the anterior margin of the tracheal pit (white line in B). Stripe of drm expression lies immediately posterior to hh-lacZ (brown staining in C). (F-I) Expression of drm (G), sob (H), and odd (I) expands posteriorly by two to three cell diameters in daGal4>UAS CiAct. ptc expression expands similarly (F; arrow) and is activated in more cells of each parasegment. (J) Diagram of Hh target gene activation in the dorsal ectoderm. ptc expression (tan) on the anterior and posterior flanks of each hh stripe (blue) expands with ubiquitous CiAct (right; red stripes), but expression of Hh targets odd, drm and sob (brown), which in wild type (WT, left) is posterior to the dorsal portion of each hh stripe, expands only on the dorsal-posterior flank.
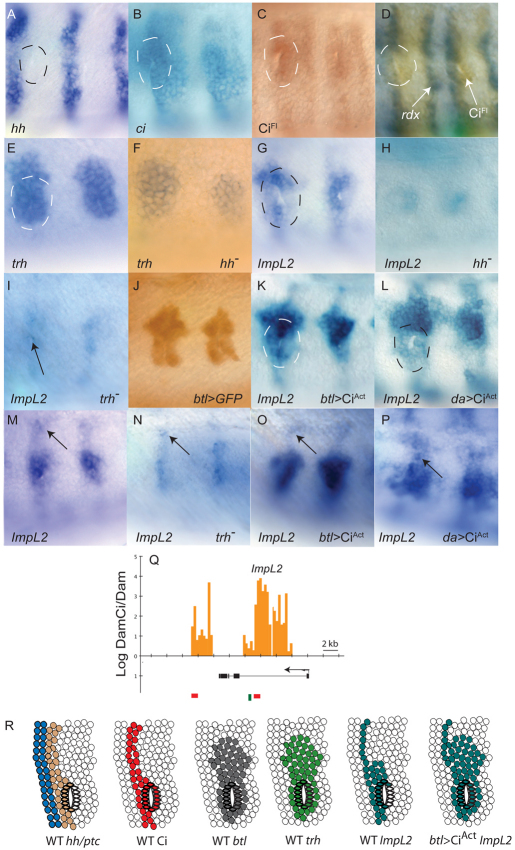
Fig. 6.
ImpL2 is a Hh target in the tracheal pit of Drosophila. (A-P) Lateral views of stage 11 embryos. Anterior is to the left. hh (A) is expressed in a stripe of cells immediately anterior to each tracheal pit (dashed circle; pit identified by gap). Pit cells express ci RNA (B), stabilize CiFL (C,D) and express rdx (D). trh is expressed in a broad region that includes the pit (E) and trh expression in the pit is not diminished in hhAC mutants (F). ImpL2 is non-uniformly expressed in the pit with highest levels in the most dorsal anterior cells (G). ImpL2 expression in the pit is severely reduced in hhAC (H) and trh1/trh2 (I; arrow). Anti-GFP in a btl>GFP embryo reveals btl expression (J). ImpL2 expression in the pit expands similarly in btl>CiAct (K) and da>CiAct (L). ImpL2 expression in ectodermal cells dorsal to the pit (M; arrow) present in trh mutants (N; arrow) and btl>CiAct (O; arrow), and elevated in da>CiAct (P; arrow). (Q) Statistically significant DamCiAct signal (orange bars) and Ci binding motif clusters (red boxes) in first intron and downstream of ImpL2; binding motif for Trh (green line) in first intron. Ci motif clusters match the motif YGSGDGGNC and are listed in Table S2 in the supplementary material. (R) Summary diagram of Hh target gene activation in the cells of the tracheal pit. Expression of hh (brown), ptc (blue), Ci (red), btl (gray), trh (light green) and Impl2 (dark green) in wild type (WT) and btl>CiAct mutants reveals the location of the tracheal pit relative to the AP compartment border (the interface of ptc and hh expressing cells), the extent of the tracheal primordium (defined by domains of btl and trh expression) and expansion of Impl2 to the entire tracheal primordium in the presence of ubiquitous CiAct.
De novo motif analysis
In addition to word searches of the Ci motifs, two programs were used to query the peaks associated with the 52 high confidence genes. All peaks neighboring the 52 genes (91 peaks, for a total of 504,561 bp) were extracted and masked for repeats using the RepeatMasker tool (http://www.repeatmasker.org/) that identifies interspersed repeats and low complexity DNA sequences. The masked sequences were then scanned for repeating patterns using default options for the MEME software (http://meme.sdsc.edu) and the oligo-analysis tool at the RSA Tools website (http://rsat.ulb.ac.be/rsat/). MEME found some local repeats (e.g. CACACACA) despite repeat masking. The RSA analysis was more successful: of the 35 most significant oligonucleotide sequences that were over-represented (occurrence E-value<10–8), most could be clustered by similarity using the RSA pattern assembly tool into seven motifs of at least three sequences. One included a portion of the Ci consensus site TGGTGGT. Other motifs included GTCCTGC, CTGCTGCC, GTCCTTG, CGGCGCG, TTTGTTT and TATTTA. We cannot distinguish whether these sequences are variations of the Ci consensus site or are alternative regulatory sequences. Otherwise, these searches, in general, identified little of note.
Expression array analysis
RNA was isolated, amplified and labeled as described previously (Klebes and Kornberg, 2008). Data were processed with Cluster and Treeview (Eisen et al., 1998), and Genepix PRO (Axon Instruments). Normalization for cluster analysis was carried out with NOMAD 2.0 (University of California, San Francisco, USA). Genes chosen had combined median intensity of >300 above background (both channels) in >80% of experiments, and above a threshold of 1.4 (0.55 of the log2-transformed ratios). The fold-change and cluster selection procedure identified 147 transcripts. Using a post hoc FDR evaluation, ∼75% of the genes represented by these transcripts were controlled at an FDR of 0.1 and 88% were controlled at a FDR of 0.2 based on P-values from a two-sided t-test across all experiments using transcripts with at least three non-missing values.
To assess the degree of similarity in expression values for array replicates, Pearson's correlation coefficient and a concordance percentage were calculated between each pair of replicates within a group and between replicates across groups. Missing values were removed from both analyses. Higher correlations (positive) or concordance percentages, with a maximum of 1.0 and 100%, respectively, indicate that the arrays are generally in agreement. Although there was some variability for replicates within a group (e.g. for the Smo group, one of the lowest correlation values was ∼0.3 between two of the replicates), the median correlation and concordance percentage were higher between replicates within the same group and were lower between different groups (see Table S3 in the supplementary material). For example, the correlation was 0.62 between replicates of the hh arrays whereas the median correlation with other arrays was 0.2.
Data access
Data for the DamID and expression arrays are in the GEO database. Results from DamID are the normalized log2-transformed ratios of DamCi/DamAlone signals. The data include three biological replicates for both DamCiAct / DamAlone (GSE23999) and DamCiRep / DamAlone (GSE24024). The gene expression array data (GSE24038) are both the log2-transformed ratios and fluorescent intensities for 14 experiments comparing expression in mutant embryos with control siblings. GSE24055 includes all of the deposited data as one superseries.
RESULTS
Specificity of Ci binding to the Drosophila genome
We used the DamID technique (van Steensel et al., 2001) to locate regions bound by Ci in the Drosophila genome. This method directs DNA methylation to sites of sequence-specific binding by expressing a fusion protein composed of a transcription factor and DNA adenine methyltransferase (Dam), and employs hybridization to DNA microarrays to detect genomic fragments for which methylation confers protection from endonuclease digestion. We generated two DamCi fusion proteins: a constitutive CiAct of Dam (DamCiAct) fused to the N terminus of a Ci mutant that activates Hh targets independently of Hh signaling (Chen et al., 1999), and Dam fused to the N terminus of Ci76 (DamCiRep) (Aza-Blanc et al., 1997). Examination of wing phenotypes after expression at high levels in wing discs (MS1096GAL4) revealed that CiAct and CiRep functionality was preserved in the respective fusion proteins (not shown).
To identify Ci targets, methylated DNA fragments were collected from stage 10-11 embryos expressing DamCiAct or DamCiRep. At this stage, organogenesis is active and Hh signaling activity is required in many areas. Three replicates were analyzed for each construct; Dam alone embryos served as a control for non-specific methylation. Expression was driven by the minimal Hsp70 promoter of the UAS vector without heat shock. We detected 2438 protected regions bound by DamCiAct and 1743 for DamCiRep (see Table S1 in the supplementary material). Median protected region length was 2133 bp for DamCiAct and 993 bp for DamCiRep. The differences in frequency and length might reflect differences in biological activity of the CiAct and CiRep constructs, different levels of expression, different affinities or other factors, but, irrespective of cause, we assume that this method has a high false positive rate (see Materials and methods for further discussion), as do other related techniques that monitor chromatin binding (Vokes et al., 2008; Zeitlinger et al., 2007). To establish the criteria that distinguish functional sites from false positives, we assessed four properties: (1) common binding by CiAct and CiRep, (2) presence of Ci binding motifs, (3) linkage to known Hh targets and (4) Hh dependence of expression levels of linked genes.
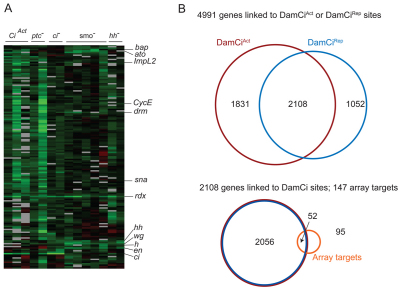
Available evidence indicates that CiAct and CiRep can bind to and function at common sites (Müller and Basler, 2000), and a significant number of the DamID-protected regions are shared. 729 out of 2439 (30%) of the DamCiAct-protected regions overlapped with DamCiRep-protected regions, with a median overlap of 1231 bases; 75% of the overlaps were between 383 and 2147 bases. The base pairs that overlapped were 8.6 times the number expected by chance. We do not understand the basis for extensive regions of nuclease protection but simply assume that the location and size of each region correlates in some way with the presence, number and affinity of binding sites, and perhaps with local chromatin conformation. Irrespective of the underlying mechanism, we analyzed the genome in the vicinity of regions bound by DamCiAct or DamCiRep to identify transcription units that contain or have adjacent upstream or downstream binding sites for both. The number of transcripts found that contain or are proximal to both DamCiAct and DamCiRep binding regions was 2108. The correlation of the transcripts that are associated with both binding regions was statistically significant (1.4× that expected by chance; P-value <2.2×10–16, Fisher's test) and they represented 67% of DamCiRep (2108 out of 3160) and 54% of DamCiAct (2108 out of 3939) binding region-associated transcripts.
We examined the DamCiRep peaks to locate sequences that conform to known Ci binding preferences. Previous work defined a consensus binding sequence for the Gli and Ci proteins (TGGGTGGTC) (Hallikas and Taipale, 2006; Kinzler and Vogelstein, 1990; Pavletich and Pabo, 1993), as well as variants at human ptc (PTCH1 – Human Gene Nomenclature database) (Kinzler and Vogelstein, 1990), Drosophila wg (Von Ohlen et al., 1997), Drosophila dpp (Müller and Basler, 2000) and mouse Foxa2 (Sasaki et al., 1997). Probing the Drosophila genome for these sequences (YGSGTGGHC and HDSSHGVHS) identified many sites, but their incidence in the DamID peaks was not significantly above that expected by chance (see Materials and methods). By probing with the consensus sequence (TGGGTGGTC), which represents an unknown fraction of Ci binding targets, we found that its frequency in the DamCiAct and DamCiRep peaks was 2.4- to 2.5-fold greater than expected by chance. Most consensus sequence motifs were present in one copy only (e.g. motifs in 89% of DamCiAct peaks were singles) and their proximity to transcription start sites was without apparent bias. We conclude that our current understanding of Ci binding specificity leaves us without the means to distinguish between bona fide binding sites and background, but that these statistics, as well as the correlation with known target genes described in the following section, suggest that the presence of consensus sequence motifs is a positive indicator for functional binding.
Next, we asked if known targets of Hh signaling were among the 2108 genes linked to DamCiAct and DamCiRep peaks. Binding regions for DamCiAct and DamCiRep were detected within 7 kb of the transcription units of 27 out of 30 known targets (Table 1). Plots of DamID protection at five representative targets, ptc, wg, rdx, bagpipe (bap) and atonal (ato), are shown in Fig. 1. At ptc, a 35 kb DamID-protected region included most of the transcription unit as well as ∼20 kb upstream. Six Ci consensus binding sites were in the protected region; the sites in the upstream sequence have been shown to have Hh-responsive enhancer activity (Alexandre et al., 1996; Forbes et al., 1993). At wg, a 2.5 kb element upstream of the transcription start site drives expression in metameric stripes (Lessing and Nusse, 1998). This region has sequences that match the degenerate consensus Ci binding site (see Table S2 in the supplementary material), and both DamCi forms conferred nuclease protection. rdx requires Hh activity for expression (Kent et al., 2006), but its regulatory sequences have not been characterized. DamCi analysis revealed binding by both DamCi forms within rdx. Enhancers responsible for Hh regulation of ato expression in eye discs and the PNS of the embryo have been identified (Sun et al., 1998). The DamID-protected regions at ato spanned the identified enhancer elements. We interpret the consistency of DamCiAct and DamCiRep binding, the presence of Ci consensus binding sites in DamCi protected regions and the tight correlation of DamID signal to known Hh targets as evidence that this DamID analysis detected Ci binding sites that function in vivo.
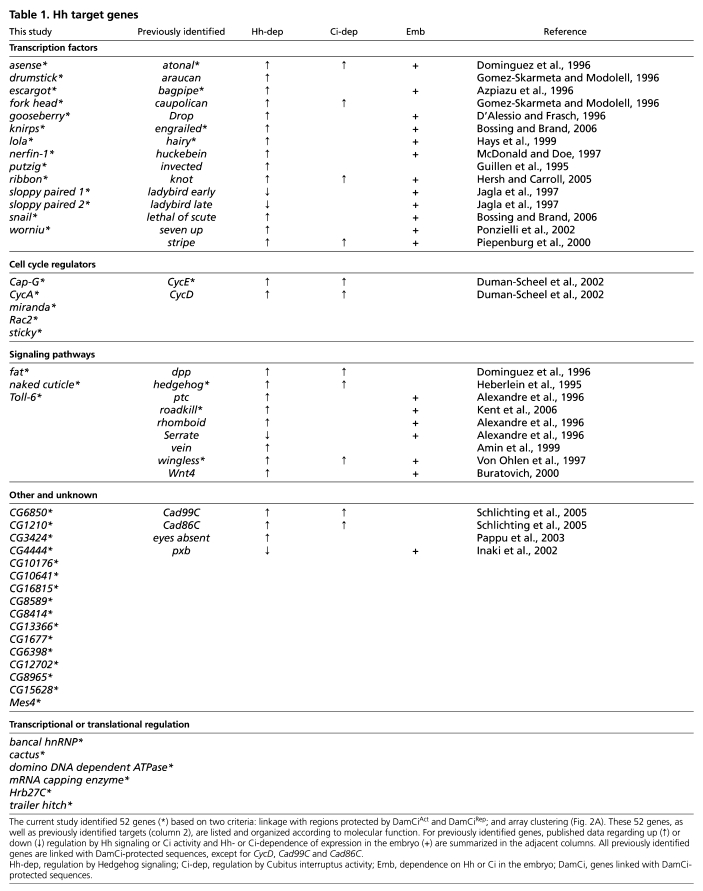
Table 1.
Hh target genes
Hedgehog responsive genes
To identify Hh-dependent transcripts, RNA extracted from five types of stage 10-11 mutant embryos was compared with controls by hybridization to whole genome expression arrays. Mutant embryos for three positive regulators of the pathway (Hh, Smo and Ci) and one negative regulator (Ptc) were tested, as well as embryos in which CiAct was ectopically expressed (daGal4>CiAct). Clustering genes according to similarity of responses yielded a group of 147 (Fig. 2A). The abundance of transcripts from these 147 genes was decreased in embryos with compromised Hh signaling (hh, smo, ci), and increased in embryos with elevated Hh signaling (ptc, daGal4>CiAct). Mutants lacking the function of Hh pathway components developed similar cuticular phenotypes, therefore, this expression array analysis shows that the cuticular phenotypes have a common mechanistic basis in a failure to regulate the Hh signaling pathway and Hh target genes.
Fig. 2.
Expression array and DamID analyses. (A) Cluster analysis of expression arrays depicted as columns representing pairwise comparisons of transcript levels of da>CiAct, ptcB98/ptc6C, ci94/ci94, smoQ/smoQ and hhAC/hh13C to controls. Each lane represents one array experiment. Genes upregulated under normal or elevated Hh signaling are represented in rows as shades of green. bap, rdx, wg and en were previously shown to respond to Hh signaling in the embryo; hh, ato, CycE, drm and h have been shown to respond to Hh signaling in non-embryonic tissues. sna and ImpL2 are characterized in this study. (B) Above: Venn diagram indicating the number of genes linked to DamCiAct and DamCiRep sites or both. Below: Overlap between expression array targets and genes linked to DamID sites.
Genes in this group of 147 might be either direct or indirect targets, and as wg is a direct target at each segment border of the embryo, many of these Hh-responsive genes could be direct targets of Wg rather than Hh. Ci binding is presumably a property that distinguishes Hh targets, and 52 genes of the 147 had regions within, or adjacent to, their transcription units that were protected by DamCiAct or DamCiRep. Published work has shown that Hh regulates 13 of the 30 known Hh targets in embryos, whereas the others are targeted at later stages or are negatively regulated (Table 1). Five of the thirteen embryo targets (atonal, bagpipe, engrailed, rdx and wg) are among the 52 candidate genes. Of the seven that are not, three (Drop, Wnt4, rhomboid) responded as expected of Hh targets in some arrays but their responses in other arrays were below our thresholds; four (ptc, huckebein, seven up, stripe) were not included in the cluster analysis because their transcripts were not represented in the hybridization probes (presumably for failure to amplify, meaning that only nine of the known embryo targets were queried in this analysis); and one (lethal of scute) is expressed in only six cells per hemi-segment and the levels of its transcripts might have been below detection. We suggest that the combination of Ci binding sites revealed by DamID analysis, together with the response to Hh signaling revealed by the expression array analysis, identifies a set of 52 probable Ci targets. In the following sections, we describe our characterization of some of these genes as well as several others that are co-regulated.
Tissue-specific responses to Ci
ptc and rdx are targets of Hh signaling that encode components of the Hh signal transduction pathway, and available evidence indicates that both are expressed in all cells that respond to Hh. By contrast, expression of all other Hh targets appears to be tissue-specific. Thirty known targets listed in Table 1 do not encode components of Hh signal transduction, and each has been shown to be expressed in tissue-specific patterns. Our analysis of many of the new candidate targets indicates that they are also expressed in only a subset of Hh-responsive tissues. Our analysis of candidate target genes expressed in the embryo dorsal ectoderm, visual primordia and tracheal placodes is described below.
Dorsal ectoderm
Among the genes that are expressed in segmentally repeated stripes in the embryo, three are members of the odd skipped family. These genes, drumstick (drm), sister of odd and bowl (sob) and odd skipped (odd), are linked on chromosome 2L and are associated with three regions of overlapping DamCiAct and DamCiRep binding (Fig. 3A). Although drm is the only candidate that fulfills both the DamCi binding and cluster analysis criteria, odd and sob expression changed in ways that are consistent with regulation by Hh signaling. drm, odd and sob were expressed in nearly identical patterns in stage 11 embryos (Fig. 3C-E). The stripes of drm, sob and odd expression were adjacent and posterior to hh-expressing cells in each parasegment (Fig. 3C; data not shown), and expression in these domains is consistent with regulation by Hh. We compared drm, sob and odd transcription in embryos that express CiAct ubiquitously (daGal4>CiAct) with controls using ptc expression as reference. In stage 11 embryos, ptc was expressed in many places, whereas in the trunk ectoderm, expression was confined to two ectodermal stripes in each parasegment, one to either side of each hh stripe (Hidalgo and Ingham, 1990). Levels of expression were higher in the more posterior stripe of each pair, and with ubiquitous CiAct, the general level of ptc expression increased; the more intensely expressing posterior stripe broadened posteriorly, and the more anterior stripe broadened anteriorly. These changes are consistent with expansion of ptc expression to all cells except for those that express En. drm, sob and odd each responded to ubiquitous CiAct with posterior expansion of each stripe by two to three cells (Fig. 3B-I). In wild-type germband-extended embryos, drm and sob were also expressed in the presumptive hindgut and odd was expressed in the presumptive foregut. We did not observe changes to these domains of expression after ubiquitous expression of CiAct, nor did we observe ectopic expression in any other tissues in which Hh signaling is also active (not shown). The patterns of drm, sob and odd expression in wild-type and ubiquitous CiAct embryos are consistent with regulation by Hh and Ci-dependent activation specifically in the trunk ectoderm.
Visual primordium
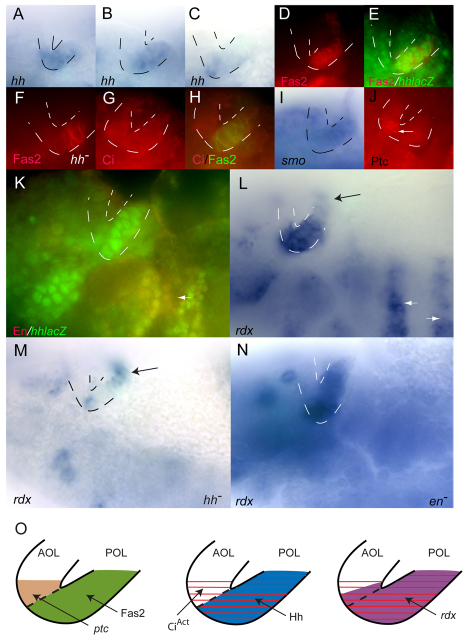
The visual primordium has anterior and posterior optic lobes (AOL and POL, respectively), and contains the precursor cells for Bolwig's organ, the larval eye. During stage 11, the placode adopts a V-like shape and has conspicuous anterior and posterior lips. hh is expressed in the visual primordium and its expression pattern changes rapidly as the embryo matures. In situ hybridization and enhancer trap expression revealed that, at early stage 11, hh expression extended from the POL into the AOL, became restricted to the POL at mid stage 11 and later localized to Bolwig's organ precursor cells in the ventral posterior lobe (Fig. 4A-C) (Chang et al., 2001). Fasciclin2 (Fas2) is expressed in the POL of stage 11 embryos where its expression coincides with hh (Fig. 4D,E) (Chang et al., 2001). Fas2 levels in the POL remained robust in hh mutants (Fig. 4F), indicating that Fas2 is not an Hh target and that at least some aspect of the POL remains in the absence of hh function. Full length Ci protein, a hallmark of active Hh signaling, was present in both the POL and adjacent AOL cells at mid stage 11 (Fig. 4G,H), and rdx expression, another hallmark of Hh signaling, had a similar pattern (Fig. 4L). smo was expressed in both lobes, although our in situ hybridization analysis was insufficient to distinguish relative levels of expression within the AOL and POL (Fig. 4I). By contrast, ptc RNA was clearly present only in AOL cells adjacent to the POL (Fig. 4J).
Fig. 4.
Hh signaling in Drosophila embryonic visual primordia. (A-N) Lips of the optic lobe (OL) placode are outlined with dotted lines. Anterior is to the left, dorsal is at the top. Patterns of hh expression in the OL change in early (A), mid (B) and late (C) stage 11 embryos. Fas2 expression (D; red) marks the posterior optic lobe (POL) and is coincident with hh-lacZ at mid-stage (E; green). Fas2 staining (F; red) marks POL in hhAC (hh–). Full-length Ci (G; red) in the AOL and in Fas2-containing POL (H; green). (I) smo expression in AOL and POL. (J) Ptc protein (arrow) in AOL. (K) En (red) is absent from the OL but is present elsewhere where hh is expressed (arrow). (L) rdx expression in POL, in adjacent AOL and in cells dorsal to the OL (black arrow) and in ectodermal stripes (white arrows). (M) rdx expression in hhAC (hh–) is largely absent from the OL but is present in more dorsal cells (arrow). (N) rdx expression in enl11/en7 is absent from ectodermal stripes but unaffected in the OL. (O) Diagram of Hh target gene activation in the optic primordium. Left: Fas2 (green) in the POL and Ptc protein in the adjacent AOL (brown). Middle: CiAct (red) in both the POL and adjacent AOL and Hh (blue) in the POL. Right: rdx (purple) in the POL and adjacent AOL.
As elevated levels of rdx expression and full length Ci protein are hallmarks of paracrine Hh signaling, their presence in the POL was unexpected. Hh-target genes are not expressed in hh-expressing cells in the wing disc or embryo trunk ectoderm, a pattern that is attributed to repression by En. En also has a role in the positive regulation of hh expression, and loss of rdx expression in the ectodermal stripes of en mutant embryos is one of the consequences (Fig. 4N). However, En is not expressed in either optic lobe (Fig. 4K), and rdx expression remained robust in the optic primordium of en mutant embryos (Fig. 4N). In situ hybridization revealed that rdx RNA was reduced in both the AOL and POL of hh mutants (Fig. 4M), showing that hh function is essential for rdx expression in both regions. The POL is present in hh mutant embryos (Fig. 4F) and expression of rdx in cells dorsal to the POL was not affected by loss of hh (Fig. 4M). In wild type, a two to three cell-wide stripe of rdx expression connected with the dorsal part of the POL (Fig. 4L). This stripe extended dorsally and curved into the anterodorsal part of the head. A similar pattern of eyes absent (eya) expression was also observed (Fig. 5B), and for both genes, this stripe was not altered in hh mutant embryos, consistent with a tissue-specific role for Hh signaling in the POL.
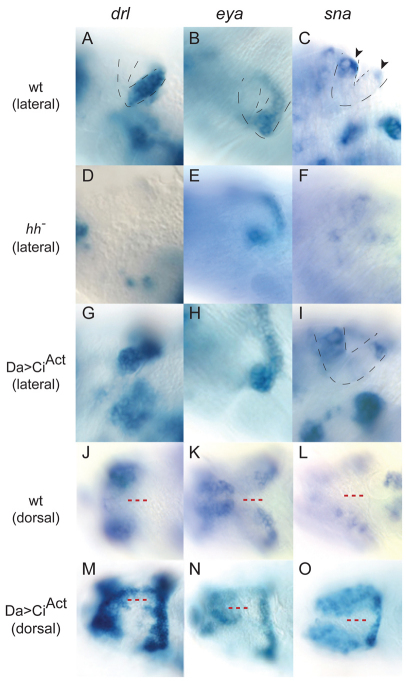
Fig. 5.
Novel Hh target genes in Drosophila embryonic visual primordia respond to CiAct in a tissue-specific manner. (A-I) Lateral view of the visual primordia. Lips of the optic lobe (OL) placode are outlined with dotted lines. Anterior is to the left. In the wild type, drl (A) and eya (B) are expressed in the POL. sna (C) is expressed in the most dorsal cells (arrowheads) of both the AOL and POL. In the hh mutant, POL expression of drl (D) is absent and expression of eya (E) and sna (F) is reduced. In daGal4>CiAct mutants, expression of drl (G), eya (H) and sna (I) is unchanged. (J-O) Dorsal view of the visual primordia. Expression of drl (J), eya (K) and sna (L) in the wild type is shown. Note the absence of expression in the dorsal head ectoderm (red dashed lines). drl (M) eya (N) and sna (O) expression in daGal4>CiAct fuses the normally bilateral fields of OL expression.
These results indicate that hh expression in the optic primordium is not dependent upon En, and that Hh signal transduction in the optic primordium is activated both in AOL cells (revealed by Ptc, Ci and rdx expression) and in hh-expressing POL cells (revealed by Ci and rdx expression). In the Discussion section, we consider the possibility that hh-expressing cells of the POL respond to Hh in an autocrine manner.
We also monitored visual primordium expression of three genes that are in the set of 2108 with linked DamCiAct and DamCiRep binding sites: snail (sna), derailed (drl) and eya. They were examined because of their similarly strong upregulation in expression arrays of embryos with ubiquitous CiAct. sna is one of the 52 genes in the hh-dependent cluster; expression of the other two changed in ways that are consistent with regulation by Hh signaling, albeit at levels that did not meet the clustering thresholds.
In situ hybridization to stage 12 embryos revealed that sna, drl and eya were expressed in many tissues, and that in the optic primordium, sna was expressed in both lobes (Fig. 5C) whereas drl and eya were expressed only in the POL (Fig. 5A,B). Expression of sna, drl and eya in the optic primordium of hh mutant embryos was severely reduced or undetectable (Fig. 5D-F). Whereas expression of sna and eya in other tissues was unaffected in hh mutants, the ectodermal stripes of drl were absent in mutant embryos (not shown). With the exception of an increase in the number of cells that express sna in the peripheral nervous system, expression of sna, drl and eya in embryos containing ubiquitous CiAct was affected only in cells associated with the optic primordium. In CiAct-expressing embryos, expression of all three genes extended from the optic primordium into the dorsal ectoderm of the head (Fig. 5M-O). These dorsal domains of expression are similar (or identical) to the Hh-induced cyclopia phenotype in ptc mutant embryos (which have upregulated Hh signaling) and in embryos that had been subjected to a heat shock-induced dose of ectopic Hh expression (Chang et al., 2001).
Tracheal placode
The tracheal system arises from twenty clusters that have approximately 90 epidermal cells each. At stage 11, the tracheal pits have begun to invaginate, hh is prominently expressed in posterior compartment epidermal cells to which the pits are juxtaposed and Hh protein accumulates in the most anterior cells of the pits (Glazer and Shilo, 2001). A role for Hh signaling in these anterior pit cells is indicated by their high levels of ptc expression and by the reduced number of cells in the pits of hh mutants (Glazer and Shilo, 2001).
In situ hybridization studies confirmed that hh is expressed in the cells immediately anterior to the pits, and known targets of Hh, such as ci, ptc and rdx were expressed in the adjacent pit cells (Fig. 3; Fig. 6A-D). ptc was expressed most prominently in the anterior portion of the pits; ci was expressed more broadly and most pit cells had elevated levels of full-length Ci protein. Expression array analysis (Fig. 2A) and the DamID assay (Fig. 6Q) led to the identification of ImpL2 as an Hh target expressed in the tracheal pit. ImpL2 lacks a known function; its expression was activated in anterior cells of the pit (Fig. 6G) and in cells immediately dorsal to the pit invagination (Fig. 6M). It was expressed in only a subset of the cells in the tracheal primordium, which we defined by btl expression (Fig. 6J). Expression of ImpL2 was severely reduced but not absent in hh null embryos (Fig. 6H), suggesting that Hh signaling is necessary for activation of ImpL2, but that ImpL2 also responds to other activators. We monitored ImpL2 expression in embryos expressing ectopic CiAct to determine the maximum range of Hh-dependent activation. btlGal4>CiAct embryos had a higher number of ImpL2-expressing cells in the tracheal primordium (Fig. 6K), generating an expression pattern in the tracheal primordium that was indistinguishable from btl. ImpL2 expression expanded similarly to the entire primordium in daGal4>CiAct. ImpL2 expression dorsal to the tracheal primordium was not affected by btlGal4>CiAct (Fig. 6O), suggesting that these cells are not part of the tracheal primordium. Expression in this region is hh-dependent, as indicated by requirement for hh (Fig. 6H) and expansion in daGal4>CiAct (Fig. 6K).
DamCiAct binds two regions at the ImpL2 locus; both contain Ci motifs (Fig. 6Q). One of these regions is in the first intron and contains three Ci motifs within a 285 bp sequence; adjacent to this cluster is a consensus Trh binding site (CTCGA). Trh is a basic helix-loop-helix (bHLH)-PAS DNA binding protein that has been implicated in tracheal development (Boube et al., 2000; Wilk et al., 1996). Our evidence suggests that Impl2 is a Trh target. In wild type, expression of trh was immediately dorsal to the tracheal pit invagination (Fig. 6E) and throughout the tracheal primordium. This pattern is similar to btl expression and to ImpL2 expression in btlGal4>CiAct embryos. To determine if Trh function is necessary for ImpL2 expression, we examined trh mutants. Although ImpL2 expression in the cells dorsal to the tracheal primordium was unaffected (Fig. 6N), it was essentially absent in the tracheal primordium (Fig. 6I,N). Based on these observations, we conclude that Trh and Ci together activate ImpL2 expression in the tracheal pit.
DISCUSSION
Because hh mutant embryos have an extensive syndrome of defects and hh is expressed in most organ systems in the embryo, we designed a global search for genes that are regulated by Hh to improve our understanding of its diverse roles. We used chromatin-binding assays to identify sequences that bind Ci in vivo, analyzed transcription profiles with expression arrays to identify genes whose expression is Hh-dependent, and monitored how several Hh-target genes are expressed and regulated. These experiments illuminate three general properties of Hh signaling: (1) targets of Hh signaling include genes that control cell behavior directly (e.g. cyclins) but a high proportion are transcription factors, with presumed indirect consequences; (2) Hh signal transduction is activated in some Hh-expressing cells where it could be autocrine; and (3) tissue-specific activation of targets requires the combinatorial activity of both Ci and non-Hh-dependent transcription factors.
Hh target genes
Chromatin binding and expression array analyses are, in essence, fractionation methods and, like all such procedures, whether they are genetic screens, biochemical purifications, or behavioral selections, they are plagued with false positives and negatives. Productive use of these methods therefore depends on additional means to extract candidates and determine relevance.
Chromatin-binding assays have a high frequency of false positives, as they detect association with many more sites than are likely to be functionally relevant. Whether binding to non-functional sites is an experimental artifact or represents the actual distribution, it complicates identification of relevant targets. Statistical measures validate the binding assays, but they do not distinguish between the sites. Association with consensus binding sequences might help, but binding to bona fide targets might not always be direct, and our understanding of how accessibility and affinity affect residency is incomplete. It is especially poor for Ci and, although we had hoped that independent identification of binding sites for CiAct and CiRep would establish whether they target the same genes or not, our results were inconclusive. Many of the protected DamID regions are shared and these overlaps are statistically significant; moreover, we show that some of these regions are linked to legitimate targets. However, these chromatin binding experiments cannot rule out the existence of functionally relevant sites that are specific to the activator or repressor forms.
Among the proteins encoded by these 52 targets, we identified six with roles in cell cycle regulation and nineteen that were transcription factors. Previous work has shown that Cyclin D and Cyclin E are Hh targets in the eye disc, that Ci binds to and regulates Cyclin E expression (Duman-Scheel et al., 2002), and that Cyclin E expression in the embryo is regulated in a tissue-specific manner (Jones et al., 2000). Our work adds Cyclin A and Sticky to the cell cycle kinases regulated by Hh. The conceptual significance of these observations is that Hh might promote cell division and growth directly by regulating such cell cycle functions.
In addition, the targets identified by our work and by previous studies show that Hh signaling also regulates cell behavior indirectly. Hh controls growth and patterning of the wing disc by employing Ci to regulate Dpp expression at the anteroposterior compartment border (Tabata et al., 1995; Zecca et al., 1995). Ci also upregulates Wg expression (Lessing and Nusse, 1998), as well as key components of the Notch and EGF signaling pathways (Table 1). Indirect regulation of cell behavior was also revealed by the high proportion of transcription factors among the targets: 19 out of 52 (37%). Transcription factors represent 40% (30 out of 74) of all targets of Drosophila Hh identified to date, and the prevalence of transcription factors is consistent with the recent tabulation of Shh targets in the vertebrate limb (Vokes et al., 2007).
Autocrine Hh signaling
Hh signaling is understood best in the wing disc and embryo, where posterior compartment cells that express En and Hh export Hh to anterior compartment cells that express neither protein. Although the paracrine response of anterior cells is well-established, posterior cells also activate Hh signaling (Ramirez-Weber et al., 2000) but the basis for their pathway activation has not been settled. Whereas one study concluded that pathway activation is not Hh-dependent (Denef et al., 2000), another concluded that it is (Ramirez-Weber et al., 2000), implying that either Hh-expressing cells respond to Hh received from neighboring cells, or to Hh that they produce themselves.
We do not know if the distinction between autocrine and paracrine signaling is important, but we are intrigued by the responsiveness of Hh-expressing cells to Hh in the optic primordium (Fig. 4). Cells in the POL express hh as well as rdx and have elevated levels of Ci, two hallmarks of pathway activation. POL cells do not express detectable levels of Ptc, an Hh receptor whose negative regulation of the Hh signal transduction pathway is relieved upon Hh binding. Although loss of Ptc function can lead to ectopic pathway activation, expression of rdx in the POL appears not to be a consequence of absent Ptc; rdx expression was markedly reduced in hh mutant embryos, indicating that activation of the pathway in the POL is Hh-dependent. One possible explanation is that some function other than Ptc keeps the Hh pathway in an `off' state when Hh is not present. Another possibility is that Ptc is expressed in these cells, but at levels too low for our methods to detect, and that, although pathway activation increases ptc expression in other contexts (for instance, in the AOL and in every known setting with paracrine signaling), hh-expressing cells of the POL keep ptc expression low. As the elevated level of Ci in the POL does not lead to high levels of ptc, either the Ci is inert or POL cells have a mechanism to override Ci-dependent activation. This mechanism cannot involve En, a negative regulator of ptc, because POL cells do not express en.
In addition to our evidence for autocrine Hh signaling in the wing disc and optic primordium, apparent autocrine signaling by sonic hedgehog has been observed. Three examples are in neural stem cells (Cai et al., 2008), large B-cell lymphoma (Singh et al., 2010) and cerebellar dysplasia in the developing brain (Wang et al., 2004). These examples indicate the potential importance of an autocrine mechanism to human development and disease.
Tissue-specificity of Hh-dependent responses
Activation of Hh targets dpp and knot in the wing disc requires participation of Ci with Vestigial (Hepker et al., 1999) or Ataxin-2 Binding Protein 1 (Usha and Shashidhara, 2010), respectively. We show here that activation of ImpL2 in the tracheal primordium requires Ci and Trh (Fig. 6). This supports the idea that cell signaling pathways such as Hh are characterized by `activator insufficiency', meaning that pathway activation is necessary, but not sufficient, to induce targets (Barolo and Posakony, 2002).
In the tracheal primordium, our genetic analysis shows that, although trh expression is not dependent on hh, ImpL2 expression depends on both hh and trh. These observations indicate that the domain of trh expression must be established independently of Hh and that ImpL2 responds only where both Hh signaling and trh expression intersect. As all cells in the tracheal primordium express trh but only a portion express ImpL2, the cells in which Hh signaling induces ImpL2 expression represent a subset of the cells that are capable of responding. ImpL2 is expressed only in cells near Hh-producing cells that, presumably, are exposed to the highest relative levels of Hh. Moreover, as ectopic activation of the pathway led to upregulation of ImpL2 expression throughout the tracheal primordium, the level of Hh signaling seems to be adjusted in normal embryos to regulate ImpL2. Because ImpL2 expression outside of the tracheal primordium is not trh-dependent, we generalize these observations and suggest that the mechanism that leads to selective responsiveness of Hh targets involves both concentration-dependent responses as well as combinatorial control by Hh-sensitive and Hh-independent transcription factors.
Supplementary Material
Acknowledgments
We thank Michael Levine, Didier Stainier and Grae Davis for advice and support; and Dave Casso, G. Ehrenkaufer, A. Klebes, S. Smolik, B. Ng, Y. N. Jan, S. Parkhurst, B. van Steensel, Richard Tabor and the late F.-A. Ramirez-Weber for advice and help. This work was supported by grants from the NIH to K.K. (AA016922) and T.B.K. (GM77407). Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.055871/-/DC1
References
- Alexandre C., Jacinto A., Ingham P. W. (1996). Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 10, 2003-2013 [DOI] [PubMed] [Google Scholar]
- Amin A., Li Y., Finkelstein R. (1999). Hedgehog activates the EGF receptor pathway during Drosophila head development. Development 126, 2623-2630 [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P., Ramirez-Weber F. A., Laget M. P., Schwartz C., Kornberg T. B. (1997). Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89, 1043-1053 [DOI] [PubMed] [Google Scholar]
- Azpiazu N., Lawrence P. A., Vincent J. P., Frasch M. (1996). Segmentation and specification of the Drosophila mesoderm. Genes Dev. 10, 3183-3194 [DOI] [PubMed] [Google Scholar]
- Barolo S., Posakony J. W. (2002). Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16, 1167-1181 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. (2001). Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279-284 [DOI] [PubMed] [Google Scholar]
- Bossing T., Brand A. H. (2006). Determination of cell fate along the anteroposterior axis of the Drosophila ventral midline. Development 133, 1001-1012 [DOI] [PubMed] [Google Scholar]
- Boube M., Llimargas M., Casanova J. (2000). Cross-regulatory interactions among tracheal genes support a co-operative model for the induction of tracheal fates in the Drosophila embryo. Mech. Dev. 91, 271-278 [DOI] [PubMed] [Google Scholar]
- Buratovich M. A. (2000). DWnt-4 and Wingless have distinct activities in the Drosophila dorsal epidermis. Dev. Genes Evol. 210, 111-119 [DOI] [PubMed] [Google Scholar]
- Cai C., Thorne J., Grabel L. (2008). Hedgehog serves as a mitogen and survival factor during embryonic stem cell neurogenesis. Stem Cells 26, 1097-1108 [DOI] [PubMed] [Google Scholar]
- Chang T., Mazotta J., Dumstrei K., Dumitrescu A., Hartenstein V. (2001). Dpp and Hh signaling in the Drosophila embryonic eye field. Development 128, 4691-4704 [DOI] [PubMed] [Google Scholar]
- Chen Y., Cardinaux J. R., Goodman R. H., Smolik S. M. (1999). Mutants of cubitus interruptus that are independent of PKA regulation are independent of hedgehog signaling. Development 126, 3607-3616 [DOI] [PubMed] [Google Scholar]
- Choksi S. P., Southall T. D., Bossing T., Edoff K., de Wit E., Fischer B. E., van Steensel B., Micklem G., Brand A. H. (2006). Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell 11, 775-789 [DOI] [PubMed] [Google Scholar]
- Chou T. B., Perrimon N. (1996). The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144, 1673-1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessio M., Frasch M. (1996). msh may play a conserved role in dorsoventral patterning of the neuroectoderm and mesoderm. Mech. Dev. 58, 217-231 [DOI] [PubMed] [Google Scholar]
- Denef N., Neubuser D., Perez L., Cohen S. M. (2000). Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell 102, 521-531 [DOI] [PubMed] [Google Scholar]
- Dominguez M., Brunner M., Hafen E., Basler K. (1996). Sending and receiving the hedgehog signal: control by the Drosophila Gli protein Cubitus interruptus. Science 272, 1621-1625 [DOI] [PubMed] [Google Scholar]
- Duman-Scheel M., Weng L., Xin S., Du W. (2002). Hedgehog regulates cell growth and proliferation by inducing Cyclin D and Cyclin E. Nature 417, 299-304 [DOI] [PubMed] [Google Scholar]
- Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863-14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes A. J., Nakano Y., Taylor A. M., Ingham P. W. (1993). Genetic analysis of hedgehog signalling in the Drosophila embryo. Development Suppl. 115-124 [PubMed] [Google Scholar]
- Glazer L., Shilo B. Z. (2001). Hedgehog signaling patterns the tracheal branches. Development 128, 1599-1606 [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta J. L., Modolell J. (1996). araucan and caupolican provide a link between compartment subdivisions and patterning of sensory organs and veins in the Drosophila wing. Genes Dev. 10, 2935-2945 [DOI] [PubMed] [Google Scholar]
- Guillen I., Mullor J. L., Capdevila J., Sanchez-Herrero E., Morata G., Guerrero I. (1995). The function of engrailed and the specification of Drosophila wing pattern. Development 121, 3447-3456 [DOI] [PubMed] [Google Scholar]
- Hallikas O., Taipale J. (2006). High-throughput assay for determining specificity and affinity of protein-DNA binding interactions. Nat. Protoc. 1, 215-222 [DOI] [PubMed] [Google Scholar]
- Hallikas O., Palin K., Sinjushina N., Rautiainen R., Partanen J., Ukkonen E., Taipale J. (2006). Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 124, 47-59 [DOI] [PubMed] [Google Scholar]
- Hays R., Buchanan K. T., Neff C., Orenic T. V. (1999). Patterning of Drosophila leg sensory organs through combinatorial signaling by hedgehog, decapentaplegic and wingless. Development 126, 2891-2899 [DOI] [PubMed] [Google Scholar]
- Heberlein U., Singh C. M., Luk A. Y., Donohoe T. J. (1995). Growth and differentiation in the Drosophila eye coordinated by hedgehog. Nature 373, 709-711 [DOI] [PubMed] [Google Scholar]
- Hepker J., Blackman R. K., Holmgren R. (1999). Cubitus interruptus is necessary but not sufficient for direct activation of a wing-specific decapentaplegic enhancer. Development 126, 3669-3677 [DOI] [PubMed] [Google Scholar]
- Hersh B. M., Carroll S. B. (2005). Direct regulation of knot gene expression by Ultrabithorax and the evolution of cis-regulatory elements in Drosophila. Development 132, 1567-1577 [DOI] [PubMed] [Google Scholar]
- Hidalgo A., Ingham P. (1990). Cell patterning in the Drosophila segment: spatial regulation of the segment polarity gene patched. Development 110, 291-301 [DOI] [PubMed] [Google Scholar]
- Inaki M., Kojima T., Ueda R., Saigo K. (2002). Requirements of high levels of Hedgehog signaling activity for medial-region cell fate determination in Drosophila legs: identification of pxb, a putative Hedgehog signaling attenuator gene repressed along the anterior-posterior compartment boundary. Mech. Dev. 116, 3-18 [DOI] [PubMed] [Google Scholar]
- Ingham P. W. (1991). Segment polarity genes and cell patterning within the Drosophila body segment. Curr. Opin. Genet. Dev. 1, 261-267 [DOI] [PubMed] [Google Scholar]
- Jagla K., Jagla T., Heitzler P., Dretzen G., Bellard F., Bellard M. (1997). ladybird, a tandem of homeobox genes that maintain late wingless expression in terminal and dorsal epidermis of the Drosophila embryo. Development 124, 91-100 [DOI] [PubMed] [Google Scholar]
- Jia J., Tong C., Jiang J. (2003). Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes Dev. 17, 2709-2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Richardson H., Saint R. (2000). Tissue-specific regulation of cyclin E transcription during Drosophila melanogaster embryogenesis. Development 127, 4619-4630 [DOI] [PubMed] [Google Scholar]
- Kechris K. J., Biehs B., Kornberg T. B. (2010). Generalizing moving averages for tiling arrays using combined P-value statistics. Stat. Appl. Genet. Mol. Biol. 9, Article 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent D., Bush E. W., Hooper J. E. (2006). Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development 133, 2001-2010 [DOI] [PubMed] [Google Scholar]
- Kinzler K. W., Vogelstein B. (1990). The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol. Cell. Biol. 10, 634-642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebes A., Kornberg T. B. (2008). Linear RNA amplification for the production of microarray hybridization probes. Methods Mol. Biol. 420, 303-317 [DOI] [PubMed] [Google Scholar]
- Lessing D., Nusse R. (1998). Expression of wingless in the Drosophila embryo: a conserved cis-acting element lacking conserved Ci-binding sites is required for patched-mediated repression. Development 125, 1469-1476 [DOI] [PubMed] [Google Scholar]
- Lum L., Zhang C., Oh S., Mann R. K., von Kessler D. P., Taipale J., Weis-Garcia F., Gong R., Wang B., Beachy P. A. (2003). Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol. Cell 12, 1261-1274 [DOI] [PubMed] [Google Scholar]
- MacArthur S., Li X. Y., Li J., Brown J. B., Chu H. C., Zeng L., Grondona B. P., Hechmer A., Simirenko L., Keranen S. V., et al. (2009). Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 10, R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Markstein P., Markstein V., Levine M. S. (2002). Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 99, 763-768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. A., Doe C. Q. (1997). Establishing neuroblast-specific gene expression in the Drosophila CNS: huckebein is activated by Wingless and Hedgehog and repressed by Engrailed and Gooseberry. Development 124, 1079-1087 [DOI] [PubMed] [Google Scholar]
- McMahon A. P., Ingham P. W., Tabin C. J. (2003). Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 53, 1-114 [DOI] [PubMed] [Google Scholar]
- Méthot N., Basler K. (1999). Hedgehog controls limb, development, by, regulating the, activities, of, distinct transcriptional activator. and repressor forms, of, Cubitus interruptus. Cell 96, 819-831 [DOI] [PubMed] [Google Scholar]
- Méthot N., Basler K. (2001). An absolute requirement for Cubitus interruptus in Hedgehog signaling. Development 128, 733-742 [DOI] [PubMed] [Google Scholar]
- Müller B., Basler K. (2000). The repressor and activator forms of Cubitus interruptus control Hedgehog target genes through common generic gli-binding sites. Development 127, 2999-3007 [DOI] [PubMed] [Google Scholar]
- O'Neill J. W., Bier E. (1994) Double-label in situ hybridization using biotin and digoxigenin-tagged RNA probes. BioTechniques 870, 874-875 [PubMed] [Google Scholar]
- Orenic T. V., Slusarski D. C., Kroll K. L., Holmgren R. A. (1990). Cloning and characterization of the segment polarity gene cubitus interruptus Dominant of Drosophila. Genes Dev. 4, 1053-1067 [DOI] [PubMed] [Google Scholar]
- Pappu K. S., Chen R., Middlebrooks B. W., Woo C., Heberlein U., Mardon G. (2003). Mechanism of hedgehog signaling during Drosophila eye development. Development 130, 3053-3062 [DOI] [PubMed] [Google Scholar]
- Pavletich N., Pabo C. (1993). Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science 261, 1701-1707 [DOI] [PubMed] [Google Scholar]
- Piepenburg O., Vorbruggen G., Jackle H. (2000). Drosophila segment borders result from unilateral repression of hedgehog activity by wingless signaling. Mol. Cell 6, 203-209 [PubMed] [Google Scholar]
- Ponzielli R., Astier M., Chartier A., Gallet A., Therond P., Semeriva M. (2002). Heart tube patterning in Drosophila requires integration of axial and segmental information provided by the Bithorax Complex genes and hedgehog signaling. Development 129, 4509-4521 [DOI] [PubMed] [Google Scholar]
- Ramirez-Weber F. A., Casso D. J., Aza-Blanc P., Tabata T., Kornberg T. B. (2000). Hedgehog signal transduction in the posterior compartment of the Drosophila wing imaginal disc. Mol. Cell 6, 479-485 [DOI] [PubMed] [Google Scholar]
- Ruel L., Rodriguez R., Gallet A., Lavenant-Staccini L., Therond P. P. (2003). Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat. Cell Biol. 5, 907-913 [DOI] [PubMed] [Google Scholar]
- Sasaki H., Hui C., Nakafuku M., Kondoh H. (1997). A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 124, 1313-1322 [DOI] [PubMed] [Google Scholar]
- Schlichting K., Demontis F., Dahmann C. (2005). Cadherin Cad99C is regulated by Hedgehog signaling in Drosophila. Dev. Biol. 279, 142-154 [DOI] [PubMed] [Google Scholar]
- Singh R. R., Kim J. E., Davuluri Y., Drakos E., Cho-Vega J. H., Amin H. M., Vega F. (2010). Hedgehog signaling pathway is activated in diffuse large B-cell lymphoma and contributes to tumor cell survival and proliferation. Leukemia 24, 1025-1036 [DOI] [PubMed] [Google Scholar]
- Smyth G. (2005). Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions using R and Bioconductor (ed. Carey V. J., Irizarry W.), pp. 397-420 New York: Springer; [Google Scholar]
- Stone D. M., Hynes M., Armanini M., Swanson T. A., Gu Q., Johnson R. L., Scott M. P., Pennica D., Goddard A., Phillips H., et al. (1996). The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384, 129-134 [DOI] [PubMed] [Google Scholar]
- Sun Y., Jan L. Y., Jan Y. N. (1998). Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development 125, 3731-3740 [DOI] [PubMed] [Google Scholar]
- Tabata T., Kornberg T. B. (1994). Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell 76, 89-102 [DOI] [PubMed] [Google Scholar]
- Tabata T., Schwartz C., Gustavson E., Ali Z., Kornberg T. B. (1995). Creating a Drosophila wing de novo, the role of engrailed, and the compartment border hypothesis. Development 121, 3359-3369 [DOI] [PubMed] [Google Scholar]
- Taipale J., Cooper M. K., Maiti T., Beachy P. A. (2002). Patched acts catalytically to suppress the activity of Smoothened. Nature 418, 892-897 [DOI] [PubMed] [Google Scholar]
- Usha N., Shashidhara L. S. (2010). Interaction between Ataxin-2 Binding Protein 1 and Cubitus-interruptus during wing development in Drosophila. Dev. Biol. 341, 389-399 [DOI] [PubMed] [Google Scholar]
- van Steensel B., Delrow J., Henikoff S. (2001). Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 27, 304-308 [DOI] [PubMed] [Google Scholar]
- Varjosalo M., Taipale J. (2008). Hedgehog: functions and mechanisms. Genes Dev. 22, 2454-2472 [DOI] [PubMed] [Google Scholar]
- Vokes S. A., Ji H., McCuine S., Tenzen T., Giles S., Zhong S., Longabaugh W. J., Davidson E. H., Wong W. H., McMahon A. P. (2007). Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development 134, 1977-1989 [DOI] [PubMed] [Google Scholar]
- Vokes S. A., Ji H., Wong W. H., McMahon A. P. (2008). A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 22, 2651-2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Ohlen T., Lessing D., Nusse R., Hooper J. (1997). Hedgehog signaling regulates transcription through cubitus interruptus, a sequence-specific DNA binding protein. Proc. Natl. Acad. Sci. USA 94, 2404-2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lin W., Popko B., Campbell I. L. (2004). Inducible production of interferon-gamma in the developing brain causes cerebellar dysplasia with activation of the Sonic hedgehog pathway. Mol. Cell. Neurosci. 27, 489-496 [DOI] [PubMed] [Google Scholar]
- Wilk R., Weizman I., Shilo B. Z. (1996). trachealess encodes a bHLH-PAS protein that is an inducer of tracheal cell fates in Drosophila. Genes Dev. 10, 93-102 [DOI] [PubMed] [Google Scholar]
- Wormald S., Hilton D. J., Smyth G. K., Speed T. P. (2006). Proximal genomic localization of STAT1 binding and regulated transcriptional activity. BMC Genomics 7, 254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M., Basler K., Struhl G. (1995). Sequential organizing activities of engrailed, hedgehog and decapentaplegic in the Drosophila wing. Development 8, 2265-2278 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Zinzen R. P., Stark A., Kellis M., Zhang H., Young R. A., Levine M. (2007). Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 21, 385-390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zhang L., Wang B., Ou C., Chien C., Jiang J. (2006). A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev. Cell 10, 719-729 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.