Abstract
Genome integrity is constantly monitored by sophisticated cellular networks, collectively termed the DNA damage response (DDR). A common feature of DDR proteins is their mobilization in response to genotoxic stress. Here, we outline how the development of various complementary methodologies has provided valuable insights into the spatiotemporal dynamics of DDR protein assembly/disassembly at sites of DNA strand breaks in eukaryotic cells. Considerable advances have also been made in understanding the underlying molecular mechanisms for these events, with post-translational modifications of DDR factors being shown to play prominent roles in controlling the formation of foci in response to DNA-damaging agents. We review these regulatory mechanisms and discuss their biological significance to the DDR.
Keywords: checkpoint, DNA breaks, DNA damage foci, post-translational modifications, repair
Genome integrity is continuously challenged by DNA lesions, many thousands of which arise in each human cell every day (Lindahl and Barnes 2000). While the majority of these lesions occur as byproducts of normal cell metabolism or DNA replication, they are also induced by radiation and toxic environmental chemicals (Friedberg et al. 2006; Jackson and Bartek 2009; Ciccia and Elledge 2010). Programmed DNA lesions also form as intermediates during developmentally regulated genome rearrangements in lymphocytes and germ cells (Jackson and Bartek 2009; Longhese et al. 2009; Tsai and Lieber 2010). DNA damage can have deleterious effects, as it interferes with DNA replication and transcription, and because it can ultimately result in mutations and chromosomal aberrations. Genome integrity is preserved by DNA damage signaling and repair machineries, which counteract the adverse consequences of DNA lesions and prevent their transmission to daughter cells (Hoeijmakers 2001; Ciccia and Elledge 2010). Defects in DNA damage signaling or repair contribute to aging and various disorders, including developmental defects, neurodegenerative diseases, and cancer (Jackson and Bartek 2009), which highlights the critical importance of an efficient DNA damage response (DDR) for cell and organism viability.
This review focuses on cellular responses to DNA single-strand breaks (SSBs) and double-strand breaks (DSBs) in eukaryotic cells, with an emphasis on mammalian systems. After a general overview of how such lesions are generated, signaled, and repaired, we outline recent work that has characterized the spatiotemporal dynamics of protein assembly/disassembly at sites of DNA breaks and the regulatory mechanisms involved. In particular, we highlight the major contribution of post-translational modifications in mediating and controlling these processes, and discuss the biological significance of DNA damage foci.
Cellular responses to DNA strand breaks
Formation and repair of DNA breaks
DNA DSBs are the most deleterious form of DNA damage because they do not leave an intact complementary strand to be used as a template for DNA repair. If left unrepaired, they can ultimately lead to chromosome breaks and translocations that are associated with developmental defects, neurodegeneration, immunodeficiency, radiosensitivity, sterility, and cancer predisposition (Jackson and Bartek 2009). DSBs are generated in response to ionizing radiation (IR) or radiomimetic drugs by free radical attack of deoxyribose, and also arise in cells treated with topoisomerase II inhibitors that prevent religation of DNA strands broken by topoisomerase II activity. DSBs can also form upon replication of DNA molecules containing other DNA lesions, such as DNA SSBs, and are produced by specific nucleases during V(D)J and class switch recombination in vertebrate lymphocytes, meiotic recombination in germ cells, mating type switching in yeast, apoptotic cell death, and retroviral integration (Friedberg et al. 2006; Jackson and Bartek 2009). Last, but not least, naturally occurring DSBs at chromosome ends are associated with human cell aging, as they are exposed when telomeres become critically short during replicative senescence (d'Adda di Fagagna et al. 2003).
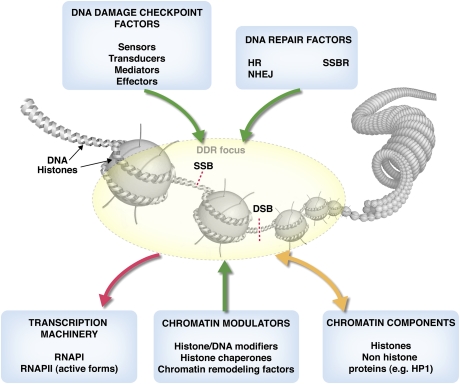
Cells rely on two major pathways to repair DSBs: homologous recombination (HR) and nonhomologous end-joining (NHEJ) (Hartlerode and Scully 2009; Pardo et al. 2009). These pathways are complementary and operate optimally under different circumstances. HR requires the presence of a homologous template, usually a sister chromatid, which allows accurate repair of post-replicative DSBs in S and G2 phases of the cell cycle (San Filippo et al. 2008; Moynahan and Jasin 2010). In contrast, NHEJ can operate throughout the cell cycle without the need for template DNA, and is often mutagenic because deletions or insertions can be induced at sites of repair (Lieber 2010). NHEJ, which is the prevalent DSB repair pathway in higher eukaryotes, essentially mediates the direct ligation of broken DNA ends, and usually involves minimal DNA end processing. In NHEJ, DNA ends are first bound by the Ku70/Ku80 heterodimer, which recruits and activates the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form the DNA-PK holoenzyme (Gottlieb and Jackson 1993). Broken DNA ends juxtaposed by DNA-PK are then acted on by factors such as the nuclease Artemis, polynucleotide kinase (PNK), Aprataxin, and APLF (Aprataxin and PNK-like factor) before being ligated by the XLF–XRCC4 (X-ray cross-complementing-4)–LigaseIV complex (Table 1; Lieber and Wilson 2010). In addition, alternative end-joining pathways sometimes operate, particularly when classical NHEJ is impeded because some NHEJ component is missing or mutated. These alternative end-joining pathways often rely on terminal microhomologies for the joining reaction, and involve certain factors that also function in HR or SSB repair (SSBR), such as the MRE11–RAD50–NBS1 (MRN) complex, poly(ADP-ribose) polymerase-1 (PARP-1), XRCC1, and DNA Ligase I or III (McVey and Lee 2008).
Table 1.
Factors involved in DNA strand break repair and damage signaling in budding yeast and mammals
An important regulatory step that determines the choice between the two main DSB repair pathways is the process of DSB resection, which is required for HR but not NHEJ. Resection comprises the 5′-to-3′ nucleolytic processing of DNA ends by the MRN complex (Rupnik et al. 2010) in conjunction with auxiliary factors including CtIP, RECQ family helicases, and the nucleases Exo1 and Dna2 (Zou and Elledge 2003; Bernstein and Rothstein 2009; Mimitou and Symington 2009; Huertas 2010; Longhese et al. 2010; You and Bailis 2010). Resulting ssDNA overhangs are then coated by the ssDNA-binding complex RPA (replication protein A) before being substituted by RAD51 proteins with the help of factors such as RAD51 paralogs, RAD52, and other proteins that comprise the FA (Fanconi anemia) pathway such as FANCD1 (FA-associated nuclease CD1)/BRCA2 (breast cancer-2, early onset) and FANCN/PALB2 (partner and localizer of BRCA2) that play key roles in detecting and repairing interstrand cross-links, particularly at sites of stalled DNA replication (Moldovan and D'Andrea 2009). The RAD51 nucleofilament, together with various other HR factors, then mediates homology search in the sister chromatid, followed by strand invasion into the homologous template. After the actions of DNA polymerases and DNA end ligation by Ligase I, DNA helicase and resolvase enzymes then mediate the cleavage and resolution of HR intermediates to yield intact, repaired DNA molecules (Table 1; Mazon et al. 2010).
Although not as harmful as DSBs, SSBs are toxic to the cell, as they can block DNA replication and transcription, and, indeed, SSBR defects are associated with several hereditary neurodegenerative diseases (Caldecott 2008). SSBs form upon oxidative attack of deoxyribose by free radicals arising from cell metabolism or through exposure to agents such as H2O2, IR, and radiomimetic drugs. They also arise as intermediates during excision repair of base damage, and upon inhibition of topoisomerase I. In most eukaryotes, SSBs are initially detected by PARP-1, whose binding to DNA breaks triggers poly-(ADP-ribosyl)ation of numerous nuclear proteins, including itself. These modifications in turn promote the binding of XRCC1, which acts as a molecular scaffold for downstream SSBR components involved in end-processing, gap filling, and ligation (Table 1).
Signaling of DNA breaks
DNA repair is tightly coordinated with cell cycle progression through the activation of orchestrated signaling pathways that are often termed DNA damage checkpoints (Harrison and Haber 2006; Harper and Elledge 2007; Lazzaro et al. 2009). In response to unrepaired DNA damage, these pathways delay or stop the cell cycle at critical stages before or during DNA replication (G1/S and intra-S checkpoints) and before cell division (G2/M checkpoint), thereby preventing duplication and segregation of damaged DNA. DNA damage signaling cascades are complex, coordinated events that require the actions of various proteins whose functions can be categorized as DNA damage sensors, transducers, mediators, and effectors. Of particular note are the MRN sensor complex that detects DSBs (Lavin 2007), and RPA that signals the accumulation of ssDNA resulting from DNA damage processing. MRN contributes to the recruitment and activation of the apical DDR kinase ATM (ataxia telangiectasia mutated) (Uziel et al. 2003; Falck et al. 2005; Lee and Paull 2005), while RPA recruits the ATR (ATM and rad3-related) kinase via its partner protein, ATRIP (ATR-interacting partner) (Cortez et al. 2001; Rouse and Jackson 2002; Zou and Elledge 2003). With the help of mediator proteins—such as MDC1 (mediator of DNA damage checkpoint), 53BP1 (p53-binding protein 1), and BRCA1 for ATM, and TopBP1 (topoisomerase-binding protein 1) and Claspin for ATR—transducer kinases activate the effector kinases Chk1 and Chk2, which then spread the signal throughout the nucleus. Ultimate targets of these signaling cascades include transcription factors, cell cycle regulators, the apoptotic machinery, and DNA repair factors (Harrison and Haber 2006; Harper and Elledge 2007; Lazzaro et al. 2009).
Responses to DNA breaks in a physiological context
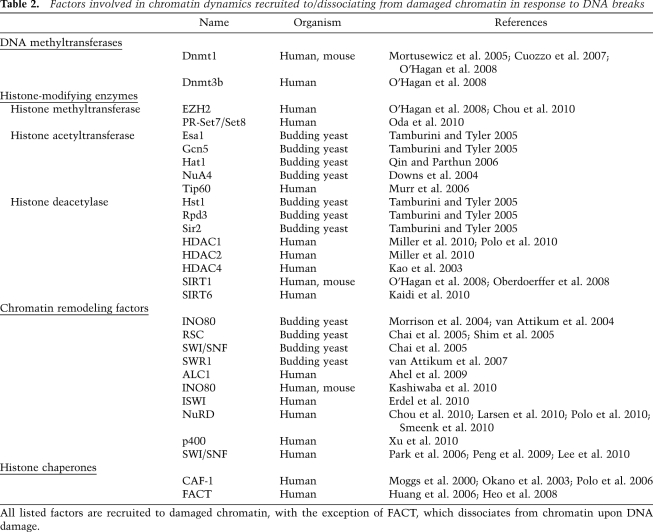
Signaling and repair of DNA breaks occur in a context where they can potentially interfere with other DNA metabolic activities such as replication and transcription. Indeed, ATM has been reported to mediate local inhibition of both RNA polymerase I- and II-dependent transcription at sites of DNA breaks in human cells (Kruhlak et al. 2007; Shanbhag et al. 2010). Such transcriptional inhibition is accompanied by the clearance of active RNA polymerases from damage sites, highlighting that protein dynamics at DNA breaks operate in both directions: While DNA damage signaling and repair factors are recruited to DNA breaks, other factors dissociate (Fig. 1).
Figure 1.
Protein dynamics to and from sites of DNA breaks. DNA damage checkpoint and repair factors and modulators of chromatin organization are recruited (green arrows) to DNA breaks (SSB and DSB), while transcription machineries are excluded from DDR foci (red arrows), and the dynamics of structural chromatin components operate in both directions (orange arrows).
When studying the DDR in its cellular context, one also has to consider that the physiological substrate for the DDR machinery in the cell nucleus is DNA wrapped around histone proteins in the form of chromatin, the basic unit of which is the nucleosome (Kornberg 1977; Luger et al. 1997). Nucleosomes can be further compacted into higher-order chromatin structures with linker histones and nonhistone components such as heterochromatin protein 1 (HP1) (McBryant et al. 2006; Fanti and Pimpinelli 2008). Although chromatin acts as a physical barrier to the detection and repair of DNA lesions, it is also a dynamic structure that can be modulated by DNA methylation (Kulis and Esteller 2010), incorporation of histone variants (Bernstein and Hake 2006), histone post-translational modifications (Kouzarides 2007), and nucleosome repositioning by ATP-dependent remodeling complexes (Clapier and Cairns 2009). Moreover, a growing body of evidence highlights the importance of such modulations of chromatin organization in the DDR (for review, see Misteli and Soutoglou 2009; van Attikum and Gasser 2009), the most prominent and widely documented being phosphorylation of the histone variant H2AX (γH2AX) identified >10 years ago (Rogakou et al. 1998). As a consequence, and as described in further detail below, it comes as no surprise that chromatin-associated proteins are also mobilized to and from DNA breaks (Fig. 1; Table 2). In particular, chromatin remodeling complexes and histone chaperones have been linked to histone mobilization (Tsukuda et al. 2005; Kent et al. 2007; Shim et al. 2007; Xu et al. 2010), histone variant replacement (Kusch et al. 2004; Papamichos-Chronakis et al. 2006; Heo et al. 2008), and new histone deposition (Polo et al. 2006; Chen et al. 2008) at sites of DNA breaks.
Table 2.
Factors involved in chromatin dynamics recruited to/dissociating from damaged chromatin in response to DNA breaks
All listed factors are recruited to damaged chromatin, with the exception of FACT, which dissociates from chromatin upon DNA damage.
Methods for studying DDR protein assembly/disassembly at DNA breaks
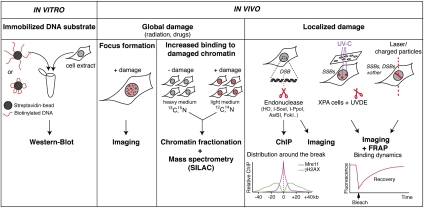
Our understanding of DDR protein dynamics at DNA breaks has been greatly advanced through the development of various methodologies for studying DDR protein assembly and disassembly at damage sites in vitro and in vivo (summarized in Fig. 2; Lukas et al. 2005; Nagy and Soutoglou 2009). Initially, the recruitment of DDR proteins to broken DNA was analyzed in vitro by biochemical assays employing various types of DNA substrates (e.g., circular vs. linear DNA), as described for the NHEJ factors Ku and DNA-PKcs, which display strong affinity for DNA ends (Gottlieb and Jackson 1993; Dynan and Yoo 1998). More recently, advanced biophysical approaches have enabled single-molecule imaging of DDR proteins such as yeast Rad54 and human RAD51 on DNA (Amitani et al. 2010).
Figure 2.
Methods for studying the recruitment of DDR proteins to DNA breaks. Scheme describing the multiple methods used to generate or mimic DNA breaks in vitro and in vivo and the techniques employed to monitor recruitment of DDR factors to such breaks.
In vivo studies of DDR protein recruitments include imaging of protein dynamics at sites of DNA breaks and biochemical approaches to analyze changes in protein binding to damaged chromatin. When DNA damage is inflicted on the whole nucleus by exposing cells to genotoxic agents such as H2O2, IR, radiomimetic drugs or topoisomerase inhibitors, protein accumulation at DNA damage sites can take the form of cytologically discernable foci, as initially observed for the proteins Rad51, BRCA1, and MRN in human cells (Haaf et al. 1995; Maser et al. 1997; Scully et al. 1997). Such foci can be visualized by indirect immunofluorescence or in real time by imaging live cells expressing fluorescently tagged proteins. Importantly, the number of focus-positive cells and the number of foci per cell each increase in a dose-dependent manner in mammalian cells (for example, 20–40 DSB foci per nucleus per Gray of radiation for a mammalian cell), and focus number and size also change over time after DNA damage induction (van Veelen et al. 2005). In this regard, it is noteworthy that similar foci can be detected at recombination sites in meiotic cells, as initially illustrated by the local accumulation of the RAD51 recombinase in vertebrate germ cells (Ashley et al. 1995). Another approach for analyzing protein dynamics in response to DNA breaks consists of monitoring increased chromatin binding of proteins in damaged versus undamaged cells by biochemical cell fractionation (Drouet et al. 2005), a method that was converted recently into high-throughput formats by the implementation of SILAC (stable isotope labeling by amino acids in cell culture)-based mass spectrometry (Chou et al. 2010; Larsen et al. 2010).
The recruitment of DDR factors to DNA breaks can also be analyzed by chromatin immunoprecipitation (ChIP) or cell imaging upon local induction of DNA damage by microirradiation or specific endonucleases. The generation of sequence-specific DSBs by selected endonucleases was pioneered with yeast homothallic (HO) endonuclease (Sugawara and Haber 2006), translated into mammalian cells by using homing endonucleases such as I-SceI (Jasin 1996) and I-PpoI (Berkovich et al. 2008), and more recently developed into genome-wide formats by combining expression of the restriction enzyme AsiSI with microarray hybridization and high-throughput sequencing (Iacovoni et al. 2010; Massip et al. 2010). Furthermore, generation of DSBs by the targeting of a nuclease to a defined locus in the genome can be attained by fusing its nuclease domain to a zinc finger protein or a Lac repressor, as was done recently with the FokI enzyme (Urnov et al. 2005; Shanbhag et al. 2010).
In addition to the use of endonucleases, several localized irradiation techniques have been developed to introduce discrete sites of DNA breaks in the cell nucleus, including partial cell volume irradiation with ultrasoft X-rays through a gridded shield (Nelms et al. 1998) or high-voltage X-rays through a microcollimator (Pataky et al. 2009). Similarly, local UV-C irradiation through micropore filters combined with the expression of UVDE (UV-specific endonuclease) in cells defective in repairing UV lesions has been used to generate localized SSBs (Okano et al. 2003). DNA breaks, together with various base alterations, can also be induced locally by laser microirradiation, with or without cell photosensitization with halogenated thymidine analogs such as bromodeoxyuridine (BrdU) or a DNA intercalating agent like Hoechst (Limoli and Ward 1993; Lukas et al. 2005). This technique has also been refined recently by the use of highly focused multiphoton lasers (Mari et al. 2006; Botchway et al. 2010). In such studies, it is important to calibrate the laser wavelength, energy output, and photosensitization method used, as these determine the type and density of DNA lesions. Other techniques such as localized cell irradiation with heavy-ion or α-particle microbeams (Jakob et al. 2005; Stap et al. 2008) elicit similar tracks of DNA breaks and base modifications in the cell nucleus. Other ways of triggering DNA DSB signaling in mammalian cells without the need for any exogenous DNA-damaging agent are telomere shortening in senescent cells and artificial telomere uncapping by inducible loss of function of a component of the Shelterin complex that protects chromosome ends, both of which trigger the formation of DDR foci at telomeres (d'Adda di Fagagna et al. 2003; Takai et al. 2003). Similar uncapping strategies have been used in yeast (Wellinger 2010). Complementary to the aforementioned methods, photobleaching techniques such as fluorescence recovery after photobleaching (FRAP) have provided valuable insights into the dynamics of DDR proteins by measuring their turnover rates at damage sites (Essers et al. 2006; Mortusewicz et al. 2008), revealing that DDR factors are not stably bound within a DDR focus but dynamically exchange with neighboring nuclear compartments. Collectively, these studies have proven critical for understanding the hierarchy and spatiotemporal dynamics of DDR factor accumulation at DNA breaks.
Spatiotemporal dynamics of DDR protein assembly/disassembly at DNA breaks
To ensure the efficient signaling and repair of DNA damage, DDR proteins must relocate to the right place at the right time. As described below, a wealth of studies have highlighted the tight spatiotemporal coordination of DDR protein assembly and disassembly at DNA breaks.
Spatial organization of DDR foci
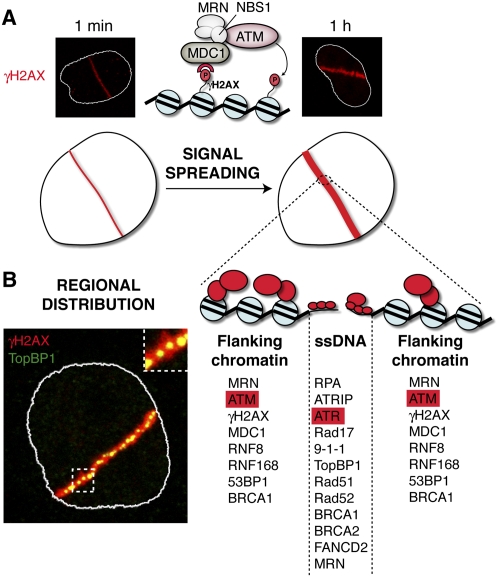
One hallmark of DDR proteins is their local accumulation at damage sites, which in many cases leads to the formation of discrete, cytologically detectable foci. Notably, however, not all DDR factors accumulate at DNA breaks in a manner that can be readily observed under the microscope. This is exemplified by NHEJ components, as described below, and the Chk1 and Chk2 effector kinases, which become phosphorylated at damage sites but then quickly dissociate and distribute throughout the nucleus (Lukas et al. 2003; Smits et al. 2006). Furthermore, factors accumulating at DSB sites do not always colocalize perfectly, with two classes of DDR proteins being readily distinguishable based on their spatial distributions (Fig. 3): those present directly at damage sites, coating ssDNA resulting from DSB resection, and those associated with DSB-flanking chromatin (Bekker-Jensen et al. 2006). In this regard, we point out that, because NHEJ components are usually confined close to the DSBs themselves and do not spread substantially into adjacent chromatin, such factors are not usually evident at radiation-induced or even laser-induced foci unless high levels of damage are used (Bekker-Jensen et al. 2006). However, they can be readily detected at DSB sites by ChIP (Zhang et al. 2007; Miller et al. 2010) or cellular fractionation methods (Drouet et al. 2005).
Figure 3.
Spatial organization of DDR protein accumulation at DNA DSBs. (A) DDR signal spreading. DDR proteins initially accumulate at DSB sites and then spread at distance via a positive feedback loop involving MDC1, which binds γH2AX, the MRN complex, and ATM kinase, which phosphorylates additional H2AX molecules further away from the break site. (B) Regional distribution of DDR proteins around DSBs. Factors involved in ATR signaling accumulate proximal to the break site on ssDNA generated by DNA end resection, while ATM signaling factors localize on flanking chromatin regions.
Strikingly, for various DSB-associated DDR factors and protein modifications, their foci increase in size over time, which reflects these factors and modifications spreading away from the DSB into adjacent chromatin (Fig. 3), as initially observed for γH2AX in mammalian cells (Rogakou et al. 1999). γH2AX spreading was also described in yeast (Downs et al. 2004; Shroff et al. 2004), although over shorter distances: up to 100 kb as opposed to several megabases in mammals (Rogakou et al. 1999; Meier et al. 2007; Iacovoni et al. 2010). Another prominent difference reported between yeast and higher eukaryotes lies in the mobility of DDR foci. In budding yeast, DDR foci are highly mobile and coalesce into repair centers (Lisby et al. 2003), which contrasts with the reported relative positional stability of broken DNA ends in mammalian cells (Nelms et al. 1998; Kruhlak et al. 2006; Soutoglou et al. 2007; Jakob et al. 2009). The reason for this apparent discrepancy is not yet clear, and might simply relate to the difference in nuclear size between these organisms. For example, it is possible that mammalian DDR foci are able to move distances similar to those traversed by foci in yeast cells, but this might not have been evident in the mammalian studies because such movements would occur only within a small fraction of the total nuclear volume. Indeed, recent studies have indicated that some DSB mobility does occur in mammalian cells to promote joining of distal DNA breaks (Difilippantonio et al. 2008; Dimitrova et al. 2008).
The dynamics of DDR foci are also dictated by chromatin organization and transcriptional activity, as they clearly differ between relaxed, highly transcribed euchromatin and more compact heterochromatin that concentrates in specific chromosomal domains such as centromeres and telomeres. For example, it has been shown that γH2AX does not propagate effectively on actively transcribed genes in human cells (Iacovoni et al. 2010). Furthermore, despite the fact that DNA breaks can be efficiently induced in heterochromatin, this chromatin compartment is generally refractory to γH2AX focus formation in yeast and mammalian cells (Cowell et al. 2007; JA Kim et al. 2007). This barrier is relieved in S phase, however, by DNA replication, and replication stress-associated γH2AX is also detected in heterochromatin regions (Chadwick and Lane 2005; Cowell et al. 2007; Rozenzhak et al. 2010; Szilard et al. 2010), suggesting that transient decompaction of heterochromatin and/or displacement of heterochromatin components upon DNA replication is required for the propagation of H2AX phosphorylation. Notably, the γH2AX foci that persist longest after damage in mammalian cells generally localize at the periphery of heterochromatin domains, suggesting that heterochromatic DSBs are refractory to repair; indeed, it has been shown that this γH2AX persistence can be alleviated by depletion of heterochromatin components (Goodarzi et al. 2008).
Temporal organization of DDR foci
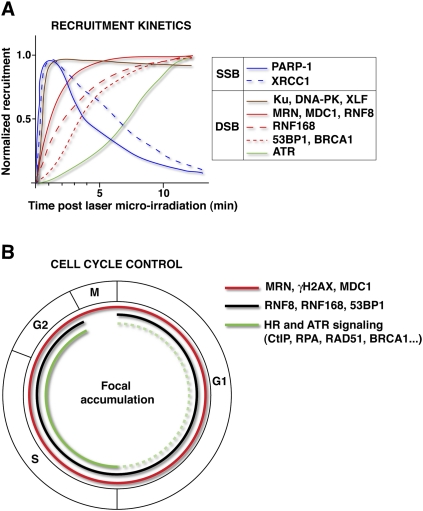
In addition to shaping our knowledge of DDR protein distributions in response to DNA breaks, live-cell imaging by time-lapse microscopy has substantially contributed to our understanding of DDR protein dynamics in both yeast and mammalian cells, especially in regard to the early kinetics of DDR focus formation. Furthermore, the use of siRNA-mediated depletion methods or mutant cell lines has been instrumental in functionally dissecting the sequential recruitment of DDR proteins to DNA breaks (e.g., Lisby et al. 2004; Mari et al. 2006; Mailand et al. 2007). Indeed, multiple lines of evidence support the idea that DDR proteins assemble in a sequential, coordinated manner at sites of DNA breaks, rather than being recruited as a preformed protein complex (Fig. 4A). For instance, the recruitment of DDR factors to SSBs is very rapid and transient (Mortusewicz et al. 2007), reaching maximal accumulation within 1–2 min, due to its dependency on poly(ADP-ribosyl)ation (PARylation), which is a relatively short-lived protein modification (Gagne et al. 2006; Hakme et al. 2008). Factors involved in DSB repair by NHEJ are also recruited within seconds upon break formation and normally dissociate within 2 h, while HR factors show delayed and persistent recruitment to DSBs, reflecting different repair kinetics between these two pathways (Kim et al. 2005; Mari et al. 2006; Mailand et al. 2007; Uematsu et al. 2007; Yano et al. 2008; Doil et al. 2009).
Figure 4.
Temporal regulation of DDR protein accumulation at DNA breaks. (A) Sequential recruitment of DDR factors to SSBs and DSBs generated by laser microirradiation. (B) Cell cycle regulation of DDR foci formation. (Solid line) Efficient focus formation; (dashed line) weak/undetectable foci.
The accumulation kinetics of factors involved in DSB signaling has also been studied in great detail, and has been described as a two-stage process in which initial recruitment occurs independently of H2AX phosphorylation, followed by sustained DDR factor retention in a γH2AX-dependent manner (Celeste et al. 2003b; Yuan et al. 2010). As discussed in the following sections, studies from multiple laboratories have built up a fairly comprehensive picture of the mechanisms involved in γH2AX-dependent focus formation. In contrast, our knowledge of the initial γH2AX-independent recruitment of DSB signaling factors is limited, although it seems to involve the MRN complex, additional histone modifications, and changes in chromatin compaction at damage sites (FitzGerald et al. 2009; Xie et al. 2009; Yuan and Chen 2010).
Importantly, responses to DSBs can be markedly influenced by cell cycle status (Fig. 4B). Thus, while focal accumulation of DDR factors such as γH2AX, MRN, and MDC1 occurs regardless of the cell cycle stage, focus formation by others—including CtIP, RPA, ATRIP–ATR, BRCA1, and RAD51—takes place effectively only in association with ssDNA formation by DNA end resection in S/G2 cells (Lisby et al. 2004; Bekker-Jensen et al. 2006; Jazayeri et al. 2006; Sartori et al. 2007). In contrast, resection has been reported in G1 at sites of IR-induced breaks in budding yeast, although the rate of resection is higher in S phase and lesions appear to become recognized by the HR machinery only upon entry into S phase (Barlow et al. 2008). Recent work has shown that cell cycle control of DNA resection and HR relies on cell cycle-regulated expression of key factors such as CtIP and BRCA1, and is also governed by cyclin-dependent kinases (Cdks) that phosphorylate evolutionarily conserved residues in CtIP (Huertas et al. 2008; Huertas and Jackson 2009; You and Bailis 2010).
In addition to regulation during interphase, recent observations have shed light on a distinct response to DSBs during mitosis, with only partial activation of the DDR taking place in this cell cycle stage (Fig. 4B). While the molecular basis of this DDR abrogation is still elusive, it appears to operate at the level of ubiquitylating enzymes that control 53BP1 and BRCA1 focus formation (Nelson et al. 2009; Giunta et al. 2010; Nakamura et al. 2010; van Vugt et al. 2010). It is speculated that early DNA damage signaling takes place in mitosis as a priming event for full DDR activation in the following G1 phase (Giunta et al. 2010).
Molecular mechanisms of DDR protein assembly/disassembly at DNA breaks
Analyzing the spatiotemporal dynamics of DDR proteins at DNA breaks has gone beyond being a descriptive approach, and has provided substantial mechanistic insights into how DDR factors assemble and disassemble at damage sites. In this section, we describe how DNA breaks are recognized by DNA damage sensors, how this drives focal recruitment of multiple downstream factors, and how protein modifications regulate these processes.
Direct recognition of DNA breaks by DDR factors
Among the first proteins recruited to DNA breaks are those able to directly recognize DNA breaks. Such factors bind broken DNA in a sequence-independent manner, and thus act as molecular sensors of DNA breaks. For example, PARP-1 and PARP-2 catalytic activity is triggered by them binding directly to SSBs and DSBs (Benjamin and Gill 1980; Ohgushi et al. 1980; de Murcia and Menissier de Murcia 1994; D'Amours et al. 1999). DNA binding is mediated via N-terminal zinc finger domains in PARP-1 (Menissier-de Murcia et al. 1989), and a basic N-terminal domain in PARP-2 (Ame et al. 1999). Similarly, the Ku70–Ku80 heterodimer and the MRN complex are DSB sensors that display direct binding to DNA ends in vitro (Mimori and Hardin 1986; de Jager et al. 2001), and are among the earliest factors to bind to DSBs in vivo (Lisby et al. 2004; Kim et al. 2005). Structural studies have shown that Ku70 and Ku80 form a ring-shaped heterodimer that encircles DNA and threads onto DNA by way of a DSB terminus (Walker et al. 2001), while the MRN complex binds DSBs via a globular head region comprised of MRE11 together with joined RAD50 ATPase domains, with MRE11 dimerization ensuring stable DNA binding, and RAD50 dimerization tethering DNA ends together (de Jager et al. 2001; Williams et al. 2008). As can be anticipated for factors binding the similar DNA structures, Ku competes with the budding yeast MRX complex (and, presumably, also with mammalian MRN) for binding to DSBs (Zhang et al. 2007; Clerici et al. 2008), and Ku also competes with PARP-1 for DNA end binding in mammalian cells (Wang et al. 2006).
Other DDR factors become recruited to processed DSBs via direct binding of evolutionarily conserved OB-fold motifs (Flynn and Zou 2010) to ssDNA generated by DNA end resection. The most extensively studied of these proteins is the ssDNA-binding complex RPA (Wold 1997), which accumulates at resected DSBs in an MRN- and CtIP-dependent manner. RPA in turn directs the recruitment of the Rad9–Rad1–Hus1 (9-1-1) complex, a heterotrimeric ring that is structurally similar to the replicative sliding clamp PCNA (proliferating cell nuclear antigen) and is loaded onto ssDNA–dsDNA junctions by an RFC (replication factor C)-like clamp loader (Parrilla-Castellar et al. 2004). The ssDNA–dsDNA junction is also a substrate for binding by helicases and nucleases involved in DNA break repair (Mimitou and Symington 2009; Bernstein et al. 2010). Recently, two novel heterotrimeric ssDNA-binding complexes have been characterized, termed sensors of ssDNA 1 (SOSS1) and SOSS2, which contain the ssDNA-binding proteins hSSB1 and hSSB2, respectively (Richard et al. 2008; Huang et al. 2009a; Li et al. 2009; Skaar et al. 2009). Unlike RPA, these proteins appear to form DDR foci independently of cell cycle stage in a CtIP-independent manner, and recent work suggests that they could function upstream of MRN (Richard et al. 2010).
DDR protein assembly mediated by protein–protein interactions
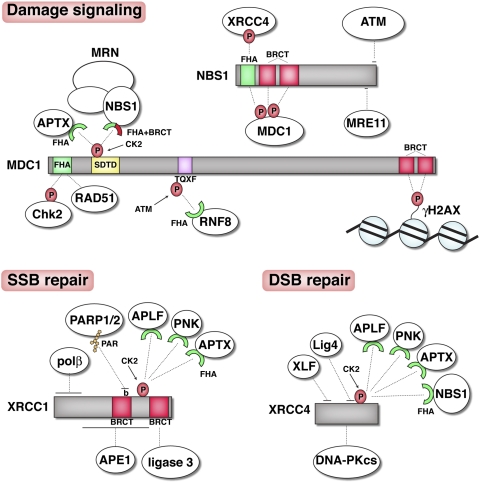
Downstream from proteins that directly sense DNA breaks, other DNA damage signaling and repair factors are sequentially recruited to DNA lesions. The critical importance of protein–protein interactions in building such DDR factor assemblies is well exemplified by MDC1, which serves as a binding platform for DNA damage checkpoint and repair proteins (Fig. 5; Jungmichel and Stucki 2010). Similarly, XRCC1 and XRCC4 act as scaffold proteins, promoting the accumulation of repair factors at SSBs and DSBs, respectively (Fig. 5; Mortusewicz et al. 2008). The pivotal role of protein–protein interactions in the sequential association of repair factors to DDR foci is also illustrated by the recruitment of HR proteins to DSBs (Huen et al. 2010; Moynahan and Jasin 2010). For instance, in human cells, BRCA1 promotes the localization of BRCA2 to damage foci through the BRCA2-binding protein PALB2 (Xia et al. 2006; Sy et al. 2009; Zhang et al. 2009a,b). Through its interaction with RAD51, BRCA2 in turn promotes RAD51 assembly onto ssDNA (Jensen et al. 2010; J Liu et al. 2010; Thorslund et al. 2010).
Figure 5.
Binding platforms at DNA breaks. NBS1, MDC1, XRCC1, and XRCC4 act as binding platforms for the recruitment of other DDR factors to DNA breaks promoting DNA damage signaling and/or repair. Dotted lines indicate protein–protein interactions, while horizontal lines at the end of the dotted lines indicate interacting regions. Some interactions involve post-translational modifications. (P) Phosphorylation; (PAR) PARylation. The red and the green semicircles represent BRCT and FHA domains, respectively. (b) Basic region at the end of the first BRCT domain of XRCC1 that interacts with poly(ADP-ribosyl)ated PARPs.
In the NHEJ pathway, the Ku70–Ku80 heterodimer plays a central role in recruiting other NHEJ components by protein–protein interactions. In particular, Ku recruits DNA-PKcs (Dvir et al. 1992; Gottlieb and Jackson 1993) via a specific interaction between DNA-PKcs and the Ku80 C terminus (Gell and Jackson 1999; Singleton et al. 1999). Ku is also involved in recruiting the downstream NHEJ complex XLF–XRCC4–LigaseIV to DNA ends in mammalian cells (Nick McElhinny et al. 2000; Calsou et al. 2003; Yano et al. 2008), with analogous interactions also taking place in budding yeast (Teo and Jackson 2000; Palmbos et al. 2008).
Regarding the apical checkpoint kinases, ATM is recruited to DSBs by the MRN/X complex (Uziel et al. 2003; Falck et al. 2005; Lee and Paull 2005) by a mechanism that involves direct and evolutionarily conserved interactions between ATM and the NBS1/Xrs2 C terminus, as shown in budding and fission yeast, Xenopus, and human cells (Nakada et al. 2003; Falck et al. 2005; You et al. 2005). In contrast, ATR is recruited to resected DSBs via its interacting partner, ATRIP (Cortez et al. 2001), which binds to RPA-coated ssDNA in human cells, a mechanism that is conserved for yeast orthologs of these factors (Rouse and Jackson 2002; Zou and Elledge 2003; Ball et al. 2007). More generally, it appears that there is a conserved mechanism for apical DDR kinase recruitment to DNA damage sites that involves similar interaction motifs in the C termini of human Ku80-, NBS1-, and ATRIP-mediating interactions with DNA-PKcs, ATM, and ATR, respectively (Falck et al. 2005).
Control of DDR protein recruitment by post-translational modifications
As can be expected for a crucial cellular process, the building of multiprotein assemblies at DNA breaks is tightly controlled. Much of this is achieved by post-translational protein modifications (Fig. 6) that promote the recruitment or dissociation of DDR factors or regulate their residence times at damage sites. While such control mechanisms rely heavily on phosphorylation, a major breakthrough in recent years has been the realization that they are also regulated by other post-translational modifications, including ubiquitylation, sumoylation, methylation, acetylation, and PARylation.
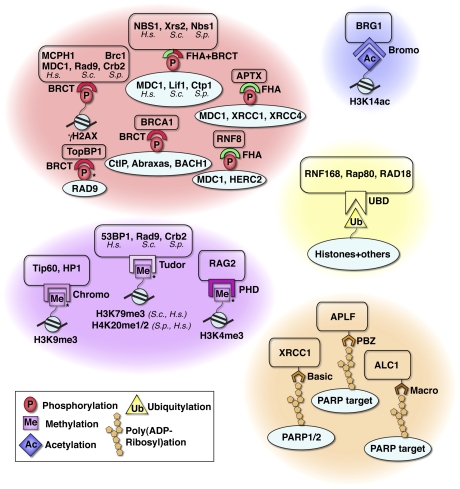
Figure 6.
Specialized binding modules for recognition of post-translational modifications (PTMs) at DNA breaks. The recruitment of DDR proteins to modified histones or other modified proteins at sites of DNA breaks is mediated by specific interactions between the post-translational modification and a dedicated binding module. BRCT and FHA domains, which are represented by red and green semicircles, bind phosphorylated serine or threonine residues; Tudor domains, chromodomains, and PDH finger domains bind methylated histones; bromodomains (Bromo) bind acetylated histones; and UBDs bind ubiquitylated proteins. The PAR-binding domain can take the form of a basic stretch of amino acids (Basic), a PAR-binding zinc finger (PBZ), or a macrodomain (Macro). Note that some of these modules are found as tandem domains and that not all post-translational modifications are damage-induced (asterisk [*] denotes constitutive modifications). The species of the proteins are indicated, unless only human proteins are listed. (H.s.) Homo sapiens; (S.c.) S. cerevisiae, (S.p.) S. pombe.
Key roles for protein phosphorylation in the assembly of DDR foci
The central role of protein kinases and the widespread importance of phosphorylation and dephosphorylation reactions in the DDR are widely acknowledged, with several hundreds of phosphorylated targets having already been identified by mass spectrometry-based screens (Matsuoka et al. 2007; Bennetzen et al. 2010; Bensimon et al. 2010). While some of these phosphorylations likely serve to directly regulate the structure and activity of DDR target proteins, in many cases they act by providing regulated docking sites for other DDR factors. In this regard, DDR proteins frequently display phospho-binding motifs such as BRCT (breast cancer C-terminal) or FHA (Forkhead-associated) domains (Bork et al. 1997; Callebaut and Mornon 1997; Durocher et al. 1999; Mohammad and Yaffe 2009) that play pivotal roles in mediating the phospho-dependent assembly of DDR protein complexes (Fig. 6).
The prime example of a DDR kinase substrate is the histone variant H2AX, which is phosphorylated on a conserved C-terminal serine residue by ATM, ATR, and DNA-PK (Rogakou et al. 1998; Downs et al. 2000; Burma et al. 2001; Ward and Chen 2001; Stiff et al. 2004). This phosphorylation directs the assembly of downstream DDR components, including checkpoint mediators such as human MDC1 and MCPH1 (Microcephalin; also named BRIT1) and their budding and fission yeast orthologs, Rad9 and Crb2, respectively (Nakamura et al. 2004; Lee et al. 2005; Stucki et al. 2005; Hammet et al. 2007; Wood et al. 2007; Sanders et al. 2010; Sofueva et al. 2010). Phosphorylated H2AX also promotes the recruitment of chromatin-modifying complexes, including yeast NuA4, INO80, and SWR1; Drosophila Tip60; and human p400 (Downs et al. 2004; Kusch et al. 2004; Morrison et al. 2004; van Attikum et al. 2004, 2007; Xu et al. 2010). Most prominent among the various factors recruited to γH2AX are MDC1 and its counterparts, which have been shown through structural and biochemical studies to bind directly to phospho-H2AX via their BRCT domains (Lee et al. 2005; Stucki et al. 2005; Hammet et al. 2007; Kilkenny et al. 2008). Notably, MDC1 binding to γH2AX can also be modulated by neighboring modifications within the H2AX C-terminal tail, such as phosphorylation on Tyr 142 by the kinase activity of WSTF (Williams-Beuren syndrome transcription factor), which prevents MDC1 focus formation and instead binds the proapoptotic kinase JNK1 (Cook et al. 2009; Xiao et al. 2009). This regulatory mechanism is thought to govern the balance between DNA damage signaling and cell death.
Once bound to γH2AX, MDC1 in turn acts as a loading platform for other DDR components, with many of these interactions also being phospho-dependent (Jungmichel and Stucki 2010). For instance, ATM-dependent phosphorylation of MDC1 on Thr–Gln–X–Phe (TQXF) motifs creates binding sites for the FHA domain of the ubiquitin E3 ligase RNF8 (Ring finger protein 8), which in turn promotes the focal accumulation of 53BP1 and BRCA1 at DSB sites (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007). Furthermore, constitutive phosphorylation of MDC1 by CK2 (casein kinase 2) on Ser–Asp–Thr–Asp (SDTD) repeat motifs mediates DSB focus formation by MRN (Fig. 5; Chapman and Jackson 2008; Melander et al. 2008; Spycher et al. 2008; Wu et al. 2008). The molecular basis for this MDC1–NBS1 interaction was unveiled recently by structural and biochemical studies that revealed the existence of a compact and evolutionarily conserved phospho-protein interaction module in NBS1 formed by its closely apposed FHA and BRCT domains (Lloyd et al. 2009; Williams et al. 2009; Hari et al. 2010). Interestingly, while this module mediates binding to MDC1 phospho-SDTD repeats in human cells, it binds the phosphorylated CtIP ortholog in fission yeast and interacts with the phosphorylated XRCC4 ortholog in budding yeast (Matsuzaki et al. 2008; Palmbos et al. 2008; Lloyd et al. 2009; Williams et al. 2009). Some additional FHA- and BRCT-mediated interactions involved in the phospho-dependent recruitment and/or retention of DDR factors at DNA breaks are illustrated in Figure 6.
Notably, in some cases, phosphorylation promotes the dissociation of proteins from sites of DNA breaks. The first characterized example of this was provided by the demonstration that DNA-PKcs autophosphorylation causes it to dissociate from Ku (Chan and Lees-Miller 1996; Merkle et al. 2002). Another example of a phospho-dependent dissociation mechanism is provided by the release from chromatin of the transcriptional cofactor KAP1, which depends on its phosphorylation by ATM (Goodarzi et al. 2008). Similarly, ATM and ATR-mediated phosphorylation of Chk1 is linked to Chk1 dissociation from chromatin in response to DNA damage in mammalian cells, presumably to allow Chk1 to access downstream target proteins in the nucleoplasm (Smits et al. 2006). In fission yeast, hyperphosphorylation of Rad9, which is part of the 9-1-1 checkpoint complex, causes it to dissociate from damaged chromatin by loosening its interaction with RPA (Furuya et al. 2010). This phospho-dependent dissociation mechanism seems critical for efficient repair of DNA damage, and thus potentially contributes to the transition from DNA damage signaling to repair. Furthermore, delocalization of the heterochromatin component HP1 from DNA damage sites has been reported to take place upon CK2-dependent phosphorylation within its chromodomain (Ayoub et al. 2008). This disrupts HP1 interactions with heterochromatin marks, and could thereby contribute to relieving the inhibitory effect of chromatin compaction on the DDR. Notably, however, HP1 recruitment to DNA breaks (and other types of DNA lesions) has also been observed, suggesting more active and dynamic roles for HP1 in the DDR (Ayoub et al. 2008, 2009; Dinant and Luijsterburg 2009; Luijsterburg et al. 2009; Zarebski et al. 2009). Future work will be necessary to fully elucidate HP1 dynamics at sites of DNA breaks, its underlying mechanisms, and its biological functions.
Ubiquitin-dependent signaling mediates DDR focus assembly
Ubiquitylation is the process whereby the 76-amino-acid polypeptide ubiquitin is covalently attached to other proteins singly (monoubiquitylation) or in the form of polyubiquitin chains (polyubiquitylation) by the concerted actions of ubiquitin E1, E2, and E3 ligase proteins (Pickart 2001). Recent studies have revealed the critical importance of such events in orchestrating the assembly of DDR proteins at DSB sites in vertebrate cells (Messick and Greenberg 2009; Al-Hakim et al. 2010), although whether similar mechanisms also operate in yeast remains to be determined. For example, various protein–ubiquitin conjugates, including ubiquitylated histones, have been detected at sites of DNA breaks in mammalian cells (Morris and Solomon 2004; Polanowska et al. 2006; Mailand et al. 2007; Doil et al. 2009; Stewart et al. 2009). While it is possible that some of these modifications target the associated protein for proteasome-mediated degradation, most do not, and instead serve as docking platforms for focal DDR–protein assembly. Consistent with ubiquitylation actively taking place at such locations, several ubiquitin ligases have been shown to accumulate at sites of DNA breaks in mammalian cells, including BRCA1, RNF8, RNF168, RAD18, HERC2, and PRC1 (Polycomb-repressive complex 1) (Scully et al. 1997; Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Wang and Elledge 2007; Doil et al. 2009; Huang et al. 2009b; Stewart et al. 2009; Watanabe et al. 2009; Wu et al. 2009a; Bekker-Jensen et al. 2010; Chou et al. 2010).
BRCA1 and its interacting partner, BARD1 (BRCA1-associated RING domain), comprise the first mammalian E3 enzyme shown to function within foci at sites of DNA breaks (Hashizume et al. 2001; Morris and Solomon 2004). While few BRCA1 ubiquitylation targets have been identified so far, one of these is CtIP, with BRCA1-mediated ubiquitylation promoting CtIP binding to damaged chromatin (Yu et al. 2006). Besides work on BRCA1/BARD1, much attention has focused on ubiquitylation of histones H2A, H2B, and H2AX, and possibly other substrates by the ubiquitin E3 ligase RNF8, which promotes the focal recruitment of various DDR factors to sites of DNA strand breaks. These factors include the checkpoint mediators 53BP1, BRCA1, and PTIP (Pax2 transactivation domain interaction protein) (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Wang and Elledge 2007; Doil et al. 2009; Gong et al. 2009; Stewart et al. 2009; Wu et al. 2009a,b); the ubiquitin ligase RAD18, which elicits post-replication repair of damaged DNA (Huang et al. 2009b); and the histone chaperone NPM1 (nucleophosmin) (Koike et al. 2010). In a parallel pathway, H2AX ubiquitylation by PRC1 also contributes to 53BP1 and BRCA1 recruitment to DSBs (Ismail et al. 2010). It will be important to clarify whether and how such PRC1-dependent events cross-talk with the RNF8-dependent pathway. RNF8-dependent ubiquitylation is further amplified by the RNF168 ubiquitin ligase, which is mutated in patients suffering from RIDDLE syndrome (radiosensitivity, immunodeficiency, dysmorphic features, and learning difficulties) (Doil et al. 2009; Pinato et al. 2009; Stewart et al. 2009). Notably, both RNF8 and RNF168 function with the ubiquitin-conjugating E2 enzyme UBC13, and the ubiquitin E3 ligase HERC2 was shown recently to promote RNF8 assembly with UBC13, as opposed to other ubiquitin E2 enzymes (Bekker-Jensen et al. 2010). Additionally, HERC2 stabilizes RNF168 protein levels, thereby regulating ubiquitin-dependent retention of 53BP1 and BRCA1 at DSB sites (Bekker-Jensen et al. 2010).
Analogous to the situation with various DDR–protein phosphorylations, chains of ubiquitin at sites of DNA breaks can be recognized by specific protein domains, termed ubiquitin-binding domains (UBDs) (Fig. 6). While such motifs have been identified in the BRCA1-associated protein RAP80 (receptor-associated protein 80) (H Kim et al. 2007; Sobhian et al. 2007; Sato et al. 2009; Sims and Cohen 2009), the ubiquitin ligase RNF168 (Doil et al. 2009; Pinato et al. 2009, 2011; Stewart et al. 2009), and the RAD18 zinc finger domain (Huang et al. 2009b), the molecular details of ubiquitin-mediated 53BP1 and PTIP recruitment to DNA damage sites remain elusive and probably involve additional post-translational modifications. Indeed, PTIP focus formation is BRCT-dependent (Manke et al. 2003), suggesting a requirement for phosphorylation, while, as described further below, 53BP1 recruitment depends on its ability to bind methylated histone residues (Huyen et al. 2004; Botuyan et al. 2006). In this respect, it is noteworthy that histone ubiquitylation and methylation can act cooperatively, since histone H4 ubiquitylation by the ubiquitin ligase BBAP (B-lymphoma and BAL-associated protein) was shown to stimulate chromatin association of an H4 methylase, thus promoting 53BP1 recruitment to damage sites (Yan et al. 2009). Besides ubiquitylation at DNA damage sites recruiting DDR components to such regions, ubiquitylation of a DDR factor can conversely serve to target this factor to DNA breaks. For instance, when DNA breaks occur during DNA replication, the FA core complex monoubiquitylates the FA proteins FANCD2 and FANCI in a manner that promotes their localization to HR foci (Alpi and Patel 2009; Moldovan and D'Andrea 2009). Ubiquitylated FA proteins in turn recruit the recently identified FAN1 nuclease to sites of DNA repair in a manner dependent on the FAN1 UBD, which might contribute to the resolution of recombination intermediates (Kratz et al. 2010; T Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010).
Sumoylation controls DDR focus assembly
Modification of proteins by SUMO (small ubiquitin-like modifier) (Creton and Jentsch 2010) has also emerged as an important regulator of the DDR (Bergink and Jentsch 2009), as demonstrated in both yeast and mammalian cells. For example, numerous DDR proteins—including human BLM, XRCC4, and RPA, and budding yeast Sgs1, Ku70, and Rad52—have been identified as sumoylation targets (Eladad et al. 2005; Zhao and Blobel 2005; Branzei et al. 2006; Sacher et al. 2006; Yurchenko et al. 2006; Dou et al. 2010). Furthermore, it was established recently that SUMO conjugation takes place at DNA break sites in mammalian cells, and that the ensuing accumulation of sumoylated proteins in these regions then promotes both protein ubiquitylation at DNA break sites and DDR focus formation (Galanty et al. 2009; Morris et al. 2009). More specifically, these studies revealed that sumoylation of 53BP1 and BRCA1 by the PIAS1 and PIAS4 SUMO E3 ligases increases the residence times of 53BP1 and BRCA1 at damage foci, and serves to enhance BRCA1/BARD1 ubiquitin E3 ligase activity. Other DDR proteins identified as sumoylation targets in response to DNA damage include budding yeast Rad52 (Sacher et al. 2006), whose sumoylation regulates its focal accumulation and recombination activity (Sacher et al. 2006; Torres-Rosell et al. 2007; Altmannova et al. 2010), and the human RPA70 subunit, whose sumoylation enhances its binding to Rad51 and facilitates Rad51 recruitment to DNA damage foci (Dou et al. 2010). Notably, sumoylation is also involved in targeting unrepaired broken DNA ends to the nuclear periphery in yeast (Nagai et al. 2008; Kalocsay et al. 2009), although it is not yet known if sumoylation promotes analogous events in mammalian cells.
Protein methylation-dependent events at DDR foci
Besides requiring histone and nonhistone protein phosphorylation and ubiquitylation, the focal recruitment of 53BP1 and its yeast orthologs to DNA breaks involves their tandem Tudor domains recognizing methylated histone residues (Fig. 6). Specifically, fission yeast Crb2 binds H4K20me2 (Sanders et al. 2004; Greeson et al. 2008) and budding yeast Rad9 binds H3K79me3 (Grenon et al. 2007), while human 53BP1 was initially reported to bind H3K79me3 and subsequently H4K20me1/2 (mono or dimethylated), which might be more physiologically relevant (Huyen et al. 2004; Botuyan et al. 2006; Oda et al. 2010). Importantly, it seems that these histone modifications are constitutive (Huyen et al. 2004; Sanders et al. 2004), which is different from H2AX phosphorylation and histone ubiquitylation that are DNA damage-induced. A proposed model is that these methylated histone residues are not normally readily accessible by 53BP1 and its counterparts, but become exposed upon DNA damage-induced changes in chromatin conformation resulting from histone ubiquitylation (Huen and Chen 2010). Nevertheless, this model is still speculative and might need to be revised, particularly in light of recent reports showing that the H4 methylase PR-Set7/Set8 is recruited to DNA damage sites, and that the H4K20me1/2 it directs is induced to some extent after DNA damage induction in human cells (Yan et al. 2009; Oda et al. 2010).
Tudor domains, PHD, and chromodomains can also recognize methylated lysine residues and target DDR factors to sites of DNA breaks (Fig. 6). For example, H3K4me3 binding at antigen receptor genes by the RAG2 recombinase PHD finger is necessary for effective V(D)J recombination, a programmed gene rearrangement that occurs during B-cell and T-cell development (Liu et al. 2007; Matthews et al. 2007). Furthermore, the chromodomain of the chromatin-modifying complex protein Tip60 binds H3K9me3 when HP1 dissociates from damage sites (Sun et al. 2009). In this case, however, binding to methylated histones acts as an allosteric regulator of Tip60 acetyltransferase function rather than directly promoting Tip60 recruitment to DSBs, which is instead mediated by MRN and ATM. Methylation of DDR factors themselves can also regulate their focal recruitment to DNA breaks. For instance, methylation of MRE11 and 53BP1 on GAR (glycine–arginine-rich) motifs promotes their focal accumulation by regulating their DNA-binding activities (Boisvert et al. 2005a,b; Dery et al. 2008).
Protein acetylation regulates DDR factor assembly
The importance of a tightly controlled histone acetylation status near DSBs is underlined by the recruitment of several histone acetyltransferases (Gcn5, Esa1, Hat1, and NuA4) and deacetylases (Rpd3, Sir2, and Hst1) to HO-induced DSBs in budding yeast (Downs et al. 2004; Tamburini and Tyler 2005; Qin and Parthun 2006). Similarly, the recruitment of the Tip60 acetyltransferase and the deacetylase enzymes HDAC1, HDAC2, HDAC4, SIRT1, and SIRT6 has been observed at DSB sites in mammalian cells (Kao et al. 2003; Murr et al. 2006; O'Hagan et al. 2008; Oberdoerffer et al. 2008; Kaidi et al. 2010; Miller et al. 2010), and several studies have suggested roles for histone acetylation in regulating the dynamics of DDR factors in the vicinity of DNA breaks. For instance, SIRT1 binding in the vicinity of a DSB promotes recruitment of NBS1 and RAD51 (Oberdoerffer et al. 2008), while SIRT6-dependent deacetylation of the CtIP protein in response to DSBs stimulates RPA and RAD51 focus formation, thus promoting ATR signaling and DSB repair by HR (Kaidi et al. 2010). On the other hand, H3K56 deacetylation by HDAC1 and HDAC2 regulates binding of NHEJ factors to DSB regions (Miller et al. 2010). Additionally, MOF (males absent on the first)-dependent acetylation of H4K16 is important for IR-induced focus formation of MDC1, 53BP1, and BRCA1 in mammalian cells, possibly through interactions between the histone H4 tail and H2AX stimulating the binding of MDC1 to γH2AX (X Li et al. 2010; Sharma et al. 2010). Along the same lines, H4 acetylation by the MOF-like protein Tip60 at an I-Sce1-induced DNA break stimulates the loading of 53BP1 and BRCA1 at such locations (Murr et al. 2006). Tip60 also acetylates H2AX, which promotes H2AX eviction from damaged chromatin, as shown in both Drosophila and mammalian cells (Kusch et al. 2004; Ikura et al. 2007). Furthermore, the acetylation status of histone proteins in the vicinity of DNA breaks can regulate the assembly of DDR factors indirectly by modulating chromatin compaction, or directly as exemplified by DNA damage-induced acetylation of histone H3 by Gcn5, which then recruits the chromatin remodeling factor BRG1 by creating a binding site for the BRG1 bromodomain (Fig. 6; Lee et al. 2010).
PARylation targets DDR proteins to DNA breaks
The covalent modification of proteins with ADP-ribose polymers, a process known as PARylation, is catalyzed by PARP enzymes (Hakme et al. 2008). Although PARP enzymes do not exist in the commonly used yeast model systems of Saccharomyces cerevisiae and Schizosaccharomyces pombe, PARP proteins comprise a large family broadly distributed among eukaryotes, several members of which have clearly identified DDR functions (Citarelli et al. 2010). Imposed by PARP1 and PARP2 enzymes in response to DNA breaks in mammalian cells, where it has been most thoroughly studied, PARylation is one of the earliest events in the DDR but is quickly removed by the action of PARG (PAR glycohydrolase) (Gagne et al. 2006; Hakme et al. 2008; Krishnakumar and Kraus 2010). Through the use of anti-PAR antibodies, PARylation can be detected locally at sites of DNA breaks in mammalian cells (Tartier et al. 2003), where it promotes recruitment of the DNA break repair factors XRCC1 (El-Khamisy et al. 2003; Okano et al. 2003) and APLF (Bekker-Jensen et al. 2007; Kanno et al. 2007; Ahel et al. 2008; Rulten et al. 2008; Eustermann et al. 2010; GY Li et al. 2010). PARylation at DNA breaks is also required for accrual of the chromatin remodeling factors ALC1 and CHD4 (Ahel et al. 2009; Gottschalk et al. 2009; Chou et al. 2010; Polo et al. 2010), the Polycomb histone-modifying complex (Chou et al. 2010), and the histone variant macroH2A (Timinszky et al. 2009). Current models envision these PAR-dependent recruitment events modulating chromatin structure locally at sites of DNA breaks in order to facilitate DNA damage signaling and/or repair. A contribution of PARylation to the early recruitment of MRN has also been reported (Haince et al. 2008). In each of the above cases, specific domains or motifs on DDR factors mediate their binding to PAR (Fig. 6; Pleschke et al. 2000; Karras et al. 2005; Ahel et al. 2008; Gagne et al. 2008; Rulten et al. 2008; Eustermann et al. 2010; Isogai et al. 2010). Interestingly, PARylation can also promote protein dissociation from DNA damage, as shown for the histone chaperone FACT (facilitates chromatin transcription), which is released from damaged chromatin upon PARylation of its Spt16 subunit (Huang et al. 2006; Heo et al. 2008).
Focus disassembly by reversion of post-translational modifications
Resumption of cell cycle progression following DNA repair requires switching off the DDR (Bartek and Lukas 2007), which involves disassembly of DDR foci. In many cases, this appears to occur mainly by reversing the post-translational modifications that led to focal DDR protein assembly. For example, PARG is responsible for erasing PARylation, as described in the preceding section (Gagne et al. 2006). Additionally, several γH2AX phosphatases have been identified, including Pph3 and PP1 in budding yeast (Keogh et al. 2006; Bazzi et al. 2010) and PP1, PP2A, PP4, PP6, and WIP1 in mammals (Nazarov et al. 2003; Chowdhury et al. 2005, 2008; Nakada et al. 2008; Cha et al. 2010; Douglas et al. 2010; Macurek et al. 2010; Moon et al. 2010). Interestingly, H2AX dephosphorylation, which plays an important role in terminating checkpoint signaling, can take place on chromatin, as reported in human cells (Chowdhury et al. 2005, 2008; Nakada et al. 2008), or after histone eviction from chromatin in yeast (Keogh et al. 2006). Furthermore, the reversal of H2AX phosphorylation also involves Tip60-dependent histone acetylation and subsequent histone eviction from damaged chromatin in Drosophila and human cells (Kusch et al. 2004; Jha et al. 2008).
Acting in a manner analogous to phosphatases, deubiquitylating enzymes (DUBs) have also been implicated in terminating DDR processes. DUBs that function in this regard include USP3 (ubiquitin-specific protease 3), which deubiquitylates histone H2A and H2B (Nicassio et al. 2007), and BRCC36 (BRCA1/BRCA2-containg complex subunit 36), which antagonizes RNF8-dependent ubiquitylation events at DSBs (Shao et al. 2009). Notably, it was shown recently that OTUB1 (OTU domain ubiquitin aldehyde binding 1) also suppresses RNF168-dependent ubiquitylation at DSBs, although in this case this occurs independently of OTUB1 catalytic activity through direct inhibition of the ubiquitin-conjugating enzyme UBC13 (Nakada et al. 2010). In addition, USP16-mediated deubiquitylation of histone H2A was shown to relieve the inhibition of RNA polymerase II transcription at DSBs (Shanbhag et al. 2010), while USP1 counteracts FA protein ubiquitylation (Nijman et al. 2005). Another DUB that regulates the DDR is USP28, which in part operates by stabilizing several DDR factors (Zhang et al. 2006). Although it remains to be demonstrated, it seems likely that desumoylating enzymes will also play important roles in promoting DDR foci disassembly after DNA repair is complete.
Intriguingly, in some cases, DDR factor automodification is coupled to its dissociation from DNA damage sites. For instance, DNA-PKcs autophosphorylation induces a significant conformational change in the protein that elicits its disassembly from Ku and the associated DNA DSB (Chan and Lees-Miller 1996; Merkle et al. 2002; Uematsu et al. 2007; Hammel et al. 2010). Similarly, auto-PARylation of PARP-1 stimulates its dissociation from DNA damage sites, which probably at least partly arise from electrostatic repulsions between DNA and the highly negatively charged PAR chains (Ferro and Olivera 1982; Mortusewicz et al. 2007). Given that acetyltransferases, ubiquitylating enzymes, and sumoylating enzymes are capable of automodifications, it is conceivable that additional examples of automodification-triggered DDR focus disassembly await identification.
Functional importance of DDR foci
The direct binding of DNA breaks by factors such as Ku and MRN is clearly crucial for the DNA repair events that they control. Similarly, the recruitment and activation of the apical DDR kinases ATM, ATR, and DNA-PKcs have well-defined roles at sites of DNA breaks and in DDR focus formation. In contrast, the functional importance of recruiting and activating downstream DDR factors is less well understood and has not been straightforward to decipher. Key reasons for this lack of understanding are the sheer complexity and diversity of downstream DDR responses, and the fact that multiple systems appear to cooperate to control the formation of DDR foci, some of which likely have overlapping and/or compensatory functions. Nevertheless, as summarized below, experimental data and evolutionary conservation indicate that proteins involved in DDR focus assembly and the activities that DDR foci control are of major importance for the maintenance of genome stability.
Evolutionary conservation and biological significance of DDR factor assembly
Many of the proteins and protein modules directing the focal assembly of DDR factors have been highly conserved throughout eukaryotic evolution. A prime example of this is illustrated by the H2AX C-terminal sequence and the tandem BRCT domains that recognize γH2AX, both of which are conserved from yeasts to humans with only a very few known exceptions (Lee et al. 2005; Stucki et al. 2005; Hammet et al. 2007; Bonner et al. 2008; Kilkenny et al. 2008). Similarly, the Tudor domains of human 53BP1 are also found in yeast 53BP1 orthologs, and associate with similar or identical methylated histone residues in all species examined (Huyen et al. 2004; Sanders et al. 2004; Botuyan et al. 2006; Grenon et al. 2007; Greeson et al. 2008; Oda et al. 2010). Other examples of strong evolutionary conservation are provided by the checkpoint kinases, which occur in virtually all eukaryotes studied and appear to rely on highly conserved mechanisms for their recruitment and activation in response to DNA breaks (Falck et al. 2005; Stracker et al. 2009). Although they have been less studied, it seems likely that the functions of chromatin-modifying proteins, acetyltransferases, deacetylases, PARP enzymes, and components of the ubiquitylation and sumoylation systems in controlling DDR focus assembly will also turn out to be highly conserved, at least among metazoans.
Not only are focus-forming DDR factors and their recruitment mechanisms evolutionarily conserved, but defects in such proteins are associated with various pathologies, in both model systems and human genetic conditions. For instance, key regulators of DDR focus formation—such as ATM, ATR, MRE11, NBS1, and RNF168—are mutated in severe genome instability disorders (Jackson and Bartek 2009; Ciccia and Elledge 2010), and MRN, RNF8, and RNF168 have been identified as prominent targets during viral infection (Carson et al. 2009; Lilley et al. 2010). In some cases, however, the ability of certain DDR factors to compensate for one another has hindered evaluation of their functional requirements. A good example of this is provided by mammalian PARP-1 and PARP-2: Single-knockout mice for PARP-1 or PARP-2 are viable, although they display increased genomic instability and hypersensitivity to DNA-damaging agents, while the double knockout is early embryonic-lethal (Wang et al. 1995; de Murcia et al. 1997; Masutani et al. 1999; Menissier de Murcia et al. 2003). Additionally, functional defects caused by DDR factor dysfunction are often not initially obvious but become apparent only under more detailed examination. For instance, while H2AX-deficient mice are viable and show only partial DDR defects, they are growth-retarded and display increased cancer predisposition in the context of oncogenic mutations (Bassing et al. 2002, 2003; Celeste et al. 2002, 2003a), and H2AX-deficient cells show increased error-prone repair of DSBs (Xie et al. 2004). Moreover, while they are viable and overtly normal, mice lacking H2AX, RNF8, or 53BP1, and, to a lesser extent, MDC1, have class switch recombination defects that would have profound impacts on animal viability in natural environments (Petersen et al. 2001; Celeste et al. 2002; Ward et al. 2004; Lou et al. 2006; L Li et al. 2010; Santos et al. 2010).
Structural and regulatory functions of DDR foci
A prominent feature of many proteins that respond to DNA breaks is their ability to concentrate into microscopically detectable foci. It is widely assumed that the focal accumulation of DDR proteins, by increasing their local concentrations, potentiates interactions between them and with the damaged DNA, thus mounting rapid and effective responses to DNA breaks, while at the same time making accidental DDR induction unlikely. Consistent with this idea, studies in yeast and mammalian systems have demonstrated that colocalization of DDR proteins rather than DNA damage per se is critical for DNA damage signaling (Bonilla et al. 2008; Soutoglou and Misteli 2008). Focus-based responses may also have evolved to allow effective DDR events to take place even under conditions in which the individual DDR components are present at fairly low levels in the cell. In this way, initial DDR events would not necessarily rely on other, potentially slower mechanisms to concentrate DDR components in the nucleus, such as by increasing their levels of expression and/or stability. It is also tempting to speculate that increasing the local concentrations of DDR factors at DNA break sites might be particularly critical in chromatin regions otherwise inhibitory to repair, such as heterochromatin (Noon et al. 2010). Another likely key facet of DDR foci is their ability to promote signal amplification through DDR factors spreading along the chromatin surrounding the break. While the importance of such mechanisms might be missed in many experimental settings, where it is common to use high levels of DNA-damaging agents, amplification of DNA damage signaling is likely to be very important in normal biological contexts. This is particularly evident when one considers that DNA damage checkpoints seem to be able to respond to very small numbers of DSBs, with some experimental data indicating that 10–20 DSBs are enough to elicit G2 arrest in human cells (Deckbar et al. 2007), while very few or even a single unrepaired DSB can be sufficient to trigger p53-dependent G1 arrest in human cells (Huang et al. 1996) or cell death in yeast (Bennett et al. 1993).
Along with concentrating DDR factors and amplifying damage signaling, DDR foci may also fulfill more structural roles by stabilizing broken DNA ends and protecting them from excessive degradation and/or illegitimate repair events (Yin et al. 2009). In line with this idea, structural maintenance of chromosome (SMC) proteins, including cohesins, are recruited to chromosomes in response to DSBs in budding yeast, where they facilitate DSB repair by HR by maintaining sister chromatids in close proximity (Strom et al. 2004; Unal et al. 2004, 2007; De Piccoli et al. 2006; Lindroos et al. 2006). Tethering of DNA ends together can also be achieved through the dimerization or oligomerization of DDR proteins, as shown for the MRN complex (de Jager et al. 2001; Moreno-Herrero et al. 2005) and 53BP1, which promotes long-range joining of deprotected telomeres and V(D)J recombination intermediates in mammalian cells (Difilippantonio et al. 2008; Dimitrova et al. 2008). Aside from such structural functions, DDR foci can also be viewed as a temporary storage site for some DDR factors, as illustrated in mitotic cells, where they mark sites of DNA damage for full DDR activation only in the following G1 phase (Giunta et al. 2010). Another example of a specialized DSB storage focus is the telomere-associated Shelterin complex, which sequesters chromosomal termini and prevents them from being recognized as damaged substrates by forming a protective complex at telomeres after their replication (Verdun et al. 2005). Another likely regulatory function for DDR foci is to contribute to the proper coordination of DNA damage signaling and repair with other DNA metabolic activities by inhibiting replication and transcription. In this regard, DNA methylation and histone modifications—including deacetylation, methylation, and ubiquitylation at sites of DNA breaks—have been proposed to contribute to silencing of damaged chromatin (O'Hagan et al. 2008; Shanbhag et al. 2010).
Conclusions and future directions
The focal accumulation of DDR proteins at sites of DNA breaks was first described >15 years ago, and the molecular mechanisms of focus formation have been the subject of intense investigation since then. As a result, we have identified many of the key protein players in these events and have a growing understanding of their biochemical and physiological functions. In particular, considerable advances have been made in elucidating the hierarchical and functional organization of DDR factor assemblies at DNA break sites, which we now know represent intricate protein and nucleic acid networks precisely coordinated in space and time. Furthermore, recent studies on the dynamics of DDR foci have revealed the prominent roles of combinatorial protein modifications in ensuring rapid, reversible, and fine-tuned regulation of DDR factor recruitment to and dissociation from sites of DNA breaks.
Nevertheless, there are many remaining challenges that will surely attract the attentions of DDR researchers for many years to come. First, while many DDR components are known, the frequent reporting of additional factors in the literature suggests that many more await identification. In this regard, ongoing genetic, proteomic, and siRNA-based screens seem set to provide many additional DDR components and regulators whose functions must then be defined. Another major challenge for the future will be to understand not only how the various post-translational modifications are imposed on DDR factors and which binding modules they specifically associate with, but also how and when they get erased to allow termination of the DDR. It will also be of great interest to further explore likely cross-talks—cooperative or antagonistic—between these post-translational modifications in coordinating the assembly/disassembly of DDR factors at DNA breaks, which we are only beginning to uncover. Further exciting challenges include determining how DNA break signaling and repair are influenced by cell physiology and disease, and how they may be differentially regulated in different cell types and tissues.
As with many areas of biological research, recent progress in the DDR field has been highly dependent on the development of new methodologies and equipment, including cutting-edge microscopy techniques. Thus, it seems that further progress in the field is also likely to be triggered by further technical improvements, such as new superresolution microscopes and associated computational tools that will allow hitherto unattainable resolution of DDR foci in both three-dimensional space and time (Schermelleh et al. 2008). Complementing such work will be the development of further biochemical and structural analyses of DDR components, together with siRNA-based and genetic manipulation techniques in cells and model organisms.
Finally, we emphasize that DNA break-associated proteins and the foci that they assemble into are of considerable medical importance, with defects in them being associated with various pathologies, particularly cancer. In this regard, it is notable that the immunodetection of γH2AX foci, which indirectly measure DSB formation and repair, is showing promise as a sensitive diagnostic tool to detect cancer cells and also monitor cancer progression and assess responses to treatment (Bonner et al. 2008; Mah et al. 2010). Moreover, the existence of many druggable protein targets in DNA break-associated events is providing exciting opportunities for developing new therapeutic agents that, by exploiting differences between normal cells and cancer cells, have the potential to markedly improve cancer management.
Acknowledgments
We apologize to those whose work we inadvertently did not cite or elected not to cite in order to keep this review to a reasonable length. We thank D. Durocher, R. Scully, K. Miller, and K. Dry for critical reading of the manuscript. Research in the Jackson laboratory is supported by grants from Cancer Research UK, the European Union (GENICA HEALTH-F2-2007-201630), the Wellcome Trust, and the BBSRC, and core infrastructure is provided by funding from Cancer Research UK and the Wellcome Trust. S.P.J. receives his salary from the University of Cambridge with a supplement from Cancer Research UK, while S.E.P. was funded by the Human Frontier Science Program Organization, followed by a fellowship from Association pour la Recherche sur le Cancer.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2021311.
References
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC 2008. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451: 81–85 [DOI] [PubMed] [Google Scholar]
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. 2009. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325: 1240–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hakim A, Escribano-Diaz C, Landry MC, O'Donnell L, Panier S, Szilard RK, Durocher D 2010. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst) 9: 1229–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpi AF, Patel KJ 2009. Monoubiquitylation in the Fanconi anemia DNA damage response pathway. DNA Repair (Amst) 8: 430–435 [DOI] [PubMed] [Google Scholar]
- Altmannova V, Eckert-Boulet N, Arneric M, Kolesar P, Chaloupkova R, Damborsky J, Sung P, Zhao X, Lisby M, Krejci L 2010. Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res 38: 4708–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G 1999. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem 274: 17860–17868 [DOI] [PubMed] [Google Scholar]
- Amitani I, Liu B, Dombrowski CC, Baskin RJ, Kowalczykowski SC 2010. Watching individual proteins acting on single molecules of DNA. Methods Enzymol 472: 261–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley T, Plug AW, Xu J, Solari AJ, Reddy G, Golub EI, Ward DC 1995. Dynamic changes in Rad51 distribution on chromatin during meiosis in male and female vertebrates. Chromosoma 104: 19–28 [DOI] [PubMed] [Google Scholar]
- Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR 2008. HP1-β mobilization promotes chromatin changes that initiate the DNA damage response. Nature 453: 682–686 [DOI] [PubMed] [Google Scholar]
- Ayoub N, Jeyasekharan AD, Venkitaraman AR 2009. Mobilization and recruitment of HP1: a bimodal response to DNA breakage. Cell Cycle 8: 2945–2950 [PubMed] [Google Scholar]
- Ball HL, Ehrhardt MR, Mordes DA, Glick GG, Chazin WJ, Cortez D 2007. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol Cell Biol 27: 3367–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Lisby M, Rothstein R 2008. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell 30: 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Lukas J 2007. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol 19: 238–245 [DOI] [PubMed] [Google Scholar]
- Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, et al. 2002. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci 99: 8173–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW 2003. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114: 359–370 [DOI] [PubMed] [Google Scholar]
- Bazzi M, Mantiero D, Trovesi C, Lucchini G, Longhese MP 2010. Dephosphorylation of γH2A by Glc7/protein phosphatase 1 promotes recovery from inhibition of DNA replication. Mol Cell Biol 30: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol 173: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Fugger K, Danielsen JR, Gromova I, Sehested M, Celis J, Bartek J, Lukas J, Mailand N 2007. Human Xip1 (C2orf13) is a novel regulator of cellular responses to DNA strand breaks. J Biol Chem 282: 19638–19643 [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, Bartek J, Lukas J, Mailand N 2010. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 12: 80–86 [DOI] [PubMed] [Google Scholar]
- Benjamin RC, Gill DM 1980. Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J Biol Chem 255: 10502–10508 [PubMed] [Google Scholar]
- Bennett CB, Lewis AL, Baldwin KK, Resnick MA 1993. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc Natl Acad Sci 90: 5613–5617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS 2010. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics 9: 1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimon A, Schmidt A, Ziv Y, Elkon R, Wang S-Y, Chen DJ, Aebersold R, Shiloh Y 2010. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci Signal 3: rs3 doi: 10.1126/scisignal.20011034 [DOI] [PubMed] [Google Scholar]
- Bergink S, Jentsch S 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458: 461–467 [DOI] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ Jr, Kastan MB 2008. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nat Protoc 3: 915–922 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Hake SB 2006. The nucleosome: a little variation goes a long way. Biochem Cell Biol 84: 505–517 [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Rothstein R 2009. At loose ends: resecting a double-strand break. Cell 137: 807–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Gangloff S, Rothstein R 2010. The RecQ DNA helicases in DNA repair. Annu Rev Genet 44: 393–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Hendzel MJ, Masson JY, Richard S 2005a. Methylation of MRE11 regulates its nuclear compartmentalization. Cell Cycle 4: 981–989 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Rhie A, Richard S, Doherty AJ 2005b. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle 4: 1834–1841 [DOI] [PubMed] [Google Scholar]
- Bonilla CY, Melo JA, Toczyski DP 2008. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell 30: 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y 2008. γH2AX and cancer. Nat Rev Cancer 8: 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV 1997. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J 11: 68–76 [PubMed] [Google Scholar]
- Botchway SW, Reynolds P, Parker AW, O'Neill P 2010. Use of near infrared femtosecond lasers as sub-micron radiation microbeam for cell DNA damage and repair studies. Mutat Res 704: 38–44 [DOI] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M 2006. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522 [DOI] [PubMed] [Google Scholar]
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 276: 42462–42467 [DOI] [PubMed] [Google Scholar]
- Caldecott KW 2008. Single-strand break repair and genetic disease. Nat Rev Genet 9: 619–631 [DOI] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP 1997. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett 400: 25–30 [DOI] [PubMed] [Google Scholar]
- Calsou P, Delteil C, Frit P, Drouet J, Salles B 2003. Coordinated assembly of Ku and p460 subunits of the DNA-dependent protein kinase on DNA ends is necessary for XRCC4-ligase IV recruitment. J Mol Biol 326: 93–103 [DOI] [PubMed] [Google Scholar]
- Carson CT, Orazio NI, Lee DV, Suh J, Bekker-Jensen S, Araujo FD, Lakdawala SS, Lilley CE, Bartek J, Lukas J, et al. 2009. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J 28: 652–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, et al. 2002. Genomic instability in mice lacking histone H2AX. Science 296: 922–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A 2003a. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114: 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A 2003b. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5: 675–679 [DOI] [PubMed] [Google Scholar]
- Cha H, Lowe JM, Li H, Lee JS, Belova GI, Bulavin DV, Fornace AJ Jr 2010. Wip1 directly dephosphorylates γ-H2AX and attenuates the DNA damage response. Cancer Res 70: 4112–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Lane TF 2005. BRCA1 associates with the inactive X chromosome in late S-phase, coupled with transient H2AX phosphorylation. Chromosoma 114: 432–439 [DOI] [PubMed] [Google Scholar]
- Chai B, Huang J, Cairns BR, Laurent BC 2005. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev 19: 1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Lees-Miller SP 1996. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem 271: 8936–8941 [DOI] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP 2008. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep 9: 795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK 2008. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ 2010. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci 107: 18475–18480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J 2005. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell 20: 801–809 [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Xu X, Zhong X, Ahmed F, Zhong J, Liao J, Dykxhoorn DM, Weinstock DM, Pfeifer GP, Lieberman J 2008. A PP4–phosphatase complex dephosphorylates γ-H2AX generated during DNA replication. Mol Cell 31: 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ 2010. The DNA damage response: making it safe to play with knives. Mol Cell 40: 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citarelli M, Teotia S, Lamb RS 2010. Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC Evol Biol 10: 308 doi: 10.1186/1471-2148-10-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP 2008. The Yku70–Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep 9: 810–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG 2009. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 458: 591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294: 1713–1716 [DOI] [PubMed] [Google Scholar]
- Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ 2007. γH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS ONE 2: e1057 doi: 10.1371/journal.pone.001057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creton S, Jentsch S 2010. SnapShot: The SUMO system. Cell 143: 848. [DOI] [PubMed] [Google Scholar]
- Cuozzo C, Porcellini A, Angrisano T, Morano A, Lee B, Di Pardo A, Messina S, Iuliano R, Fusco A, Santillo MR, et al. 2007. DNA damage, homology-directed repair, and DNA methylation. PLoS Genet 3: e110 doi: 10.1371/journal.pgen.0030110 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194–198 [DOI] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342: 249–268 [PMC free article] [PubMed] [Google Scholar]
- de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell 8: 1129–1135 [DOI] [PubMed] [Google Scholar]
- de Murcia G, Menissier de Murcia J 1994. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem Sci 19: 172–176 [DOI] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, et al. 1997. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci 94: 7303–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckbar D, Birraux J, Krempler A, Tchouandong L, Beucher A, Walker S, Stiff T, Jeggo P, Lobrich M 2007. Chromosome breakage after G2 checkpoint release. J Cell Biol 176: 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piccoli G, Cortes-Ledesma F, Ira G, Torres-Rosell J, Uhle S, Farmer S, Hwang JY, Machin F, Ceschia A, McAleenan A, et al. 2006. Smc5–Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat Cell Biol 8: 1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dery U, Coulombe Y, Rodrigue A, Stasiak A, Richard S, Masson JY 2008. A glycine–arginine domain in control of the human MRE11 DNA repair protein. Mol Cell Biol 28: 3058–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, et al. 2008. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T 2008. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant C, Luijsterburg MS 2009. The emerging role of HP1 in the DNA damage response. Mol Cell Biol 29: 6335–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446 [DOI] [PubMed] [Google Scholar]
- Dou H, Huang C, Singh M, Carpenter PB, Yeh ET 2010. Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol Cell 39: 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Zhong J, Ye R, Moorhead GB, Xu X, Lees-Miller SP 2010. Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates γ-H2AX. Mol Cell Biol 30: 1368–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Lowndes NF, Jackson SP 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell 16: 979–990 [DOI] [PubMed] [Google Scholar]
- Drouet J, Delteil C, Lefrancois J, Concannon P, Salles B, Calsou P 2005. DNA-dependent protein kinase and XRCC4-DNA ligase IV mobilization in the cell in response to DNA double strand breaks. J Biol Chem 280: 7060–7069 [DOI] [PubMed] [Google Scholar]
- Durocher D, Henckel J, Fersht AR, Jackson SP 1999. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell 4: 387–394 [DOI] [PubMed] [Google Scholar]
- Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS 1992. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci 89: 11920–11924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan WS, Yoo S 1998. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res 26: 1551–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eladad S, Ye TZ, Hu P, Leversha M, Beresten S, Matunis MJ, Ellis NA 2005. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum Mol Genet 14: 1351–1365 [DOI] [PubMed] [Google Scholar]
- El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW 2003. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res 31: 5526–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, Schubert T, Marth C, Langst G, Rippe K 2010. Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites. Proc Natl Acad Sci 107: 19873–19878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, Houtsmuller AB, Kanaar R 2006. Analysis of DNA recombination and repair proteins in living cells by photobleaching microscopy. Methods Enzymol 408: 463–485 [DOI] [PubMed] [Google Scholar]
- Eustermann S, Brockmann C, Mehrotra PV, Yang JC, Loakes D, West SC, Ahel I, Neuhaus D 2010. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose). Nat Struct Mol Biol 17: 241–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434: 605–611 [DOI] [PubMed] [Google Scholar]
- Fanti L, Pimpinelli S 2008. HP1: a functionally multifaceted protein. Curr Opin Genet Dev 18: 169–174 [DOI] [PubMed] [Google Scholar]
- Ferro AM, Olivera BM 1982. Poly(ADP-ribosylation) in vitro. Reaction parameters and enzyme mechanism. J Biol Chem 257: 7808–7813 [PubMed] [Google Scholar]
- FitzGerald JE, Grenon M, Lowndes NF 2009. 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans 37: 897–904 [DOI] [PubMed] [Google Scholar]
- Flynn RL, Zou L 2010. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit Rev Biochem Mol Biol 45: 266–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T 2006. DNA repair and mutagenesis, 2nd ed ASM Press, New York [Google Scholar]
- Furuya K, Miyabe I, Tsutsui Y, Paderi F, Kakusho N, Masai H, Niki H, Carr AM 2010. DDK phosphorylates checkpoint clamp component Rad9 and promotes its release from damaged chromatin. Mol Cell 40: 606–618 [DOI] [PubMed] [Google Scholar]
- Gagne JP, Hendzel MJ, Droit A, Poirier GG 2006. The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr Opin Cell Biol 18: 145–151 [DOI] [PubMed] [Google Scholar]
- Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG 2008. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res 36: 6959–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP 2009. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462: 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell D, Jackson SP 1999. Mapping of protein–protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res 27: 3494–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta S, Belotserkovskaya R, Jackson SP 2010. DNA damage signaling in response to double-strand breaks during mitosis. J Cell Biol 190: 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Cho YW, Kim JE, Ge K, Chen J 2009. Accumulation of Pax2 transactivation domain interaction protein (PTIP) at sites of DNA breaks via RNF8-dependent pathway is required for cell survival after DNA damage. J Biol Chem 284: 7284–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA 2008. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 31: 167–177 [DOI] [PubMed] [Google Scholar]
- Gottlieb TM, Jackson SP 1993. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72: 131–142 [DOI] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, et al. 2009. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci 106: 13770–13774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greeson NT, Sengupta R, Arida AR, Jenuwein T, Sanders SL 2008. Di-methyl H4 lysine 20 targets the checkpoint protein Crb2 to sites of DNA damage. J Biol Chem 283: 33168–33174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenon M, Costelloe T, Jimeno S, O'Shaughnessy A, Fitzgerald J, Zgheib O, Degerth L, Lowndes NF 2007. Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast 24: 105–119 [DOI] [PubMed] [Google Scholar]
- Haaf T, Golub EI, Reddy G, Radding CM, Ward DC 1995. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci 92: 2298–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG 2008. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem 283: 1197–1208 [DOI] [PubMed] [Google Scholar]
- Hakme A, Wong HK, Dantzer F, Schreiber V 2008. The expanding field of poly(ADP-ribosyl)ation reactions. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep 9: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, et al. 2010. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem 285: 1414–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammet A, Magill C, Heierhorst J, Jackson SP 2007. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep 8: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari FJ, Spycher C, Jungmichel S, Pavic L, Stucki M 2010. A divalent FHA/BRCT-binding mechanism couples the MRE11–RAD50–NBS1 complex to damaged chromatin. EMBO Rep 11: 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ 2007. The DNA damage response: ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Harrison JC, Haber JE 2006. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 40: 209–235 [DOI] [PubMed] [Google Scholar]
- Hartlerode AJ, Scully R 2009. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J 423: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T 2001. The RING heterodimer BRCA1–BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 276: 14537–14540 [DOI] [PubMed] [Google Scholar]
- Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W 2008. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell 30: 86–97 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH 2001. Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374 [DOI] [PubMed] [Google Scholar]
- Huang LC, Clarkin KC, Wahl GM 1996. Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G1 arrest. Proc Natl Acad Sci 93: 4827–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Chen WH, Chang YL, Wang HT, Chuang WT, Lee SC 2006. Modulation of nucleosome-binding activity of FACT by poly(ADP-ribosyl)ation. Nucleic Acids Res 34: 2398–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gong Z, Ghosal G, Chen J 2009a. SOSS complexes participate in the maintenance of genomic stability. Mol Cell 35: 384–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J 2009b. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol 11: 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Chen J 2010. Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem Sci 35: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, Chen J 2010. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol 11: 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P 2010. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol 17: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284: 9558–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432: 406–411 [DOI] [PubMed] [Google Scholar]
- Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G 2010. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J 29: 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, et al. 2007. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol 27: 7028–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, Andrin C, McDonald D, Hendzel MJ 2010. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol 191: 45–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai S, Kanno S, Ariyoshi M, Tochio H, Ito Y, Yasui A, Shirakawa M 2010. Solution structure of a zinc-finger domain that binds to poly-ADP-ribose. Genes Cells 15: 101–110 [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob B, Rudolph JH, Gueven N, Lavin MF, Taucher-Scholz G 2005. Live cell imaging of heavy-ion-induced radiation responses by beamline microscopy. Radiat Res 163: 681–690 [DOI] [PubMed] [Google Scholar]
- Jakob B, Splinter J, Taucher-Scholz G 2009. Positional stability of damaged chromatin domains along radiation tracks in mammalian cells. Radiat Res 171: 405–418 [DOI] [PubMed] [Google Scholar]
- Jasin M 1996. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet 12: 224–228 [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC 2010. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467: 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Shibata E, Dutta A 2008. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol 28: 2690–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S, Stucki M 2010. MDC1: The art of keeping things in focus. Chromosoma 119: 337–349 [DOI] [PubMed] [Google Scholar]
- Kaidi A, Weinert BT, Choudhary C, Jackson SP 2010. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science 329: 1348–1353 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kalocsay M, Hiller NJ, Jentsch S 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell 33: 335–343 [DOI] [PubMed] [Google Scholar]
- Kanno S, Kuzuoka H, Sasao S, Hong Z, Lan L, Nakajima S, Yasui A 2007. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J 26: 2094–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao GD, McKenna WG, Guenther MG, Muschel RJ, Lazar MA, Yen TJ 2003. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J Cell Biol 160: 1017–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG 2005. The macro domain is an ADP-ribose binding module. EMBO J 24: 1911–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwaba S, Kitahashi K, Watanabe T, Onoda F, Ohtsu M, Murakami Y 2010. The mammalian INO80 complex is recruited to DNA damage sites in an ARP8 dependent manner. Biochem Biophys Res Commun 402: 619–625 [DOI] [PubMed] [Google Scholar]
- Kent NA, Chambers AL, Downs JA 2007. Dual chromatin remodeling roles for RSC during DNA double strand break induction and repair at the yeast MAT locus. J Biol Chem 282: 27693–27701 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, et al. 2006. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature 439: 497–501 [DOI] [PubMed] [Google Scholar]
- Kilkenny ML, Dore AS, Roe SM, Nestoras K, Ho JC, Watts FZ, Pearl LH 2008. Structural and functional analysis of the Crb2-BRCT2 domain reveals distinct roles in checkpoint signaling and DNA damage repair. Genes Dev 22: 2034–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K 2005. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J Cell Biol 170: 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X 2007. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316: 1202–1205 [DOI] [PubMed] [Google Scholar]
- Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE 2007. Heterochromatin is refractory to γ-H2AX modification in yeast and mammals. J Cell Biol 178: 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike A, Nishikawa H, Wu W, Okada Y, Venkitaraman AR, Ohta T 2010. Recruitment of phosphorylated NPM1 to sites of DNA damage through RNF8-dependent ubiquitin conjugates. Cancer Res 70: 6746–6756 [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD 1977. Structure of chromatin. Annu Rev Biochem 46: 931–954 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavo E, Sartori AA, Hengartner MO, Jiricny J 2010. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 142: 77–88 [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL 2010. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell 39: 8–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, McNally JG, Bazett-Jones DP, Nussenzweig A 2006. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol 172: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M, Crouch EE, Orlov M, Montano C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R 2007. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 447: 730–734 [DOI] [PubMed] [Google Scholar]
- Kulis M, Esteller M 2010. DNA methylation and cancer. Adv Genet 70: 27–56 [DOI] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR III, Abmayr SM, Washburn MP, Workman JL 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306: 2084–2087 [DOI] [PubMed] [Google Scholar]
- Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Danielsen JM, Menard P, Sand JC, Stucki M, et al. 2010. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol 190: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF 2007. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 26: 7749–7758 [DOI] [PubMed] [Google Scholar]
- Lazzaro F, Giannattasio M, Puddu F, Granata M, Pellicioli A, Plevani P, Muzi-Falconi M 2009. Checkpoint mechanisms at the intersection between DNA damage and repair. DNA Repair (Amst) 8: 1055–1067 [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT 2005. ATM activation by DNA double-strand breaks through the Mre11–Rad50–Nbs1 complex. Science 308: 551–554 [DOI] [PubMed] [Google Scholar]
- Lee MS, Edwards RA, Thede GL, Glover JN 2005. Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the γ-H2AX histone tail. J Biol Chem 280: 32053–32056 [DOI] [PubMed] [Google Scholar]
- Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J 2010. A cooperative activation loop among SWI/SNF, γ-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J 29: 1434–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bolderson E, Kumar R, Muniandy PA, Xue Y, Richard DJ, Seidman M, Pandita TK, Khanna KK, Wang W 2009. HSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J Biol Chem 284: 23525–23531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GY, McCulloch RD, Fenton AL, Cheung M, Meng L, Ikura M, Koch CA 2010. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc Natl Acad Sci 107: 9129–9134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Halaby MJ, Hakem A, Cardoso R, El Ghamrasni S, Harding S, Chan N, Bristow R, Sanchez O, Durocher D, et al. 2010. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J Exp Med 207: 983–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Corsa CA, Pan PW, Wu L, Ferguson D, Yu X, Min J, Dou Y 2010. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol 30: 5335–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79: 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Wilson TE 2010. SnapShot: Nonhomologous DNA end joining (NHEJ). Cell 142: 496–496 e491 [DOI] [PubMed] [Google Scholar]
- Lilley CE, Chaurushiya MS, Boutell C, Landry S, Suh J, Panier S, Everett RD, Stewart GS, Durocher D, Weitzman MD 2010. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J 29: 943–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli CL, Ward JF 1993. A new method for introducing double-strand breaks into cellular DNA. Radiat Res 134: 160–169 [PubMed] [Google Scholar]
- Lindahl T, Barnes DE 2000. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol 65: 127–133 [DOI] [PubMed] [Google Scholar]
- Lindroos HB, Strom L, Itoh T, Katou Y, Shirahige K, Sjogren C 2006. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol Cell 22: 755–767 [DOI] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R 2003. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol 5: 572–577 [DOI] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S 2007. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen–receptor–gene rearrangement. Immunity 27: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD 2010. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol 17: 1260–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J 2010. FAN1 acts with FANCI–FANCD2 to promote DNA interstrand cross-link repair. Science 329: 693–696 [DOI] [PubMed] [Google Scholar]
- Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ 2009. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell 139: 100–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Bonetti D, Guerini I, Manfrini N, Clerici M 2009. DNA double-strand breaks in meiosis: checking their formation, processing and repair. DNA Repair (Amst) 8: 1127–1138 [DOI] [PubMed] [Google Scholar]
- Longhese MP, Bonetti D, Manfrini N, Clerici M 2010. Mechanisms and regulation of DNA end resection. EMBO J 29: 2864–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. 2006. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell 21: 187–200 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, et al. 2009. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol 185: 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J 2003. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 5: 255–260 [DOI] [PubMed] [Google Scholar]
- Lukas C, Bartek J, Lukas J 2005. Imaging of protein movement induced by chromosomal breakage: tiny ‘local’ lesions pose great ‘global’ challenges. Chromosoma 114: 146–154 [DOI] [PubMed] [Google Scholar]
- MacKay C, Declais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. 2010. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 142: 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macurek L, Lindqvist A, Voets O, Kool J, Vos HR, Medema RH 2010. Wip1 phosphatase is associated with chromatin and dephosphorylates γH2AX to promote checkpoint inhibition. Oncogene 29: 2281–2291 [DOI] [PubMed] [Google Scholar]
- Mah LJ, El-Osta A, Karagiannis TC 2010. γH2AX as a molecular marker of aging and disease. Epigenetics 5: 129–136 [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB 2003. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636–639 [DOI] [PubMed] [Google Scholar]
- Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, et al. 2006. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci 103: 18597–18602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Monsen KJ, Nelms BE, Petrini JH 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol 17: 6087–6096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massip L, Caron P, Iacovoni JS, Trouche D, Legube G 2010. Deciphering the chromatin landscape induced around DNA double strand breaks. Cell Cycle 9: 2963–2972 [DOI] [PubMed] [Google Scholar]
- Masutani M, Nozaki T, Nishiyama E, Shimokawa T, Tachi Y, Suzuki H, Nakagama H, Wakabayashi K, Sugimura T 1999. Function of poly(ADP-ribose) polymerase in response to DNA damage: gene-disruption study in mice. Mol Cell Biochem 193: 149–152 [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Matsuzaki K, Shinohara A, Shinohara M 2008. Forkhead-associated domain of yeast Xrs2, a homolog of human Nbs1, promotes nonhomologous end joining through interaction with a ligase IV partner protein, Lif1. Genetics 179: 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. 2007. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450: 1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazon G, Mimitou EP, Symington LS 2010. SnapShot: Homologous recombination in DNA double-strand break repair. Cell 142: 646, 646 e641 [DOI] [PubMed] [Google Scholar]
- McBryant SJ, Adams VH, Hansen JC 2006. Chromatin architectural proteins. Chromosome Res 14: 39–51 [DOI] [PubMed] [Google Scholar]
- McVey M, Lee SE 2008. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet 24: 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier A, Fiegler H, Munoz P, Ellis P, Rigler D, Langford C, Blasco MA, Carter N, Jackson SP 2007. Spreading of mammalian DNA-damage response factors studied by ChIP–chip at damaged telomeres. EMBO J 26: 2707–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J 2008. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol 181: 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menissier-de Murcia J, Molinete M, Gradwohl G, Simonin F, de Murcia G 1989. Zinc-binding domain of poly(ADP-ribose)polymerase participates in the recognition of single strand breaks on DNA. J Mol Biol 210: 229–233 [DOI] [PubMed] [Google Scholar]
- Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, et al. 2003. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J 22: 2255–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle D, Douglas P, Moorhead GB, Leonenko Z, Yu Y, Cramb D, Bazett-Jones DP, Lees-Miller SP 2002. The DNA-dependent protein kinase interacts with DNA to form a protein–DNA complex that is disrupted by phosphorylation. Biochemistry 41: 12706–12714 [DOI] [PubMed] [Google Scholar]
- Messick TE, Greenberg RA 2009. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol 187: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP 2010. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 17: 1144–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2009. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci 34: 264–272 [DOI] [PubMed] [Google Scholar]
- Mimori T, Hardin JA 1986. Mechanism of interaction between Ku protein and DNA. J Biol Chem 261: 10375–10379 [PubMed] [Google Scholar]
- Misteli T, Soutoglou E 2009. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 10: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moggs JG, Grandi P, Quivy JP, Jonsson ZO, Hubscher U, Becker PB, Almouzni G 2000. A CAF-1–PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol 20: 1206–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad DH, Yaffe MB 2009. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst) 8: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, D'Andrea AD 2009. How the fanconi anemia pathway guards the genome. Annu Rev Genet 43: 223–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SH, Lin L, Zhang X, Nguyen TA, Darlington Y, Waldman AS, Lu X, Donehower LA 2010. Wild-type p53-induced phosphatase 1 dephosphorylates histone variant γ-H2AX and suppresses DNA double strand break repair. J Biol Chem 285: 12935–12947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C 2005. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature 437: 440–443 [DOI] [PubMed] [Google Scholar]
- Morris JR, Solomon E 2004. BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet 13: 807–817 [DOI] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. 2009. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462: 886–890 [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X 2004. INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119: 767–775 [DOI] [PubMed] [Google Scholar]
- Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H 2005. Recruitment of DNA methyltransferase I to DNA repair sites. Proc Natl Acad Sci 102: 8905–8909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortusewicz O, Ame JC, Schreiber V, Leonhardt H 2007. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res 35: 7665–7675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortusewicz O, Leonhardt H, Cardoso MC 2008. Spatiotemporal dynamics of regulatory protein recruitment at DNA damage sites. J Cell Biochem 104: 1562–1569 [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 11: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z 2006. Histone acetylation by Trrap–Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol 8: 91–99 [DOI] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Soutoglou E 2009. DNA repair: easy to visualize, difficult to elucidate. Trends Cell Biol 19: 617–629 [DOI] [PubMed] [Google Scholar]
- Nakada D, Matsumoto K, Sugimoto K 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev 17: 1957–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada S, Chen GI, Gingras AC, Durocher D 2008. PP4 is a γH2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep 9: 1019–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, O'Donnell L, Kumakubo A, Munro M, Sicheri F, et al. 2010. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466: 941–946 [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Du LL, Redon C, Russell P 2004. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol 24: 6215–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura AJ, Rao VA, Pommier Y, Bonner WM 2010. The complexity of phosphorylated H2AX foci formation and DNA repair assembly at DNA double-strand breaks. Cell Cycle 9: 389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarov IB, Smirnova AN, Krutilina RI, Svetlova MP, Solovjeva LV, Nikiforov AA, Oei SL, Zalenskaya IA, Yau PM, Bradbury EM, et al. 2003. Dephosphorylation of histone γ-H2AX during repair of DNA double-strand breaks in mammalian cells and its inhibition by calyculin A. Radiat Res 160: 309–317 [DOI] [PubMed] [Google Scholar]
- Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH 1998. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280: 590–592 [DOI] [PubMed] [Google Scholar]
- Nelson G, Buhmann M, von Zglinicki T 2009. DNA damage foci in mitosis are devoid of 53BP1. Cell Cycle 8: 3379–3383 [DOI] [PubMed] [Google Scholar]
- Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP, et al. 2007. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol 17: 1972–1977 [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA 2000. Ku recruits the XRCC4–ligase IV complex to DNA ends. Mol Cell Biol 20: 2996–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R 2005. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 17: 331–339 [DOI] [PubMed] [Google Scholar]
- Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA 2010. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol 12: 177–184 [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. 2008. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135: 907–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D 2010. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell 40: 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan HM, Mohammad HP, Baylin SB 2008. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet 4: e1000155 doi: 10.1371/journal/pgen.1000155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgushi H, Yoshihara K, Kamiya T 1980. Bovine thymus poly(adenosine diphosphate ribose) polymerase. Physical properties and binding to DNA. J Biol Chem 255: 6205–6211 [PubMed] [Google Scholar]
- Okano S, Lan L, Caldecott KW, Mori T, Yasui A 2003. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol 23: 3974–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmbos PL, Wu D, Daley JM, Wilson TE 2008. Recruitment of Saccharomyces cerevisiae Dnl4–Lif1 complex to a double-strand break requires interactions with Yku80 and the Xrs2 FHA domain. Genetics 180: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Krebs JE, Peterson CL 2006. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev 20: 2437–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B, Gomez-Gonzalez B, Aguilera A 2009. DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol Life Sci 66: 1039–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J 2006. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting γ-H2AX induction. EMBO J 25: 3986–3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrilla-Castellar ER, Arlander SJ, Karnitz L 2004. Dial 9-1-1 for DNA damage: the Rad9–Hus1–Rad1 (9-1-1) clamp complex. DNA Repair (Amst) 3: 1009–1014 [DOI] [PubMed] [Google Scholar]
- Pataky K, Villanueva G, Liani A, Zgheib O, Jenkins N, Halazonetis DJ, Halazonetis TD, Brugger J 2009. Microcollimator for micrometer-wide stripe irradiation of cells using 20–30 keV X rays. Radiat Res 172: 252–259 [DOI] [PubMed] [Google Scholar]
- Peng G, Yim EK, Dai H, Jackson AP, Burgt I, Pan MR, Hu R, Li K, Lin SY 2009. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol 11: 865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, et al. 2001. AID is required to initiate Nbs1/γ-H2AX focus formation and mutations at sites of class switching. Nature 414: 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM 2001. Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Pinato S, Scandiuzzi C, Arnaudo N, Citterio E, Gaudino G, Penengo L 2009. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol Biol 10: 55 doi: 10.1086/1471-2199-10-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinato S, Gatti M, Scandiuzzi C, Confalonieri S, Penengo L 2011. UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway. Mol Cell Biol 31: 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR 2000. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem 275: 40974–40980 [DOI] [PubMed] [Google Scholar]
- Polanowska J, Martin JS, Garcia-Muse T, Petalcorin MI, Boulton SJ 2006. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. EMBO J 25: 2178–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Roche D, Almouzni G 2006. New histone incorporation marks sites of UV repair in human cells. Cell 127: 481–493 [DOI] [PubMed] [Google Scholar]
- Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP 2010. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J 29: 3130–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Parthun MR 2006. Recruitment of the type B histone acetyltransferase Hat1p to chromatin is linked to DNA double-strand breaks. Mol Cell Biol 26: 3649–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard DJ, Bolderson E, Cubeddu L, Wadsworth RI, Savage K, Sharma GG, Nicolette ML, Tsvetanov S, McIlwraith MJ, Pandita RK, et al. 2008. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature 453: 677–681 [DOI] [PubMed] [Google Scholar]
- Richard DJ, Savage K, Bolderson E, Cubeddu L, So S, Ghita M, Chen DJ, White MF, Richard K, Prise KM et al. 2010. hSSB1 rapidly binds at the sites of DNA double-strand breaks and is required for the efficient recruitment of the MRN complex. Nucleic Acids Res. doi: 10.1093/nar/gkq1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868 [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 146: 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP 2002. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol Cell 9: 857–869 [DOI] [PubMed] [Google Scholar]
- Rozenzhak S, Mejia-Ramirez E, Williams JS, Schaffer L, Hammond JA, Head SR, Russell P 2010. Rad3 decorates critical chromosomal domains with γH2A to protect genome integrity during S-Phase in fission yeast. PLoS Genet 6: e1001032 doi: 10/1371/journal.pgen.1001032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Cortes-Ledesma F, Guo L, Iles NJ, Caldecott KW 2008. APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol Cell Biol 28: 4620–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupnik A, Lowndes NF, Grenon M 2010. MRN and the race to the break. Chromosoma 119: 115–135 [DOI] [PubMed] [Google Scholar]
- Sacher M, Pfander B, Hoege C, Jentsch S 2006. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat Cell Biol 8: 1284–1290 [DOI] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119: 603–614 [DOI] [PubMed] [Google Scholar]
- Sanders SL, Arida AR, Phan FP 2010. Requirement for the phospho-H2AX binding module of Crb2 in double-strand break targeting and checkpoint activation. Mol Cell Biol 30: 4722–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H 2008. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257 [DOI] [PubMed] [Google Scholar]
- Santos MA, Huen MS, Jankovic M, Chen HT, Lopez-Contreras AJ, Klein IA, Wong N, Barbancho JL, Fernandez-Capetillo O, Nussenzweig MC, et al. 2010. Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8. J Exp Med 207: 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP 2007. Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S 2009. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J 28: 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, et al. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320: 1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM 1997. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90: 425–435 [DOI] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA 2010. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141: 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA 2009. The Rap80–BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8–Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci 106: 3166–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, Bhadra U, Pandita RK, Porteus MH, Chen DJ, et al. 2010. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol 30: 3582–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Ma JL, Oum JH, Yanez Y, Lee SE 2005. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol Cell Biol 25: 3934–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE 2007. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol 27: 1602–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol 14: 1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JJ, Cohen RE 2009. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell 33: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton BK, Torres-Arzayus MI, Rottinghaus ST, Taccioli GE, Jeggo PA 1999. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol Cell Biol 19: 3267–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Richard DJ, Saraf A, Toschi A, Bolderson E, Florens L, Washburn MP, Khanna KK, Pagano M 2009. INTS3 controls the hSSB1-mediated DNA damage response. J Cell Biol 187: 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H 2010. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol 190: 741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits VA, Reaper PM, Jackson SP 2006. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr Biol 16: 150–159 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, et al. 2010. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell 39: 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316: 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Du LL, Limbo O, Williams JS, Russell P 2010. BRCT domain interactions with phospho-histone H2A target Crb2 to chromatin at double-strand breaks and maintain the DNA damage checkpoint. Mol Cell Biol 30: 4732–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T 2008. Activation of the cellular DNA damage response in the absence of DNA lesions. Science 320: 1507–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T 2007. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol 9: 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M 2008. Constitutive phosphorylation of MDC1 physically links the MRE11–RAD50–NBS1 complex to damaged chromatin. J Cell Biol 181: 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stap J, Krawczyk PM, Van Oven CH, Barendsen GW, Essers J, Kanaar R, Aten JA 2008. Induction of linear tracks of DNA double-strand breaks by α-particle irradiation of cells. Nat Methods 5: 261–266 [DOI] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434 [DOI] [PubMed] [Google Scholar]
- Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA 2004. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res 64: 2390–2396 [DOI] [PubMed] [Google Scholar]
- Stracker TH, Usui T, Petrini JH 2009. Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 8: 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom L, Lindroos HB, Shirahige K, Sjogren C 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell 16: 1003–1015 [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Sugawara N, Haber JE 2006. Repair of DNA double strand breaks: in vivo biochemistry. Methods Enzymol 408: 416–429 [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD 2009. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol 11: 1376–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Chen J 2009. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci 106: 7155–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilard RK, Jacques PE, Laramee L, Cheng B, Galicia S, Bataille AR, Yeung M, Mendez M, Bergeron M, Robert F, et al. 2010. Systematic identification of fragile sites via genome-wide location analysis of γ-H2AX. Nat Struct Mol Biol 17: 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T 2003. DNA damage foci at dysfunctional telomeres. Curr Biol 13: 1549–1556 [DOI] [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK 2005. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol 25: 4903–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartier L, Spenlehauer C, Newman HC, Folkard M, Prise KM, Michael BD, Menissier-de Murcia J, de Murcia G 2003. Local DNA damage by proton microbeam irradiation induces poly(ADP-ribose) synthesis in mammalian cells. Mutagenesis 18: 411–416 [DOI] [PubMed] [Google Scholar]
- Teo SH, Jackson SP 2000. Lif1p targets the DNA ligase Lig4p to sites of DNA double-strand breaks. Curr Biol 10: 165–168 [DOI] [PubMed] [Google Scholar]
- Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC 2010. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol 17: 1263–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, et al. 2009. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol 16: 923–929 [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M 2007. The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol 9: 923–931 [DOI] [PubMed] [Google Scholar]
- Tsai AG, Lieber MR 2010. Mechanisms of chromosomal rearrangement in the human genome. BMC Genomics 11: S1 doi: 10.1186/1471-2164-11-S1-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA 2005. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438: 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ 2007. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol 177: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell 16: 991–1002 [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Koshland D 2007. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317: 245–248 [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC 2005. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651 [DOI] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 22: 5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM 2009. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 19: 207–217 [DOI] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Hohn B, Gasser SM 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119: 777–788 [DOI] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Gasser SM 2007. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J 26: 4113–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veelen LR, Cervelli T, van de Rakt MW, Theil AF, Essers J, Kanaar R 2005. Analysis of ionizing radiation-induced foci of DNA damage repair proteins. Mutat Res 574: 22–33 [DOI] [PubMed] [Google Scholar]
- van Vugt MA, Gardino AK, Linding R, Ostheimer GJ, Reinhardt HC, Ong SE, Tan CS, Miao H, Keezer SM, Li J, et al. 2010. A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol 8: e1000287 doi: 10.1371/journal.pbio.1000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Crabbe L, Haggblom C, Karlseder J 2005. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell 20: 551–561 [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412: 607–614 [DOI] [PubMed] [Google Scholar]
- Wang B, Elledge SJ 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci 104: 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF 1995. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev 9: 509–520 [DOI] [PubMed] [Google Scholar]
- Wang M, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G 2006. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 34: 6170–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Chen J 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 276: 47759–47762 [DOI] [PubMed] [Google Scholar]
- Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, et al. 2004. 53BP1 is required for class switch recombination. J Cell Biol 165: 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Iwabuchi K, Sun J, Tsuji Y, Tani T, Tokunaga K, Date T, Hashimoto M, Yamaizumi M, Tateishi S 2009. RAD18 promotes DNA double-strand break repair during G1 phase through chromatin retention of 53BP1. Nucleic Acids Res 37: 2176–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RJ 2010. When the caps fall off: responses to telomere uncapping in yeast. FEBS Lett 584: 3734–3740 [DOI] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. 2008. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. 2009. Nbs1 flexibly tethers Ctp1 and Mre11–Rad50 to coordinate DNA double-strand break processing and repair. Cell 139: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold MS 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 66: 61–92 [DOI] [PubMed] [Google Scholar]
- Wood JL, Singh N, Mer G, Chen J 2007. MCPH1 functions in an H2AX-dependent but MDC1-independent pathway in response to DNA damage. J Biol Chem 282: 35416–35423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Luo K, Lou Z, Chen J 2008. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci 105: 11200–11205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huen MS, Lu LY, Ye L, Dou Y, Ljungman M, Chen J, Yu X 2009a. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol Cell Biol 29: 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Prindle MJ, Dressler GR, Yu X 2009b. PTIP regulates 53BP1 and SMC1 at the DNA damage sites. J Biol Chem 284: 18078–18084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22: 719–729 [DOI] [PubMed] [Google Scholar]
- Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, et al. 2009. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 457: 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Puget N, Shim I, Odate S, Jarzyna I, Bassing CH, Alt FW, Scully R 2004. Control of sister chromatid recombination by histone H2AX. Mol Cell 16: 1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Kwok A, Scully R 2009. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol 16: 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD 2010. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol 191: 31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Dutt S, Xu R, Graves K, Juszczynski P, Manis JP, Shipp MA 2009. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol Cell 36: 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ 2008. Ku recruits XLF to DNA double-strand breaks. EMBO Rep 9: 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin B, Savic V, Juntilla MM, Bredemeyer AL, Yang-Iott KS, Helmink BA, Koretzky GA, Sleckman BP, Bassing CH 2009. Histone H2AX stabilizes broken DNA strands to suppress chromosome breaks and translocations during V(D)J recombination. J Exp Med 206: 2625–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Bailis JM 2010. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol 20: 402–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P 2005. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol 25: 5363–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Fu S, Lai M, Baer R, Chen J 2006. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev 20: 1721–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Chen J 2010. MRE11–RAD50–NBS1 complex dictates DNA repair independent of H2AX. J Biol Chem 285: 1097–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Adamski R, Chen J 2010. Focus on histone variant H2AX: to be or not to be. FEBS Lett 584: 3717–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenko V, Xue Z, Sadofsky MJ 2006. SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol Cell Biol 26: 1786–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarebski M, Wiernasz E, Dobrucki JW 2009. Recruitment of heterochromatin protein 1 to DNA repair sites. Cytometry A 75: 619–625 [DOI] [PubMed] [Google Scholar]
- Zhang D, Zaugg K, Mak TW, Elledge SJ 2006. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 126: 529–542 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE 2007. Role of Dnl4–Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol 14: 639–646 [DOI] [PubMed] [Google Scholar]
- Zhang F, Fan Q, Ren K, Andreassen PR 2009a. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res 7: 1110–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X 2009b. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol 19: 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Blobel G 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci 102: 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ 2003. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]