Abstract
Differential responses to stimuli can affect how cells succumb to disease. In yeast, DNA damage can create heterogeneous responses. To delineate how a response contributes to a cell's future behavior, we constructed a transcription-based memory circuit that detects DNA repair to isolate subpopulations with heritable damage responses. Strongly responsive cells show multigenerational effects, including growth defects and iron-associated gene expression. Less-responsive cells exhibit increased mutation frequencies but resume wild-type behavior. These two subpopulations remain distinct for multiple generations, indicating a transmissible memory of damage. Collectively, this work demonstrates the efficacy of using synthetic biology to define how environmental exposure contributes to distinct cell fates.
Keywords: synthetic circuit, bistable, DNA damage, heterogeneous, cell fate
A single cell encounters numerous environmental cues throughout its lifetime that can direct it toward specific pathways of behavior. One such stimulus is DNA damage. While initial population-level damage responses are well-characterized in the yeast Saccharomyces cerevisiae (Fu et al. 2008), transcriptional responses exhibit cell-to-cell variability due to factors such as unequal exposure, epigenetic elements, cell cycle, and cell age (Kale and Jazwinski 1996; Avery 2006). Significant single-cell variability is masked in population-based studies, preventing exploration of differential responses within cellular subsets (Bishop et al. 2007). As a result, little is understood about the heritability of heterogeneous damage responses and their contribution to a cell's future. We wished to determine whether a cell's history—its response to past damage—might influence its fate.
One method of detecting heritable, differential responses within a population is to employ a persistent autoregulatory positive feedback circuit. In this network design, feedback is only self-sustainable when the input exceeds the circuit's threshold for feedback. Such an input allows a circuit to switch from an OFF steady state to an alternative ON steady state that is heritable. Examples of this biological memory mechanism in nature include phage λ, the cell cycle, and cell differentiation (Burrill and Silver 2010). Using natural memory circuits as guides, a variety of synthetic memory pathways have been engineered in bacteria, yeast, and mammalian cells (Burrill and Silver 2010).
The bistable nature of memory circuits allows for identification of cell subpopulations that are responsive to specific events, and tracking of these cells during their progression through the cellular response (Ajo-Franklin et al. 2007). Cells that respond strongly to a stimulus can exceed a circuit's bistable threshold, while more weakly responsive cells do not activate autofeedback. In such a situation, two cell populations coexist, and their progeny can be followed over time to determine how their distinct responses affect their respective future fates. Thus, bistable circuits can be applied to the study of heritable DNA damage responses within whole-cell populations to determine how a cell's personal history affects its later behavior.
The application of a synthetic memory circuit toward studying a biological phenomenon would present a significant bioengineering advancement. Many previously designed circuits were used as proof-of-principle models to both demonstrate synthetic biological design approaches and elucidate the mathematical considerations required for devices with predictable behavior (Khali and Collins 2010). These circuits have laid the foundation for synthetic tools capable of addressing specific biological questions. Here we present a complex gene circuit designed to investigate heterogeneous responses to DNA damage in a eukaryotic system. Our work demonstrates the utility of applying engineered genetic devices to the study of biology.
Results and Discussion
Synthetic detection of DNA damage
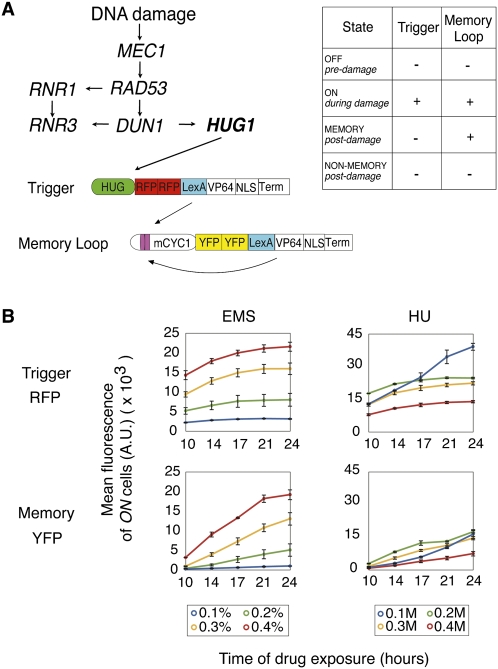
To identify yeast cell subpopulations that differ in initial and long-term memory of damage response, we used a novel transcription-based marker system that permits isolation and tracking of cells that were exposed to DNA damage. This marker system is a genetic positive feedback device (Ajo-Franklin et al. 2007) employing the promoter of the Mec1-mediated repair protein Hug1 (Basrai et al. 1999) to activate an RFP-expressing trigger transactivator in response to damage (Fig. 1A). The transactivator, composed of the transcription factor LexA and activator VP64, subsequently initiates transcription of a YFP-expressing memory loop that produces an identical transactivator (Ajo-Franklin et al. 2007). If the trigger activates the loop above the bistable threshold for feedback, the loop continues to self-activate its CYC1 promoter post-damage over many generations (Fig. 1A; Ajo-Franklin et al. 2007). This characteristic permits use of the memory device as a detector of cells derived from a parent cell that responded to a stimulus strongly enough to overcome the bistable threshold. For our purposes, the subpopulation of cells that maintain this memory loop activity post-damage can be identified as descendents of cells that experienced above-threshold HUG1-mediated repair activity in initial response to damage.
Figure 1.
Synthetic detection of DNA damage. (A) Schematic of bistable DNA damage memory device, composed of trigger and memory loop genes. (B) Induction of trigger (RFP) and memory loop (YFP) genes over time in response to varying EMS or HU concentrations. Induction was measured by FACS to determine mean RFP and YFP levels in ON cells. Values represent mean ± SD, n = 3.
Synthetic memory of DNA damage
HUG1 memory device activation was tested using the toxins ethyl methanesulfonate (EMS) and hydroxyurea (HU). EMS is an alkylating agent causing specific point mutations and double-strand breaks (Sega 1984). HU inhibits DNA synthesis by depleting dNTPs, resulting in S-phase arrest, genomic rearrangements, and deletions (Galli and Schiestle 1996). Both drugs resulted in dose- and time-dependent responses in trigger and memory loop induction, as measured by fluorescence-activated cell sorting (FACS) (Fig. 1B; Supplemental Fig. S1). While EMS exposure resulted in linear dose response curves that stabilized after 24 h of damage, HU caused a nonlinear dose response on device induction. This difference is likely due to HU causing dNTP synthesis malfunction at higher concentrations, interfering with the normal function of our transcription-based device. The memory loop is specifically responsive to trigger activation, as demonstrated by negligible loop activity in the absence of the trigger gene (Supplemental Fig. S2).
Memory loop persistence was measured by quenching exposed cells of toxins and using FACS to measure YFP levels at post-damage time points. A single cell was considered a memory cell if it expressed only YFP above background level, meaning the trigger had turned off and the memory loop maintained activity. A cell was considered a nonmemory cell if it did not express RFP or YFP above background levels. It should be noted that the designation of memory cells may be specific to experimental and biological conditions. The formation and defining characteristics of memory and nonmemory populations could vary depending on cell state, damaging agents, length of exposure, and time of recovery. Our goal was to identify whether a protracted biological response indeed distinguishes memory and nonmemory cell populations after cellular insult.
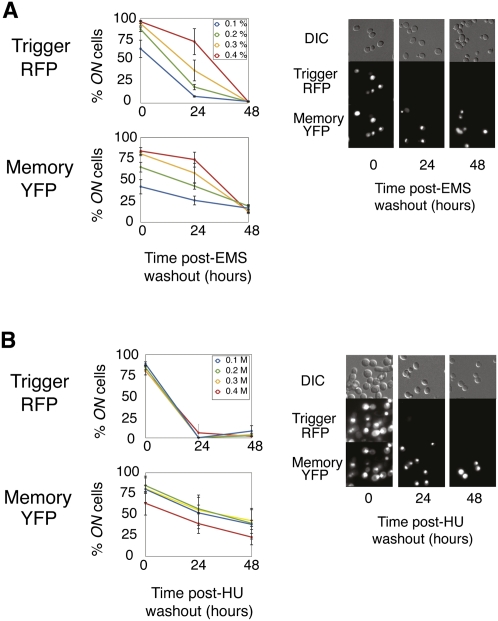
Toward this goal, we determined that memory loop expression was sustained in the absence of trigger activity for at least 48 h post-damage in a small subset of cells after EMS or HU exposure (Fig. 2). We compared this behavior with that of the previously designed GAL1/10 memory device (Ajo-Franklin et al. 2007) that is identical to the HUG1 circuit except that its promoter is galactose-inducible. The GAL1/10 circuit's behavior was quite different: Cells were exposed to galactose for 24 h, and memory loop expression persisted in at least 85% of cells for 72 h post-galactose (Supplemental Fig. S3). The difference in response is likely due to the more heterogeneous transcriptional response caused by DNA damage. When trigger versus memory loop induction is plotted, the uniform response to galactose contrasts with the more heterogeneous response to EMS (Supplemental Fig. S1). HUG1 is not uniformly activated within a cell population, resulting in variable trigger and memory loop activation between cells. The memory circuit thus permits the isolation of two distinct post-damage subpopulations—one that responds strongly with HUG1 activation, and one that responds more weakly—a distinction that could impact immediate and long-term cell identity.
Figure 2.
Synthetic memory of DNA damage. (Left) Trigger (RFP) and memory loop (YFP) expression post-EMS damage (A) and post-HU damage (B) was assessed via FACS by measuring the percentage of ON cells within a population. Zero hours post represents cells after 24 h exposure to EMS or HU. Twenty-four hours post and 48 h post represent cells after 24 h and 48 h of recovery from DNA damage. Values represent means ± SD; n = 3. (Right) Further analysis was done by fluorescence microscopy of the whole population after exposure to 0.3% EMS (top) or 0.3 M HU (bottom).
Sorting of memory and nonmemory cells reveals two distinct populations
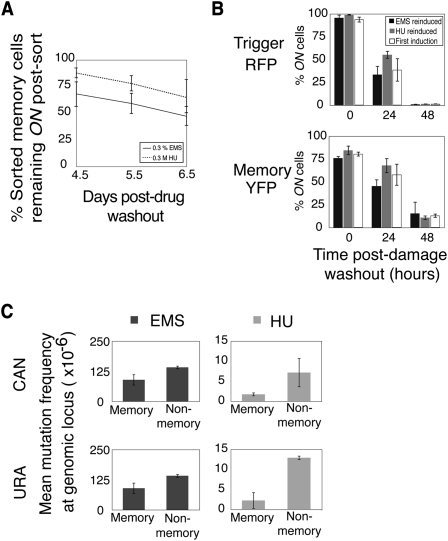
Our bistable circuit allows for isolation of memory and nonmemory cell populations post-damage via memory loop YFP expression. We exposed cells to EMS or HU for 24 h and then sorted memory and nonmemory cells after a 36-h recovery period using FACS (Supplemental Fig. S1). After sorting, the two populations were cultured, and YFP persistence in the sorted memory population was measured over multiple days. More than half of cells maintained YFP expression 6.5 d after EMS or HU exposure (Fig. 3A). This result suggested that the proportion of memory loop cells decreased post-damage due to a growth defect, resulting in the memory population being diluted over time by the more rapidly growing nonmemory cells.
Figure 3.
Sorting of memory and nonmemory cells reveals two distinct populations. (A) Cells that recovered for 36 h from DNA damage were sorted based on memory loop expression, and the percentage of persistent ON memory cells was analyzed by FACS. (B) Nonmemory cells previously exposed to EMS were reinduced with EMS or HU, and the percentage of trigger or memory loop ON cells was measured by FACS. (C) Mutation frequencies were measured at CAN and URA loci in sorted populations. Values in A–C represent means ± SD; n = 3. Statistical significance was determined in C using a paired two-tail t-test.
To investigate this issue, we measured the growth rate of sorted memory cells versus sorted nonmemory cells after 36 h of recovery from EMS or HU. Memory cells grew 1.4 times or 1.27 times slower, respectively, than their nonmemory cell counterparts, which resumed normal growth rate (Supplemental Fig. S4). DNA damage can induce slower growth to allow time for repair (Bartek and Lukas 2007). Additionally, it has been shown in bacteria that growth retardation may occur due to the additional transcription and translation required for positive feedback loop expression (Tan et al. 2009). Both factors likely contribute to the observed decrease in the percentage of memory cells post-damage when the two populations remain unsorted (Fig. 2).
We verified that memory loop dysfunction was not a contributing factor to the memory population decrease by measuring the potential of nonmemory cells to be reinduced by DNA damage. Dysfunction could result, for example, from mutations in the DNA encoding the memory loop. Cells were exposed to EMS or HU for 24 h, and nonmemory cells were sorted after a 36-h recovery period. The sorted nonmemory cells were cultured for 12 h and then re-exposed to EMS or HU (Fig. 3B represents data from cells first exposed to EMS; Supplemental Fig. S5 represents data from cells first exposed to HU). Memory loop behavior was comparable with that observed after the first induction. Nearly all cells reactivated the trigger and memory loop genes, and a similar number of cells had persistent post-damage memory loop expression. Thus, the HUG1 memory device detects two distinct populations based on differential responses to damage.
Memory population has a lower mutation frequency
Having isolated these two subpopulations, we next determined whether they differed in their long-term responses to DNA damage. Differences would indicate that a cell's individual repair history contributes to an inherited stress response in its progeny. Cells were exposed to EMS or HU for 24 h and recovered for 36 h, and memory and nonmemory cells were sorted using FACS. Mutational analysis of the two populations at the CAN (Whelan et al. 1979) and URA (Crouse 2000) loci revealed a lower nuclear genomic mutation (NGM) frequency in memory cells (Fig. 3C; Supplemental Figs. S6, S7A): 1.6 times (0.02 < P < 0.025) and 4.0 times (0.05 < P < 0.10) lower at the CAN locus after EMS or HU exposure, and 3.1 times (0.005 < P < 0.01) and 5.6 times (0.02 < P < 0.025) lower at the URA locus after EMS or HU exposure. These results are consistent with the memory device's input being activation of HUG1-mediated repair: Cells that maintained memory loop expression likely experienced HUG1 expression levels that exceeded the bistable threshold for positive feedback. Strong activation of HUG1-mediated repair—and, by proxy, the likely up-regulation of other repair pathways—resulted in fewer NGMs. This result does not necessarily reflect the current cell state—i.e., vulnerability toward mutation accumulation—but rather reflects a state that occurred in response to damage and was carried through many generations. Thus, we demonstrated that DNA damage has a sustained impact, as evidenced by differential mutation frequencies persisting for multiple cell divisions.
Memory population exhibits gene expression associated with iron starvation response
Transcriptional profiling of memory and nonmemory cells was next performed to interrogate each population's unique response to DNA damage at the expression level. Although initial transcriptional responses have been identified in yeast (Fu et al. 2008), little is known about the sustained effects on gene expression (Dubacq et al. 2006; Davies et al. 2009), particularly with respect to isolated, damaged subpopulations. To define the long-term effects of DNA damage on the gene expression of subpopulations, cells were exposed to 0.3% EMS for 24 h, and memory and nonmemory cells were isolated after 36 h of recovery. The two populations were cultured for another 12 h (a total of 48 h of recovery), at which point transcriptional profiling was performed. We determined that 48 h post-EMS exposure, the two separable populations continued to have distinct expression signatures: 66 genes were up-regulated and 15 were down-regulated in cells with sustained memory as compared with those without memory of DNA damage (fold-induction [FI] ≥ 1.5, coefficients of variation [CV] ≤ 1.5) (Supplemental File S1). Bioinformatics analysis identified the transcriptional pattern of iron starvation in memory cells (Fig. 4A). Up-regulated genes were enriched for iron-related activities: siderophore (P = 1.4e-10), siderophore-iron (P = 1.68e-06), metal ion transport (P = 1.68e-06), and iron assimilation (P = 9.85e-06). Homeostasis genes were also up-regulated: the vacuole (P = 6.23e-05), iron ion (P = 6.91e-05), and chemical (P = 5.01e-04) homeostasis, as well as general homeostatic processes (P = 8.21e-04). Enrichments were validated by real-time PCR analysis (Supplemental Figs. S8, S9).
Figure 4.
Characterization of memory and nonmemory cells. (A) Gene ontology enrichment. (B) Difference in gene expression between HUG1 memory cells and GAL1/10 memory cells. (C) Profiling validation using GFP-tagged yeast strain of the differentially expressed gene AIM19. (D) Respiration was visualized by fluorescence microscopy in memory cells. Values represent means ± SD; n = 3. Statistical significance was determined using a paired two-tail t-test. (E) Model of the heterogeneous response to EMS exposure and sustained effects in subpopulations.
We determined that the memory cell expression profile was independent of effects caused by the synthetic device itself. HUG1 memory cell gene expression was compared with that of GAL1/10 memory cells by isolating each population after a 36-h recovery period from EMS or galactose exposure, respectively. Cells were cultured for an additional 12 h (a total of 48 h of DNA damage recovery), and expression differences were measured by real-time PCR (Fig. 4B). Expression was quantified for six genes that were up-regulated in the original transcriptional profiling of HUG1 memory cells: FIT1, FIT2, FIT3 and ZRT1 are involved in the iron starvation response, and XBP1 and AIM19 are stress response genes. All six genes were up-regulated in HUG1 memory cells as compared with GAL1/10 memory cells. XBP1 and AIM19 had the lowest difference in expression between the two strains, suggesting that raffinose-to-galactose carbon source switching or sustained positive feedback loop activity incurs some background stress on cell physiology.
To establish that DNA damage truly causes sustained up-regulation of XBP1 and AIM19 above the influence of positive feedback loop activity, strains containing GFP fusions of XBP1 and AIM19 were used to assess whether expression remained active in a subpopulation of cells post-DNA damage (these strains do not contain a memory device) (Fig. 4C; Supplemental Fig. S10; Huh et al. 2003). This result would be expected, based on our observation that these genes remain up-regulated in memory cells. Cells were exposed to EMS or galactose for 24 h and recovered for 48 h. In comparison with galactose-regulated expression of XBP1 and AIM19, EMS produced a lasting stress response for at least 48 h post-damage in a subpopulation of cells. This stress response is induced by DNA damage and contributes to the up-regulation of XBP1 and AIM19 in damaged-induced memory cells. The results indicate that the identified gene profile is largely due to the long-term effects of DNA damage and not expression of a positive feedback loop. In sum, EMS-mediated DNA damage produces a heritable transcriptional response that is maintained through multiple rounds of DNA replication and cell division.
We next compared the transcriptional profile of HUG1 memory cells with other DNA damage- or iron-related data sets. A comparison of our data with that acquired immediately after exposure to MMS (Benton et al. 2006), an alkylating agent similar to EMS, determined a 24.2% overlap of up-regulated genes (data not shown). This indicates that primary damage response pathways were partially active 48 h post-damage in memory cells. Comparison with the expression profile of iron-starved cells (Puig et al. 2005) revealed a stronger intersection of 38.1% of up-regulated genes (data not shown). Thus, damage-induced expression of iron-related genes is activated and sustained post-damage.
The iron starvation response is a well-characterized program in S. cerevisiae that tightly regulates cellular iron uptake and utilization to prevent toxicity caused by iron overload. It is induced primarily by defective iron metabolism, dysfunctional mitochondrial iron sulfur cluster (ISC) biogenesis, and DNA damage (Lee and Wei 2005; Veatch et al. 2009; Berthelet et al. 2010). DNA repair proteins commonly require iron as a cofactor, and this essential role may endow iron with protective effects against DNA damage (Berthelet et al. 2010). Memory cells might continue to recruit iron for generations post-damage to facilitate DNA repair protein production, or even as a protective advantage against future damage. Iron may also be needed to compensate for loss of ISC biogenesis as a result of damaged mitochondria. Indeed, a comparison of the transcriptional profile of memory cells with that obtained 27 h after mitochondrial loss of function (Veatch et al. 2009) revealed a significant overlap of 45.5% of up-regulated genes (data not shown). These observations indicate that defective iron metabolism in memory cells may be part of a sustained response to DNA damage and its impact on mitochondria.
Memory population exhibits increased levels of respiration
We further probed the concept of a prolonged damage response program by comparing respiration in memory and nonmemory cells. Prolonged DNA repair responses coincide with elevated respiratory activity in yeast, perhaps to meet energy requirements for DNA damage repair (Schaus et al. 2001). Furthermore, studies suggest that damage can result in the eventual biogenesis of new mitochondria with the respiratory capacity to provide metabolic energy and replenish ISC pools for repair proteins (Gasch et al. 2001; Aguilaniu et al. 2003; Lee and Wei 2005; Rasbach and Schnellmann 2007; Thorsen 2009).
Using a reduced Red MitoTracker dye (Invitrogen) that fluoresces in the presence of respiring mitochondria, we determined respiration levels in memory and nonmemory populations. Forty-eight hours post-damage, the number of respiring memory cells exceeded that of nonmemory cells by 4.1-fold (0.025 < P < 0.05) (Fig. 4D; Supplemental Fig. S7B). We believe this phenomenon is due to a sustained DNA damage response. Using the GAL1/10 memory device to control for the influence of positive feedback activity on respiration, we observed that galactose memory cells did not demonstrate a behavior similar to our DNA damage memory cells (Supplemental Fig. S11). This suggests that loop expression did not contribute to observed respiratory differences. In sum, the memory population exhibits a sustained state of damage response that is partially characterized by up-regulated respiration, perhaps to provide energy or ISCs for DNA repair.
Collectively, our data provide evidence that activation of the yeast DNA damage response strongly influences the state of a cell for many generations (Fig. 4E). Cells experiencing a greater damage response will elevate repair pathways, resulting in reduced growth and fewer nuclear mutations. Additionally, these cells exhibit sustained up-regulation of the iron starvation response and respiratory activity post-damage, suggesting that these are two components of a prolonged program of damage response. Cells experiencing a lower level of cellular insult instead respond with minimal repair, greater NGM accumulation, and the eventual resumption of normal growth. Since population identities were defined numerous generations post-damage, we propose that these responses are transmissible through DNA replication and cell division. Further studies will determine how one population maintains a specific cell state (e.g., chromatin remodeling or protein modification) and whether it is beneficial: The differential responses may translate into increased vulnerability to further damage or effects on cell aging, or the notion that regulation of iron metabolism provides a protective advantage. By isolating distinct DNA damage responses within a population and tracking cell identity over time, we illuminated how a cell's unique history contributes to its future behavior.
Materials and methods
Strains and media
The HUG1 promoter was amplified from S. cerevisiae genomic DNA. Trigger and memory loop genes were integrated into PSY580A (MATα, ura3-52, trp1Δ63, and leu2Δ1) using Sikorski vectors; strain DBY2 contained only memory loop (Ajo-Franklin et al. 2007). The GAL1/10 memory strain from Ajo-Franklin et al. (2007) was used as control for circuit behavior; it is identical to the HUG1 strain except for its promoter being galactose-inducible. Yeast were rotated at 30°C in 2% raffinose synthetic dropout (SD) media (leu−, ura−) and maintained in log phase for all experiments.
FACS and fluorescence microscopy
For FACS, cells were run on an LSRII (BD Biosciences) with 488-nm and 568-nm lasers (Harvard Systems Biology). Cells (2 × 104) were analyzed for RFP and YFP fluorescence and gated based on cell size and granularity. The uninduced HUG1 strain controlled for basal expression, so single cells were defined as ON or OFF for both fluorophores (Supplemental Fig. S1). Reported fluorescence units are mean values of ON or OFF cells; intensities were normalized so the uninduced strain was identical between experiments. For sorting, cells were run on a Coulter High-Speed Sorter (MoFlo) with 488-nm and 647-nm lasers (Dana Farber) and analyzed as described above. Cells were imaged on an Eclipse TE2000-E with a 60× objective, Orca 285 CCD camera, Metamorph software, and HcRed (RFP) and JP2 (YFP) filters.
Device induction
Yeast were exposed to 0.1%–0.4% EMS or 0.1–0.4 M HU for 24 h. Aliquots were sampled over a 24-h period and toxins were quenched: HU-exposed aliquots were washed three times in SD media, and EMS-exposed aliquots were washed twice in SD media, twice in 5% sodium thiosulfate, and again twice in SD media. Aliquots were fixed in 3.7% paraformaldehyde, stored in Tris-EDTA for 1–2 d at 4°C, and analyzed by FACS and fluorescence microscopy. Experiments were repeated in biological triplicate.
Memory loop sustainability
Yeast were exposed to 0.1–0.4% EMS or 0.1–0.4 M HU for 24 h. Aliquots were washed and fixed as described above for FACS analysis. Washed aliquots were also used to inoculate fresh SD media and every 24 h post-damage, and aliquots were fixed and analyzed by FACS and fluorescence microscopy to detail memory gene expression for 48 h. Experiments were repeated in biological triplicate.
A similar procedure was performed using the GAL1/10 memory strain. Cells were grown to log phase in 2% raffinose SD media, exposed to 2% galactose for 24 h, and then cultured in 2% raffinose SD media for analysis of memory sustainability.
Sustainability and reinducibility of sorted memory populations
Yeast were exposed to 0.3 M HU or 0.3% EMS for 24 h, and aliquots were washed as described above and used to inoculate fresh media. Cultures were maintained in log phase for 36 h, and samples were sorted based on YFP expression. Five-hundred-thousand cells of each population were sorted and resuspended into SD media and kept on a rotator at 37°C. Aliquots were fixed every 24 h at subsequent time points and analyzed by FACS and fluorescence microscopy. Growth rates of two sorted populations were measured with a Genesys spectrophotometer. Experiments were repeated in biological triplicate.
Mutation assay
Yeast were exposed to 0.3 M HU or 0.3% EMS for 24 h, recovered for 36 h, and sorted based on YFP expression. One million to 2 million YFP+ and YFP− cells were sorted, and equal numbers were plated on mutation plates (mutation frequency) and nonselective plates (viability). Undamaged yeast were similarly sorted and plated as a background control. Cells grew on nonselective plates for 3 d, on CANR plates for 3 d, and on 5-FOAR plates for 4 d. CANR plates were as follows: SD media (leu− ura− arg−) and 60 μg/mL L-canavanine. 5-FOAR plates were as follows: SD media (leu− ura−), 1 g/L 5-FOA, and 50 mg/L uracil. Experiments were repeated in biological triplicate. Statistics were determined with paired two-tail t-test, assuming equal variance; DF = 2, α cutoff = 0.10.
cDNA microarray
Yeast were exposed to 0.3% EMS for 24 h in biological triplicate. After 36 h of recovery, 10 million memory and nonmemory cells were sorted and cultured in SD media for 12 h (total recovery period of 48 h). cDNA was extracted with phenol/chloroform and used as a template for random priming with BioPrime DNA labeling kits (Invitrogen) and 3 μL of 1 nM Cy5-dUTP or Cy3-dUTP (PerkinElmer). In two biological replicates, memory cells were labeled with Cy3-dUTP and nonmemory cells were labeled with Cy5-dUTP; in one biological replicate, reverse fluorophore orientation accounted for dye bias. Samples were hybridized to 6.4Kv6 double-spotted yeast ORF microarrays (University Health Networks, Toronto).
Microarray data analysis
Microarrays were scanned using Axon Genepix 4000B (BioPolymer Facility, Harvard University). Individual arrays underwent background subtraction and log transformation. Arrays were global median-normalized, intensities across arrays were averaged, and CV and FI were calculated. Genes with CV ≤ 1.5 and FI ≥ 1.5 were considered differentially expressed and analyzed for gene ontology enrichment via GoStat (http://gostat.wehi.edu.au) with Benjamini correction for multiple hypothesis testing; enrichment was considered significant with P ≤ 0.001.
Up-regulated genes were compared with other profiling data: In Benton et al. (2006), genes were compared if FI ≥ 1.5 in both data sets; in Puig et al. (2005), genes were compared if FI ≥ 2.0 in both data sets; and in Veatch et al. (2009), genes were compared if FI ≥ 1.5 in both data sets.
Real-Time PCR
Differentially expressed genes were validated by real-time PCR using 0.5 μg of RNA of sorted memory and nonmemory cells. Primers amplify ∼100 base pairs near the 3′ end of the ORF. Differential expression was normalized to the control gene ADH1.
GFP fusion assay
Strains (Huh et al. 2003) contained GFP fused to YIL087C (AIM19), YIL101C (XBP1), or YOL086C (ADH1). PSY580A controlled for background fluorescence, and ADH1 controlled for expression changes. Yeast were grown in 2% raffinose SC media, exposed to 0.3% EMS for 24 h, and imaged 0 h, 24 h, and 48 h post-damage. Experiments were repeated in biological triplicate; 200 cells were counted per replicate.
Respiration assay
MitoTracker Red CMXRos (Invitrogen) selectively stained mitochondria, and CM-H2XRos (Invitrogen) was a reduced version that fluoresced when oxidized by respiring mitochondria. HUG1 yeast were exposed to 0.3% EMS for 24 h; GAL1/10 yeast were exposed to 2% galactose for 24 h. Yeast recovered for 48 h and were resuspended in SD media containing 1 μM CM-XRos or 3 μM CM-H2XRos. Cells were incubated for 30 min at 30°C while rotating and then analyzed by FACS or fluorescence microscopy. Experiments were repeated in duplicate. Statistics were determined with paired two-tail t-test, assuming equal variance; DF = 2, α cutoff = 0.10.
Acknowledgments
We thank D. Drubin, M. Inniss, B. Afonso, E. Wintermute, P. Boyle, and C. Agapakis for all their advice and time; the entire Silver laboratory for all their support; R. Ward, F. Winston, and the Walker laboratory for helpful discussion; and J. Moore, the Dana Farber Flow Cytometry Facility, and the Biopolymers Facility for technical assistance. D.R.B. was supported by NSF SYNBERC (520.45321.148830.377227.0402.66376), and P.A.S. was supported by DOD W81XWH-08-1-0608 and NIHGM36373-22.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1994911.
Supplemental material is available for this article.
References
- Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 299: 1751–1753 [DOI] [PubMed] [Google Scholar]
- Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EPS, Landgraf D, Phillips I, Silver PA 2007. Rational design of memory in eukaryotic cells. Genes Dev 21: 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery SV 2006. Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol 4: 577–587 [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J 2007. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol 19: 238–245 [DOI] [PubMed] [Google Scholar]
- Basrai MA, Velculescu VE, Kinzler KW, Hieter P 1999. NORF5/HUG1 is a component of the MEC1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol Cell Biol 19: 7041–7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MG, Somasundaram S, Glasner JD, Palecek SP 2006. Analyzing the dose- dependence of the Saccharomyces cerevisiae global transcriptional response to methyl methanesulfonate and ionizing radiation. BMC Genomics 7: 305–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelet S, Usher J, Shulist K, Hamza A, Maltez N, Johnston A, Fong Y, Harris LJ, Baetz K 2010. Functional genomics analysis of the Saccharomyces cerevisiae iron responsive transcriptional factor Aft1 reveals iron-independent functions. Genetics 185: 1111–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AL, Rab FA, Sumner ER, Avery SV 2007. Phenotypic heterogeneity can enhance rare-cell survival in ‘stress sensitive’ yeast populations. Mol Microbiol 63: 507–520 [DOI] [PubMed] [Google Scholar]
- Burrill DR, Silver PA 2010. Making cellular memories. Cell 140: 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse GF 2000. Mutagenesis assays in yeast. Methods 22: 116–119 [DOI] [PubMed] [Google Scholar]
- Davies BW, Kohanski MA, Simmons LA, Winkler JA, Collins JJ, Walker GC 2009. Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli. Mol Cell 36: 845–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubacq C, Chevalier A, Courbeyrette R, Petat C, Gidrol X, Mann C 2006. Role of the iron mobilization and oxidative stress regulons in the genomic response of yeast to hydroxyurea. Mol Genet Genomics 275: 114–124 [DOI] [PubMed] [Google Scholar]
- Fu Y, Pastushok L, Xiao W 2008. DNA damage-induced gene expression in Saccharomyces cerevisiae. FEMS Microbiol Rev 32: 908–926 [DOI] [PubMed] [Google Scholar]
- Galli A, Schiestle RH 1996. Hydroxyurea induces recombination in dividing but not in G1 or G2 cell cycle arrested yeast cells. Mutat Res 354: 69–75 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell 12: 2987–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Kale SP, Jazwinski SM 1996. Differential response to UV stress and DNA damage during the yeast replicative life span. Dev Genet 18: 154–160 [DOI] [PubMed] [Google Scholar]
- Khali AS, Collins JJ 2010. Synthetic biology: applications come of age. Nat Rev Genet 11: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Wei YH 2005. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol 37: 822–834 [DOI] [PubMed] [Google Scholar]
- Puig S, Askeland E, Thiele DJ 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120: 99–110 [DOI] [PubMed] [Google Scholar]
- Rasbach KA, Schnellmann RG 2007. Signaling of mitochondrial biogenesis following oxidant injury. J Biol Chem 282: 2355–2362 [DOI] [PubMed] [Google Scholar]
- Schaus SE, Cavalieri D, Myers AG 2001. Gene transcription analysis of Saccharomyces cerevisiae exposed to neocarzinostatin protein–chromophore complex reveals evidence of DNA damage, a potential mechanism of resistance, and consequences of prolonged exposure. Proc Natl Acad Sci 98: 11075–11080 doi: 10.1073/pnas.191340698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sega GA 1984. A review of the genetic effects of ethyl methanesulfonate. Mutat Res 134: 113–142 [DOI] [PubMed] [Google Scholar]
- Tan C, Marguet P, You L 2009. Emergent bistability by a growth-modulating positive feedback circuit. Nat Chem Biol 5: 842–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen M 2009. Genetic basis of arsenite and cadmium tolerance in Saccharomyces cerevisiae. BMC Genomics 10: 105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch JR, McMurray MA, Nelson ZW, Gottschling DE 2009. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell 137: 1247–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan WL, Gocke E, Manney TR 1979. The CAN1 locus of Saccharomyces cerevisiae: fine-structure analysis and forward mutation rates. Genetics 91: 35–51 [DOI] [PMC free article] [PubMed] [Google Scholar]