Abstract
Duplex formation between the branch point-binding region (BBR) of U2 snRNA and the branch point sequence (BPS) in the intron is essential for splicing. Both the BBR and BPS interact with the U2 small nuclear ribonucleoprotein (snRNP)-associated SF3b complex, which is the target of the anti-tumor drug E7107. We show that E7107 blocks spliceosome assembly by preventing tight binding of U2 snRNP to pre-mRNA. E7107 has no apparent effect on U2 snRNP integrity. Instead, E7107 abolishes an ATP-dependent conformational change in U2 snRNP that exposes the BBR. We conclude that SF3b is required for this remodeling, which exposes the BBR for tight U2 snRNP binding to pre-mRNA.
Keywords: small molecule splicing inhibitor, spliceosome assembly, pladienolide derivative E7107, 17S U2 snRNP, SF3b, branch point-binding region, anti-tumor drug
Most pre-mRNAs in higher eukaryotes contain multiple introns that are excised by the spliceosome to generate mature mRNA (Konarska and Query 2005; Smith et al. 2008; Wahl et al. 2009). The spliceosome, composed of five snRNAs (U1, U2, U4, U5, and U6) and many proteins, undergoes a series of dynamic changes during the splicing pathway (Staley and Guthrie 1998; Smith et al. 2008; Wahl et al. 2009). In the early steps, U1 small nuclear ribonucleoprotein (snRNP) binds to the pre-mRNA, and U1 snRNA base-pairs to the 5′ splice site to form the ATP-independent early (E) complex (Reed 1996; Smith et al. 2008; Wahl et al. 2009). U2 snRNP is also recruited to the E complex, but is not tightly bound (Das et al. 2000; Donmez et al. 2004, 2007). Subsequently, in an ATP-dependent step, the A complex is assembled, in which U2 snRNP binds tightly. At this time, a duplex is formed between the branch point sequence (BPS) in the intron and the branch point-binding region (BBR) located near the 5′ end of U2 snRNA (Konarska and Query 2005; Smith et al. 2008; Wahl et al. 2009). This tight binding of U2 snRNP to the pre-mRNA and formation of the BBR–BPS duplex are critical early steps in establishing the catalytic center of the spliceosome. In later steps, multiple complex rearrangements occur, resulting in binding of U4/U5/U6 snRNPs to form the B complex, and finally the catalytically active C complex.
Although a great deal has been learned about spliceosome assembly and splicing, it has been difficult to achieve a detailed understanding of the numerous steps in this pathway without the aid of small molecule inhibitors. Recently, an anti-tumor drug, E7107 (E7), was identified that blocks splicing by targeting the U2 snRNP-associated complex SF3b (Kotake et al. 2007). SF3b associates with both the 5′ end of U2 snRNA and the BPS region of the intron (Gozani et al. 1996; Kramer et al. 1999; Will et al. 2001; Dybkov et al. 2006). Thus, SF3b is positioned at the critical site of the spliceosome, where the BBR–BPS duplex forms during establishment of the catalytic center of the spliceosome. However, the precise function of SF3b is not known. Here, we show that E7 results in formation of a defective spliceosome in which U2 snRNP fails to bind tightly to pre-mRNA. Our data indicate that E7 does not disrupt U2 snRNP or its association with SF3b, but does block an ATP-dependent remodeling event that exposes the BBR of U2 snRNA. We conclude that SF3b is required to remodel U2 snRNP to expose the BBR, and this event may allow the SF3b- and ATP-dependent tight binding of U2 snRNP to the pre-mRNA. These results provide important new insights into the mechanism of action of E7, and should aid in elucidating the novel anti-tumor properties of this drug.
Results and Discussion
E7 results in formation of a defective, labile spliceosome
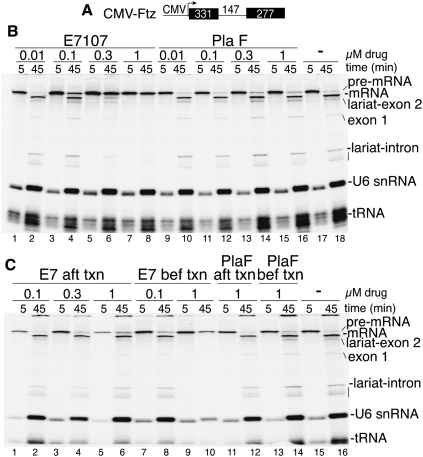
To determine how E7 blocks splicing, we used a coupled RNA polymerase II (RNAP II) transcription/splicing system (Das et al. 2006). When a CMV-Ftz DNA template encoding one intron and two exons (Fig. 1A) was incubated in the presence of E7, transcription occurred normally, but splicing was blocked in a dose-dependent manner (Fig. 1B, lanes 1–8). Splicing was not affected by a negative control compound, pladienolide F (PlaF), and was the same as with no drug treatment (Fig. 1B, lanes 9–18). Significantly, however, when CMV-Ftz was first transcribed by RNAP II, followed by addition of the drug, splicing was not affected by E7 (or PlaF) (Fig. 1C). Previous work showed that the spliceosome assembles immediately on pre-mRNA when synthesized in the coupled system (Das et al. 2006). Thus, the data in Figure 1C indicate that E7 inhibits spliceosome assembly. We obtained the same results with CMV-AdML, indicating that the effects of E7 are general (Supplemental Fig. 1).
Figure 1.
E7 blocks splicing when added prior to but not after RNAP II transcription. (A) Schematic of CMV-Ftz DNA template showing the CMV promoter and sizes of the exons and intron. (B) CMV-Ftz DNA was incubated in coupled RNAP II transcription/splicing reaction mixtures for 5 min in the presence of the indicated amounts of E7, PlaF, or no drug. α-Amanitin was then added, and incubation was continued for 45 min. (C) CMV-Ftz DNA was incubated for 5 min, followed by addition of α-amanitin. (Lanes 1–6,11,12) After transcription, the indicated amounts of E7 or PlaF were added, and incubation was continued for 45 min. In lanes 7–10, 13, and 14, the indicated amounts of E7 or PlaF were added at the beginning of the coupled reaction as in B. No drug was added in lanes 15 and 16. Total RNA was fractionated on a 5% denaturing polyacrylamide gel and detected by PhosphorImager. The splicing intermediates and products and endogenous U6 snRNA and tRNA are indicated.
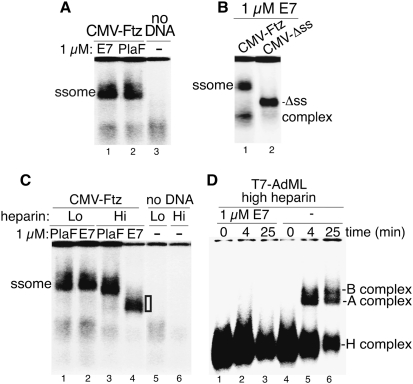
We next directly assayed for the spliceosome by transcribing CMV-Ftz in the coupled system and analyzing the reactions on a native gel. Surprisingly, the spliceosome had the same mobility whether assembled in E7 or PlaF (Fig. 2A). Moreover, the spliceosome assembled in E7 was splice site-dependent, as it did not form on a CMV-Δss transcript, which lacks functional splice sites and assembles into a nonspecific hnRNP complex (Fig. 2B; Das et al. 2006). We conclude that E7 results in formation of a complex that requires splicing signals and comigrates with the spliceosome, but is incapable of splicing. To determine why the E7 spliceosome is defective, we examined it using different amounts of heparin, which affects spliceosome stability. Significantly, in the presence of high heparin, the E7 spliceosome was disrupted, whereas the PlaF spliceosome had similar mobility in low and high heparin (Fig. 2C). These data indicate that the E7 spliceosome is an aberrant, labile complex.
Figure 2.
E7 results in formation of a labile spliceosome and blocks the A complex assembly. (A) CMV-Ftz was incubated in coupled transcription/splicing mixtures in the presence of E7 or PlaF for 8 min. CMV-Ftz was omitted in lane 3. Heparin (0.5 μL) was added to the reaction mixtures, and then an aliquot was fractionated on a 1.2% native low-melting agarose gel and detected by PhosphorImager. The spliceosome (ssome) is indicated. (B) Same as A, except using CMV-Ftz and CMV-Δss DNA templates. The spliceosome and nonspecific Δss complexes are indicated. (C) Same as A, except 0.5 μL and 1.5 μL of heparin was added. The white box to the right of lane 4 indicates the heparin-sensitive E7 spliceosome. (D) T7-AdML was incubated under standard splicing conditions in E7 or no drug for the indicated times, heparin was added, and spliceosomal complexes were separated on a 1.5% native low-melting agarose gel.
To determine when along the spliceosome assembly pathway E7 exerts its effect, we made use of the uncoupled splicing system and a T7 transcript, because the kinetics of spliceosome assembly are so fast with the coupled system that the different spliceosomal complexes (e.g., A and B) are not detected. When T7 AdML was incubated in an uncoupled splicing reaction for 0, 4, or 25 min, the nonspecific H, A, and B complexes were detected with no drug (Fig. 2D) or PlaF (data not shown). In contrast, only the H complex was detected with E7 (Fig. 2D). We conclude that E7 blocks assembly of the spliceosomal A complex.
E7 results in weak binding of U2 snRNP to pre-mRNA
As E7 targets U2 snRNP, and this snRNP first binds tightly in the A complex, we next asked whether U2 snRNP was present in the defective, heparin-sensitive E7 spliceosome. To do this, we inactivated U2 snRNP using the endogenous RNase H in the nuclear extract with an oligonucleotide complementary to the 5′ portion of U2 snRNA (Fig. 3A). U2 snRNA was efficiently cleaved in the presence of this oligo, whereas a negative control oligo (cntl) had no effect (Fig. 3B). When CMV-Ftz was incubated in U2 snRNA or cntl extracts in the coupled system, the spliceosome was detected in the PlaF-treated cntl extract, but was disrupted in the U2 snRNA extract (Fig. 3C, cf. lanes 1 and 2). In contrast, the heparin-disrupted E7 spliceosome had the same mobility in cntl and U2 snRNA extracts (Fig. 3C, lanes 3,4). These data indicate that U2 snRNP is not a stable component of the heparin-disrupted, E7 spliceosome. In contrast, in low heparin, both the E7 and normal (PlaF) spliceosomes migrated with faster mobility in the U2 snRNA extract (Fig. 3D, lanes 2,4) versus the cntl extract (Fig. 3D, lanes 1,3), indicating that U2 snRNP is a component of the low-heparin E7 (and PlaF) spliceosome.
Figure 3.
E7 results in weak binding of U2 snRNP to pre-mRNA. (A) Schematic of the 5′ end of U2 snRNA. The line indicates the 20-nucleotide (nt) oligonucleotide complementary to U2 snRNA used for RNase H cleavage. See the Materials and Methods for sequences of the U2 oligonucleotide and the cntl. (B) Analysis of RNase H-cleaved U2 snRNA. Total RNA from U2 snRNA or cntl extracts was fractionated on a 5% denaturing polyacrylamide gel, and RNA was detected by ethidium bromide. The snRNAs and cleaved U2 snRNA are indicated. (C,D) CMV-Ftz DNA template was incubated in coupled RNAP II transcription/splicing reaction mixtures containing either PlaF (lanes 1,2) or E7 (lanes 3,4) for 5 min in U2 snRNA (ΔU2) or cntl extracts. An aliquot of each sample was mixed with 1.5 μL (Fig. 2C) or 0.5 μL (Fig. 2D) of heparin and fractionated on a 1.2% native agarose gel. (E) Chase assay showing that nonfunctional U2 snRNP remains loosely bound to pre-mRNA after E7 treatment. CMV-Ftz DNA template was transcribed for 5 min in the coupled transcription/splicing system containing 1 μM E7 (lanes 1,2) or PlaF (lanes 3,4). An aliquot (2 μL) of each sample in lanes 1 and 2 was diluted into a new 25-μL reaction mixture in the presence of α-amanitin and incubated for an additional 40 min. For cntl samples in lanes 5–8, CMV-Ftz was transcribed for 5 min in the presence of 100 nM E7107 (lanes 5,6) or 100 nM PlaF (lanes 7,8), α-amanitin was added, and incubation was continued for an additional 40 min. Total RNA was fractionated on a 5% denaturing polyacrylamide gel and detected by PhosphorImager. (F) CMV-Ftz DNA template was incubated in coupled transcription/splicing reactions for 5 min in the presence of E7 or PlaF. Immunoprecipitations were carried out in low salt (250 mM) using a monoclonal antibody to the U2 snRNP B″ protein, a rabbit polyclonal to SF3b155, and monoclonal or polyclonal antibodies to nonrelated proteins as negative controls. Total RNA was run on a 5% denaturing polyacrylamide gel, and CMV-Ftz pre-mRNA was detected by PhosphorImager.
To further verify that the low-heparin E7 spliceosome contains loosely bound U2 snRNP, we next asked whether this snRNP could be competed off of the pre-mRNA in the presence of normal nuclear extract. To do this, we carried out a chase assay in which 1 μM E7 or PlaF was added prior to transcription for 5 min to allow formation of the E7 or PlaF spliceosome, followed by a 10-fold dilution of these reactions into normal nuclear extract. At this dilution (100 nM), E7 has no effect on splicing (Fig. 3E, lanes 5–8). However, when the E7 spliceosome was chased in the normal extract, splicing was inefficient (Fig. 3E, lanes 1,2). In contrast, splicing was efficient when the PlaF spliceosome was chased in normal nuclear extract (Fig. 3E, lanes 3,4). We conclude that the E7 spliceosome contains nonfunctional U2 snRNP that is loosely bound to pre-mRNA but cannot exchange for the U2 snRNP in the normal nuclear extract. Finally, antibodies to the U2 snRNP protein B″ or an SF3b protein (SF3b155) specifically immunoprecipitate both the E7 and PlaF spliceosomes assembled on the CMV-Ftz transcript (Fig. 3F), providing further evidence that SF3b/U2 snRNP is associated with the defective E7 spliceosome. We conclude that E7 causes the formation of a labile spliceosome containing weakly bound U2 snRNP. In recent work, another small molecule splicing inhibitor (spliceostatin A), which also targets SF3b (Kaida et al. 2007), was shown to inhibit spliceosome assembly (Roybal and Jurica 2010). Moreover, in a study published in this issue of Genes & Development, Corrionero et al. (2011) found that spliceostatin A, like E7, inhibits tight binding of U2 snRNP to pre-mRNA by interfering with mechanisms that ensure proper base-pairing interactions between U2 snRNA and the pre-mRNA. Together, these results show that both drugs disrupt stable U2 snRNP recruitment, indicating that this step in spliceosome assembly is a key target for anti-tumor drugs.
E7 blocks ATP-dependent remodeling of U2 snRNP that exposes the BBR
We next sought to determine why E7 inhibits tight binding of U2 snRNP to pre-mRNA. We first examined the integrity of U2 snRNP, which exists as a 17S particle containing a 12S core U2 snRNP particle associated with SF3b, and another multiprotein complex known as SF3a (Wahl et al. 2009). When U2 snRNP was immunoprecipitated from nuclear extract after treatment with E7, PlaF, or no drug, the full set of 17S U2 snRNP proteins, including the SF3a/b proteins, was detected on a Coomassie-stained gel and by mass spectrometry of total proteins (Supplemental Fig. 2). Consistent with these results, Western analyses showed that the levels of SF3a/b components were the same in immunopurified E7-U2 snRNP and PlaF-U2 snRNP (Supplemental Fig. 2). Together, these data indicate that E7 has no apparent effect on the integrity of U2 snRNP or its association with SF3a/b.
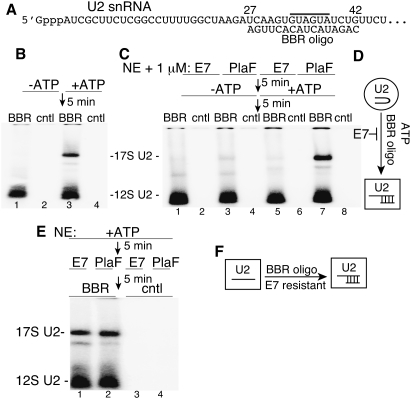
We next asked whether E7 alters U2 snRNP conformation. The 5′ region of U2 snRNA, which associates with SF3b, plays a critical role early in spliceosome assembly (Kramer et al. 1999; Dybkov et al. 2006). As E7 targets SF3b, we focused on this region of U2 snRNP. Using native gels, we assayed for binding of a 32P-labeled 2′OMethyl oligonucleotide complementary to nucleotides 27–42 of U2 snRNA (designated BBR oligo) (Fig. 4A). Consistent with previous work in yeast (Abu Dayyeh et al. 2002), the BBR oligonucleotide (Fig. 4B, lanes 1,3), but not a cntl (Fig. 4B, lanes 2,4), binds to 17S U2 snRNP in an ATP-dependent manner. In striking contrast, when nuclear extract is treated with E7, ATP-dependent binding of the BBR oligonucleotide to 17S U2 snRNP is abolished (Fig. 4C, lanes 5–8). We conclude that E7 potently inhibits an ATP-dependent conformational change that exposes the BBR of U2 snRNA (Fig. 4D).
Figure 4.
E7 blocks ATP-dependent remodeling of U2 snRNP to expose the BBR. (A) Schematic of the 5′-end region of U2 snRNA. The 2′ OMethyl BBR oligonucleotide is complementary to nucleotides 27–42 of U2 snRNA. The gray line above the U2 snRNA sequence indicates the BBR. (B) Nuclear extracts were incubated in the presence or absence of ATP for 5 min, followed by a 5-min incubation with 32P-labeled BBR or cntl 2′ OMethyl oligonucleotide. Samples were fractionated on a 1.2% agarose gel and detected by PhosphorImager. The 17S and 12S U2 snRNPs are indicated. (C) Same as B, except that nuclear extract was preincubated with 1 μM E7 or PlaF for 5 min. (D) Model showing that E7 blocks the ATP-dependent remodeling of U2 snRNP that exposes the BBR. The circle indicates the ATP-independent form of U2 snRNP, and the curved line within the circle indicates the inaccessible BBR in U2 snRNA. The square indicates remodeled U2 snRNP, and the straight lines indicate U2 snRNA base-paired to the BBR oligonucleotide. (E) Same as C, except that nuclear extract was preincubated in ATP for 5 min, followed by addition of the drugs for 5 min, and then the BBR or cntl oligonucleotide for 5 min. The 17S and 12S U2 snRNPs are indicated. (F) Model showing that the BBR oligonucleotide binds to remodeled 17S U2 snRNP in an E7-resistant manner. The square on the left indicates remodeled U2 snRNP, and the line within the square indicates U2 snRNA with an exposed BBR. In the square on the right, the lines indicate U2 snRNA and U2 snRNA base-paired to the BBR oligonucleotide.
To further investigate the remodeling of U2 snRNP, we carried out an order-of-addition experiment in which nuclear extract was first preincubated in ATP, and then E7 or PlaF was added. Significantly, binding of the BBR oligonucleotide to 17S U2 snRNP was now completely resistant to E7 (Fig. 4E). These data, together with the data in Figure 4C, suggest that, once the ATP-dependent, SF3b-dependent remodeling of U2 snRNP exposes the BBR (Fig. 4D), then binding of the BBR oligonucleotide can occur in an SF3b-independent manner (i.e., E7-resistant manner) (Fig. 4F). We conclude that SF3b and ATP, but not the BBR oligonucleotide, are required to trigger remodeling of U2 snRNP to expose the BBR.
In this study, we showed that E7 results in formation of defective spliceosomes in which U2 snRNP cannot bind tightly to pre-mRNA. Consistent with this conclusion, our data show that E7 abolishes the A complex assembly, in which U2 first binds tightly to pre-mRNA. We did not detect any effect of E7 on the integrity of U2 snRNP, but showed that E7 does potently block an ATP-dependent remodeling event that exposes the BBR of U2 snRNA. As E7 targets SF3b, our data lead to a model in which SF3b is required for the ATP-dependent remodeling of U2 snRNP to expose the BBR, and this event may be required for the SF3b- and ATP-dependent tight binding of U2 snRNP to pre-mRNA. Previous studies in yeast suggested that splicing factor Prp5p, which is an ATPase/helicase, facilitates an ATP-dependent alteration in U2 structure that is required for the BBR of U2 snRNA to become accessible to the BPS region in the intron (Ruby et al. 1993; Abu Dayyeh et al. 2002). Our work showing that E7 blocks ATP-dependent remodeling of U2 snRNP to expose the BBR strongly suggests that E7 exerts its effects at this Prp5p-dependent remodeling step. Another study showed that, during the A complex assembly, SF3a/b components bind tightly to the pre-mRNA at a region known as the anchoring site, which is located immediately upstream of the BPS (Gozani et al. 1996). More recently, Query and colleagues (Newnham and Query 2001) showed that the anchoring site mediates the ATP requirement for stable U2 snRNP binding, and suggested that this could be due to remodeling of SF3a/b. Our studies using E7 provide direct evidence for this proposal, showing that SF3b is required for tight binding of U2 snRNP to pre-mRNA and for remodeling U2 snRNP.
The identification of specific roles for SF3b via the use of E7 and spliceostatin A, as well as other recent work using small molecules to examine splicing, underscores the utility of these compounds for elucidating specific functions of the highly complex, dynamic factors in the spliceosome (Pilch et al. 2001; Muraki et al. 2004; Kaida et al. 2007; Kotake et al.2007; O'Brien et al. 2008; Stoilov et al. 2008; Sumanasekera et al. 2008; Kuhn et al.2009; Younis et al. 2010). Although the binding affinity of SF3b to E7 is highly correlated with its cell growth inhibition activity (Kotake et al. 2007), the mechanistic basis for the anti-tumor effect of E7 is not known. If the effect is due to splicing inhibition, then E7 may, for example, differentially affect splicing of pre-mRNAs containing weak splicing signals, and these pre-mRNAs may encode proteins involved in tumorogenesis. Our insights into the mechanism of action of E7 and its effects on SF3b/U2 snRNP will contribute to establishing the therapeutic potential of the drug, as well as the development of second-generation derivatives.
Materials and methods
Plasmids
The plasmid encoding the 755-nucleotide (nt) CMV-Ftz transcript was described (Das et al. 2006). The plasmid encoding the 695-nt CMV-Δss transcript was a generous gift from H. Lei and encodes the naturally intronless HSBP3 gene. CMV-DNA templates used in the coupled RNAP II transcription/splicing system were amplified by PCR using a forward primer (5′-TGGAGGTCGCTGAGTAGTGC-3′) and reverse primer (5′-TAGAAGGCACAGTCGAGGCT-3′). AdML pre-mRNA was transcribed with T7 RNA polymerase from a PCR product using the same primers and the DoA plasmid (Das et al. 2006).
Coupled transcription/splicing
In vitro RNAP II transcription/splicing reactions were performed at 30°C in 25-μL reaction mixtures containing 200 ng of CMV-DNA template, 1 μL of α-32P-UTP (800 Ci/mmol; Perkin Elmer Life Sciences catalog no. BLU507x), 0.5 mM ATP, 3.2 mM MgCl2, 20 mM creatine phosphate (di-Tris salt), and 15 μL of HeLa nuclear extract (Das et al. 2006). The indicated amounts of E7 or PlaF were added to the reaction mixtures before or after RNAP II transcription, as designated. α-Amanitin (10 ng/25 μL) was added after transcription, and incubation was continued for the indicated times.
Spliceosome assays
For spliceosome assembly, CMV-DNA templates were incubated under RNAP II transcription/splicing conditions for 8 min. G-50 columns (GE Healthcare Life Sciences) were used to remove unincorporated α-32P-UTP. For low- or high-heparin samples, 0.5 or 1.5 μL of heparin (6.5 g/L), respectively, was added to 10 μL of G-50 column-purified reactions prior to loading on 1.2% low-melting-point agarose gels (Das and Reed 1999). For assembly of spliceosomal complexes using the uncoupled system, AdML pre-mRNA (2 ng) was incubated for 0, 4, and 25 min under standard splicing conditions (0.5 mM ATP, 3.2 mM MgCl2, 20 mM creatine phosphate [di-Tris salt]) in a 25-μL reaction mixture containing 7.5 μL of nuclear extract and 7.5 μL of Splicing Dilution Buffer (20 mM Hepes at pH 7.6, 100 mM KCl). No drug or 1 μM E7 was included in the reactions. Aliquots (10 μL) of each reaction were mixed with 1 μL of heparin (6.5 g/L) and run on 1.5% low-melting-point agarose gel. For preparation of U2 snRNA and control extracts, 250 ng of oligonucleotide targeting U2 snRNA (5′-CCAAAAGGCCGAGAAGCGAT-3′) or cntl (5′-GGGGTGAATTCTTTGCCAA-3′) was added to 15 μL of nuclear extract, followed by incubation for 5 min at 30°C for endogenous RNase H cleavage of the snRNA. Total RNA was prepared from these samples and fractionated on a 5% denaturing gel, and snRNAs were detected by ethidium bromide. The ΔU2 and cntl extracts were used for spliceosome assembly assays as described above.
U2 snRNP conformation assay
The BBR 2′OMethyl oligonucleotide complementary to the BBR (5′-mCmAmGmAmUmAmCmUmAmAmCmAmCmUmUmGmA-3′) and the cntl (5′-mAmCmUmGmUmAmCmUmAmAmCmUmGmAmCmUmG-3′) were 32P-labeled using γ-ATP (6000 Ci/mmol) and T4 polynucleotide kinase. For all assays, ATP was depleted from nuclear extract by incubation for 20 min at room temperature. Reaction mixtures (25 μL) containing the depleted nuclear extract (15 μL) were incubated in the presence or absence of 0.5 mM ATP, 3.2 mM MgCl2, and 20 mM creatine phosphate (di-Tris salt) for the indicated times. BBR or control 2′OMethyl oligonucleotide (34 ng of each) and 1 μM E7 or PlaF were added to the reaction mixtures in the orders indicated in Figure 4. For reactions carried out in the absence of ATP, water was used to bring the volume up to 25 μL.
Acknowledgments
We are grateful to R. Das, K. Dufu, C.S. Lee, and members of the Reed laboratory for useful discussions, and to H. Lei and K. Dufu for critical comments on the manuscript. HeLa cells were obtained from the National Cell Culture Center (Minneapolis, MN). This work was supported by a post-doctoral fellowship to E.F. from Eisai Pharmaceuticals and an NIH grant to R.R.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2009411.
Supplemental material is available for this article.
References
- Abu Dayyeh BK, Quan TK, Castro M, Ruby SW 2002. Probing interactions between the U2 small nuclear ribonucleoprotein and the DEAD-box protein, Prp5. J Biol Chem 277: 20221–20233 [DOI] [PubMed] [Google Scholar]
- Corrionero A, Miñana B, Valcárcel J 2011. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev (this issue). doi: 10.1101/gad.2014311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Reed R 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5: 1504–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Zhou Z, Reed R 2000. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol Cell 5: 779–787 [DOI] [PubMed] [Google Scholar]
- Das R, Dufu K, Romney B, Feldt M, Elenko M, Reed R 2006. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev 20: 1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G, Hartmuth K, Luhrmann R 2004. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA 10: 1925–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G, Hartmuth K, Kastner B, Will CL, Luhrmann R 2007. The 5′ end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol Cell 25: 399–411 [DOI] [PubMed] [Google Scholar]
- Dybkov O, Will CL, Deckert J, Behzadnia N, Hartmuth K, Luhrmann R 2006. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol Cell Biol 26: 2803–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Feld R, Reed R 1996. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev 10: 233–243 [DOI] [PubMed] [Google Scholar]
- Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. 2007. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 3: 576–583 [DOI] [PubMed] [Google Scholar]
- Konarska MM, Query CC 2005. Insights into the mechanisms of splicing: more lessons from the ribosome. Genes Dev 19: 2255–2260 [DOI] [PubMed] [Google Scholar]
- Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y 2007. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol 3: 570–575 [DOI] [PubMed] [Google Scholar]
- Kramer A, Gruter P, Groning K, Kastner B 1999. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J Cell Biol 145: 1355–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn AN, van Santen MA, Schwienhorst A, Urlaub H, Luhrmann R 2009. Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation. RNA 15: 153–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraki M, Ohkawara B, Hosoya T, Onogi H, Koizumi J, Koizumi T, Sumi K, Yomoda J, Murray MV, Kimura H, et al. 2004. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem 279: 24246–24254 [DOI] [PubMed] [Google Scholar]
- Newnham CM, Query CC 2001. The ATP requirement for U2 snRNP addition is linked to the pre-mRNA region 5′ to the branch site. RNA 7: 1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien K, Matlin AJ, Lowell AM, Moore MJ 2008. The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. J Biol Chem 283: 33147–33154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilch B, Allemand E, Facompre M, Bailly C, Riou JF, Soret J, Tazi J 2001. Specific inhibition of serine- and arginine-rich splicing factors phosphorylation, spliceosome assembly, and splicing by the antitumor drug NB-506. Cancer Res 61: 6876–6884 [PubMed] [Google Scholar]
- Reed R 1996. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev 6: 215–220 [DOI] [PubMed] [Google Scholar]
- Roybal GA, Jurica MS 2010. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res 38: 6664–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby SW, Chang TH, Abelson J 1993. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev 7: 1909–1925 [DOI] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM 2008. ‘Nought may endure but mutability’: spliceosome dynamics and the regulation of splicing. Mol Cell 30: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL 2008. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc Natl Acad Sci 105: 11218–11223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanasekera C, Watt DS, Stamm S 2008. Substances that can change alternative splice-site selection. Biochem Soc Trans 36: 483–490 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Will CL, Schneider C, MacMillan AM, Katopodis NF, Neubauer G, Wilm M, Luhrmann R, Query CC 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J 20: 4536–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis I, Berg M, Kaida D, Dittmar K, Wang C, Dreyfuss G 2010. Rapid-response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Mol Cell Biol 30: 1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]