Abstract
Spliceostatin A (SSA) is a stabilized derivative of a Pseudomonas bacterial fermentation product that displays potent anti-proliferative and anti-tumor activities in cancer cells and animal models. The drug inhibits pre-mRNA splicing in vitro and in vivo and binds SF3b, a protein subcomplex of U2 small nuclear ribonucleoprotein (snRNP), which is essential for recognition of the pre-mRNA branch point. We report that SSA prevents interaction of an SF3b 155-kDa subunit with the pre-mRNA, concomitant with nonproductive recruitment of U2 snRNP to sequences 5′ of the branch point. Differences in base-pairing potential with U2 snRNA in this region lead to different sensitivity of 3′ splice sites to SSA, and to SSA-induced changes in alternative splicing. Indeed, rather than general splicing inhibition, splicing-sensitive microarray analyses reveal specific alternative splicing changes induced by the drug that significantly overlap with those induced by knockdown of SF3b 155. These changes lead to down-regulation of genes important for cell division, including cyclin A2 and Aurora A kinase, thus providing an explanation for the anti-proliferative effects of SSA. Our results reveal a mechanism that prevents nonproductive base-pairing interactions in the spliceosome, and highlight the regulatory and cancer therapeutic potential of perturbing the fidelity of splice site recognition.
Keywords: spliceostatin A, U2 snRNP, branch point, splicing fidelity, alternative splicing
Excision of intronic sequences from pre-mRNAs (pre-mRNA splicing) is an obligatory step for the expression of the majority of higher eukaryotic genes and requires the function of a complex molecular machinery, the spliceosome, which is composed of five small nuclear ribonucleoprotein (snRNP) complexes and >200 proteins (for review, see Wahl et al. 2009). The function of the spliceosome relies on an intricate network of protein–protein, protein–RNA, and RNA–RNA interactions that undergo significant conformational and compositional rearrangements to facilitate intron excision (for review, see Smith et al. 2008; Wahl et al. 2009). These interactions can be modulated to allow the generation of alternative patterns of splicing, a phenomenon reported to occur in the majority of human genes (for review, see Chen and Manley 2009; Nilsen and Graveley 2010). Alternative splicing plays important roles in the development of multicellular organisms and in numerous pathologies, including cancer (for review, see Cooper et al. 2009; David and Manley 2010).
The efficiency and precision required for intron removal is in contrast to the limited sequence consensus at intron/exon boundaries, a feature likely connected to the establishment of mechanisms of splice site regulation mediated by a diverse array of auxiliary sequences and cognate factors (for review, see Wang and Burge 2008; Barash et al. 2010). In higher eukaryotes, the 3′ end of the intron is characterized by a polypyrimidine-rich sequence (the pY-tract) followed by a conserved AG as the intron 3′-most dinucleotide. Upstream of the pY-tract is the branch point, which functions optimally when it coincides with the invariant yeast sequence UACUAAC (Zhuang et al. 1989), but which corresponds to the loose consensus YUNAY in higher eukaryotes (Y represents pyrimidines, R represents purines, N represents any nucleotide, and the underlined A represents the invariant adenosine that engages in a 2′–5′ phosphodiester bond with the 5′ end of the intron after the first catalytic step of the splicing reaction) (Gao et al. 2008; for review, see Wahl et al. 2009). Initial recognition of the 3′ splice site sequences involves cooperative binding of the branch point-binding protein (BBP/SF1) to the branch point, and of the U2 snRNP auxiliary factor (U2AF) 65-kDa and 35-kDa subunits to the pY-tract and the AG dinucleotide, respectively (Ruskin et al. 1988; Zamore et al. 1992; Berglund et al. 1997; Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999; Liu et al. 2001; Selenko et al. 2003). RNA–protein and protein–protein contacts involving these factors are believed to enforce a unique RNA structure that triggers subsequent events in 3′ splice site recognition (Kent et al. 2003). These interactions, together with recognition of the 5′ splice site by U1 snRNP and possibly other contacts across the intron, establish the earliest ATP-independent (E) complex that commits the pre-mRNA to the splicing pathway (Michaud and Reed 1991, 1993).
The next step in 3′ splice site recognition is the recruitment of U2 snRNP, which is composed of U2 snRNA and several polypeptides, including two protein subcomplexes (SF3a and SF3b), which are part of the 17S U2 snRNP complex (for review, see Wahl et al. 2009). SF3a is composed of three subunits of 60, 66, and 120 kDa, while SF3b is composed of at least eight subunits of 10, 14a, 14b, 49, 125, 130, 140, and 155 kDa (for review, see Wahl et al. 2009). U2 snRNP recruitment involves at least two separable steps. U2 snRNP has been detected in E complexes, but U2 binding in this complex is not stable and does not require the presence of a branch point (Hong et al. 1997; Das et al. 2000; Dönmez et al. 2004, 2007). Stable binding of U2 snRNP to form prespliceosomal complex A requires ATP and depends on base-pairing interactions between the branch point and a region of U2 snRNA known as the branch point recognition sequence (bprs) (Parker et al. 1987; Nelson and Green 1989; Wu and Manley 1989; Zhuang and Weiner 1989). Base-pairing between the branch point and the bprs bulges out the branch point adenosine, which facilitates the first catalytic step (Query et al. 1994; Smith et al. 2009). It has been proposed recently that initial interaction of the bprs with the branch point requires a stem–loop structure in U2 snRNA, which is subsequently disrupted to facilitate stable base-pairing between the branch point and the bprs (Perriman and Ares 2010). This rearrangement of RNA–RNA interactions could be mediated by Prp5, a member of the RNA-dependent DExH/D-box family of ATPases/helicases involved in fidelity of splice site recognition and other key spliceosomal transitions (for review, see Konarska et al. 2006), because the ATPase activity of Prp5 has been linked to kinetic proofreading of branch point/bprs base-pairing interactions (Xu and Query 2007).
Stable U2 snRNP recruitment also relies on additional RNA–protein and protein–protein interactions. Components of the SF3a and SF3b subcomplexes bind tightly to pre-mRNA sequences 5′ (anchoring site [AS]) or 3′ of the branch point, apparently in a sequence-independent manner (Gozani et al. 1996). Interestingly, SF3b 155, the most conserved SF3b subunit, cross-links to sequences both 5′ and 3′ of the branch point (Gozani et al. 1998), suggesting an important role for this factor in tethering U2 snRNP in the proximity of the branch point. A central function for SF3b 155 in U2 snRNP recruitment and splicing catalysis is also suggested by its interaction with U2AF65 (Gozani et al. 1998; Spadaccini et al. 2006; Thickman et al. 2006); its interaction with p14, a protein known to interact directly with the branch point adenosine (Query et al. 1996; Will et al. 2001; Dybkov et al. 2006); and its phosphorylation concomitant with catalytic activation of the spliceosome (Wang et al. 1998).
U2 snRNP binding serves as a platform for additional RNA–RNA and protein interactions, leading to the recruitment of the tri-snRNP U4/U5/U6 and formation of mature spliceosome complexes, within which numerous RNA rearrangements and changes in protein composition facilitate splicing catalysis (for review, see Smith et al. 2008; Wahl et al. 2009).
Investigation of the function of the ribosome, another complex ribonucleoprotein machinery, has been greatly assisted by the identification of families of antibiotics and other small molecules that interfere with specific steps of the translation process (for review, see Ogle and Ramakrishnan 2005). In contrast, only a handful of small molecules have been identified that affect the function of the spliceosome (Kaida et al. 2007; Kotake et al. 2007; O'Brien et al. 2008; Stoilov et al. 2008; Kuhn et al. 2009; Roybal and Jurica 2010). Spliceostatin A (SSA) is a stabilized methyl ketal derivative of FR901464, a fermentation product of Pseudomonas bacteria that displays potent cytotoxic activity against various solid tumor cell lines at nanomolar IC50 and prolongs the life of tumor-bearing mice (Nakajima et al. 1996; Kaida et al. 2007). Both SSA and pladienolide, a structurally related anti-cancer compound from Streptomyces, have been found to bind to protein components of SF3b (Kaida et al. 2007; Kotake et al. 2007) and inhibit pre-mRNA splicing in vitro and in cultured cells (Kaida et al. 2007, 2010; Kotake et al. 2007; Furumai et al. 2010; Roybal and Jurica 2010).
In this study, we investigated the mechanism by which SSA inhibits pre-mRNA splicing. Our results indicate that SSA prevents the interaction of SF3b 155 with pre-mRNA sequences, concomitant with the establishment of base-pairing interactions between U2 snRNA and sequences 5′ of the branch point. This leads to unstable, nonproductive recruitment of U2 snRNP, and the potential of these sequences to base-pair with U2 snRNA establishes differences in the sensitivity of 3′ splice sites to the drug. Differences in SSA sensitivity between competing 3′ splice sites underlie changes in alternative splicing, and we report that such changes have an impact on the expression of key regulators of cell division. We propose that SSA influences the fidelity of 3′ splice site recognition, leading to changes in alternative splicing of cell cycle regulators that can contribute to the anti-proliferative activity of the drug.
Results
Effects of SSA on spliceosome assembly
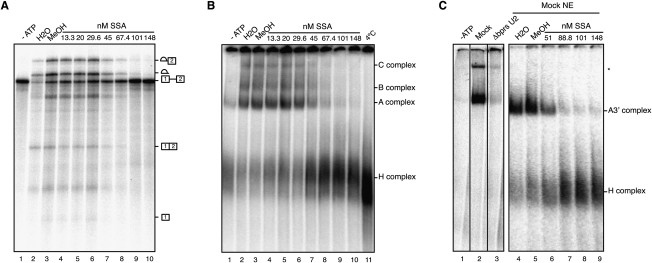
To study the mechanism of splicing inhibition by SSA, a radioactively labeled model pre-mRNA substrate from adenovirus (adenovirus major late promoter [AdML]) was incubated with HeLa nuclear extracts under splicing conditions in the absence or presence of increasing concentrations of SSA. ATP-dependent accumulation of splicing intermediates and products was inhibited at concentrations starting at 45 nM, with essentially complete inhibition at 100 nM (Fig. 1A). Fully assembled splicing complexes contain five snRNP particles (U1, U2, and the tri-snRNP U4/U5/U6) (Wahl et al. 2009). Distinct splicing-related subcomplexes can be resolved by native agarose–polyacrylamide gel electrophoresis after treatment of pre-mRNA/nuclear extract incubation mixes with heparin (Konarska and Sharp 1986; Krämer 1988): Complex A contains U2 snRNP bound to the 3′ splice site region of the pre-mRNA, while complexes B and catalytically active C contain in addition U4/U5/U6 snRNPs. Heterogeneous complexes of pre-mRNA with hnRNP proteins (H complex) display higher electrophoretic mobility in these assays. SSA was found to inhibit U2 snRNP-containing A and subsequent complexes at concentrations that parallel those leading to splicing inhibition (Fig. 1B). Complex A assembly on an RNA corresponding to the 3′ half of the intron and downstream exon was also inhibited at equivalent concentrations, indicating that a 5′ splice site is not required for SSA-mediated inhibition (Fig. 1C).
Figure 1.
SSA inhibits U2 snRNP-containing A complex assembly. (A) In vitro splicing assays using AdML RNA in the presence of the indicated concentrations of SSA or its carrier (methanol), analyzed by denaturing polyacrylamide gel electrophoresis. Pre-mRNA, splicing intermediates, and products are indicated. (B) Spliceosome assembly assays corresponding to in vitro splicing mixes as in A in the presence of heparin (5 μg/μL), analyzed by native agarose–polyacrylamide gel electrophoresis. The positions of heterogeneous nuclear RNP (H) and spliceosomal A, B, and C complexes are shown. (C) Spliceosome assembly assays as in B using AdML 3′ RNA containing the 3′ 40 nt of AdML intron 1 and exon 2. The positions of the A3′ and H complexes are indicated.
SSA destabilizes U2 snRNP assembly
Previous work (Das et al. 2000; Dönmez et al. 2004, 2007) established that initial, more labile, association of U2 snRNP with the pre-mRNA can be detected in the absence of heparin. In the absence of heparin, we found that SSA alters the electrophoretic mobility in low-melting agarose gels but does not inhibit formation of ATP-dependent, U2 snRNP-containing complexes (Fig. 2A, lanes 1–7). Indeed, using this assay system, the drug inhibits U2 snRNP complexes only when heparin (5 mg/mL) was added after complex formation previous to electrophoresis (Fig. 2B, cf. lanes 1–6 and 7–12). These observations suggest that SSA weakens the interaction of U2 snRNP with the pre-mRNA, thus facilitating that the RNA mimetic properties of the highly negatively charged heparin molecules displace U2 snRNP from the pre-mRNA.
Figure 2.
SSA destabilizes U2 snRNP assembly. (A) Spliceosome assembly assays as in Figure 1C in the absence of heparin, analyzed by low-melting agarose-gel electrophoresis, using either mock-treated or nuclear extracts in which the bprs of U2 snRNA was inactivated by RNase H-mediated degradation (ΔbprsU2) in the absence or presence of SSA at the indicated concentrations. (B) Spliceosome assembly assays as in A in the presence of different concentrations of the indicated nuclear extracts with or without SSA, and in the absence or presence (1.67 and 5 μg/μL) of heparin. (C) Spliceosome assembly assays as in A using wild-type AdML (−40), a mutant in which the branch point region was deleted (−19) or a mutant in which the branch point region (ACUUAU, where the underlined adenosine is the branch site) was mutated to GTCCTC (−40 X). (D) Spliceosome assembly assays were performed as in A and Supplemental Figure S2 on AdML or Fas RNAs. After electrophoresis, complexes were extracted from the gel, the RNA was purified, and the presence of U2 snRNA in the complex was analyzed by primer extension using 5′ 32P-labeled oligonucleotides complementary to U2 snRNA nucleotides 13–28 (top panel) (expected length of the primer extension product: 28 nt) or 171–187 (bottom panel) (expected lengths of primer extension products: 187 nt for full-length U2 snRNA and 145 nt for Δbprs U2 snRNA).
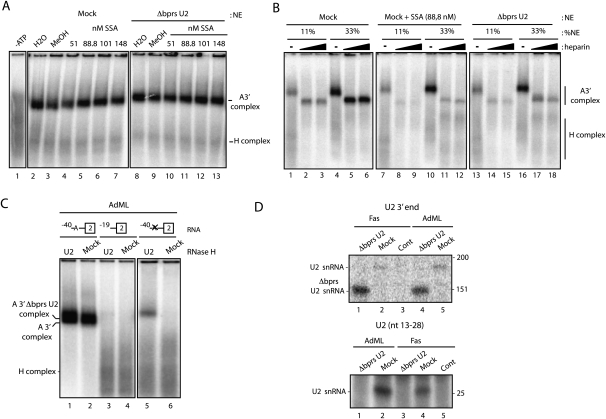
U2 snRNP binding involves base-pairing between sequences in the pre-mRNA branch point region and the bprs in U2 snRNA. RNase H-mediated degradation of the bprs is commonly used to inactivate U2 snRNP (ΔbprsU2). Unexpectedly, however, a complex was detected between Δbprs U2 snRNP and the 3′ splice site region in the absence of heparin (Fig. 2A, lane 8; Supplemental Fig. 1A), indicating that base-pairing between the pre-mRNA branch point and U2 snRNA bprs is not essential for initial U2 snRNP binding. Neither the amount nor the mobility of this complex was affected by SSA treatment (Fig. 2A, lanes 8–13).
Similar to the complex formed with intact U2 snRNP in the presence of SSA, the ΔbprsU2 complex was heparin-sensitive (Fig. 2B, lanes 13–18) and showed hallmarks of functional U2 snRNP recruitment, including (1) dependence on ATP, (2) dependence on the presence of a branch point (Fig. 2C), (3) dependence on U2AF (Supplemental Fig. 1B,C), and (4) sensitivity to exon definition effects (Supplemental Fig. 2). Indeed, the presence of U2 snRNA in the complex was confirmed by primer extension (Fig. 2D, top panel) and RNase H-mediated degradation (Fig. 3C). Interestingly, U2 snRNA sequences corresponding to the 5′ digestion product could not be detected (Fig. 2D, bottom panel), indicating that the complex lacks stem–loop I of U2 snRNA.
Figure 3.
SSA treatment and U2 snRNA bprs inactivation alter RNA–protein interactions important for U2 snRNP recruitment but do not inhibit U2 snRNA base-pairing. (A) UV light-induced cross-linking, followed by immunoprecipitation of SF3b155, SF3a60, and U2AF65 in the presence of SSA (88 nM) or methanol, incubated with mock or ΔbprsU2 nuclear extracts. Asterisk indicates a cross-linked protein coimmunoprecipitated with the SF3a60 antibody, possibly SF3a66 (Brosi et al. 1993). (B) Psoralen-induced cross-linking was analyzed as in A using either mock or Δbprs U2 snRNA nuclear extracts, in the presence or absence of SSA (53 or 267 nM) or methanol. The positions of cross-linked adducts (CXL) formed with wild-type and ΔbprsU2 are labeled as T (top) and B (bottom), respectively. (C, top) Schematic representation of U2 snRNA secondary structure domains and mapping of U2 snRNA/pre-mRNA interactions by RNase H-mediated degradation of psoralen-induced cross-links between radioactively labeled AdML 3′ RNA and either U2 snRNA (Mock) or ΔbprsU2 snRNA, analyzed by denaturing gel electrophoresis. As expected, the position of the cross-linked RNA is affected by RNase H-mediated degradation of U2 snRNA at sequences 5′ (Loop I) or 3′ (Sm site) of the bprs in mock extracts, while cross-linking of the ΔbprsU2 snRNA occurs 3′ of the bprs, as indicated by altered electrophoretic mobility of the RNA–RNA cross-link upon degradation of the 3′ Sm site, but not of the 5′ loop I.
Taken together, our observations indicate that complexes between U2 snRNP and the pre-mRNA formed in the presence of SSA have lower stability and share properties with similarly labile complexes formed in the absence of U2 bprs (see also below).
SSA leads to altered RNA–protein and RNA–RNA interactions in U2 snRNP recruitment
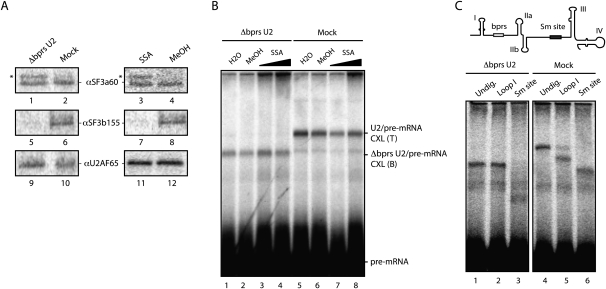
To further characterize these complexes, the interactions of protein and RNA components of U2 snRNP with the pre-mRNA were analyzed. Ultraviolet (UV) light irradiation and immunoprecipitation experiments were used to test the interaction of 3′ splice site-recognizing proteins with radioactively labeled pre-mRNA in the presence or absence of SSA. While interactions with the pre-mRNA of U2AF65 or SF3a60 were not affected by SSA, cross-linking of SF3b 155 was significantly decreased in the presence of the drug (Fig. 3A, right panel). The presence of an additional cross-linked species immunoprecipitated by anti-SF3a 60 antibodies, which may correspond to the 66-Kda subunit of the SF3a complex (Bennett and Reed 1993; Brosi et al. 1993), further points to SSA-induced conformational rearrangements of RNA–protein contacts between U2 snRNP components and the pre-mRNA. Adding to the similarities mentioned above between U2 snRNP complexes assembled in the presence of SSA and upon degradation of U2 snRNA bprs, cross-linking of SF3b 155, but not of U2AF65 or SF3a 60, was significantly decreased in ΔbprsU2 nuclear extracts (Fig. 3A, left panel).
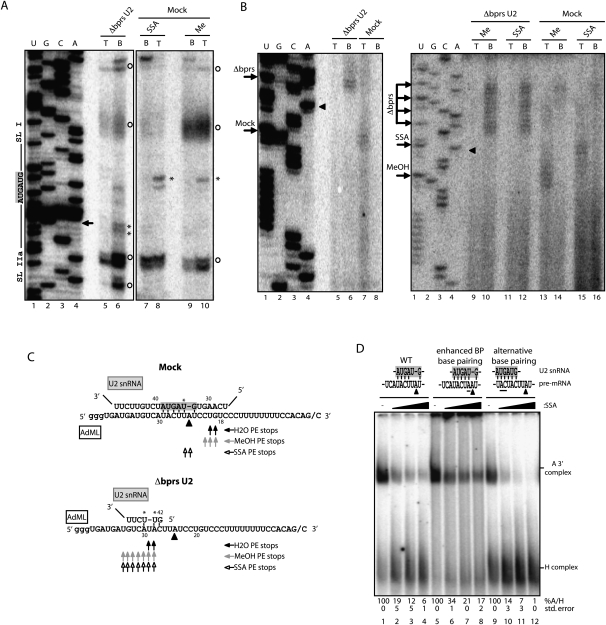
Base-pairing interactions between U2 snRNA and the pre-mRNA were monitored using psoralen-mediated, long-wave UV light-induced cross-linking (Wassarman and Steitz 1992). Cross-linking between U2 snRNA and the pre-mRNA was largely unaffected by SSA treatment (Fig. 3B, lanes 5–8). Surprisingly, cross-linking could also be detected between ΔbprsU2 snRNA and the pre-mRNA, and this cross-linking was not affected by SSA either (Fig. 3B, lanes 1–4). RNase H-mediated digestion of different regions of U2 snRNA indicated that cross-linking involved residues 3′ of the bprs in the ΔbprsU2 snRNA: RNase H digestion using an oligonucleotide complementary to the Sm site affected the electrophoretic mobility of the RNA–RNA adduct (Fig. 3C, cf. lanes 1 and 3), while digestion using an oligonucleotide complementary to stem–loop I did not (Fig. 3C, lane 2). As expected, both oligonucleotides affected the mobility of adducts formed in mock-inactivated nuclear extracts (Fig. 3C, lanes 4–6). To precisely map the cross-linked residues in U2 snRNA and the pre-mRNA, primer extension assays were carried out on the purified RNA–RNA adduct. Using an oligonucleotide complementary to the Sm site of U2 snRNA, similar primer extension products were obtained in the presence or absence of SSA (Fig. 4A, cf. lanes 8 and 10): Specific primer extension stops (Fig. 4A, labeled with an asterisk) correspond to nucleotides within the bprs, as expected for standard base-pairing of U2 snRNA with the pre-mRNA. Consistent with the RNase H degradation results of Figure 3C, nucleotides 3′ from the bprs were involved in cross-linking of the ΔbprsU2 RNA (Fig. 4A, lane 6). Using an oligonucleotide complementary to the pre-mRNA 3′ exon, primer extension stops mapped the sites of cross-linking just 3′ of the branch point adenosine in the absence of SSA, as expected, while, in the presence of the drug, cross-linking was mapped to nucleotides further upstream in the pre-mRNA (Fig. 4B [lane 13 vs. 15, arrows labeled MeOH and SSA, respectively], C). These observations suggest that SSA induces altered base-pairing between U2 snRNA and the pre-mRNA. Remarkably, in both the absence or presence of SSA, cross-linking between ΔbprsU2 snRNA and the pre-mRNA was mapped to multiple nucleotides significantly 5′ distant from the branch point (Fig. 4B [lane 6 vs. 7, lanes 10,12 vs. 13], C). Taken together, the results indicate that SSA treatment or U2 snRNA bprs inactivation alters the pattern of U2 snRNA base-pairing with the pre-mRNA, allowing interactions of U2 snRNA with regions upstream of the branch point (summarized in Fig. 4C) that occur concomitant with loss of SF3b 155 interaction with the pre-mRNA (Fig. 3A).
Figure 4.
SSA induces altered base-pairing between U2 snRNA and the pre-mRNA. (A) Mapping of U2 snRNA–AdML cross-links by primer extension. U2 snRNA–AdML adducts from either mock or Δbprs U2 nuclear extracts and methanol-treated or SSA-treated (88 nM) nuclear extracts were gel-extracted and purified. To assess the presence of background levels of U2 snRNA in the lanes, gel slices corresponding to top (T) and bottom (B) positions defined as in Figure 3B were analyzed in parallel for each sample. Primer extension was carried out using a 32P-labeled oligonucleotide complementary to the Sm site sequence in U2 snRNA. Lanes 1–4 correspond to dideoxy sequencing of U2 snRNA using the same primer. The bprs (5′-GUAGUA-3′) and boundaries of the flanking stem–loops I and IIa are indicated. Asterisks indicate specific primer extension stops. Circles indicate nonspecific stops observed in primer extension of U2 snRNA in the absence of psoralen cross-linking. The arrow indicates the 5′ boundary of the antisense oligonucleotide used for RNase H treatment. (B) Mapping of AdML pre-mRNA cross-linked residues by primer extension as in A using a 32P-labeled oligonucleotide primer complementary to AdML exon 2. Lanes 1–4 correspond to dideoxy sequencing reactions of AdML DNA using the same primer. Arrows indicate primer extension stops. Triangles indicate the position of the branch point adenosine. (C) Schematic representation of base-pairing interactions between U2 snRNA and the AdML branch point region in mock or Δbprs U2 nuclear extracts in the presence of SSA, methanol, or water. Asterisks indicate U2 snRNA cross-linked nucleotides as mapped in A. Triangles indicate the branch point adenosine, and residues in the gray box indicate the bprs. Arrows summarize primer extension (PE) stops observed in B using different conditions as indicated. (D) Spliceosome assembly assays (as in Fig. 1C) using AdML wild type and mutants harboring either a consensus branch point (enhanced BP base-pairing) or a sequence located 5′ of the branch point region that is fully complementary to U2 snRNA bprs (alternative base-pairing). Mutated residues are underlined. Quantification of the A/H complex ratio for a minimum of three independent experiments (with standard deviation) is shown. The ratio in methanol-treated samples was considered as 100% in each case.
To assess whether the potential of sequences 5′ of the branch point to form alternative base-pairing interactions with U2 snRNA can modulate the effects of SSA, mutant pre-mRNAs were generated that either increase the potential of base-pairing interactions of the natural branch point region or offer an alternative site of perfect base-pairing (Fig. 4D, top). Enhancing base-pairing with the proper branch site region reduced the sensitivity of the 3′ splice site region to SSA (Fig. 4D, cf. lanes 1–4 and 5–8). Reciprocally, stronger inhibition was observed in a mutant that improves the base-pairing potential of the region 5′ of the branch point (Fig. 4D, cf. lanes 1–4 and 9–12), possibly due to steric hindrance between U2 particles competing for recruitment at the overlapping sites. These results are consistent with the possibility that the splicing inhibitory effects of SSA involve the establishment of alternative, nonproductive base-pairing between U2 snRNA and the pre-mRNA in the presence of the drug.
SSA leads to changes in alternative splicing
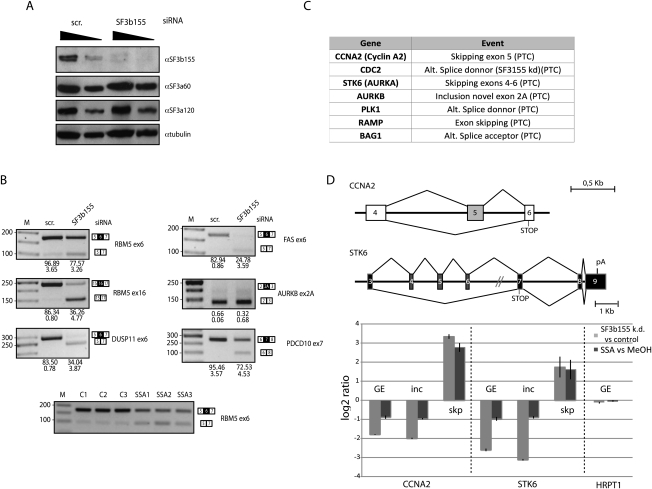
Given the differential sensitivity of 3′ splice sites to SSA, it is conceivable that the drug can lead to altered splice site selection in alternatively spliced transcripts. To test this possibility, splicing-sensitive microarrays (Muñoz et al. 2009) were used to evaluate the impact of SSA in alternative splicing decisions. The results of these experiments revealed that treatment of HeLa cells with 100 ng/mL SSA for 3 h affected 11% of the alternative splicing events monitored by the array in 21% of the genes, with a validation rate by quantitative RT–PCR of 72% (Supplemental Figs. 3–6). Gene expression changes were also observed in 10% of the genes, 72% of which were concomitant with changes in alternative splicing, often coupled to the generation of premature translation termination codons (PTCs) and, consequently, subject to nonsense-mediated mRNA degradation (AS-NMD) (Lareau et al. 2007). In agreement with the observation that SSA prevents interaction of SF3b 155 with the pre-mRNA (Fig. 3A), a sample of validated SSA-induced alternative splicing events showed similar changes upon SF3b 155 knockdown (Fig. 5A,B). Furthermore, splicing-sensitive microarray analyses showed that 30% of the SSA-induced alternative splicing events were also observed in microarray analyses of RNAs isolated after knockdown of SF3b 155 using two different siRNAs (Supplemental Fig. S5), which represents a highly significant statistical overlap (P < 0.001, Fischer exact test). We conclude that, under our experimental conditions, SSA does not cause a general inhibition of splicing, but rather induces changes in alternative splicing that significantly overlap with changes induced by SF3b 155 knockdown, consistent with the inactivating effect of SSA on SF3b155 function.
Figure 5.
SSA induces changes in alternative splicing and expression of genes important for cell cycle control. (A) Western blot analysis of the effects of SF3b 155 RNAi on the levels of SF3b 155 and other protein components of U2 snRNP (as well as α-tubulin as loading control) demonstrates specific depletion of SF3b 155. (B) Semiquantitative RT–PCR analysis of alternative splicing changes upon SF3b 155 knockdown from a selection of alternative splicing events induced by SSA. Genes and alternative splicing events are indicated, as well as the position of the alternatively spliced products. The numbers below each lane indicate percentage of exon inclusion (average and standard deviation for a minimum of three independent experiments). Alternative splicing changes common to those induced by SSA are observed for all except AURKB, which displays essentially quantitative exon skipping. The bottom panel shows changes in RBM5 exon 6 induced by SSA treatment: C1–C3 show results from three control samples, while SSA1–SSA3 correspond to three independent RNA samples from cells treated with SSA. (C) Genes encoding important cell cycle regulators whose expression is down-regulated by SSA and knockdown of SF3b 155 using two independent siRNAs. Down-regulation is coupled to the generation of alternatively spliced isoforms containing PTCs and, consequently, AS-NMD. (D, top) Schematic representation of alternative splicing events leading to NMD in the cyclin A2 (CCNA2) and Aurora Kinase A (STK6) genes, activated by both SSA treatment and SF3b 155 knockdown; indicated are PTCs associated with the alternative splicing patterns leading to NMD. (Bottom) Quantitative RT–PCR analysis using primers that monitor general gene expression in a constitutive exon (GE) or primers specific for splice junctions corresponding to exon inclusion (inc) or skipping (skp). Also shown is the absence of effects of the treatments on expression of a housekeeping gene (HRPT1).
Both SSA treatment and SF3b 155 knockdown reduced cell proliferation (Kaida et al. 2007; data not shown), and significant down-regulation by AS-NMD of genes that encode important regulators of cell proliferation was observed in both situations (Fig. 5C). For example, in addition to the previously reported inhibition of p27 splicing (Kaida et al. 2007; data not shown), AS-NMD was observed for the CCNA2 (cyclin A2) and STK6 (Aurora A kinase) genes by quantitative RT–PCR using primer pairs that monitor gene expression (by amplification of contitutive exons) or alternative exon inclusion and skipping (by including specific splice junction primers in the amplification reactions) (Fig. 5D). The mRNA levels of other intron-containing genes like HRPT1 were not affected under the same conditions (Fig. 5D). Cyclin A2 and Aurora A kinase are important for cell cycle transitions similar to those blocked by SSA (Mitra and Enders 2004; Saeki et al. 2009). Taken together, the results reveal that SSA treatment and SF3b 155 inactivation induce specific changes in alternative splicing and gene expression, including changes in key regulators of cell division that can contribute to explaining the anti-proliferative properties of the drug.
Decoy branch points in the AS determine SSA/SF3b 155-mediated alternative splicing
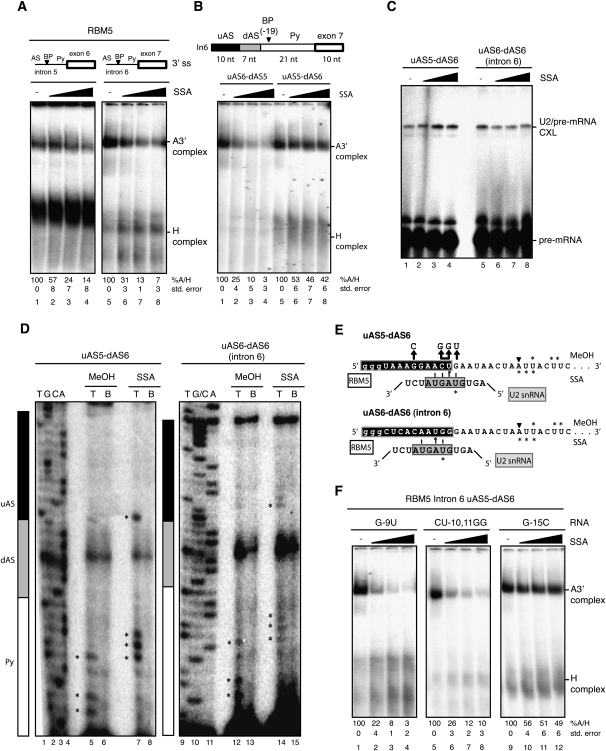
To address the mechanism of alternative splicing changes induced by SSA, we focused on an alternative splicing event in RBM5, a putative tumor suppressor gene encoding a splicing regulator implicated in the control of cell division and apoptosis (Oh et al. 2006; Bonnal et al. 2008; Fushimi et al. 2008). Both SSA treatment and SF3b 155 knockdown induce skipping of exon 6 (Fig. 5B; Supplemental Fig. 3D), and the competing 3′ splice site regions proved amenable for biochemical studies (Fig. 6). Spliceosome assembly assays showed that recruitment of U2 snRNP to RNAs containing the competing 3′ regions displays differential sensitivity to SSA (Fig. 6A, cf. lanes 1–4 and 5–8). Contrary to expectation, however, the 3′ splice site of intron 5 displays higher apparent resistance to SSA than the 3′ splice site of intron 6, while splicing to the intron 5 3′ splice site is repressed and splicing to the intron 6 3′ splice site remains active in the presence of the drug. This counterintuitive pattern of differential sensitivity was shared by another tested set of alternative 3′ splice sites involved in similar SSA-mediated alternative splicing effects (Supplemental Fig. 7). Of relevance, the 3′ splice site regions that are more affected by SSA are also more efficient in complex A assembly in the absence of the drug (Fig. 6A, cf. the ratio between A and H complexes in lanes 1 and 5; Supplemental Fig. 7). These results suggest that differential sensitivity to SSA does not simply reflect the relative efficiency of the splice sites to engage U2 snRNP.
Figure 6.
Differential sensitivity to SSA is modulated by sequences upstream of the branch point. (A) Spliceosome assembly assays were carried out as in Figure 1C using RNAs corresponding to RBM5 introns 5 and 6 3′ splice site regions, including the polypyrimidine tract (Py), branch point (BP), and AS. (B) Spliceosome assembly assays (as in A) using chimeric RBM5 intron 6 RNAs in which the indicated sequences have been replaced by the corresponding fragments of intron 5. The length of the different regions of the RNAs used is indicated. (C) Psoralen cross-linking assays (as in Fig. 2B) using the indicated RNAs. (D) Mapping of U2 snRNA/RBM5 intron 6 psoralen-induced cross-links by primer extension in the absence or presence of SSA, as in Figure 2C. Lanes 1–4 and 9–11 correspond to dideoxy sequencing reactions of the corresponding DNA products using the same primer. Asterisks indicate primer extension stops. Black, gray, and white boxes designate the different segments of the 3′ splice site region, as in B. (E) Schematic representation of potential U2 snRNA/RBM5 pre-mRNA base-pairing interactions induced by SSA. Asterisks mark the cross-linked nucleotides mapped in C in methanol- and SSA-treated samples. Nucleotides in the gray box represent the U2 snRNA bprs. (F) Spliceosome assembly assays (as in A) using RNAs harboring mutations (also indicated in E) in the residues proposed to base-pair with U2 snRNA bprs (or control residues) of the uAS5–dAS6 chimeric RNA.
Given the contribution of sequences 5′ of the branch site to the differential sensitivity of 3′ splice site regions to SSA (Fig. 4D), we tested whether such sequences also contribute to the differential effects of SSA on the alternative 3′ splice sites of RBM5. A segment of 17 nucleotides (nt) 5′ of the branch point (within the region known as the AS) (Gozani et al. 1998) of RBM5 intron 6 was divided in two subregions (upstream AS [uAS] and downtream AS[dAS]), and each of them was independently replaced by sequences located at equivalent positions in the 3′ splice site region of RBM5 intron 5. While replacement of the dAS sequence did not alter the sensitivity of the 3′ splice site region to SSA (Fig. 6, cf. lanes 5–8 in A and lanes 1–4 in B), replacement of the uAS sequence significantly reduced the effects of SSA on this 3′ splice site (Fig. 6B, lanes 5–8). These results reinforce the concept that differential sensitivity to SSA can be modulated by sequences 5′ of the branch point. Interestingly, replacement of the uAS sequence resulted in enhanced psoralen-mediated cross-linking with U2 snRNA in the presence of SSA, while this was not the case for the wild-type intron 6 3′ splice site region (Fig. 6C, cf. lanes 1–4 and 5–8). These results suggest that the uAS sequence actively facilitates U2 snRNP binding, concomitant with higher apparent resistance to SSA.
To determine whether SSA induces relocalization of U2 snRNP interactions, as observed for AdML pre-mRNA, the residues involved in psoralen-mediated cross-linking were determined by primer extension, as in Figure 4B. As observed for AdML pre-mRNA, mapping of U2 snRNA/pre-mRNA interactions revealed that SSA induces interactions of U2 snRNA with regions 5′ of the branch point (Fig. 6D [cf. asterisk-labeled bands in lanes 5,7,12,14], summarized in E). Also similar to the observations made with the AdML substrate (Fig. 3A), the changes in the location of U2 snRNA interaction were concomitant with reduced SF3b 155 (but not SF3a60 or U2AF65) cross-linking to the pre-mRNA (Supplemental Fig. 8). Significantly, the novel sites of U2 interaction included a relatively distant cross-linking site located in the uAS region (Fig. 6D), which confers SSA resistance (Fig. 6B) and enhanced U2 interaction (Fig. 6C). This region has substantial base-pairing potential with U2 snRNA (Fig. 6E), and mutation of residues involved in this base-pairing scheme strongly increased sensitivity to SSA (Fig. 6, cf. lanes 1–8 in F and lanes 5–8 in B), while mutation of another nucleotide not involved in this base-pairing scheme did not (Fig. 6, cf. lanes 9–12 in F and lanes 5–8 in B). We conclude that SSA induces base-pairing of U2 snRNA to sequences substantially 5′ of the branch point, and that the base-pairing potential of these regions with U2 snRNA influences the differential effects of the drug on alternative 3′ splice site regions.
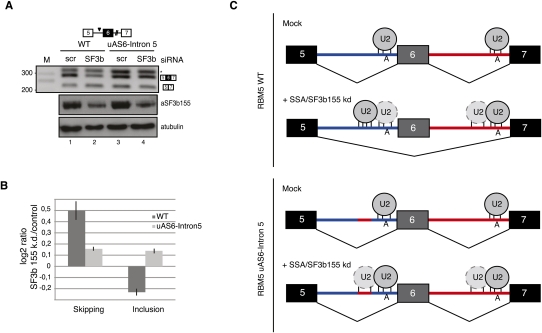
To test whether such sequences can indeed influence splice site choice, we transiently transfected HeLa cells with minigenes that include the genomic region of RBM5 between exons 5 and 7. Cotransfection with siRNAs against SF3b 155 resulted in increased levels of exon 6 skipping (Fig. 7, cf. inclusion/skipping ratio between lanes 1,2 in A and results of quantitative PCR using splice junction primers in B), as observed for the endogenous gene upon SSA treatment or SF3b 155 knockdown (Fig. 5B). Based on the U2 snRNP recruitment and mapping results above, we predicted that the competitive disadvantage of the RBM5 intron 5 3′ splice site under conditions of SSA treatment or SF3b 155 knockdown would be attenuated if the sequence acting as a decoy site for U2 snRNA binding was replaced by the corresponding sequence of intron 6 (mutant uAS6–intron 5). Consistent with this prediction, SF3b 155 knockdown did not induce exon 6 skipping in this construct (Fig. 7, cf. inclusion/skipping ratio between lanes 3,4 in A and quantitative PCR in B). We conclude that the presence of sequences harboring strong base-pairing potential with U2 snRNA 5′ of the branch point can modulate the competitiveness of 3′ splice sites in the presence of SSA, and thus explain alternative splicing changes induced by the drug or knockdown of SF3b 155 (Fig. 7C).
Figure 7.
Base-pairing potential with U2 snRNA of sequences 5′ of the branch point can modulate SSA-induced alternative splicing. (A) RT–PCR analysis of RBM5 RNA isoforms expressed from plasmids expressing the alternatively spliced RBM5 region (exons 5–7), either wild type or a mutant, that replaces the upstream region of the intron 5 AS with the corresponding sequence from intron 6 (uAS6–intron 5) in control cells or in cells in which expression of SF3b 155 has been knocked down. The asterisk indicates an aberrant amplification product caused by primer hybridization in intron 5, as determined by sequencing. The bottom panel shows Western blot analysis of the levels of SF3b 155 and α-tubulin in control and SF3b 155 knockdown cells. (B) Analysis of RBM5 exon 6 skipping and inclusion of RNAs analyzed in A in SF3b 155 knockdown versus control cells by quantitative RT–PCR, using splice junction primers specific for exon inclusion or skipping as one of the oligonucleotides used in the PCR amplification reactions. (C) Mechanistic model for alternative splicing regulation induced by SSA or SF3b155 knockdown. Under normal conditions, both of the branch point regions of RBM5 introns 5 and 6 are occupied by U2 snRNP, and the alternatively spliced exon 6 is preferentially included in mature transcripts. SSA or SF3b knockdown inactivates a fidelity mechanism that prevents U2 snRNP binding to decoy branch point-like sequences 5′ of the functional sites. The presence of a decoy site with strong base-pairing complementarity with U2 snRNA leads to preferential recruitment of the snRNP to this site, inhibition of the intron 5 3′ splice site, and exon skipping. Consistent with this hypothesis, replacement of the decoy sequence by a sequence in intron 6 (uAS6–intron 5) with lower base-pairing potential with U2 snRNA attenuates the effects of SSA/SF3b 155 knockdown. Exons, introns, and branch point adenosines are indicated. The relative base-pairing potential of U2 snRNA with different pre-mRNA sequences is represented by the number of vertical lines.
Discussion
Small molecules that target the ribosome, including antibiotics that promote inaccurate base-pairing interactions, have been immensely useful over the past 60 years for both mechanistic studies of the translation process and treatment of human disease (Ogle and Ramakrishnan 2005). Clearly lagging behind is the identification of compounds that target the spliceosome. A few small molecules have been identified that stall spliceosome assembly at different steps of the pathway (Kuhn et al. 2009) or act as inhibitors of SR protein factors (Soret et al. 2005; Bakkour et al. 2007; Keriel et al. 2009) or SR protein kinases (Muraki et al. 2004; Fukuhara et al. 2006). Such compounds hold the promise of novel insights into the dynamic transitions required for spliceosome assembly and catalysis. Based on the findings presented in this study, we propose that the anti-tumor drug SSA modulates alternative splicing by compromising the fidelity of branch point recognition by U2 snRNP. We also report alternative splicing changes induced by the drug that affect the expression of key regulators of cell division, which in turn can contribute to the anti-proliferative properties of this compound.
Mechanisms of 3′ splice site fidelity
A recent report by Roybal and Jurica (2010) concluded that SSA inhibits spliceosome assembly subsequent to prespliceosome formation, impeding the transition between complex A and B, implying a role for SF3b in maturation steps of the spliceosome after its established function in early U2 snRNP recruitment. Our results indicate that complex A formation is compromised by SSA when spliceosome assembly mixes are treated with heparin (5 mg/mL) previous to electrophoretic fractionation of the splicing complexes. We also found that, in the absence of heparin (or likely in the presence of low heparin concentrations like those used by Roybal and Jurica [2010], 0.125 mg/mL), U2 snRNP-containing complexes that form in the presence of SSA lack critical RNA–protein interactions between SF3b 155 and the pre-mRNA. In the absence of SSA, these interactions provide contacts between SF3b 155 and regions 5′ and 3′ of the branch point (Gozani et al. 1998), contributing to U2 snRNP tethering at the branch point region. Our results indicate that SSA interferes with this early function of SF3b in U2 snRNP recruitment, and suggest that the absence of contacts between SF3b 155 and the pre-mRNA enables alternative patterns of base-pairing of U2 snRNA with pre-mRNA sequences 5′ of the branch point, as observed in the presence of the drug (Figs. 4, 6). In this model, SSA inactivates a normal function of SF3b 155 (and possibly other U2 snRNP components) that prevents the establishment of nonproductive base-pairing interactions between U2 snRNA and (partially) complementary sequences in the neighborhood of the functional branch point. SSA would thus act by disabling a mechanism that ensures fidelity in branch point recognition. Such a fidelity mechanism is particularly important considering the limited base-pairing potential with the bprs of the very diverse sequences observed at natural branch points (Gao et al. 2008). More generally, while snRNA/snRNA and snRNA/pre-mRNA base-pairing interactions play key roles in the dynamic architecture of the spliceosome, base-paired regions are usually short and complementarity is often imperfect, raising the question of how specific recognition of particular sequences (e.g., splice sites vs. decoy neighboring sequences) can be achieved. The abnormal base-pairing schemes established in the presence of SSA may interfere with normal branch point activation by sterically competing with formation of productive branch point/bprs interactions (Fig. 4D). Alternatively, they may act as decoy sites for U2 snRNP binding that engage in nonproductive interactions with other spliceosome components, leading to splice site repression or alternative splice site selection (Figs. 6, 7C). The latter scenario could be at the basis of some of the intriguing results from Roybal and Jurica (2010), arguing for additional effects of SSA on subsequent steps of spliceosome assembly.
Recent work by Perriman and Ares (2010) supports the existence of a stem–loop structure in U2 snRNA that helps to establish initial interactions between the bprs (located mainly in the loop) and the branch point. Upon initial recognition, the stem–loop structure is remodeled to allow stable U2 snRNP recruitment (Perriman and Ares 2010), a remodeling step that may be guided by the DEAD-box helicase/ATPase Prp5, which is known to kinetically proofread U2 snRNP assembly by monitoring U2 bprs/branch point base-pairing (Xu and Query 2007). These insights into early 3′ splice site recognition, obtained in budding yeast, are likely to be conserved in mammalian cells, and it is therefore conceivable that SSA influences the efficiency or the proofreading properties of this U2 snRNA remodeling step, thereby allowing formation of the alternative base-pairing schemes observed in the presence of the drug. Consistent with this possibility, the related anti-tumor drug E7107, which also binds SF3b, specifically blocks an ATP-dependent remodeling event that exposes the bprs of U2 snRNA (EG Folco, KE Coil, and R Reed, pers. comm.).
Splicing fidelity, alternative splicing, and the control of cell proliferation
Given its binding to a general splicing factor, it has been assumed that SSA causes a general inhibition of pre-mRNA splicing (Kaida et al. 2007, 2010; Furumai et al. 2010). General splicing inhibition is likely to compromise cell viability and, consequently, the utility of SSA as an anti-tumor drug. Our splicing-sensitive microarray results indicate, in contrast, that, under the conditions of the experiment, SSA induces alternative splicing changes in living cells, revealing differential sensitivity of alternative splice sites to the action of the drug. A very significant overlap (P-value Fischer exact test <0.001) was observed between these changes and those induced by knockdown of SF3b 155, consistent with the inactivating effect of SSA on SF3b 155 interaction with the pre-mRNA. Differences in experimental setup (e.g., in the time required to observe effects of the drug and efficiently deplete SF3b 155) likely contribute to the differences in alternative splicing changes observed in the two conditions. SF3b 155 was involved previously in alternative splicing regulation of the Bcl-X gene, but this activity of SF3b 155 was linked to its binding to a ceramide-responsive regulatory sequence (Massiello et al. 2006), rather than to its activity as a general splicing factor.
Our analysis of RBM5 alternative splicing suggests a mechanism by which inactivation or depletion of SF3b 155 can lead to differential effects on competing 3′ splice sites (Fig. 7C). Reducing the fidelity of U2 snRNP recruitment is expected to have stronger inhibitory effects on 3′ splice sites that harbor better alternative sites for U2 snRNA base-pairing, because such decoy sites will more efficiently prevent productive U2 binding to the functional branch point. The results of the biochemical and RNAi experiments of Figures 6 and 7 are consistent with this hypothesis: A region 5′ of the branch point in intron 5 that can establish base-pairing interactions with U2 snRNA confers differential sensitivity to SSA, and its replacement antagonizes the exon-skipping effects that SSA or SF3b 155 depletion induces in the wild-type RNA. Given the multiple proofreading mechanisms that operate to ensure splice site recognition and splicing catalysis, our results suggest that a variety of novel mechanisms affecting splicing fidelity can contribute to alternative splicing regulation.
The cytotoxic and tumor inhibitory effects of SSA have been linked to cell cycle arrest and angiogenesis inhibition due to the production of a more stable C-terminally truncated form of the p27 cyclin-dependent kinase inhibitor and down-regulation of the angiogenesis inducer VEGF (Kaida et al. 2007; Furumai et al. 2010). These changes have been interpreted in the context of a general inhibitory effect of the drug on the pre-mRNA splicing. Our results indicate that SSA induces changes in alternative splicing, including changes that lead to the generation of transcripts containing PTCs in genes encoding important regulators of cell cycle progression like Cyclin A2 and Aurora kinase A. These transcripts are subject to degradation by the NMD pathway (Lareau et al. 2007), and therefore lead to down-regulation of these factors, which can contribute to the anti-proliferative effects of the drug (Mitra and Enders 2004; Saeki et al. 2009). These observations raise the intriguing possibility that physiological variations in the levels or activity of SF3b 155 modulate alternative splicing programs linked to the control of cell proliferation.
Alternative splicing networks linked to cell cycle control, apoptosis, and cancer progression are indeed emerging (for review, see David and Manley 2010; Moore et al. 2010). Thus, the classical splicing regulatory factor SRSF1 can act as a potent oncogene (Karni et al. 2007) by inducing alternative splicing changes that affect key signaling pathways involved in tumor suppression, activation of the mTORC1 pathway, and apoptosis control (Karni et al. 2007, 2008; Moore et al. 2010). Misregulation of alternative splicing of the pyruvate kinase M gene in tumor cells confers a competitive metabolic and proliferative advantage (Christofk et al. 2008), and is triggered by myc-induced increased expression of hnRNP proteins A1/A2 and I (Clower et al. 2010; David et al. 2010). These examples illustrate the significant impact of alternative splicing on tumor progression, and suggest that modulation of splice site selection can provide novel therapeutic approaches. Collectively, our results indicate that inhibition of the function of SF3b by SSA leads to relaxed fidelity in branch point recognition, and this in turn leads to alternative splicing changes that affect the expression of regulators of cell division such as Cyclin A2 and Aurora kinase A. These observations open the possibility that additional compounds that target the fidelity of the splicing process could engross a new generation of anti-cancer drugs, a general principle with analogies to the function of a class of antibiotics that promote inaccurate base-pairing interactions in the ribosome.
Materials and methods
In vitro splicing assays
Splicing assays were performed in 9-μL mixtures containing 3 μL of HeLa nuclear extracts (CilBiotech) and 10 fmol of in vitro transcribed 32P-labeled AdML pre-mRNA under standard conditions (3 mM MgCl2, 1.1 mM ATP, 22 mM creatine phosphate, 1.67% polyvinyl alcohol). After incubation for 1 h at 30°C, RNA was isolated by treatment with proteinase K and ethanol precipitation. Pre-mRNA precursor, splicing intermediates, and products were resolved by electrophoresis on denaturing 13% polyacrylamide gels and analyzed by a PhosphorImager.
Spliceosome assembly
Splicing mixtures were assembled as above using 32P-uridine-labeled pre-mRNA or fragments corresponding to the 3′ splice site region and downstream exon. After 15 min of incubation at 30°C, 5 μg/μL heparin was added to the mix, kept for 10 min at room temperature, and resolved by electrophoresis on 4% acrylamide:bisacrylamide (80:1) and 0.5% agarose gels in 50 mM Tris base and 50 mM glycine buffer. The gels were dried and exposed to a PhosphorImager screen. Alternatively, 2.2 μL of 6× DNA loading dye (20 mM Tris-HCl at pH 7.5, 0.25% bromophenol blue, 0.25% xylene cyanol, 30% glycerol) was added to the reaction directly after incubation at 30°C, and mixtures were then resolved by electrophoresis in 1.5% low-melting agarose gels (Ecogen, catalog no. AG-420) in 50 mM Tris base and 50 mM glycine buffer. The gels were fixed in 10% methanol and 10% acetic acid for 10 min at room temperature, dried, and exposed to a PhosphorImager screen.
UV cross-linking and immunoprecipitation
32P-labeled RNAs were incubated under in vitro splicing conditions in a final volume of 27 μL. Mixtures were incubated for 30 min at 30°C and irradiated with UV light (254 nm, 0.4 J). After further incubation with 1 mg/mL RNase A for 30 min at 37°C, immunoprecipitation of U2AF65 and U2 snRNP components was carried out by addition of anti-U2AF65 MC3 monoclonal antibody hybridoma supernatant (Gama-Carvalho et al. 1997) or anti-SF3b 155 or anti-SF3a 60 polyclonal antibodies (kind gifts of Professor Angela Krämer, University of Geneva). After incubating for 60 min at 4°C, 30 μL of a 50% slurry of protein A/G sepharose beads (GE Healthcare, catalog nos. 17-5280-01 and 17-0618-01, respectively) was added and the mixture was incubated under rotation for another 60 min. Beads were sedimented by centrifugation and washed four times with 800 μL of high-salt buffer (500 mM NaCl, 50 mM Tris-Hcl at pH 8.0, 1% NP-40) and once with the same buffer, except that the concentration of NaCl was 150 mM. Sedimented beads were resuspended in 4× SDS loading dye and boiled, and the supernatant was resolved by electrophoresis on 8%–10% SDS polyacrylamide gels. Gels were dried and exposed to a PhosphorImager screen.
Psoralen-mediated RNA–RNA cross-linking
Fifteen-microliter reactions containing 80 fmol of 32P-uridine-labeled 3′ half RNAs were incubated for 20 min at 30°C in the presence of 5 μL of HeLa nuclear extracts and 22 μg/mL 4′-aminomethyltrioxsalen hydrochloride (Sigma, catalog no. A4330). The mixture was irradiated with a 365-nm wavelength UV light for 10 min. RNA was then treated with proteinase K, ethanol-precipitated, and analyzed by electrophoresis on a denaturing 6% polyacrylamide gel. RNase H-mediated (Invitrogen, catalog no. 18021-014) inactivation of U2 snRNP was performed by incubating the nuclear extracts with an oligodeoxynucleotide complementary to positions 28–42 of U2 snRNA. RNase H–mediated digestion after cross-linking was performed similarly using the specified oligodeoxynucleotides.
Mapping of psoralen cross-linked nucleotides by primer extension
Psoralen-mediated cross-linked RNAs and splicing lariat intermediates were gel-purified, and primer extension was performed using the indicated 5′ 32P-labeled oligodeoxynucleotide and SuperScript II (Invitrogen, catalog no. 18064014) following the manufacturer's protocol. Primer extension products were resolved by electrophoresis on denaturing polyacrylamide gels in parallel with a sequencing reaction performed using the same primer and fmol DNA Cycle Sequencing System (Promega, catalog no. Q4100).
SSA treatments
Nuclear extract
Nuclear extracts (3 μL for splicing reactions and 5 μL for psoralen cross-linking assays) were treated for 30 min at 4°C with SSA previous to their use in subsequent assays. The indicated amounts correspond to the final concentration in the reaction mixtures.
Tissue culture
Subconfluent HeLa cells were treated with SSA (at a final concentration of 100 ng/mL, 260 nM) for 3 h. Control cells were treated with methanol, the solvent in which the SSA stock was dissolved.
RNA isolation
Total RNA was isolated using the Qiagen RNeasy minikit (catalog no. 74104) with DNase digestion on column before elution using the RNase-free DNase set (catalog no. 79254). RNA quality was assessed by Agilent Bioanalyzer nano assay and nanodrop spectrophotometry.
Microarray analyses
Cy3- and Cy5-labeled cRNAs were generated from 500 ng of total RNA using the Quick-Amp Labeling kit, No Dye (catalog no. 5190-0447) and Cy3 and Cy5-CTP from Perkin-Elmer (NEL580 and NEL581). Seven micrograms of each cRNA was hybridized into our custom oligo microarray platform (Agilent) in an Agilent rotating oven for 18 h at 60°C. After hybridization, arrays were washed and scanned images were analyzed. Statistical analyses were carried out with LIMMA package (Bioconductor) as described previously (Muñoz et al. 2009). Microarray results have been deposited in the Gene Expression Omnibus (GEO) database (GSE22614 and GSE22952). Microarray data as well as a detailed explanation of data analysis using this platform can be downloaded anonymously from the following Web site: http://davinci.crg.es/splicingarrays (user: jvlab; password: Web01).
Real-time quantitative PCR analyses
Primers were designed using Primer 3 Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) using the reference and the alternative RefSeq accession numbers. Primers used are listed in Supplemental Table 1. The primers were designed such that their annealing temperature was 60°C, generating single-amplification products in the range of 60- to 200-base-pairs (bp) long. For analysis of alternatively spliced isoforms, one of the primers was chosen as overlapping an exon junction, such that only upon hybridization to the correctly spliced mRNA was the primer able to produce an amplification product. First strand cDNA synthesis was carried out using 1 μg of RNA, 50 pmol of oligo-dT (Sigma-Aldrich), 75 ng of random hexamers (Roche, 11034731001), and Reverse Transcriptase SuperScript II (Invitrogen, catalog no. 18064-014) following the manufacturer's instructions. PCR amplification was carried out with 1 μL of the 1:10 diluted reverse transcription sample with 5 μL of 2× SYBR Green Master Mix (Roche, catalog no. 4309155) and 4 pmol of specific gene primer pairs to a 10-μL total volume in 384-well microtiter plates. PCR reactions were run in triplicates on a LightCycler 480 system (Roche). Relative copy number (RCC) or fold change ratio was calculated according to Pfaffl (2001). A list of the primers used for RT–PCR analyses using this platform can be downloaded anonymously from the following Web site: http://davinci.crg.es/splicingarrays (user: jvlab; password: Web01).
Semiquantitative RT–PCR
Total RNA was isolated 72 h after SF3b 155 siRNA transfection using the GE Healthcare RNAspin minikit (catalog no. 25-0500-72) and was analyzed by RT–PCR. One microgram of RNA was reverse-transcribed using random primers and SuperScript II (Invitrogen, catalog no. 18064014). One-fourth of the cDNA was amplified by PCR using the appropriate primers. After 24 cycles of 1 min at 93°C, 45 sec at 56°C, and 1 min at 72°C, the products were analyzed on 2% agarose gels.
RNAi
To knock down SF3b 155, cells were transfected with Lipofectamine 2000 following the manufacturer's protocol with three different siRNAs against SF3b155 (Invitrogen): siRNA #3, 5′-GACAGCAGAUUUGCUGGAUACGUGA-3′; siRNA #4, 5′-GCUUGGUCAGAAGAAGCCAGGAUAU-3′; siRNA #5, 5′-CCCUGUGGCAUUGCUUAAUGAUAU-3′. siRNA #4 was used in initial knockdown experiments, siRNAs #3 and #5, which achieved higher levels of knockdown, were used for independent microarray analyses.
Acknowledgments
We thank Robin Reed for communicating results prior to publication, and Professors M. Yoshida and A. Krämer for their kind gifts of SSA and antibodies, respectively. We thank members of our department for support and helpful discussions. This work was supported by grants from Fundación Marcelino Botín, Fundación Alicia Koplowicz, AICR, EURASNET, Ministerio de Ciencia e Innovación, and Consolider-Ingenio RNAREG. A.C. was supported by an FPI fellowship from the Spanish Ministerio de Ciencia e Innovación.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2014311.
Supplemental material is available for this article.
References
- Bakkour N, Lin YL, Maire S, Ayadi L, Mahuteau-Betzer F, Nguyen CH, Mettling C, Portales P, Grierson D, Chabot B, et al. 2007. Small-molecule inhibition of HIV pre-mRNA splicing as a novel antiretroviral therapy to overcome drug resistance. PLoS Pathog 3: 1530–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ 2010. Deciphering the splicing code. Nature 465: 53–59 [DOI] [PubMed] [Google Scholar]
- Bennett M, Reed R 1993. Correspondence between a mammalian spliceosome component and an essential yeast splicing factor. Science 262: 105–108 [DOI] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89: 781–787 [DOI] [PubMed] [Google Scholar]
- Bonnal S, Martinez C, Forch P, Bachi A, Wilm M, Valcarcel J 2008. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol Cell 32: 81–95 [DOI] [PubMed] [Google Scholar]
- Brosi R, Groning K, Behrens SE, Luhrmann R, Kramer A 1993. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science 262: 102–105 [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC 2008. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452: 230–233 [DOI] [PubMed] [Google Scholar]
- Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR 2010. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci 107: 1894–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G 2009. RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Zhou Z, Reed R 2000. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol Cell 5: 779–787 [DOI] [PubMed] [Google Scholar]
- David CJ, Manley JL 2010. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev 24: 2343–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL 2010. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463: 364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dönmez G, Hartmuth K, Lührmann R 2004. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA 10: 1925–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dönmez G, Hartmuth K, Kastner B, Will CL, Lührmann R 2007. The 5′ end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol Cell 25: 399–411 [DOI] [PubMed] [Google Scholar]
- Dybkov O, Will CL, Deckert J, Behzadnia N, Hartmuth K, Luhrmann R 2006. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol Cell Biol 26: 2803–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara T, Hosoya T, Shimizu S, Sumi K, Oshiro T, Yoshinaka Y, Suzuki M, Yamamoto N, Herzenberg LA, Hagiwara M 2006. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc Natl Acad Sci 103: 11329–11333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumai R, Uchida K, Komi Y, Yoneyama M, Ishigami K, Watanabe H, Kojima S, Yoshida M 2010. Spliceostatin A blocks angiogenesis by inhibiting global gene expression including VEGF. Cancer Sci 101: 2483–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi K, Ray P, Kar A, Wang L, Sutherland LC, Wu JY 2008. Up-regulation of the proapoptotic caspase 2 splicing isoform by a candidate tumor suppressor, RBM5. Proc Natl Acad Sci 105: 15708–15713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Carvalho M, Krauss RD, Chiang L, Valcárcel J, Green MR, Carmo-Fonseca M 1997. Targeting of U2AF65 to sites of active splicing in the nucleus. J Cell Biol 137: 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K, Masuda A, Matsuura T, Ohno K 2008. Human branch point consensus sequence is yUnAy. Nucleic Acids Res 36: 2257–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Feld R, Reed R 1996. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev 10: 233–243 [DOI] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, Reed R 1998. A potential role for U2AF–SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol 18: 4752–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Bennett M, Xiao Y, Feld Kramer R, Wang C, Reed R 1997. Association of U2 snRNP with the spliceosomal complex E. Nucleic Acids Res 25: 354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. 2007. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 3: 576–583 [DOI] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 468: 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR 2007. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol 14: 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R, Hippo Y, Lowe SW, Krainer AR 2008. The splicing-factor oncoprotein SF2/ASF activates mTORC1. Proc Natl Acad Sci 105: 15323–15327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent OA, Reayi A, Foong L, Chilibeck KA, MacMillan AM 2003. Structuring of the 3′ splice site by U2AF65. J Biol Chem 278: 50572–50577 [DOI] [PubMed] [Google Scholar]
- Keriel A, Mahuteau-Betzer F, Jacquet C, Plays M, Grierson D, Sitbon M, Tazi J 2009. Protection against retrovirus pathogenesis by SR protein inhibitors. PLoS ONE 4: e4533 doi: 10.1371/journal.pone.0004533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Sharp PA 1986. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell 46: 845–855 [DOI] [PubMed] [Google Scholar]
- Konarska MM, Vilardell J, Query CC 2006. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell 21: 543–553 [DOI] [PubMed] [Google Scholar]
- Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y 2007. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol 3: 570–579 [DOI] [PubMed] [Google Scholar]
- Krämer A 1988. Presplicing complex formation requires two proteins and U2 snRNP. Genes Dev 2: 1155–1167 [DOI] [PubMed] [Google Scholar]
- Kuhn A, van Santen M, Schwienhorst A, Urlaub H, Luhrmann R 2009. Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation. RNA 15: 153–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE 2007. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv Exp Med Biol 623: 190–211 [DOI] [PubMed] [Google Scholar]
- Liu Z, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Krämer A, Sattler M 2001. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294: 1098–1102 [DOI] [PubMed] [Google Scholar]
- Massiello A, Roesser JR, Chalfant CE 2006. SAP155 Binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J 20: 1680–1682 [DOI] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841 [DOI] [PubMed] [Google Scholar]
- Michaud S, Reed R 1991. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev 5: 2534–2546 [DOI] [PubMed] [Google Scholar]
- Michaud S, Reed R 1993. A functional association between the 5′ and 3′ splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev 7: 1008–1020 [DOI] [PubMed] [Google Scholar]
- Mitra J, Enders GH 2004. Cyclin A/Cdk2 complexes regulate activation of Cdk1 and Cdc25 phosphatases in human cells. Oncogene 23: 3361–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA 2010. An alternative splicing network links cell-cycle control to apoptosis. Cell 142: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz MJ, Perez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano JJ 2009. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell 137: 708–720 [DOI] [PubMed] [Google Scholar]
- Muraki M, Ohkawara B, Hosoya T, Onogi H, Koizumi J, Koizumi T, Sumi K, Yomoda J, Murray MV, Kimura H 2004. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem 279: 24246–24254 [DOI] [PubMed] [Google Scholar]
- Nakajima H, Hori Y, Terano H, Okuhara M, Manda T, Matsumoto S, Shimomura K 1996. New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J Antibiot 49: 1204–1211 [DOI] [PubMed] [Google Scholar]
- Nelson KK, Green MR 1989. Mammalian U2 snRNP has a sequence-specific RNA-binding activity. Genes Dev 3: 1562–1571 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien K, Matlin AJ, Lowell AM, Moore MJ 2008. The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. J Biol Chem 283: 33147–33154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V 2005. Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177 [DOI] [PubMed] [Google Scholar]
- Oh JJ, Razfar A, Delgado I, Reed RA, Malkina A, Boctor B, Slamon DJ 2006. 3p21.3 tumor suppressor gene H37/Luca15/RBM5 inhibits growth of human lung cancer cells through cell cycle arrest and apoptosis. Cancer Res 66: 3419–3427 [DOI] [PubMed] [Google Scholar]
- Parker R, Siliciano PG, Guthrie C 1987. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell 49: 229–239 [DOI] [PubMed] [Google Scholar]
- Perriman R, Ares M Jr 2010. Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Mol Cell 38: 416–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29: e45. doi: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query CC, Moore MJ, Sharp PA 1994. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev 8: 587–597 [DOI] [PubMed] [Google Scholar]
- Query CC, Strobel SA, Sharp PA 1996. Three recognition events at the branch-site adenine. EMBO J 15: 1392–1402 [PMC free article] [PubMed] [Google Scholar]
- Roybal GA, Jurica MS 2010. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res 38: 6664–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B, Zamore P, Green MR 1988. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52: 207–219 [DOI] [PubMed] [Google Scholar]
- Saeki T, Ouchi M, Ouchi T 2009. Physiological and oncogenic Aurora-A pathway. Int J Biol Sci 5: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P, Gregorovic G, Sprangers R, Stier G, Rhani Z, Krämer A, Sattler M 2003. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol Cell 11: 965–976 [DOI] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM 2008. ‘Nought may endure but mutability’: spliceosome dynamics and the regulation of splicing. Mol Cell 30: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Konarska MM, Query CC 2009. Insights into branch nucleophile positioning and activation from an orthogonal pre-mRNA splicing system in yeast. Mol Cell 34: 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soret J, Bakkour N, Maire S, Durand S, Zekri L, Gabut M, Fic W, Divita G, Rivalle C, Dauzonne D 2005. Selective modification of alternative splicing by indole derivatives that target serine-arginine-rich protein splicing factors. Proc Natl Acad Sci 102: 8764–8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadaccini R, Reidt U, Dybkov O, Will C, Frank R, Stier G, Corsini L, Wahl MC, Lührmann R, Sattler M 2006. Biochemical and NMR analyses of an SF3b155–p14–U2AF–RNA interaction network involved in branch point definition during pre-mRNA splicing. RNA 12: 410–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL 2008. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc Natl Acad Sci 105: 11218–11223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thickman KR, Swenson MC, Kabogo JM, Gryczynski Z, Kielkopf CL 2006. Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein-protein interactions among pre-mRNA splicing factors. J Mol Biol 356: 664–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R 1998. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev 12: 1409–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman DA, Steitz JA 1992. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science 257: 1918–1925 [DOI] [PubMed] [Google Scholar]
- Will CL, Schneider C, MacMillan AM, Katopodis NF, Neubauer G, Wilm M, Lührmann R, Query CC 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J 20: 4536–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Manley JL 1989. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev 3: 1553–1561 [DOI] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835 [DOI] [PubMed] [Google Scholar]
- Xu YZ, Query CC 2007. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell 28: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355: 609–614 [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM 1989. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev 3: 1545–1552 [DOI] [PubMed] [Google Scholar]
- Zhuang YA, Goldstein AM, Weiner AM 1989. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Nat Acad Aci 86: 2752–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402: 835–838 [DOI] [PubMed] [Google Scholar]