Abstract
Autophagy is a catabolic pathway used by cells to support metabolism in response to starvation and to clear damaged proteins and organelles in response to stress. We report here that expression of a H-rasV12 or K-rasV12 oncogene up-regulates basal autophagy, which is required for tumor cell survival in starvation and in tumorigenesis. In Ras-expressing cells, defective autophagosome formation or cargo delivery causes accumulation of abnormal mitochondria and reduced oxygen consumption. Autophagy defects also lead to tricarboxylic acid (TCA) cycle metabolite and energy depletion in starvation. As mitochondria sustain viability of Ras-expressing cells in starvation, autophagy is required to maintain the pool of functional mitochondria necessary to support growth of Ras-driven tumors. Human cancer cell lines bearing activating mutations in Ras commonly have high levels of basal autophagy, and, in a subset of these, down-regulating the expression of essential autophagy proteins impaired cell growth. As cancers with Ras mutations have a poor prognosis, this “autophagy addiction” suggests that targeting autophagy and mitochondrial metabolism are valuable new approaches to treat these aggressive cancers.
Keywords: autophagy, p62, Ras, cancer, metabolism, mitochondria
The products of essential autophagy genes (atg)—atg5, atg6/beclin1, atg7, and lc3/atg8, among others—respond to starvation, forming double-membrane phagophores that engulf cellular proteins, lipids, and organelles, capturing them in autophagosomes that are degraded in lysosomes (Levine and Kroemer 2008). This catabolic cellular self-degradation provides a mechanism to produce building blocks for macromolecular synthesis and maintain energy homeostasis through intracellular recycling in periods of nutrient limitation (Rabinowitz and White 2010). In established tumors, autophagy is up-regulated in hypoxic tumor regions where it promotes survival, indicating that cancer cells use the catabolic function of autophagy to tolerate stress (Degenhardt et al. 2006; Mathew et al. 2007a,b, 2009). As autophagy primarily generates substrates for oxidative metabolism, the means by which autophagy promotes survival in hypoxia are not clear.
Autophagy also mitigates cellular damage by preventing the toxic buildup of unfolded and aggregation-prone proteins, lipids, and damaged organelles. This waste removal function of autophagy may be important for both cell survival and tumor suppression (Mathew et al. 2007a; Levine and Kroemer 2008; White et al. 2010). Autophagy defects cause accumulation of p62 and ubiquitin (Ub)-containing protein aggregates and damaged organelles, particularly mitochondria (Hara et al. 2006; Komatsu et al. 2006, 2007; Mathew et al. 2009). This is associated with production of reactive oxygen species (ROS), genome damage, and spontaneous tumor development, suggesting that autophagy suppresses chronic tissue damage and cancer initiation (Mathew et al. 2009). Since autophagy both mitigates damage and maintains energy homeostasis, we hypothesized that autophagy has opposing roles in cancer initiation and in established tumors. Whereas damage mitigation may be important for suppressing tumor initiation, in aggressive cancers, growth in a stressed microenvironment may instead cause dependency on autophagy for survival. We envisioned that the tumor-promoting role of autophagy is twofold: providing energy substrates during periods of nutrient limitation, and preserving organelle function required for cell growth. To test this hypothesis, we examined the levels of autophagy in models of aggressive cancers, focusing on those with activating mutations in H-ras and K-ras. Indeed, introduction of activated Ras promoted tumorigenesis but also up-regulated basal autophagy that was required for maintenance of mitochondrial metabolism, cell growth, and cellular viability in vitro and in vivo in tumors. Since human cancer cell lines with active Ras can similarly depend on autophagy, this suggests that targeting tumor cell metabolism by autophagy inhibition may be a valuable approach to therapy for aggressive cancers with Ras mutations.
Results
Activated Ras-expressing cells are dependent on autophagy to survive starvation
We tested the hypothesis that activation of a strong cell growth-promoting oncogene such as H-rasV12 or K-rasV12 would alter the requirement for autophagy. Specifically, we hypothesized that cells expressing activated Ras would be less able to reduce metabolic expenditure during starvation; accordingly, they might be more dependent on autophagy. H-rasV12 or K-rasV12 genes were introduced into immortal, nontumorigenic baby mouse kidney epithelial (iBMK) cells (Supplemental Figs. S1A, S2A). The level of Ras expression in iBMK cell lines was comparable with that in human cancer cell lines with activating Ras mutations (see below).
Autophagy was measured by the frequency of cells displaying membrane translocation of the autophagosome component reporter LC3, and by assessing proteolytic processing of endogenous LC3-I to LC3-II. In iBMK cell lines without Ras, the level of basal autophagy was low (0%–5%) in nutrient-replete conditions and was up-regulated >10-fold by starvation (Hank's balanced salt solution [HBSS]) (Fig. 1A,B; Supplemental Figs. S1D, S2B,C). In contrast, expression of H-rasV12 or K-rasV12 (Ras) increased basal autophagy 10-fold and limited starvation-induced autophagy (Fig. 1A,B; Supplemental Figs. S1B,D, 2B,C). Isogenic iBMK cell lines deficient for the essential autophagy genes atg5 or atg7 were completely defective for autophagy (Fig. 1A,B, Supplemental Figs. S1D, S2B,C), and allelic loss of the essential autophagy gene beclin1 produced a partial autophagy defect (Supplemental Fig. S1B,C). Accumulation of the autophagy substrate p62 was observed only when autophagy was genetically impaired in vitro (Supplemental Figs. S1A, S2A) or in vivo (see below), suggesting that overexpression of Ras does not interfere with autophagic flux. This high level of autophagy in Ras-expressing cells occurred despite active mTOR, which suppresses autophagy and is activated by Ras, as levels of phospho-S6 were comparable between Ras-expressing cells and control cells (Supplemental Fig. S1E). This suggests that the high basal autophagy caused by Ras is not due to down-regulation of mTOR and must result from an mTOR-independent mechanism.
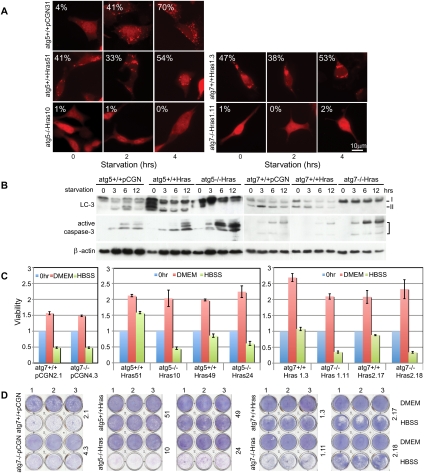
Figure 1.
H-rasV12-expressing cells are dependent on autophagy to survive starvation. (A) Autophagy-competent (atg5+/+ and atg7+/+) or authophagy-deficient (atg5−/− and atg7−/−) cells expressing H-rasV12 or vector were transiently transfected with the fluorescent autophagosome marker p-tFL-LC3 and subjected to starvation. Representative images depict RFP-LC3 localization. Numbers indicate percentage of cells with LC3 translocation to autophagosomes (punctate localization). (B) Evaluation for processing of endogenous LC3-I to LC3-II indicative of autophagy induction and activation of caspase-3 (apoptosis). (C) Ras-expressing cells were treated with HBSS for 12–20 h, and cell viability was examined by a trypan blue exclusion-based cell viability analyzer and normalized to untreated cells at the time of initiation of starvation. (D) Cells treated as in C were allowed to recover in normal medium for 2 d and assessed for clonogenic survival.
In nutrient-replete conditions, autophagy deficiency had no effect on proliferation or survival of Ras-expressing cells (Fig. 1C,D; Supplemental Fig. S2C,E). In starvation, Ras-expressing, autophagy-competent cells survived, whereas autophagy-deficient cells did not, and loss of viability was accompanied by activation of caspase-3 (Fig. 1B; Supplemental Fig. S2D). Starvation sensitivity of autophagy-deficient cells was Ras-dependent, since autophagy status did not dramatically affect survival of cells without Ras (Fig. 1C,D; Supplemental Fig. S1F). Survival impairment in starvation correlated with the severity of the autophagy defect, as Ras-expressing atg5−/− and atg7−/− cells were more sensitive to starvation (Fig. 1C,D; Supplemental Figs. S1F, S2D,E) than those with allelic loss of beclin1 (Supplemental Fig. S1G,H). Autophagy defects similarly sensitized to cell death in ischemia (glucose deprivation and 1% oxygen) (data not shown) but not to the proapoptotic ATP-competitive nonselective kinase inhibitor staurosporine (Supplemental Fig. S1I), suggesting that autophagy-dependent survival was specific to metabolic stress. Apoptosis of Ras-expressing, autophagy-defective cells was not rescued by methyl-pyruvate (Supplemental Fig. S1J) or ROS scavengers (Supplemental Fig. S1K), nor was ROS production apparent (Supplemental Fig. S1L), suggesting that survival impairment was not due to mitochondrial substrate limitation or elevated oxidative stress. Autophagy defects did not alter the level of GTP-bound Ras (Supplemental Fig. S1M), indicating that autophagy promotes survival independently of Ras activity. Thus, Ras expression promotes cellular survival in an autophagy-dependent manner when nutrients are limiting.
Autophagy supports activated Ras-mediated tumorigenesis
To assess the role of autophagy in Ras-mediated tumorigenesis, nontumorigenic iBMK cells transduced with Ras, which dramatically promotes tumor formation (Fig. 2A,D; Supplemental Fig. S2F; Degenhardt et al. 2006), were grown in nude mice. Ras-expressing atg5−/− and atg7−/− cells displayed reduced tumor growth (Fig. 2 A,B,D,E; Supplemental Fig. S2F,G) and tumors displayed abnormal histology, active caspase-3, and p62 and Ub accumulation (Fig. 2C,F; Supplemental Fig. S2H). The defect in tumorigenesis was more pronounced in atg5−/− and atg7−/− cells than those with beclin1+/−, which caused impaired tumor growth only in the context of high Ras expression (beclin1+/+ Hras2-15 and beclin1+/−Hras2) (Supplemental Fig. S3 A–C). Thus, a complete autophagy defect was more effective at compromising tumorigenesis by Ras. Facilitation of tumorigenesis by autophagy is Ras-specific, since growth of tumors without Ras is not reduced by autophagy deficiency (Degenhardt et al. 2006; Mathew et al. 2007b, 2009). Autophagy-competent and autophagy-defective Ras-expressing tumors stably expressing the autophagy reporter LC3 showed punctate LC3 distribution indicative of autophagosome formation in an atg5-dependent manner (Supplemental Fig. S3D), demonstrating that autophagy was active in Ras-driven tumors.
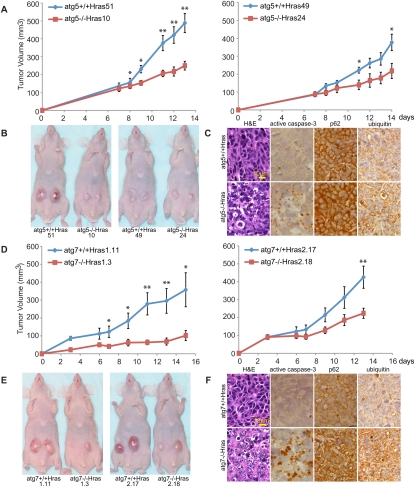
Figure 2.
Autophagy supports Ras tumorigenesis. (A) Tumor growth of Ras-expressing atg5+/+ and atg5−/− cells. Error bars represent standard errors. (*) P < 0.05; (**) P < 0.01 (t-test). (B) Representative tumor-bearing mice at day 13 (51 and 10) or day 15 (49 and 24) post-injection from A. (C) Histology (H&E) and immunohistochemistry for active caspase-3, p62, or Ub in tumors from A. (D–F) Ras-expressing atg7−/− tumors show reduced growth, elevated apoptosis, and accumulation of p62 and Ub. Error bars represent standard errors. (*) P < 0.05; (**) P < 0.01 (t-test).
p62 is required for efficient tumorigenesis by Ras
The autophagy cargo receptor p62 binds LC3 and Ub on modified proteins, including those on organelles such as depolarized mitochondria, thereby targeting cargo to autophagosomes for degradation (Pankiv et al. 2007; Geisler et al. 2010). Interestingly, deficiency in p62 impairs spontaneous lung adenocarcinoma development in mice upon activation of an oncogenic K-ras allele (Duran et al. 2008). We tested the hypothesis that p62 deficiency impairs cargo delivery to autophagosomes, thereby compromising Ras tumorigenesis by the same mechanism as deficiency in atg5 or atg7. Ras-expressing p62−/− iBMK cells (Fig. 3A) had reduced viability in starvation (Fig. 3B,C; Supplemental Fig. S4A) and decreased tumorigenicity in comparison with p62+/+ and p62-reconstituted controls (Fig. 3D,E,G,H). Ras-expressing p62−/− tumors showed abnormal histology, apoptosis (active caspase-3), and Ub accumulation (Fig. 3F,I), indistinguishable from Ras-expressing atg5−/− and atg7−/− tumors. Thus, interfering with either autophagosome cargo delivery or autophagosome formation has the common feature of impeding Ras-dependent tumorigenesis.
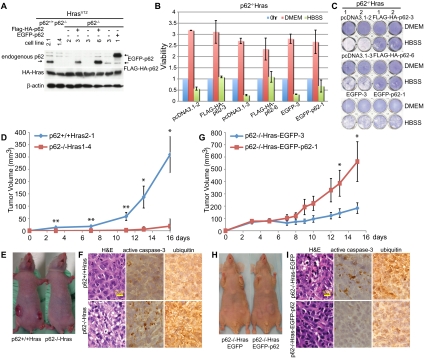
Figure 3.
p62 is required for efficient tumorigenesis by Ras. (A) HA-tagged H-rasV12 was stably expressed in p62+/+ and p62−/− iBMK cells in which p62 expression was reconstituted. Western blot shows the protein level of HA-HrasV12, Flag-HA-p62, EGFP-p62, and endogenous p62. (B,C) p62−/−-HrasV12 cells expressing vector or p62 were treated with HBSS for 16 h and then analyzed for viability (B) and clonogenic survival (C) as described in Figure 1, C and D. (D) Tumorigenesis of Ras-expressing, p62+/+, or p62−/− iBMK cells. Error bars represent standard errors. (*) P < 0.05; (**) P < 0.01 (t-test). (E) Mice at day 16 post-injection from D. (F) H&E and immunohistochemistry for activated caspase-3 and Ub in tumors from D. (G) Tumor growth of p62−/−-HrasV12 iBMK cells expressing EGFP or EGFP-p62. Error bars represent standard errors. (*) P < 0.05; (**) P < 0.01 (t-test). (H) Mice from G at day 15 post-injection. (I) H&E and immunohistochemistry of tumors from G.
High basal autophagy in human cancer cell lines with Ras mutations
To further confirm that autophagy plays a role in the growth and survival of human cancer cell lines with activating mutations in Ras, we evaluated the consequence of autophagy inhibition. We assessed the requirement of autophagy for growth and survival in T24 (bladder carcinoma cell line, H-rasG12V mutation), H1299 (lung carcinoma cell line, N-rasG12C mutation), H460 (large-cell lung cancer cell line, K-rasQ61H mutation), PANC-1 (pancreatic adenocarcinoma cell line, K-rasG12V mutation), HCT-116 (colorectal carcinoma cell line, K-rasG13D mutation), and PC-3 (prostate carcinoma cell line, wild-type Ras). The level of activated Ras expression in the human cancer cell lines was comparable with the iBMK cell lines where activated Ras expression was engineered (Fig. 4A). Except for PC3, the human cancer cell lines have high phospho-ERK (Fig. 4A), consistent with high flux through the Ras pathway. Autophagy was assessed in two ways: by the frequency of cells displaying membrane translocation of the autophagosome marker tandem-tagged p-tFL-LC3 via immunofluorescence, and by examining proteolytic processing of endogenous LC3-I to LC3-II (Fig. 4B,C). In nutrient-replete conditions, basal autophagy was up-regulated in the human cancer cells, with the exception of H460. Where H460 had few LC3 puncta and predominant LC3-I, the other human cancer cell lines had a severalfold increase in LC3 puncta and predominant LC3-II, indicative of high basal autophagy (Fig. 4B,C).
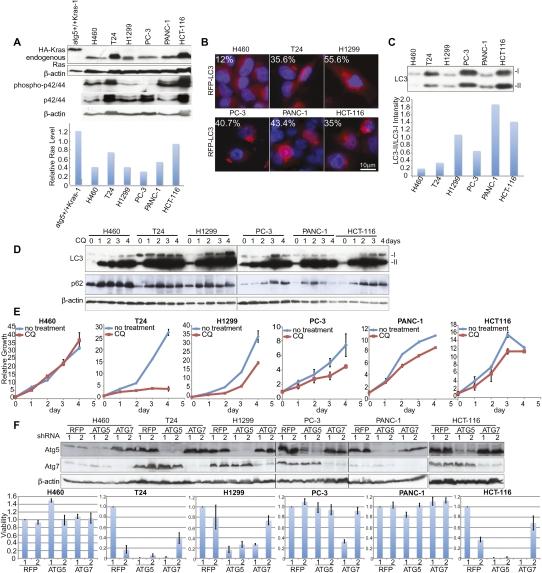
Figure 4.
Sensitivity of human cancer cell lines with activating Ras mutations to autophagy inhibition. (A, top panel) Western blot shows the protein level of HA-K-rasV12 in an iBMK cell line, endogenous Ras, phospho-p42/44, and p42/44 in human cancer cell lines. The bottom graph shows quantification of Ras levels relative to β-actin. (B) Human cancer cells were transiently transfected with the fluorescent autophagosome marker p-tFL-LC3 and cultured in nutrient-replete conditions. Representative images depict RFP-LC3 localization. Numbers indicate percentage of cells with LC3 translocation to autophagosomes (punctate localization). (C, top panel) Human cancer cells were cultured under nutrient-replete conditions and collected at 30% of confluence to evaluate processing of endogenous LC3-I to LC3-II. (Bottom panel) The ratio of LC3-II to LC3-I expression is shown quantitatively as a graph. (D) Evaluation of processing of endogenous LC3-I to LC3-II of human cancer cell lines under CQ (30 μM) treatment to block flux through the autophagy pathway. Human cancer cells were cultured under nutrient-replete conditions, treated with CQ when cells were 25%–30% confluent, and assessed in comparison with cells at the start of CQ administration. (E) Growth curve of six different cancer cell lines treated with 30 μM CQ from D. (F, top panel) Western blot shows expression of Atg5 and Atg7. (Bottom panel) Cell viability of six different cancer cell lines in response to lentiviral shRNA knockdown of essential autophagy regulators Atg5 and Atg7.
Autophagosome accumulation can be due to induction of autophagy or inhibition of flux through the autophagy pathway. To assess flux, the autophagy substrates LC3 and p62 were monitored in the presence of chloroquine (CQ), which blocks lysosome acidification, degradation of autophagosome contents, and flux through the pathway. All cell lines accumulated LC3-II and p62 in the presence of CQ, indicative of high autophagic flux, although H460 did so with slower kinetics, as expected (Fig. 4D). Thus, the high level of autophagosome formation in human cancer cell lines is due to high basal autophagy.
Autophagy facilitates growth and survival of human cancer cell lines with active Ras
To test if high basal autophagy was required for growth or survival in human cancer cell lines with activated Ras, viability was monitored in the presence of autophagy inhibitor CQ. Among cell lines with high basal autophagy, CQ dramatically suppressed growth of T24, and attenuated growth of H1299, PC-3, PANC-1, and HCT116 (Fig. 4E). In contrast, CQ did not affect proliferation of H460, which has low basal autophagy (Fig. 4E).
To further probe the significance of autophagy in human cancer cell lines with Ras mutations, the functional consequence of lentiviral knockdown of essential autophagy regulators Atg5 and Atg7 was assessed. Atg5 and Atg7 expression was suppressed by targeted shRNAs, and this was associated with a dramatic decrease in cell survival of the T24, H1299, and HCT116 cell lines (which were also sensitive to CQ) but not the others (Fig. 4F). Taken together, our results demonstrate that a subset of human cancer cell lines with activating mutations in Ras rely on autophagy for normal growth and survival.
Autophagy supports cancer cell survival through the maintenance of mitochondrial metabolic function and energy levels
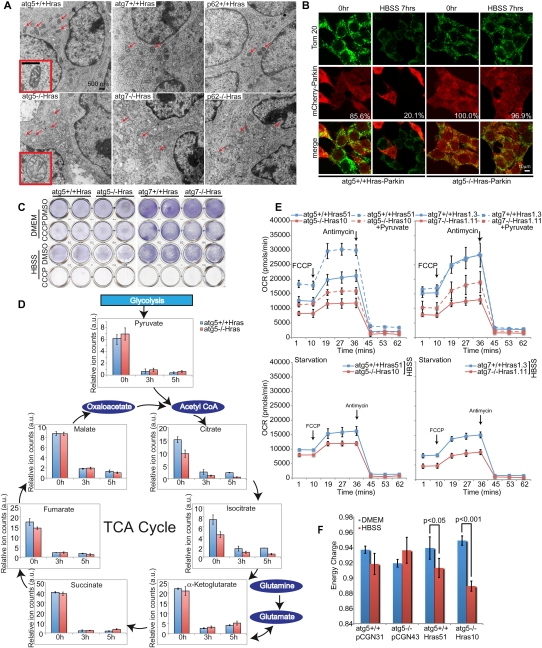
To begin to address the mechanism by which autophagy defects impair survival and tumorigenesis when Ras is activated, isogenic iBMK tumors that were either wild-type or defective for autophagy were examined by electron microscopy. In Ras-expressing atg5−/−, atg7−/−, beclin1+/−, and p62−/− tumors, there was striking accumulation of abnormal, swollen mitochondria consistent with defective mitophagy (Fig. 5A; Supplemental Fig. S5A). Mitophagy is triggered by mitochondrial depolarization, translocation of the E3 ligase Parkin to mitochondria, and ubiquitination of mitochondrial proteins, causing p62 binding and autophagosome targeting (Wild and Dikic 2010). To assess mitophagy initiation in starvation, Ras-expressing atg5+/+ and atg5−/− cells were transfected with a tagged Parkin expression vector and examined pre- and post-starvation for mitochondrial translocation (Narendra et al. 2008). Parkin translocation occurred at low levels in Ras-expressing atg5+/+ and atg5−/− cells under nutrient-replete conditions, and was induced by either the mitochondrial uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) or starvation, independently of autophagy functional status (Supplemental Fig. S5B). atg5 deficiency impaired clearance of Parkin following starvation, consistent with defective mitophagy (Fig. 5B). The failure of autophagy-defective cells to engage mitophagy-mediated degradation of mitochondria in starvation was also evident from the sustained high levels of the mitochondrial protein Tom20 (Fig. 5B; Supplemental Fig. S5C). To test if mitochondrial function was required for viability in starvation, mitochondria were depolarized with CCCP and survival in starvation was assessed. While mitochondrial depolarization had no effect on cell survival in nutrient-replete conditions, viability in starvation was abrogated independently of autophagy status (Fig. 5C; Supplemental Fig. S6A). Thus, mitochondrial function is critical for cellular survival in starvation.
Figure 5.
Autophagy maintains mitochondrial oxidative metabolism. (A) Representative EMs show the accumulation of dysmorphic mitochondria (arrows) of Ras-expressing, autophagy-defective tumors. Insets show typical mitochondrial morphology. (B) mCherry-Parkin-expressing cells were treated with HBSS for 7 h, then fixed and stained with the mitochondrial marker Tom20. Representative images show mitochondrial clearance in atg5+/+ cells during starvation (reduction in Parkin- and Tom 20-positive cells). Numbers indicate percentage of Parkin-expressing cells. (C) Autophagy-competent or autophagy-deficient cells expressing Ras were pretreated with CCCP (20 μM) for 4 h to induce mitochondrial uncoupling, and then subjected to HBSS + CCCP treatment for another 4 h. Cells were allowed to recover for 20 h and assessed for clonogenic survival. DMSO was used for CCCP control. (D) Pool sizes of major TCA cycle intermediates in atg5+/+-Hras and atg5−/−-Hras cells under nutrient-replete and starvation conditions. Graphs showing cell number normalized relative pool sizes (arbitrary unit [a.u.]) of major TCA metabolites by LC-MS in HBSS for the indicated times. (E) Oxygen consumption rates in cells under nutrient-replete conditions, following addition of FCCP (1.5 μM) to establish maximum respiratory capacity and complex III inhibitor anti-mycin (20 μM) to inhibit mitochondrial respiration. Measurements were done in DMEM without and with sodium pyruvate (0.11 g/L) (top panels) or in HBSS (bottom panel). (F) EC ([ATP] + [0.5ADP])/([ATP] + [ADP] + [AMP]) in atg5+/+ and atg5−/− cells without or with Ras under nutrient-replete and starvation conditions.
To test if autophagy defects impair mitochondrial function, maintenance of mitochondrial membrane potential was examined. When starved, autophagy-defective cells more readily lost membrane potential, and the severity of the effect correlated with the level of autophagy deficiency, consistent with reduced mitochondrial functionality (Supplemental Fig. S5D). To test if stress-induced accumulation of abnormal mitochondria impacted mitochondrial metabolism, relative pool sizes of tricarboxylic acid (TCA) cycle intermediates in Ras-expressing atg5+/+ and atg5−/− cells were examined by LC-MS under nutrient-replete (pyruvate-free DMEM) and starvation (HBSS) conditions. All TCA metabolites showed appreciable basal levels in both atg5+/+ and atg5−/− cells, which were dramatically reduced under starvation (Fig. 5D). Importantly, TCA metabolites that are exclusively produced in the mitochondria (citrate, aconitate, and isocitrate) showed reduced levels in Ras-expressing atg5−/− cells compared with atg5+/+ cells under both basal and starvation conditions (Fig. 5D; data not shown), with citrate depleted to almost undetectable levels during starvation of atg5−/− Hras cells. This was not the case for TCA metabolites that can also be produced in the cytosol from amino acid carbon skeletons (α-ketoglutarate from glutamine, and fumarate and malate from aspartate) (Fig. 5D). This citrate and isocitrate depletion was not observed in autophagy-deficient cells when tumorigenesis was rendered with an oncogene other than Ras (Bcl-2), suggesting Ras-specific effect (Supplemental Fig. S6B). Pyruvate, the final product of glycolysis, which can be taken up by mitochondria to produce acetyl-CoA and thus citrate, was actually slightly elevated in the atg5−/− cells. Thus, autophagy deficiency depletes mitochondrially produced TCA cycle metabolites, likely by impairing mitochondrial conversion of pyruvate and/or fatty acids into acetyl-CoA and citrate under Ras expression. As Ras is known to promote conversion of pyruvate to lactate and to suppress oxidation of fatty acids (Kim and Dang 2006), this may increase the requirement for autophagy to supply substrates for mitochondrial metabolism and preserve mitochondrial function.
To test if autophagy deficiency also resulted in impaired mitochondrial respiration, oxygen consumption (OCR) was examined without and with an uncoupler (p-trifluoromethoxy carbonyl cyanide phenyl hydrazone [FCCP]) to assess the spare respiratory capacity (SRC) (Choi et al. 2009), an indicator of the potential reserve capacity for bioenergetic function in cells (Dranka et al. 2010). Under nutrient-replete conditions, Ras-expressing autophagy-competent cells showed appreciable OCR, which was elevated by FCCP and pyruvate and reduced under starvation (Fig. 5E; Supplemental Fig. S6C). Thus, the autophagy-competent cells effectively tailored mitochondrial respiration to ATP needs and substrate availability. In contrast, Ras-expressing, autophagy-deficient cells showed reduced OCR levels and a blunted FCCP and pyruvate response, as well as a blunted response to starvation. Thus, distinct from any shortage of metabolic substrates, the Ras-expressing, autophagy-defective cells are fundamentally impaired in mitochondrial respiration (Fig. 5E; Supplemental S6C).
Given that autophagy deficiency in Ras-expressing tumor cells resulted in defective mitochondrial respiration, the levels of ATP, ADP, AMP, and energy charge (EC) was examined. Basal EC ([ATP + half-ADP]/[ATP + ADP + AMP]), a measure of the overall energy status of the cell, was comparable in both autophagy-competent and autophagy-defective cells with and without Ras. Ras expression resulted in a drop in EC in atg5+/+ cells upon starvation, which was decreased further by autophagy deficiency (Fig. 5F). Decreased EC in Ras-expressing, autophagy-defective cells was accompanied by a decreased ATP/ADP ratio and increased AMP levels (Supplemental Fig. S6D). Although mitochondrial ROS production has been associated with Ras-driven tumorigenesis (Weinberg et al. 2010), these data regarding EC implicate the energy-generating function of mitochondria. Ras expression amplifies energy depletion in starvation, rendering cells autophagy-dependent to buffer this demand by preservation of mitochondrial function and possibly also by production of catabolically derived metabolic substrates. In autophagy-defective cells, this metabolic insufficiency in starvation produces an acute energy crisis, leading to cell death.
Discussion
Autophagy deficiency in mice produces tissues with swollen mitochondria and reduced respiration associated with ATP depletion, but also failure of protein and lipid homeostasis, and it is not often clear which contributes to disease manifestation (Kuma et al. 2004; Komatsu et al. 2005, 2006, 2007; Hara et al. 2006; Mathew et al. 2009; Wu et al. 2009). In post-mitotic tissues, the accumulation of autophagy protein substrates may be particularly toxic, resulting in tissue damage and disease. In contrast, tumors can lessen damaged protein accumulation through cell division, whereas damaged mitochondria may be more deleterious due to proliferation in a stressed microenvironment. Importantly, functional mitochondria are required for viability of Ras-transformed cells in starvation (Fig. 5C) and in vivo (Weinberg et al. 2010), suggesting that mitochondrial quality control by mitophagy is critical for maintaining oxidative metabolism and energy homeostasis when nutrients are limiting. Thus, autophagy suppresses cancer initiation while enabling growth of aggressive cancers. In the former, autophagy prevents tissue damage that can promote cancer initiation and progression of early stage cancers. In the latter, autophagy maintains mitochondrial metabolic function important for growth of aggressive cancers, creating “autophagy addiction.”
Tumors have different levels of mitochondrial bioenergetic function (Wu et al. 2007), attributable to oncogenic mutations and activation of signaling pathways that control metabolism (Vander Heiden et al. 2009) or mutations in TCA cycle enzymes (Gottlieb and Tomlinson 2005) that promote aerobic glycolysis (Warburg effect) (Warburg 1956). As some oncogenic events suppress autophagy (White and DiPaola 2009), direct mitochondrial dysfunction derived from substrate limitation and defective mitophagy may also contribute to the Warburg effect. In the former, genetic reprogramming of metabolic pathways by mutation promotes aerobic glycolysis necessary for generation of anabolic precursors for biosynthetic pathways required for generation of new cells (Vander Heiden et al. 2009). In the latter, failure of mitochondrial quality control may prevent tumor cells from relying on oxidative metabolism to maintain cellular bioenergetics. By necessity, this may increase or select for reliance on glycolysis to support metabolism as Warburg (1956) originally described.
Cancers (such as lung, pancreatic, and colon) with a high prevalence of activating mutations in Ras have the unfortunate distinction of a particularly poor prognosis. We found that many human cancer cell lines with activating mutations in Ras have high basal autophagy, and some depend on autophagy for normal growth (T24, H1299, and HCT116) (Fig. 4E). The variance in autophagy dependence among human cancer cell lines could be due to differing additional mutations. Another intriguing possibility is that autophagy dependency may be related to high Ras activity and flux through the Ras pathway that occurs in more aggressive cancers and that is mimicked in the iBMK cells transduced with Ras. These findings suggest that these cancers with high levels of mutant Ras are most likely to respond to inhibition of autophagy, mitophagy, or mitochondrial metabolism. Indeed, autophagy is elevated in pancreatic cancer cell lines and tumors that display sensitivity to autophagy inhibition (Yang et al. 2011), consistent with the autophagy addiction model. Agents such as hydroxychloroquine (HCQ) that interfere with lysosome function at the terminal step of autophagic degradation have entered the clinic to test if autophagy inhibition can improve cancer treatment efficacy (White and DiPaola 2009). These data now inform and support a more individualized approach to assess for activating mutations in Ras in clinical trials of autophagy inhibitors. However, it is important to note that HCQ may not function equivalently to Atg5-shRNA- or Atg7-shRNA-mediated autophagy inhibition that block initiation of autophagosome formation. Development of specific autophagy inhibitors and determination of the optimal point in the autophagy pathway to compromise cancer cell survival is clearly warranted. Moreover, tumors with defective mitochondria may be particularly sensitive to agents that interfere with glucose utilization while being resistant to treatments that rely on toxic mitochondrial ROS production for cell killing.
Materials and methods
Cell culture and reagents
Baby mice iBMK cells were cultured in DMEM (GIBCO/Invitrogen) containing 10% fetal bovine serum (FBS) and 1% Pen-Strep at 38.5°C with 8.5% CO2. For human cancer cell lines, H460 and PC-3 cells were cultured in RPM1640 with 10% FBS and 1% Pen-Strep; T24 and H1299 cells were grown in 10% DMEM with 10% FBS and 1% Pen-Strep; HCT116 cells were cultured in McCoy's 5A medium containing 10% FBS and 1% Pen-Strep; and PANC-1 cells were cultured in DMEM with 10% FBS, 1% sodium pyruvate, and 1% Pen-Strep. All human cancer cells were cultured at 37°C with 5% CO2. To assess the basal autophagy level, human cancer cells were cultured in nutrient-replete conditions and collected at 30% of confluence. For starvation conditions, cells growing in multiwell plates were washed twice with phosphate-buffered saline (PBS; GIBCO/Invitrogen) and placed in HBSS (GIBCO/Invitrogen). Cell viability was assessed by a trypan blue exclusion-based cell viability analyzer (Vi-CELL, Beckman Coulter, Inc.) and normalized to untreated cells at the time of initiation of starvation. Clonogenic survival assays were performed following normal or starvation conditions by restoring normal growth medium (DMEM + 10% FBS), and the recovered cells were then visualized by staining the plates with Giemsa (Sigma-Aldrich). N-Acetyl-L-cysteine (NAC), methyl-pyruvate, staurosporine, CCCP, bafilomycin A1, anti-mycin, and FCCP were purchased from Sigma-Aldrich.
Constructs and protein expression
pEGFP-p62 and pEGFP expression vectors were gifts from Dr. Terje Johansen, the pmCherry-Parkin expression vector was generously provided by Dr. Richard Youle, and the p-tFL-LC3 tandem-tagged LC3 expression vector was gift from Dr. Tamotsu Yoshimori. Flag-HA-tagged p62 was generated by fusion of human full-length p62 to the N-terminal epitope tag and cloned into pCDNA3.1 (+) (Invitrogen). To generate rabbit polyclonal anti-p62 antibodies, the full-length human p62 cDNA was cloned into pMal-C2x (NEB) at EcoRI and SalI enzyme-cutting sites. MBP-p62 fusion protein was expressed in BL21 (DE3) Escherichia coli by IPTG induction, affinity-purified using an amylose column, and then eluted with maltose. Rabbit polyclonal anti-p62 antibody was generated by Cocalico Biologicals, Inc., using the MBP-p62 fusion protein as an antigen.
Generation of cell lines
Immortal cell lines were generated as described previously (Degenhardt et al. 2002a,b). To obtain cells stably expressing H-rasV12 or K-rasV12, atg5+/+ and atg5−/− iBMK cell lines were cotransfected with the selection marker plasmid pcDNA3.1/Zeo (Invitrogen) and pCGN-HA-HrasV12, pCGN-HA-KrasV12, or control vector pCGN followed by zeocin (Invitrogen) selection; atg7+/+, atg7−/−, beclin1+/+, and beclin1+/− iBMK cell lines were transfected with pCGN-HA-HrasV12 or control vector pCGN followed by hygromycin B (Sigma-Aldrich) selection; p62+/+ and p62−/− iBMK cell lines were cotransfected by the selection marker plasmid pCMV-BSD (Invitrogen) with pCGN-HA-HrasV12 or with control vector pCGN followed by blasticidin (GIBCO/Invitrogen) selection. p62 reconstituted cell lines were derived from p62−/−Hras cells engineered to stably express EGFP-p62 or Flag-HA-p62 by cotransfecting with pcDNA3.1/Zeo and pEGFP-p62 or pcDNA3.1(+)-Flag-HA-p62 followed by zeocin selection. atg5+/+Hras-mCherry-Parkin and atg5−/−Hras-mCherry-Parkin iBMK cell lines were derived from atg5+/+Hras or atg5−/−Hras cell lines transfected with pmCherry-Parkin and followed by geneticin (GIBCO/Invitrogen) selection. atg5+/+Hras-GFP-RFP-LC3 and atg5−/−Hras-GFP-RFP-LC3 cell lines were derived from atg5+/+Hras or atg5−/−Hras cells by transfecting with p-tFL-LC3 and followed by geneticin selection. pLKO.1-derived vectors with shRNAs targeting human atg5 or atg7 were purchased from Sigma-Aldrich (TRCN0000151963, TRCN0000330394 TRCN0000007584, and TRCN0000007587). Virus was produced using a second-generation packaging system in 293T cells as described (Root et al. 2006). Cells were selected in the presence of puromycin for 72 h prior to collection for analysis or experimental usage.
Antibodies
The following antibodies were used for Western blotting, immunohistochemistry, and immunofluorescence: p62 (antisera raised against MBP full-length human p62 described above or that purchased from Enzo Life Sciences); Beclin1 (H-300), and Tom20 (FL-145) (Santa Cruz Biotechnology, Inc.); ATG5-ATG12 (Cosmo Bio Co); HA (12CA5) (Roche Applied Science); β-actin and Ras (Sigma-Aldrich); LC3 and Ub (Ubi-1) (Novus Biologicals); and active caspase-3 (Asp175), phospho-S6, S6, phospho-p42/44, and p42/44 (Cell Signaling Technology, Inc.). Western blotting, immunohistochemistry, and immunofluorescence procedures were performed as described previously (Degenhardt et al. 2002b; Nelson et al. 2004; Mathew et al. 2009).
Electron microscopy
Tumors from atg5−/−, atg7−/−, beclin1+/−, p62−/−, or wild-type control iBMK cells expressing H-rasV12 or K-rasV12 were excised at day 11 post-injection and fixed in 0.1 M cacodylate buffer with 2.5% glutardehyde, 4% paraformaldehyde, and 8 μM CaCl2 and analyzed by a JOEL 1200EX electron microscope as described previously (Degenhardt et al. 2006).
Autophagy and mitophagy assessment
For quantification of autophagy induction, cells were transiently transfected (Amaxa, Lonza) with the p-tFL-LC3 reporter and allowed to recover overnight. Cells were treated with 1 nM bafilomycin A1 to facilitate visualization of autophagosomes and incubated with HBSS for the indicated times prior to fixation and mounting. Images were analyzed at 60×. A minimum of 200 cells was scored for each condition in three replicate experiments. The percentage of cells expressing RFP-LC3 puncta is indicated. The percentage of cells with mCherry-Parkin translocation was quantified by determining the number of cells displaying punctate (rather than diffuse) fluorescence out of a population of 100 fluorescent cells replicated in three independent experiments.
Tumor growth assays
Tumor formation assays and in vivo LC3 translocation were performed as described previously (Degenhardt et al. 2002b; Nelson et al. 2004; Mathew et al. 2009). Briefly, three mice per cell line and two independently derived cell lines per genotype were used to assess tumor formation. Cells (107) (106 in Fig. 3D; Supplemental Fig. S2F, right panel) were subcutaneously injected in nude mice and growth was monitored at the indicated time points. Statistical significance was calculated by t-test. Tumor formation assays were performed using Institutional Animal Care and Use Committee-approved protocols.
Histology
For paraffin sections, tumors were fixed overnight in 10% buffered formalin solution (Formalde-Fresh, Fisher Scientific) and sectioned. For fluorescent localization of p-tFL-LC3 tandem-tagged LC3 in vivo, tumors were fixed overnight in 10% buffered formalin solution followed by dehydration—first in 15% for 6 h, and then in 30% sucrose solution overnight—and sectioned.
OCR rate measurement
OCR rates were measured using Seahorse Biosciences extracellular flux analyzer (XF24) as described previously (Wu et al. 2007). Briefly, cells were seeded at 2.3 × 104 cells per well (0.32 cm2) in XF24 plates in 250 μL of DMEM (10% FBS, 1% Pen-Strep) and incubated for 20–24 h at 38.5°C and 8.5% CO2 prior to XF assay. Basal OCR measurements were made in HBSS and DMEM without and with sodium pyruvate (Invitrogen). SRC (maximal respiratory capacity − basal respiration rate) and total reserve capacity (Choi et al. 2009; Dranka et al. 2010) were measured under these conditions by injecting the mitochondrial uncoupler FCCP (1.5 μM), and Complex III inhibitor anti-mycin (20 μM), respectively, from XF24 reagent ports as indicated. All measurements were normalized to OCR per million cells.
Metabolomic analysis by LC-MS
Pool size measurements in major TCA cycle intermediates by LC-MS were performed as described previously (Munger et al. 2006). Briefly, cells were cultured in complete media (DMEM, 10% dialyzed FBS, 1% Pen-Strep) for 20–24 h, after which media was replaced with HBSS for the indicated time. Metabolism was quenched by removing the media and adding 4 mL of methanol:water (80:20) at −80°C. Extracted metabolites were dried under nitrogen flow, reconstituted in 350 μL of water, and measured by LC-MS. Major TCA metabolites were selected and cell number-normalized pool sizes were calculated as an average of three replicate measurements using MatLab. For ATP, ADP, and AMP measurements, individual pool sizes were normalized to cell number and to known amounts of uniformly 15N-labeled ATP (4.89 nmol/mL) and 13C-labeled ADP (2.27 nmol/mL), and AMP (0.35 nmol/mL) added to the extraction media. EC was calculated using the formula EC = (ATP + half-ADP)/(ATP + ADP + AMP). Statistical significance was calculated by two-way ANOVA with Bonferroni post-test.
Assessment of mitochondrial membrane potential, ROS levels, and Ras activity
Live cells were stained with 25 nM MitoTracker-Red CMXRos (Molecular Probes/Invitrogen) and 25 nM MitoTracker-Green FM (Molecular Probes/Invitrogen) fluorescence dyes for 30 min under growth conditions, washed with growth medium, and analyzed by flow cytometry (BD influx cell sorter, BD Biosciences). ROS levels were determined using 10 μM 2′-7′-dichlorodihydrofluorescene diacetate (DCF- DA, Molecular Probes) as described previously (Mathew et al. 2009). The Ras activity assay was performed as the manufacturer recommended (Thermo Fisher Scientific, Inc.). Briefly, 80 μg of GST-Raf1-Ras-binding domain (RBD) was incubated in 500 mg of cell lysate together with glutathione resin for 1 h at 4°C. After incubation, the protein beads were retrieved, washed, resolved on 12% SDS-PAGE, and Western-blotted with an anti-Ras mouse monoclonal antibody.
Acknowledgments
We thank M. Komatsu for the atg7−/− mice, T. Ishii for the p62−/− mice, R. Youle for the pmCherry-Parkin, T. Yoshimori for the p-tFL-LC3, and T. Johansen for the p62 expression vectors. We also thank R. Patel for the EM, and A. Roberts and the CINJ Flow Cytometry and Tissue Analytical Services and DNA sequencing core facilities. This work was supported by grants from the NIH (R37 CA53370 and RO1 CA130893 to E.W.; RC1 CA147961 to E.W., H.A.C., and J.D.R.; and R00 CA133181 to V.K.), the New Jersey Commission on Cancer Research (09-1083-CCR-EO to E.W., C.G., H.C., J.D.R., and R.S.D.), and the DOD (W81XWH06-1-0514 and W81XWH05 to E.W. and R.S.D).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.2016311.
Supplemental material is available for this article.
References
- Choi SW, Gerencser AA, Nicholls DG 2009. Bioenergetic analysis of isolated cerebrocortical nerve terminals on a microgram scale: spare respiratory capacity and stochastic mitochondrial failure. J Neurochem 109: 1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Chen G, Lindsten T, White E 2002a. BAX and BAK mediate p53-independent suppression of tumorigenesis. Cancer Cell 2: 193–203 [DOI] [PubMed] [Google Scholar]
- Degenhardt K, Sundararajan R, Lindsten T, Thompson C, White E 2002b. Bax and Bak independently promote cytochrome C release from mitochondria. J Biol Chem 277: 14127–14134 [DOI] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. 2006. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10: 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranka BP, Hill BG, Darley-Usmar VM 2010. Mitochondrial reserve capacity in endothelial cells: the impact of nitric oxide and reactive oxygen species. Free Radic Biol Med 48: 905–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J 2008. The signaling adaptor p62 is an important NF-κB mediator in tumorigenesis. Cancer Cell 13: 343–354 [DOI] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W 2010. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12: 119–131 [DOI] [PubMed] [Google Scholar]
- Gottlieb E, Tomlinson IP 2005. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer 5: 857–866 [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885–889 [DOI] [PubMed] [Google Scholar]
- Kim JW, Dang CV 2006. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res 66: 8927–8930 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880–884 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131: 1149–1163 [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N 2004. The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036 [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G 2008. Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E 2007a. Role of autophagy in cancer. Nat Rev Cancer 7: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E 2007b. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes & Dev 21: 1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell 137: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD 2006. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog 2: e132 doi: 10.1371/journal.ppat.0020132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DA, Tan TT, Rabson AB, Anderson D, Degenhardt K, White E 2004. Hypoxia and defective apoptosis drive genomic instability and tumorigenesis. Genes & Dev 18: 2095–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Rabinowitz JD, White E 2010. Autophagy and metabolism. Science 330: 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM 2006. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods 3: 715–719 [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O 1956. On respiratory impairment in cancer cells. Science 124: 269–270 [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS 2010. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci 107: 8788–8793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, DiPaola RS 2009. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res 15: 5308–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, Karp C, Strohecker AM, Guo Y, Mathew R 2010. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol 22: 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Dikic I 2010. Mitochondria get a Parkin' ticket. Nat Cell Biol 12: 104–106 [DOI] [PubMed] [Google Scholar]
- Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, Teich J, et al. 2007. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol 292: C125–C136 doi: 10.1152/ajpcell.00247.2006 [DOI] [PubMed] [Google Scholar]
- Wu JJ, Quijano C, Chen E, Liu H, Cao L, Fergusson MM, Rovira II, Gutkind S, Daniels MP, Komatsu M, et al. 2009. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging 1: 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell'Antonio G, et al. 2011. Pancreatic cancers require autophagy for tumor growth. Genes Dev (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]