Abstract
Amplification of 8p11-12 in human breast cancers is associated with increased proliferation and tumor grade and reduced metastasis-free patient survival. We identified Zeppo1 (zinc finger elbow-related proline domain protein 1) (FLJ14299/ZNF703) within this amplicon as a regulator of cell adhesion, migration, and proliferation in mammary epithelial cells. Overexpression of Zeppo1 reduces cell–cell adhesion and stimulates migration and proliferation. Knockdown of Zeppo1 induces adhesion and lumen formation. Zeppo1 regulates transcription, complexing with Groucho and repressing E-cadherin expression and Wnt and TGFβ reporter expression. Zeppo1 promotes expression of metastasis-associated p120-catenin isoform 1 and alters p120-catenin localization upon cell contact with the extracellular matrix. Significantly, Zeppo1 overexpression in a mouse breast cancer model increases lung metastases, while reducing Zeppo1 expression reduces both tumor size and the number of lung metastases. These results indicate that Zeppo1 is a key regulator of breast cancer progression.
Keywords: Znf703, breast cancer, metastasis, p120-catenin, E-cadherin, Wnt
Chromosomal rearrangements of the short arm of chromosome 8 are relatively common in epithelial cancers (Birnbaum et al. 2003). These rearrangements are often complicated (Pole et al. 2006), and involve loss, amplification, and inversion of chromosomal DNA. Amplification of 8p11-12, in particular, is seen frequently in both familial (Melchor et al. 2007) and sporadic (Garcia et al. 2005; Prentice et al. 2005) breast cancers, and may occur in as many as 24% of sporadic cases. Amplification in breast tumors is associated with a high proliferative index, a high tumor grade, and reduced metastasis-free survival (Gelsi-Boyer et al. 2005), suggesting that this region contains a tumor/metastasis-promoting gene or genes. A minimal 8p12 amplicon and the genes that lie within it have been identified (Garcia et al. 2005).
Tumor metastasis is the major cause of poor prognosis in breast cancer. It is a multistage process involving changes in tumor cell migration and invasiveness. These properties, in turn, depend on alterations in cell–cell and cell–extracellular matrix (ECM) adhesion that are highly dynamic and regulated processes (Hynes 2002; Gumbiner 2005). E-cadherin is the primary adhesive molecule in epithelial cell–cell adhesion. It interacts with members of the catenin family of proteins to form a protein complex at sites of cell–cell contact known as adherens junctions (AJs). p120-catenin (p120) binds to the juxtamembrane domain of the E-cadherin cytoplasmic tail and stabilizes E-cadherin membrane localization (Ishiyama et al. 2010). β-Catenin binds to the cytoplasmic tail of E-cadherin and recruits α-catenin to facilitate the connection between E-cadherin and the actin cytoskeleton (Nelson 2008). Cadherins and catenins also interact with receptor tyrosine kinases such as the epidermal growth factor receptor (EGFR) at the cell membrane (Qian et al. 2004). This allows AJs to integrate and coordinate growth factor signaling with cell adhesion, migration, and gene transcription.
The Wnt and TGFβ signaling pathways integrate cell signaling with changes in cell adhesion. The Wnt pathway, in particular, is strongly implicated in this process because it requires β-catenin to mediate extracellular signals from the cell membrane to the nucleus (Clevers 2006). Activation of the canonical Wnt pathway leads to nuclear accumulation of β-catenin and the formation of a protein complex between β-catenin and members of the Lef/Tcf family of transcription factors. This protein complex can activate or repress gene transcription, depending on context (MacDonald et al. 2009). Significantly, in mammals, E-cadherin expression is repressed through a combination of Wnt and TGFβ signaling (Jamora et al. 2003; Medici et al. 2006; Nawshad et al. 2007) in epithelial-to-mesenchymal transition (EMT).
In this study, we sought to identify the gene that accounts for the heightened metastatic properties of the 8p11-12 amplicon. We cloned and characterized Zeppo1 (zinc finger elbow-related proline domain protein 1; Zpo1), the mouse ortholog of ZNF703/FLJ14299, a gene that lies within the amplicon and is frequently overexpressed in breast cancers.
Results
Identification of mouse Zpo1 and Zeppo2 (Zpo2)
Within the 8p12 amplicon, the gene FLJ14299/ZNF703 has been identified repeatedly as being amplified and significantly overexpressed in sporadic and familial breast cancers (Garcia et al. 2005; Adelaide et al. 2007; Melchor et al. 2007). Using the ZNF703 human sequence, we identified a mouse Znf703 ortholog and another closely related family member (Znf503) from the NCBI database. We screened a BAC library to clone the mouse genes in their genomic context. Both genes consisted of two exons separated by a short intron. The exons for each gene were amplified by PCR and ligated to generate full-length cDNAs.
The mouse and human genes are 96% identical at the amino acid level. Homology searches identified the genes as orthologs of the Drosophila zinc finger gene elbow (Dorfman et al. 2002). We therefore named Znf703 and Znf503 as Zeppo1 (Zinc finger elbow-related proline domain protein 1) and Zeppo2, respectively. Zpo1 and Zpo2 are also orthologs of the zebrafish Nlz1 (Runko and Sagerstrom 2003) and Nlz2 (Runko and Sagerstrom 2004) genes, respectively. Zpo1 and Zpo2 proteins are 54% identical. Comparison of the protein sequences from Drosophila, zebrafish, mice, and humans identified several conserved domains, including an N-terminal Sp domain, a C2H2 zinc finger, and a C-terminal proline–tyrosine-rich domain (Fig. 1A). In this study, we focus on understanding Zpo1.
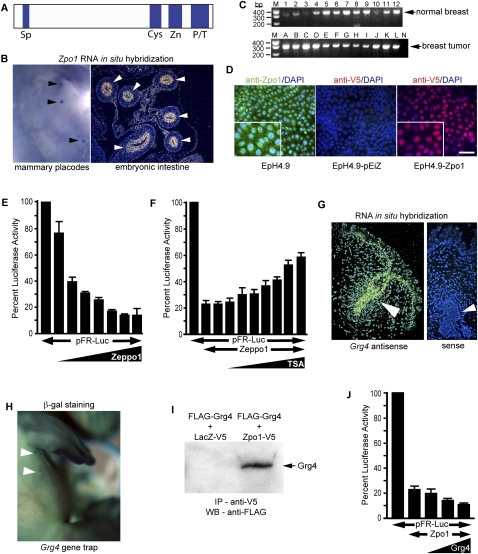
Figure 1.
Zpo1 is a transcriptional repressor expressed in mammary epithelium. (A) Schematic diagram of Zpo1 protein showing domains conserved with Drosophila elbow. (Sp) Sp box; (Cys) cysteine-rich domain; (Zn) C2H2 zinc finger; (P/T) proline- and tyrosine-rich domain. (B) Whole-mount RNA in situ hybridization demonstrating Zpo1 expression in the mammary placodes (black arrowheads) of E12.5 mouse embryo. Zpo1 expression (yellow) in the developing intestinal epithelium (white arrowheads) by RNA in situ hybridization. (C) RT–PCR demonstrating human ZPO1 expression (arrow) in a panel of normal (samples 1–12) and tumor (samples A–L) human breast epithelium. (Lane N) Negative water control. (D) Localization of endogenous Zpo1 protein (green) using rabbit anti-Zpo1 antibody in mouse EpH4.9 epithelial cells counterstained with DAPI (blue). Localization of V5-tagged Zpo1 protein (red) in EpH4.9-pEiZ and EpH4.9-Zpo1 cells using an anti-V5 antibody. Zpo1-V5 protein is detected predominantly in the nucleus. Bar, 50 μm. (E) 3T3 cells were transfected with 400 ng of the GAL4 luciferase reporter construct pFR-Luc and increasing quantities of either a Zpo1-GAL4 expression construct or vector control and assayed for luciferase activity. Firefly luciferase activity was normalized to Renilla luciferase, and activity from Zpo1-GAL4-transfected cells is expressed relative to control. Data represent the mean of three separate experiments with each experiment performed in triplicate. Error bars represent ±SEM. (F) 3T3 cells were transfected with 400 ng of pFR-Luc and 20 ng of Zpo1-GAL4 expression construct or vector control. Increasing concentrations of TSA (5–800 nM) were added to cell cultures after 24 h, and luciferase activity was measured and normalized to Renilla after a further 24 h. Error bars represent ±SEM. (G) Grg4 expression (yellow) in E13.5 mouse embryonic mammary epithelium (arrowhead) by RNA in situ hybridization. (H) LacZ staining of E12.5 Grg4 gene trap mouse embryo demonstrating Grg4 expression in the mammary placodes (arrowheads). (I) Coimmunoprecipitation of Zpo1 and Grg4 from 3T3 cells transfected with Flag-Grg4 and Zpo1-V5 or LacZ-V5 expression plasmids. Lysates were immunoprecipitated with anti-Flag antibody and blotted with anti-V5 antibody. (J) 3T3 cells were transfected with 400 ng of pFR-Luc, 20 ng of Zpo1-GAL4, and increasing quantities of a Grg4 expression plasmid or a vector control. Luciferase activity was measured and normalized to Renilla after 48 h. Activity from Grg4 transfected cells is expressed relative to vector control. Error bars represent ±SEM.
Zpo1 is expressed in the mammary epithelium
We first asked if Zpo1 is expressed in the normal mouse mammary gland. Zpo1 was expressed in the developing mammary placodes from embryonic day 11.5 (E11.5) onward (Fig. 1B). Interestingly, Zpo1 was also strongly expressed in the developing intestinal epithelium (Fig. 1B), a tissue in which the 8p11-12 amplification is also associated with tumor formation (Nakao et al. 2004; Pole et al. 2006). We also found Zpo1 in the adult mouse and human mammary epithelium (Fig. 1C; data not shown).
Using a rabbit polyclonal antibody directed against a peptide in the C-terminal region of the Zpo1 protein, we observed that endogenous Zpo1 protein was localized predominantly in the nucleus in nontumorigenic EpH4.9 mouse mammary epithelial cells (MECs), despite the lack of an identifiable nuclear localization signal, and was also present in the cytoplasm (Fig. 1D). We obtained a similar result using an anti-V5 tag antibody in cells infected with a C-terminal V5-tagged Zpo1 (Zpo1-V5) lentiviral expression construct (Fig. 1D).
Zpo1 functions as a transcriptional repressor
We then generated a plasmid expressing full-length Zpo1 fused to the GAL4 DNA-binding domain (Zpo1-GAL4) and determined its effect on the expression of a GAL4-luciferase reporter construct. Increasing levels of Zpo1-GAL4 repressed transcription of the reporter gene (Fig. 1E). Transcriptional repression is frequently achieved through recruitment of histone deacetylase (HDAC) to gene promoters (Yang and Seto 2008). Trichostatin A (TSA), a specific inhibitor of HDAC function, partially inhibited Zpo1-mediated repression (Fig. 1F).
Drosophila elbow and zebrafish Nlz1 proteins interact in vitro with the transcriptional corepressor Groucho (Dorfman et al. 2002; Runko and Sagerstrom 2003). There are at least five members of the Groucho-related gene (Grg) family in mice (Gasperowicz and Otto 2005). We observed that Grg4 was expressed in the mammary epithelium at the same developmental time points as Zpo1 (Fig. 1G). Using embryos from Grg4 gene trap mice expressing β-geo under the control of the endogenous Grg4 promoter, we detected expression in mammary placodes from E11.5 (Fig. 1H). Immunoprecipitation of lysates from cells expressing V5-tagged Zpo1 and Flag-tagged Grg4 with a V5 antibody, followed by Western blotting with an anti-Flag antibody, demonstrated coimmunoprecipitation of Grg4 with Zpo1, but not with V5-tagged lacZ (Fig. 1I). This interaction suggests that Grg4 may affect Zpo1-mediated transcriptional repression. To test this directly, we coexpressed increasing amounts of Grg4 with Zpo1 in luciferase transcription assays. Indeed, increasing levels of Grg4 led to enhanced repression of the luciferase reporter (Fig. 1J).
Overexpression of Zpo1 in MECs reduces cell–cell adhesion and increases cell invasion
Amplification of 8p12 is associated with metastasis, a process that involves changes in cell adhesion and migration. Therefore, we generated a polyclonal EpH4.9 cell line overexpressing Zpo1 (Supplemental Fig. 1) at levels comparable with those seen in human tumor samples (Garcia et al. 2005; Haverty et al. 2008). We then assessed the effect of Zpo1 overexpression on EpH4.9 cell migration using a wound healing assay. Zpo1-overexpressing cells not only migrated a greater distance than control cells (EpH4.9-pEiZ) infected with empty vector (Fig. 2A), but also migrated individually with reduced cell–cell adhesion at the wound edge (Supplemental Movies 1, 2). We also found that cells overexpressing Zpo1 were significantly more invasive through a cell culture insert coated with Matrigel than control cells (Fig. 2B).
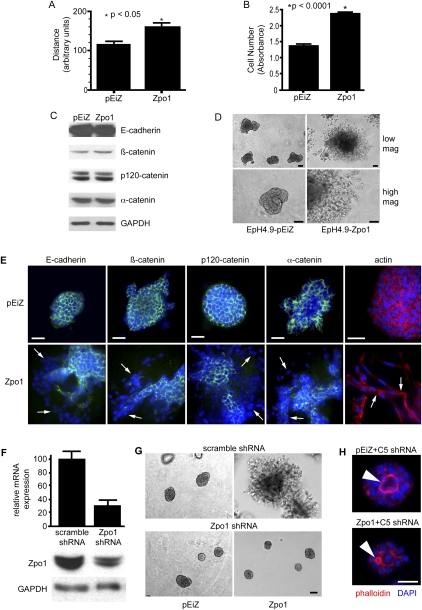
Figure 2.
Zpo1 overexpression promotes epithelial migration and invasion. (A) Migration of control (pEiZ) and Zpo1-overexpressing (Zpo1) EpH4.9 cells in wound healing assays. Distance traveled is averaged from six separate assays per cell line and three separate measurements per assay; P < 0.05. (B) Quantification of Matrigel invasion of control (pEiZ) and Zpo1-overexpressing (Zpo1) EpH4.9 cells. Experiments were performed in triplicate; P < 0.05. (C) Lysates from EpH4.9-pEiZ and EpH4.9-Zpo1 cells analyzed by Western blot for AJ proteins. (D) Bright-field images of EpH4.9-pEiZ and EpH4.9-Zpo1 cells cultured in 3D Matrigel for 4 d. Bar: top panels, 500 μm; bottom panels, 50 μm. (E) Immunofluorescent localization of E-cadherin, β-catenin, p120-catenin, and α-catenin (green) in EpH4.9-pEiZ and EpH4.9-Zpo1 cells cultured in 3D Matrigel. F-actin localization is shown in red using Alexa Fluor 568-conjugated phalloidin. Cells were counterstained with DAPI (blue). Arrows point to regions of reduced and/or relocalized protein expression. Bar, 50 μm. (F) Quantification of Zpo1 expression in EpH4.9 cells stably infected with control or Zpo1-specific shRNA (C5) by Western blot and qPCR using GAPDH as a control. (G) Bright-field images of EpH4.9-pEiZ and EpH4.9-Zpo1 cells stably infected with scramble or Zpo1-specific shRNA lentivirus and cultured in 3D Matrigel. Bar, 100 μm. (H) EpH4.9-pEiZ and EpH4.9-Zpo1 cells stably expressing a Zpo1 shRNA (C5) cultured in 3D Matrigel and stained with phalloidin demonstrating lumen formation (arrowhead). Bar, 50 μm.
Control and Zpo1-overexpressing cells showed similar levels of E-cadherin and α-, β-, and p120-catenin protein, the major components of AJ complexes (Fig. 2C). In two dimensions (2D) on tissue culture plastic, all four AJ proteins were membrane-localized (Supplemental Fig. 2). When we repeated these experiments with cells cultured in three dimensions (3D) within Matrigel, we observed that EpH4.9-Zpo1 cells formed numerous large filopodia/invadopodia-like structures at the periphery of the aggregates and migrated out from the body of the aggregates to form long chains of single cells (Fig. 2D). The cells within the chains showed minimal or a complete absence of cell–cell adhesion. Time-lapse imaging revealed the rapid formation and retraction of membrane protrusions (Supplemental Movies 3, 4). In contrast, EpH4.9-pEiZ cells formed tightly adherent aggregates (Fig. 2D).
AJ proteins in cells within the main body of both EpH4.9-pEiZ and EpH4.9-Zpo1 aggregates in 3D were localized along the cell membrane (Fig. 2E). In migratory cells of EpH4.9-Zpo1 aggregates, AJ proteins were present at significantly reduced levels and demonstrated cytoplasmic localization. Similarly, actin stained diffusely throughout the cytoplasm in these cells (Fig. 2E), unlike the cortical localization in cells within the aggregate bodies. These results suggest that Zpo1 overexpression inhibits or disrupts AJ formation at sites of cell interactions with the microenvironment, consequently reducing cell–cell adhesion. As a control, we knocked down Zpo1 expression in EpH4.9-pEiZ and EpH4.9-Zpo1 cells using a Zpo1-specific shRNA. Knockdown of Zpo1 expression was confirmed by quantitative PCR (qPCR) and Western blot (Fig. 2F). Reduced Zpo1 expression in both EpH4.9-pEiZ and EpH4.9-Zpo1 cells in 3D culture induced the formation of tightly adherent aggregates containing small lumens and an absence of migratory cell chains (Fig. 2G,H). This confirms that loss of cell adhesion and increased migration were due to Zpo1 overexpression. These results demonstrate that Zpo1 levels directly affect not only epithelial cell–cell adhesion, but also cell polarization and multicellular organization.
Overexpression of Zpo1 in MECs increases cell proliferation in 3D culture
Increased ZPO1 expression in human breast cancers is associated with an increased proliferative index. In 2D culture, there was no difference in cell proliferation between Zpo1-overexpressing and control cells (Fig. 3A). In contrast, aggregates from Zpo1-overexpressing cells cultured in 3D were significantly larger compared with control cells (Fig. 2D). When we assessed the number of cells in the aggregates that were positive for phosphorylated histone H3, we observed that ∼1% of EpH4.9-pEiZ cells were positive, while >3% of EpH4.9-Zpo1 cells were positive (P < 0.001) (Fig. 3B). This confirms that cell proliferation was increased in Zpo1-overexpressing cells in 3D.
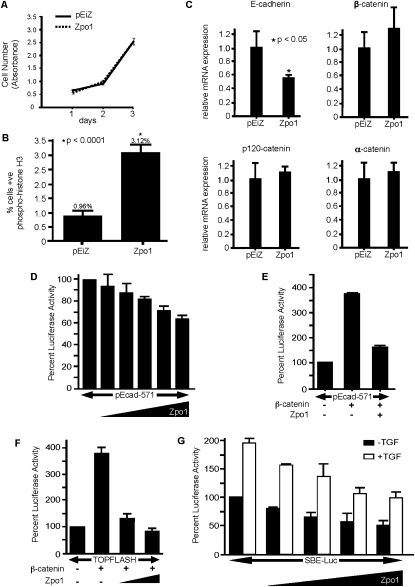
Figure 3.
Zpo1 promotes increased proliferation and represses E-cadherin expression. (A) Proliferation of control (solid line) and Zpo1-overexpressing (dotted line) EpH4.9 cells in 2D culture. Cells in three wells were counted for each time point. (B) Quantification of EpH4.9-pEiZ and EpH4.9-Zpo1 nuclei positive for phospho-histone H3 when cultured in 3D Matrigel for 48 h. Error bars represent ±SEM. (C) mRNA quantification by qPCR of AJ proteins from EpH4.9-pEiZ and EpH4.9-Zpo1 cells normalized to GAPDH expression. Error bars represent mean ± SD. (D) 3T3 cells were transfected with 400 ng of pEcad-Luc reporter plasmid and increasing quantities of Zpo1-V5 expression plasmid and assayed for luciferase activity. (E) 3T3 cells were transfected with pEcad-Luc, and the effect of β-catenin and Zpo1 expression was determined. (F) 3T3 cells were transfected with TOPFLASH reporter and increasing quantities of Zpo1-V5. (G) 3T3 cells were transfected with a TGFβ reporter plasmid and increasing quantities of Zpo1-V5, with or without exogenous TGFβ.
Zpo1 represses E-cadherin gene expression
The reduction in cell adhesion caused by Zpo1 overexpression in 3D culture suggested that Zpo1 may regulate the expression of genes involved in cell–cell adhesion. Zpo1 overexpression had no effect on α-, β- and p120-catenin gene expression levels in EpH4.9 cells, but reduced E-cadherin mRNA by ∼45% (Fig. 3C). To determine if Zpo1 repressed transcription, we cloned a 570-base-pair (bp) fragment of the human E-cadherin promoter immediately upstream of the ATG transcription start site, 5′ to a luciferase reporter gene. Transfection of the reporter construct into 3T3 cells resulted in significant luciferase activity that was reduced by cotransfection of a Zpo1 expression plasmid (Fig. 3D).
E-cadherin gene expression is reported to be negatively regulated by the Wnt pathway (Jamora et al. 2003; Yook et al. 2005). Surprisingly, we found that cotransfection of a β-catenin expression plasmid significantly increased expression from the E-cadherin reporter construct (Fig. 3E). However, cotransfection of Zpo1 with the β-catenin expression plasmid abrogated transcriptional activation (Fig. 3E). We then repeated these experiments using the TOPFLASH reporter plasmid that contains multiple copies of a Lef/Tcf-binding site upstream of a luciferase reporter gene. Increased β-catenin activity significantly increased luciferase activity as expected; however, cotransfection of Zpo1 again reduced β-catenin-mediated transcriptional activation (Fig. 3F).
Since both Wnt and TGFβ signaling negatively regulate E-cadherin expression, we used a luciferase reporter plasmid containing Smad-binding elements to determine if Zpo1 also regulates TGFβ-responsive transcription. Coexpression of Zpo1 was sufficient to repress transcription from this reporter by ∼50% in both the absence and presence of exogenous TGFβ (Fig. 3G).
Zpo1 overexpression induces an EMT
Zpo1-overexpressing cells at the invasive edge of 3D aggregates clearly differed from those in the center, as evidenced by reduced E-cadherin expression, remodeling of the cytoskeleton, and increased cell migration. These are hallmarks of EMT and a more progenitor or stem cell-like phenotype (Mani et al. 2008). To validate this observation, we evaluated other EMT markers. Zpo1-overexpressing cells within the body of the aggregates were negative for vimentin, a marker of mesenchymal cells, as were control cells; however, Zpo1-overexpressing cells at the interface with the microenvironment were clearly vimentin-positive (Fig. 4A). All EpH4.9-Zpo1 cells were positive for Snail and N-cadherin, while control cells were negative. Interestingly, N-cadherin staining was mainly plasma membrane-localized in the body of the Zpo1-overexpressing aggregates, but in the cytoplasm in peripheral cells (Fig. 4A). Both control and Zpo1-overexpressing cells were positive for cytokeratin (Fig. 4A).
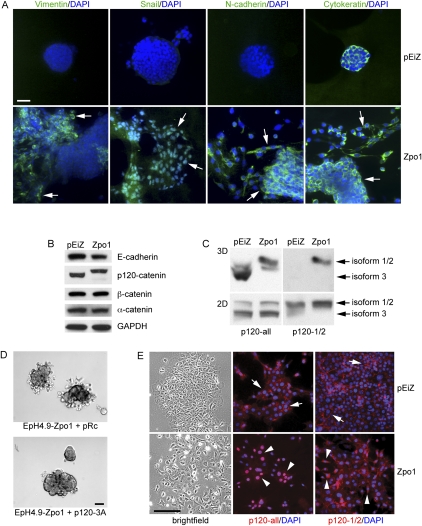
Figure 4.
Zpo1 overexpression promotes EMT and p120-catenin isoform expression and localization. (A) Immunofluorescent localization of vimentin, Snail, N-cadherin, and cytokeratin (green) in EpH4.9-pEiZ and EpH4.9-Zpo1 cells in 3D Matrigel culture. Arrows in vimentin staining of EpH4.9-Zpo1 cells point to vimentin-positive migratory cells. Cells were counterstained with DAPI (blue). Bar, 50 μm. (B) Western blot of AJ proteins from 3D cultures of control and Zpo1-overexpressing EpH4.9 cells after 6 d in culture. (C) Western blot showing p120-catenin isoform expression in 2D and 3D cultured EpH4.9-pEiZ and EpH4.9-Zpo1 cells using antibodies recognizing all p120-catenin isoforms (p120-all) or, specifically, isoforms 1 and 2 (p120-1/2). (D) Bright-field image of EpH4.9-Zpo1 cells stably transfected with control or p120-3A expression constructs and cultured in 3D Matrigel for 48 h. Bar, 100 μm. (E, left panels) Bright-field image of EpH4.9-pEiZ and EpH4.9-Zpo1 cells cultured on 2D Matrigel. Immunolocalization of all p120-catenin isoforms (p120-all) or isoforms 1/2 only (p120-1/2) demonstrates nuclear localization of p120 isoform 3 only in Zpo1-overexpressing cells (red). Bar, 100 μm.
Zpo1 overexpression promotes a switch to a promigratory p120-catenin isoform in 3D culture
To gain mechanistic insight into how Zpo1 mediates the differences between 2D and 3D culture conditions, we next investigated another promigratory molecule. p120 exists as multiple isoforms due to alternative splicing (Mo and Reynolds 1996; Keirsebilck et al. 1998) and four alternative transcriptional start sites. The most commonly expressed isoforms are 1 and 3. Using an antibody recognizing all p120 isoforms, we observed that the banding pattern from EpH4.9-Zpo1 cells differed from controls (Fig. 4B). An antibody that detects isoform 1 but not isoform 3 showed that EpH4.9-Zpo1 cells in 3D culture predominantly express isoform 1, while controls predominantly express isoform 3 (Fig. 4C). In contrast, both isoforms 1 and 3 were expressed in 2D culture (Fig. 4C), with isoform 3 as the predominant isoform in both control and Zpo1-overexpressing cells.
If the relative levels of p120 isoforms affect the adhesive and/or migratory phenotype of MECs, then increasing the level of p120 isoform 3 in EpH4.9-Zpo1 cells should reduce the invasive phenotype in 3D culture. Accordingly, we developed stable cell lines from EpH4.9-Zpo1 cells carrying either a p120 isoform 3 expression construct or the empty expression vector as a control. Cell aggregates generated from EpH4.9-Zpo1 cells overexpressing isoform 3 and cultured in 3D Matrigel demonstrated reduced cell migration compared with control cells (Fig. 4D), consistent with our hypothesis.
Zpo1 regulates ECM-dependent intracellular p120 isoform distribution
A striking phenotype of the Zpo1-overexpressing cells cultured in 3D was that redistribution of AJ proteins appeared to be restricted to cells on the periphery of the aggregates that are in contact with the ECM (Fig. 2E). This suggested that redistribution might be a result of cell–ECM interactions. To test this, we cultured EpH4.9-pEiZ and EpH4.9-Zpo1 cells on top of a thin 2D layer of Matrigel and determined AJ protein localization. EpH4.9-pEiZ cells formed compact epithelial colonies, whereas EpH4.9-Zpo1 cells were more scattered with reduced cell–cell contact (Fig. 4E). All AJ proteins, with the exception of p120, were membrane-localized in control and Zpo1-overexpressing cells (data not shown). EpH4.9-Zpo1 cells showed a predominantly nuclear localization of p120 with only weak cytoplasmic and membrane staining using a pan-isoform antibody (Fig. 4E); however, with the isoform 1/2-specific antibody, we saw protein only at the cell membrane and in the cytoplasm, but not in the nucleus (Fig. 4E). These data suggest that isoform 3 specifically localizes to the nucleus.
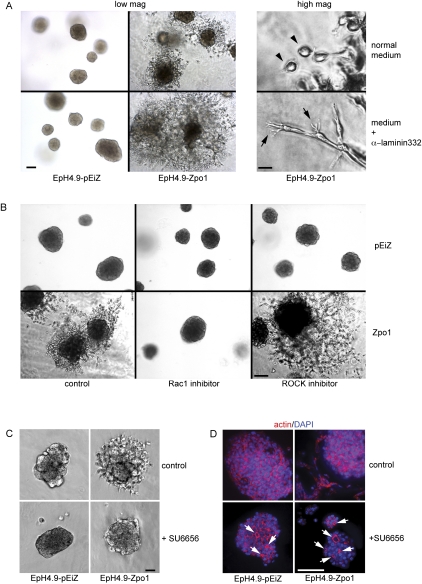
Based on our results, we propose that cell–ECM interactions mediate the effects of Zpo1 overexpression. To test this hypothesis, we used a function-inhibiting antibody directed against laminin 332 (also called laminin 5) to perturb cell–ECM interactions. Laminin 332 is not present in Matrigel, but is expressed endogenously by EpH4 cells (Maschler et al. 2005). Laminin 332 negatively regulates the number of actin-rich invadopodial protrusions (Liu et al. 2010) that facilitate ECM degradation and invasion (Albiges-Rizo et al. 2009). Addition of the anti-laminin 332 antibody to the growth medium of 3D cultures had no visible effect on control EpH4.9-pEiZ cells (Fig. 5A); however, in EpH4.9-Zpo1 cell cultures, migrating cells were more elongated, possessed longer membrane protrusions, and invaded further into the Matrigel compared with cells grown in control medium (Fig. 5A). These results suggest that loss of cell–laminin 332 interactions promotes epithelial cell invasion in Zpo1-overexpressing cells.
Figure 5.
Zpo1-mediated changes in cell adhesion and migration are regulated by the ECM and require Rho GTPase and SFK activity. (A) Low-magnification bright-field images of EpH4.9-pEiZ and EpH4.9-Zpo1 cells cultured for 5 d in the absence or presence of the anti-laminin 332 antibody, showing increased EpH4.9-Zpo1 cell migration in the presence of the antibody. Bar, 100 μm. High-magnification bright-field images of EpH4.9-Zpo1 cells cultured in the presence of the anti-laminin 332 antibody, showing elongated cells with long, membrane protrusions (arrows). Bar, 10 μm. (B) Bright-field images of EpH4.9-pEiZ and EpH4.9-Zpo1 cells cultured in the presence of the ROCK inhibitor Y-27632 (5 μM) and Rac1 inhibitor (100 μM). Bar, 100 μm. (C) Bright-field images of EpH4.9-pEiZ and EpH4.9-Zpo1 cells cultured in 3D Matrigel in the presence of the SFK inhibitor SU6656 (5 μM). Bar, 100 μm. (D) F-actin staining of EpH4.9-pEiZ and EpH4.9-Zpo1 cells cultured in 3D in the presence of 5 μM SU6656 showing lumen formation (arrows). Bar, 100 μm.
Rac1 activity and inhibition of RhoA is required for the Zpo1 overexpression phenotype
p120 can promote cell migration via interactions with Rho GTPases, increasing Rac1 activity and inhibiting RhoA. Isoform 1 specifically promotes increased cell migration compared with other p120 isoforms due to its increased affinity for, and inhibition of, RhoA (Yanagisawa et al. 2008). Addition of Rac1 inhibitor significantly inhibited EpH4.9-Zpo1 cell migration and invasion, but had no obvious effect on control cells (Fig. 5B). We then inhibited ROCK, a downstream mediator of RhoA that becomes activated upon RhoA activity. Addition of a ROCK inhibitor enhanced migration and invasion of Zpo1-overexpressing cells, but had no obvious effect on control cells (Fig. 5B).
Epithelial cell–ECM interactions and binding of p120 to RhoA (Castano et al. 2007) are both mediated through activity of the Src family of kinases (SFK). Therefore, inhibition of SFK activity would be expected to alter the Zpo1-mediated increase in MEC migration and invasion. EpH4.9-pEiZ cells cultured in 3D in the presence of the SFK inhibitor SU6656 (Fig. 5C) or PP1 (data not shown) showed no obvious effect. However, EpH4.9-Zpo1 cells in the presence of SFK inhibitors had a rounder shape, formed significantly fewer membrane protrusions, and, for the most part, remained attached to the aggregate (Fig. 5C). Sectioning of aggregates from both EpH4.9-pEiZ and EpH4.9-Zpo1 cells showed the presence of small lumens (Fig. 5D).
Zpo1 overexpression promotes tumor metastasis
To determine directly if Zpo1 overexpression is tumor- and/or metastasis-promoting, we transplanted EpH4.9-pEiZ or EpH4.9-Zpo1 cells into the mammary fat pad of syngeneic mice. Parental EpH4.9 cells are nontumorigenic; however, Zpo1 overexpression alone was not sufficient to induce tumor formation, even 12 mo after transplantation (data not shown).
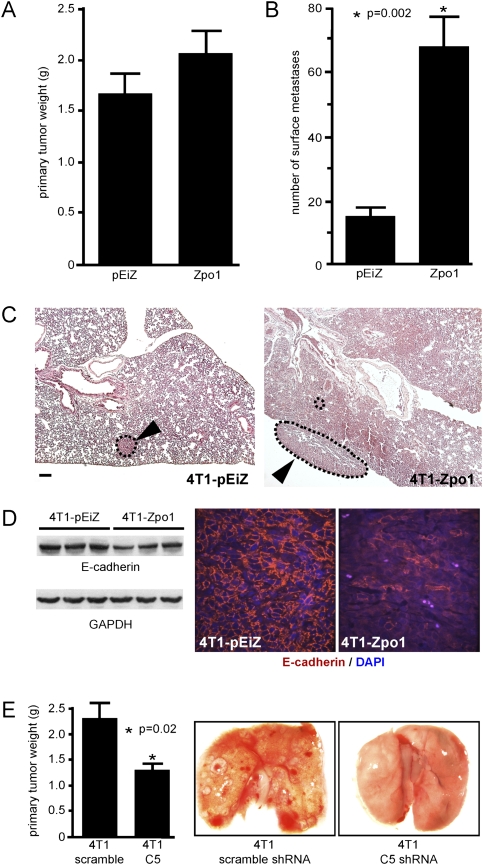
We then asked if Zpo1 overexpression affected tumor metastasis. The MEC line 4T1 is both tumorigenic and metastatic to the lung, liver, bone, and brain when transplanted into the mammary fat pad of immunocompetent mice (Aslakson and Miller 1992). We generated Zs-green-labeled polyclonal 4T1 cells overexpressing Zpo1 (4T1-Zpo1), and control 4T1 cells infected with empty vector (4T1-pEiZ), and transplanted them into the mammary fat pads of syngeneic BALB/c mice. After 3 wk, the weight of tumors from mice transplanted with Zpo1-overexpressing cells was slightly increased compared with controls (Fig. 6A). Significantly, the number of metastatic lung nodules was dramatically increased in mice transplanted with Zpo1-overexpressing cells compared with controls (Fig. 6B,C). Furthermore, the 4T1-Zpo1 primary tumors clearly showed a reduction in E-cadherin protein levels compared with control tumors (Fig. 6D). We next determined if reducing Zpo1 expression in 4T1 cells would affect tumor progression. We generated polyclonal 4T1 cell lines stably expressing either a control scrambled shRNA or a Zpo1-specific shRNA that reduced Zpo1 expression by ∼60% (data not shown) and repeated the transplant experiments. After 3 wk, both the size of the primary tumors and the number of lung metastases were significantly reduced in Zpo1 knockdown cells compared with controls (Fig. 6E). These results suggest that Zpo1 is the driver gene for increased tumor metastases in patients with the 8p12 amplicon.
Figure 6.
Zpo1 overexpression increases breast cancer metastasis. (A) Graph showing the weight of primary tumors formed by 4T1-pEiZ and 4T1-Zpo1 cells 3 wk post-transplantation. (B) Graph showing the number of lung surface metastases formed by 4T1-pEiZ and 4T1-Zpo1 cells 3 wk post-transplantation. Error bars represent ±SEM. (C) Representative hematoxylin-eosin-stained section of lungs from mice transplanted with 4T1-pEiZ and 4T1-Zpo1 cells containing lung metastases (dotted lines). Bar, 200 μm. (D) Western blot of E-cadherin protein from three primary tumor lysates isolated from 4T1-pEiZ and 4T1-Zpo1 tumors. Immunofluorescence staining of E-cadherin protein in sections of primary tumors formed from 4T1-pEiZ and 4T1-Zpo1 cells. Bar, 50 μm. (E) Graph showing weight of primary tumors formed by 4T1 cells expressing either a scrambled or Zpo1-specific (C5) shRNA 3 wk post-transplantation. Representative images of lungs from mice transplanted with shRNA-expressing 4T1 cells showing a reduction in lung surface metastases from Zpo1 knockdown cells.
Discussion
In this study, we identified Zpo1/Znf703, a critical gene in the 8p12 breast cancer-associated amplicon, as one of the earliest genes expressed during mammary gland development, and elucidated its functions as a transcriptional repressor and a regulator of cell motility and invasion. Together with the observation that Zpo1 increases metastasis, we propose that Zpo1 is a metastasis promoter that is responsible for poor breast cancer outcome.
Zeppo1 and transcriptional regulation
Zpo1 is a member of the NET (Noc/Nlz, Elbow, and Tlp-1) family of proteins (Nakamura et al. 2004) that function in embryonic development in zebrafish (Hoyle et al. 2004; Brown et al. 2009), Drosophila (Dorfman et al. 2002), and Caenorhabditis elegans (Zhao et al. 2002). We show here that Zpo1 functions as a transcriptional repressor. While some of Zpo1's repressor function requires HDAC activity, it also demonstrates HDAC-independent repressor activity, possibly as a result of its interactions with other corepressors such as Grg4. These results are consistent with studies showing that, while Nlz1 activity is associated with HDACs in vitro (Runko and Sagerstrom 2004), HDAC activity accounts for only part of the repression (Nakamura et al. 2008).
We showed that increased Zpo1 expression represses endogenous mouse E-cadherin gene transcription and inhibits transcription from a human E-cadherin promoter fragment. Interestingly, Zpo1 also inhibited β-catenin-mediated transcriptional activation of both the human E-cadherin and the TOPFLASH promoters. Since β-catenin competes with Grg proteins for binding to Lef/Tcf transcription factors (Daniels and Weis 2005), one possible mechanism of Zpo1 function may be to promote binding of Grg proteins to Lef/Tcf. An alternative possibility is that Zpo1 functions to convert β-catenin from a transcriptional activator into a repressor. β-Catenin can function as part of a repression complex in conjunction with either members of the Lef/Tcf family (Jamora et al. 2003; Theisen et al. 2007) or other transcription factors (Olson et al. 2006), and β-catenin-mediated repression requires binding of corepressors and recruitment of HDAC activity (Olson et al. 2006; Iguchi et al. 2007). Nonetheless, Zpo1-mediated repression of E-cadherin transcription is significant, since E-cadherin is a tumor and metastasis suppressor, and reduced gene expression, protein levels, or changes in protein localization are found in the majority of metastatic carcinomas, including breast cancers (Cowin et al. 2005). Repression of E-cadherin gene expression is also regulated by transcription factors such as Snail1/2, ZEB1/2, and E12/47, and, like Zpo1, they are frequently up-regulated during tumorigenesis (Peinado et al. 2007).
Interestingly, while Zpo1 was able to repress E-cadherin gene transcription, E-cadherin protein levels were not reduced in 2D culture (Fig. 2C) and were only minimally reduced in 3D culture. This is likely a consequence of the high stability and low turnover of E-cadherin protein under the conditions of stable cell–cell adhesion that we see in 2D. In contrast, E-cadherin is rapidly degraded or recycled upon loss of cell–cell contact (Daniel and Reynolds 1999); however, since this phenotype is restricted to cells on the outer edge of the 3D aggregates, overall protein levels are not greatly affected.
Zeppo1 and EMT
Zpo1 overexpression promoted EMT and the expression of the EMT markers vimentin, Slug, and N-cadherin. Interestingly, Slug and N-cadherin expression was induced in all cells, while vimentin expression was restricted to migratory cells at the interface with the microenvironment. This suggests that expression of these genes is differentially regulated, and that cell–ECM interactions specifically regulate Zpo1-mediated vimentin expression.
Loss of E-cadherin in EMT not only reduces cell–cell adhesion, but also induces a loss of cell polarity and acquisition of a more progenitor-like state. EpH4.9 cells cultured in 3D Matrigel form poorly polarized, adherent cell aggregates. We showed that knockdown of endogenous Zpo1 expression in EpH4.9 cell aggregates induced the formation of small lumens lined with cells rich in apical actin, suggesting polarization, while overexpression of Zpo1 induced the cytoplasmic localization of AJ proteins. Interestingly, we obtained a similar phenotype when we treated cell aggregates with SFK inhibitors. Src activation has long been known as a tumor promoter (Frame 2002) and a regulator of cell polarity (Timpson et al. 2001; Yamana et al. 2006; Grande-Garcia et al. 2007; Wang et al. 2009). The inhibition of the Zpo1 phenotype by SFK inhibitors and the formation of lumens by either Zpo1 knockdown or SFK inhibition suggest that Zpo1 may function, in part, through SFK activity.
p120 in cell migration and cancer
p120 is a multifunctional protein that localizes to the cell membrane, cytoplasm, and nucleus. At the cell membrane, p120 binds to E-cadherin, promoting clustering (Yap et al. 1998) and stabilization of E-cadherin by inhibiting protein turnover (Davis et al. 2003). In the cytoplasm, p120 interacts with members of the Rho family of GTPases to regulate the actin cytoskeleton and, consequently, several actin-dependent cellular processes, including cell shape, adhesion, migration, and polarity (Anastasiadis 2007). Nuclear p120 function is less well understood, but it can interact with the transcription factor Kaiso (Daniel and Reynolds 1999) to inhibit Kaiso-mediated transcriptional repression (Daniel et al. 2002). The combined regulation of both cell–cell adhesion and actin dynamics provides an important role for p120 in regulating the motile state of cells.
p120 may function as both a tumor suppressor and a metastasis promoter (Reynolds and Roczniak-Ferguson 2004), and its expression is altered in the majority of human cancers at the level of either gene expression or protein localization (van Hengel and van Roy 2007). In one study, 88% of lobular human breast carcinomas had cytoplasmic localization of p120 with a corresponding absence of E-cadherin (Sarrio et al. 2004).
p120 exists as multiple isoforms due to alternative splicing of a single gene (Reynolds and Roczniak-Ferguson 2004). The most commonly expressed isoforms are isoforms 1 and 3. Isoform 1 is predominantly expressed in motile cells such as fibroblasts and in epithelial tumors, while isoform 3 is the predominant isoform in sessile epithelial cells. p120 expression is an independent prognosticator of breast cancer survival, and isoform 1 expression predicts metastatic disease (Talvinen et al. 2010). Isoform 1 expression and metastasis are also significantly correlated in both lung (Miao et al. 2009) and renal (Yanagisawa et al. 2008) carcinoma.
Our studies show that EpH4.9 MECs preferentially express p120 isoform 3 when cultured on 2D plastic. Culturing EpH4.9 cells within a laminin-rich 3D matrix, however, further reduces isoform 1 expression. Overexpression of Zpo1 in these cells had no effect on p120 isoform expression in normal 2D culture; however, when cultured within an ECM, increased Zpo1 expression induced an isoform switch such that isoform 1 was preferentially expressed. Interestingly, transcriptional repression of E-cadherin in combination with isoform switching of p120 from isoform 3 to isoform 1 is seen in MDCK cells overexpressing the EMT-inducing transcriptional repressors Snail, Slug, or E47 (Ohkubo and Ozawa 2004; Sarrio et al. 2004).
Zpo1 overexpression also promoted relocalization of p120 protein away from the cell membrane that was also dependent on cell–ECM contact. In 2D Matrigel culture, Zpo1 overexpression induced nuclear localization of isoform 3, while isoform 1 was restricted to the cell membrane and cytoplasm. In 3D culture, both isoforms were localized to the cell membrane in cells in the body of the aggregates, away from the ECM, but were localized to the cytoplasm in migratory cells in contact with the ECM. These results provide further evidence that Zpo1 function is regulated by cell–ECM interactions.
A model for Zpo1-mediated increase in tumor metastasis
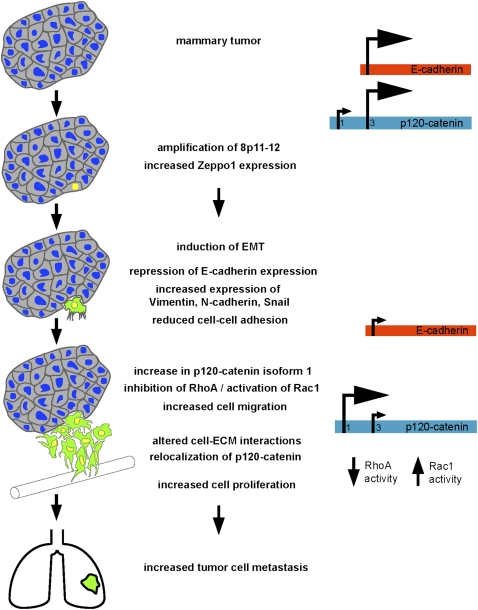
The metastatic process requires cells to alter their normal interactions with neighboring cells and the extracellular environment (Gupta and Massague 2006). Understanding how these processes are regulated is of major importance for the development of therapies that might interfere with metastasis formation. Based on the data presented in this study, we propose a model for Zpo1 function (Fig. 7) to explain how overexpression can lead to increased tumor metastasis. The 8p12 amplification and the overexpression of multiple genes within the amplicon result in increased Zpo1 gene expression. Increased levels of Zpo1 protein repress E-cadherin expression, resulting in a decrease in cell–cell adhesion and relocalization of p120 from the cell membrane to the cytoplasm. Concomitantly, increased Zpo1 expression induces a switch in p120 isoforms from isoform 3 to isoform 1 that leads to increased binding of p120 to RhoA, decreasing RhoA activity. Zpo1 expression also increases cell proliferation through an as-yet-unknown mechanism. In combination, these events reduce tumor cell adhesion and cell polarity while increasing tumor cell migration and proliferation, thus promoting a higher tumor grade and an increase in tumor metastases.
Figure 7.
Model of Zpo1 overexpression and metastasis promotion. Amplification of chromosome 8p11-12 in a mammary tumor cell leads to overexpression and increased activity of Zpo1. As a result, E-cadherin expression is reduced with a consequent reduction in tumor cell adhesion. Loss of E-cadherin at the cell membrane reduces membrane-localized p120-catenin and increases the pool of cytoplasmic p120-catenin that is able to interact with Rho GTPases. At the same time, Zpo1 overexpression also promotes a switch in p120-catenin isoform expression from isoform 3 to isoform 1 that promotes increased cell migration by binding to and inhibiting RhoA while increasing Rac1 activity. Zpo1 overexpression also reduces tumor cell interactions with the ECM and increases tumor cell migration through unknown mechanisms. In combination, these effects lead to a more proliferative tumor with increased migratory and invasive behavior, leading to increased tumor metastasis.
Role of other genes within the 8p11-12 amplicon
Overexpression of Zpo1 in nontumorigenic EpH4.9 cells was insufficient to induce tumor formation, suggesting that Zpo1 overexpression by itself may not be a tumor-initiating event. It is possible that an unrelated genetic event is responsible for tumor formation, and that amplification of 8p12 occurs later in the process. Alternatively, since 8p12 amplification affects the expression of multiple genes within the locus, altered expression of more than one gene may be responsible for tumor formation and metastatic progression. Several genes within this locus have been studied with regard to their oncogenic capabilities; however, none have been convincingly identified as classical oncogenes. One such gene, RCP (also known as RAB11FIP1), was identified recently as a candidate oncogene associated with breast cancer (Miao et al. 2009); however, overexpression of RCP in tumorigenic MECs has no effect on tumor cell growth, invasion, or metastasis in vivo. RCP overexpression in MCF10A cells did result in soft agar colony formation, reduced requirement for growth factors, and altered cell migration. Interestingly, similar results were obtained when three other genes located at 8p11-12 (LSM1, BAG4, and C8orf4) were overexpressed individually or in combination in MCF10A cells (Yang et al. 2006). It is therefore possible that amplification and overexpression of some combination of these genes are required for the full 8p11-12 amplification phenotype seen in human breast cancer. However, a study that profiled 1172 primary breast cancers also has identified ZNF703 as the sole 8p12 candidate responsible for poor prognosis in ER+ tumors (Holland et al. 2011).
In conclusion, we show that Zpo1 overexpression plays an important role in regulating MEC adhesion, migration, and polarity, and can significantly increase the number of mammary tumor metastases. Our data strongly implicate Zpo1 overexpression as a metastasis driver in breast carcinogenesis.
Materials and methods
RNA in situ hybridization of mouse tissues
Gene expression on paraffin sections of mouse embryos by RNA in situ hybridization was carried out as described previously (Lum et al. 2007). Whole-mount embryo in situ hybridizations were performed on CD1 mouse embryos as described previously (Cross et al. 1995).
The Zpo1 riboprobe was generated from a 635-bp BamHI fragment from exon 2. The Grg4 riboprobe was generated from a PCR fragment amplified using primers 5′-TCCAACTCCACGCATCAAGG-3′ and 5′-TGCTATGAGGAGGAGTCCAGTTTTG-3′.
Cell culture
Mouse EpH4.9 cells are a subclone derived from EpH4 cells. EpH4.9 cells grown on tissue culture plastic were cultured in DME-H21 medium supplemented with 5% (v/v) fetal bovine serum (FBS), insulin (5 μg/mL), 100 IU of penicillin, and 100 μg/mL streptomycin. EpH4.9 cells to be cultured in 3D Matrigel were maintained in low-adhesion plates overnight to form aggregates. The aggregates were separated from remaining single cells by gravity, resuspended in ice-cold Matrigel, and plated in 25-μL volumes. The Matrigel/cell mix was allowed to solidify for 1 h at 37°C, and was cultured in serum-free DMEM:F12 medium supplemented with 1× ITS (Invitrogen), 50 ng/mL EGF (Invitrogen), 100 IU of penicillin, and 100 μg/mL streptomycin. SFK inhibitor SU6656 (Sigma-Aldrich) was added to EpH4.9 growth medium at 5–10 μM. Mouse 3T3 and 4T1 cells and human 293T cells were cultured in DME H-21 medium supplemented with 10% (v/v) FBS, 100 IU of penicillin, and 100 μg/mL streptomycin.
Cell proliferation
Cell proliferation in 2D cultures was quantified using a colorimetric assay (CellTiter 96, Promega) to determine the number of viable cells, according to the manufacturer's instructions. Proliferation of EpH4.9 cells cultured in 3D for 24 h was determined by counting the number of cells positive for phospho-histone H3. This was achieved by staining 50-μm frozen sections of cultured cells using a phospho-histone H3 antibody (Cell Signaling Technology) and taking serial stack images of individual aggregates by confocal microscopy. The total number of cells visualized by DAPI staining and the number of phospho-histone H3-positive cells in the fifth and tenth images of each stack was counted to determine the percent of proliferating cells.
Invasion assay
Transwell cell migration assays were performed using the Millipore Cell Invasion Assay kit following the manufacturer's instructions. Briefly, 3 × 105 cells were plated onto ECM-coated cell culture inserts and incubated for 48 h. Cells that had invaded through the ECM to the lower surface of the insert were stained and quantified by colorimetric analysis.
Wound healing assay
EpH4.9 cells were grown to confluence in a 24-well plate, and were wounded using a 200-μL sterile pipette tip. Movies of wound healing were generated using an inverted microscope over a 24-h time period. The distance traveled by cells at the migrating edge after 15 h was calculated by measuring three separate points within each field of view, from at least three separate experiments.
Antibodies and immunofluorescence
Cells cultured in 2D were fixed in 4% paraformaldehyde and incubated with primary antibody, followed by an appropriate secondary antibody conjugated to Alexa Fluor 568 or 647 (Invitrogen). The actin cytoskeleton was stained with phalloidin (Invitrogen) and cell nuclei with DAPI (Invitrogen). Immunofluorescence images were obtained using confocal microscopy and Imaris imaging software (Bitplane Scientific Software). The primary antibodies used were specific for p120-catenin (all isoforms), V5 epitope, and GAL4 DNA-binding domain (Invitrogen); Flag tag, α-catenin, p120-catenin (isoforms 1 and 2), and vimentin (Sigma-Aldrich); N-cadherin, Snail, and phospho-histone H3 (Cell Signaling Technology); and E-cadherin (BD Biosciences) and β-catenin (Abcam).
Lentiviral production and shRNA knockdown
Lentiviral production was carried out using calcium phosphate-mediated transfection of HEK 293T cells as described previously (Welm et al. 2008). Expression Arrest GIPZ lentiviral shRNA constructs were purchased from Open Biosystems. Cells were transduced with lentivirus according to the manufacturer's instructions, and cells were selected in puromycin for at least 5 d.
Quantitative real-time PCR
Total RNA was isolated from cells using the RNeasy minikit (Qiagen). cDNA was synthesized from 1 μg of total RNA using the SuperScript III Reverse Transcriptase First Strand synthesis kit (Invitrogen). Quantitative PCR was performed using FastStart Universal SYBR Green master mix (Roche Applied Science) in an Eppendorf Mastercycler realplex machine, and samples were run in quadruplicate. Ct values were normalized to actin, and relative expression was calculated using the 2−DDCt method. Statistical analysis was performed using Student's t-test, and P < 0.05 was considered to be significant. All statistical values indicate mean ± standard deviation. Primer sequences are detailed in Supplemental Table 1.
Transcription assays
3T3 cells were seeded at 5 × 104 cells per well in 48-well plates and transfected with 400 ng of pFR-Luc luciferase reporter construct (Stratagene), 5 ng of pRL-TK (Promega) internal control plasmid, and the appropriate test plasmids. Cell lysates were obtained 48 h post-transfection, and luciferase reporter activity was determined using the Promega Dual-Luciferase Reporter assay system following the manufacturer's protocol. Firefly luciferase activity was normalized to the Renilla luciferase internal control and expressed relative to control plasmid. The data presented are pooled from three replicate experiments in which each assay condition was performed in triplicate. The E-cadherin promoter luciferase reporter construct was generated by amplifying human genomic DNA using the primers 5′-TTGCTAGCAAAAGAAAAAAAAAATTAGCCTGGCGTGG-3′ and 5′-AAACTCGAGCGGGCTGGAGCGGGC-3′. Amplified DNA was digested with NheI/XhoI and cloned into the pGL3-basic reporter plasmid (Promega). The β-catenin expression plasmid was a kind gift from Dr. Thaddeus Allen (University of California at San Francisco, San Francisco, CA). TSA (Calbiochem) was added to 3T3 growth medium at various concentrations as described in the Results section.
Tumorigenesis and metastasis assays
Cells were harvested, counted, and resuspended in PBS. Cells (1 × 106) were injected into the fat pad of the inguinal (#4) mammary gland of syngeneic 5-wk-old BALB/c mice. Primary tumors were dissected, weighed, and measured after 3 wk. The number of lung metastases was determined by counting the number of Zs-Green-positive areas visible on the lung surface.
Additional Materials and Methods can be found in the Supplemental Material.
Acknowledgments
We thank Heidi Savage and Ying Yu for excellent technical assistance. We are grateful to Dr. Monique Aumailley (Center for Biochemistry, Cologne, Germany) for the gift of the rabbit anti-laminin 332 antibody. This study was supported by funds from the UCSF Program for Breakthrough Biomedical Research, the National Cancer Institute (CA057621, CA129523, and CA129523S1), the Human Frontiers Science Program (RG 0051/1999-M), and a Stand up to Cancer-American Association for Cancer Research Dream Team Translational Cancer Research Grant (SU2C-AACR-DT0409). E.M.S. was funded by fellowships from The Wellcome Trust and the California Breast Cancer Research Program. J.C. was funded by the Medical Scientist Training Program and a predoctoral fellowship from the California Breast Cancer Research Program.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1998111.
Supplemental material is available for this article.
References
- Adelaide J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, Charafe-Jauffret E, Cervera N, Desplans J, Parzy D, et al. 2007. Integrated profiling of basal and luminal breast cancers. Cancer Res 67: 11565–11575 [DOI] [PubMed] [Google Scholar]
- Albiges-Rizo C, Destaing O, Fourcade B, Planus E, Block MR 2009. Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J Cell Sci 122: 3037–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiadis PZ 2007. p120-ctn: a nexus for contextual signaling via Rho GTPases. Biochim Biophys Acta 1773: 34–46 [DOI] [PubMed] [Google Scholar]
- Aslakson CJ, Miller FR 1992. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res 52: 1399–1405 [PubMed] [Google Scholar]
- Birnbaum D, Adelaide J, Popovici C, Charafe-Jauffret E, Mozziconacci MJ, Chaffanet M 2003. Chromosome arm 8p and cancer: a fragile hypothesis. Lancet Oncol 4: 639–642 [DOI] [PubMed] [Google Scholar]
- Brown JD, Dutta S, Bharti K, Bonner RF, Munson PJ, Dawid IB, Akhtar AL, Onojafe IF, Alur RP, Gross JM, et al. 2009. Expression profiling during ocular development identifies 2 Nlz genes with a critical role in optic fissure closure. Proc Natl Acad Sci 106: 1462–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano J, Solanas G, Casagolda D, Raurell I, Villagrasa P, Bustelo XR, Garcia de Herreros A, Dunach M 2007. Specific phosphorylation of p120-catenin regulatory domain differently modulates its binding to RhoA. Mol Cell Biol 27: 1745–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H 2006. Wnt/β-catenin signaling in development and disease. Cell 127: 469–480 [DOI] [PubMed] [Google Scholar]
- Cowin P, Rowlands TM, Hatsell SJ 2005. Cadherins and catenins in breast cancer. Curr Opin Cell Biol 17: 499–508 [DOI] [PubMed] [Google Scholar]
- Cross JC, Flannery ML, Blanar MA, Steingrimsson E, Jenkins NA, Copeland NG, Rutter WJ, Werb Z 1995. Hxt encodes a basic helix–loop–helix transcription factor that regulates trophoblast cell development. Development 121: 2513–2523 [DOI] [PubMed] [Google Scholar]
- Daniel JM, Reynolds AB 1999. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol 19: 3614–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JM, Spring CM, Crawford HC, Reynolds AB, Baig A 2002. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res 30: 2911–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Weis WI 2005. β-Catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol 12: 364–371 [DOI] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB 2003. A core function for p120-catenin in cadherin turnover. J Cell Biol 163: 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman R, Glazer L, Weihe U, Wernet MF, Shilo BZ 2002. Elbow and Noc define a family of zinc finger proteins controlling morphogenesis of specific tracheal branches. Development 129: 3585–3596 [DOI] [PubMed] [Google Scholar]
- Frame MC 2002. Src in cancer: deregulation and consequences for cell behaviour. Biochim Biophys Acta 1602: 114–130 [DOI] [PubMed] [Google Scholar]
- Garcia MJ, Pole JC, Chin SF, Teschendorff A, Naderi A, Ozdag H, Vias M, Kranjac T, Subkhankulova T, Paish C, et al. 2005. A 1 Mb minimal amplicon at 8p11-12 in breast cancer identifies new candidate oncogenes. Oncogene 24: 5235–5245 [DOI] [PubMed] [Google Scholar]
- Gasperowicz M, Otto F 2005. Mammalian Groucho homologs: redundancy or specificity? J Cell Biochem 95: 670–687 [DOI] [PubMed] [Google Scholar]
- Gelsi-Boyer V, Orsetti B, Cervera N, Finetti P, Sircoulomb F, Rouge C, Lasorsa L, Letessier A, Ginestier C, Monville F, et al. 2005. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res 3: 655–667 [DOI] [PubMed] [Google Scholar]
- Grande-Garcia A, Echarri A, de Rooij J, Alderson NB, Waterman-Storer CM, Valdivielso JM, del Pozo MA 2007. Caveolin-1 regulates cell polarization and directional migration through Src kinase and Rho GTPases. J Cell Biol 177: 683–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM 2005. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol 6: 622–634 [DOI] [PubMed] [Google Scholar]
- Gupta GP, Massague J 2006. Cancer metastasis: building a framework. Cell 127: 679–695 [DOI] [PubMed] [Google Scholar]
- Haverty PM, Fridlyand J, Li L, Getz G, Beroukhim R, Lohr S, Wu TD, Cavet G, Zhang Z, Chant J 2008. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer 47: 530–542 [DOI] [PubMed] [Google Scholar]
- Holland D, Burleigh A, Git A, Goldgraben MA, Perez-Mancera PA, Chin S-F, Hurado A, Bruna A, Ali R, Greenwood W, et al. 2011. ZNF703 is a common Luminal B breast cancer oncogene that differentially regulates luminal and basal progenitors in human mammary epithelium. EMBO Mol Med doi: 10.1002/emmm.201100122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle J, Tang YP, Wiellette EL, Wardle FC, Sive H 2004. nlz gene family is required for hindbrain patterning in the zebrafish. Dev Dyn 229: 835–846 [DOI] [PubMed] [Google Scholar]
- Hynes RO 2002. Integrins: bidirectional, allosteric signaling machines. Cell 110: 673–687 [DOI] [PubMed] [Google Scholar]
- Iguchi H, Urashima Y, Inagaki Y, Ikeda Y, Okamura M, Tanaka T, Uchida A, Yamamoto TT, Kodama T, Sakai J 2007. SOX6 suppresses cyclin D1 promoter activity by interacting with β-catenin and histone deacetylase 1, and its down-regulation induces pancreatic β-cell proliferation. J Biol Chem 282: 19052–19061 [DOI] [PubMed] [Google Scholar]
- Ishiyama N, Lee SH, Liu S, Li GY, Smith MJ, Reichardt LF, Ikura M 2010. Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell–cell adhesion. Cell 141: 117–128 [DOI] [PubMed] [Google Scholar]
- Jamora C, DasGupta R, Kocieniewski P, Fuchs E 2003. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature 422: 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keirsebilck A, Bonne S, Staes K, van Hengel J, Nollet F, Reynolds A, van Roy F 1998. Molecular cloning of the human p120ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics 50: 129–146 [DOI] [PubMed] [Google Scholar]
- Liu S, Yamashita H, Weidow B, Weaver AM, Quaranta V 2010. Laminin-332–β1 integrin interactions negatively regulate invadopodia. J Cell Physiol 223: 134–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum DH, Tan J, Rosen SD, Werb Z 2007. Gene trap disruption of the mouse heparan sulfate 6-O-endosulfatase gene, Sulf2. Mol Cell Biol 27: 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X 2009. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell 17: 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. 2008. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maschler S, Wirl G, Spring H, Bredow DV, Sordat I, Beug H, Reichmann E 2005. Tumor cell invasiveness correlates with changes in integrin expression and localization. Oncogene 24: 2032–2041 [DOI] [PubMed] [Google Scholar]
- Medici D, Hay ED, Goodenough DA 2006. Cooperation between snail and LEF-1 transcription factors is essential for TGF-β1-induced epithelial–mesenchymal transition. Mol Biol Cell 17: 1871–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchor L, Garcia MJ, Honrado E, Pole JC, Alvarez S, Edwards PA, Caldas C, Brenton JD, Benitez J 2007. Genomic analysis of the 8p11-12 amplicon in familial breast cancer. Int J Cancer 120: 714–717 [DOI] [PubMed] [Google Scholar]
- Miao Y, Liu N, Zhang Y, Liu Y, Yu JH, Dai SD, Xu HT, Wang EH 2009. p120ctn isoform 1 expression significantly correlates with abnormal expression of E-cadherin and poor survival of lung cancer patients. Med Oncol 27: 880–886 [DOI] [PubMed] [Google Scholar]
- Mo YY, Reynolds AB 1996. Identification of murine p120 isoforms and heterogeneous expression of p120cas isoforms in human tumor cell lines. Cancer Res 56: 2633–2640 [PubMed] [Google Scholar]
- Nakamura M, Runko AP, Sagerstrom CG 2004. A novel subfamily of zinc finger genes involved in embryonic development. J Cell Biochem 93: 887–895 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Choe SK, Runko AP, Gardner PD, Sagerstrom CG 2008. Nlz1/Znf703 acts as a repressor of transcription. BMC Dev Biol 8: 108 doi: 10.1186/1471-213X-8-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao K, Mehta KR, Fridlyand J, Moore DH, Jain AN, Lafuente A, Wiencke JW, Terdiman JP, Waldman FM 2004. High-resolution analysis of DNA copy number alterations in colorectal cancer by array-based comparative genomic hybridization. Carcinogenesis 25: 1345–1357 [DOI] [PubMed] [Google Scholar]
- Nawshad A, Medici D, Liu CC, Hay ED 2007. TGFβ3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2–Smad4–LEF1 transcription complex. J Cell Sci 120: 1646–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ 2008. Regulation of cell–cell adhesion by the cadherin–catenin complex. Biochem Soc Trans 36: 149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo T, Ozawa M 2004. The transcription factor Snail downregulates the tight junction components independently of E-cadherin downregulation. J Cell Sci 117: 1675–1685 [DOI] [PubMed] [Google Scholar]
- Olson LE, Tollkuhn J, Scafoglio C, Krones A, Zhang J, Ohgi KA, Wu W, Taketo MM, Kemler R, Grosschedl R, et al. 2006. Homeodomain-mediated β-catenin-dependent switching events dictate cell-lineage determination. Cell 125: 593–605 [DOI] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A 2007. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7: 415–428 [DOI] [PubMed] [Google Scholar]
- Pole JC, Courtay-Cahen C, Garcia MJ, Blood KA, Cooke SL, Alsop AE, Tse DM, Caldas C, Edwards PA 2006. High-resolution analysis of chromosome rearrangements on 8p in breast, colon and pancreatic cancer reveals a complex pattern of loss, gain and translocation. Oncogene 25: 5693–5706 [DOI] [PubMed] [Google Scholar]
- Prentice LM, Shadeo A, Lestou VS, Miller MA, Deleeuw RJ, Makretsov N, Turbin D, Brown LA, Macpherson N, Yorida E, et al. 2005. NRG1 gene rearrangements in clinical breast cancer: identification of an adjacent novel amplicon associated with poor prognosis. Oncogene 24: 7281–7289 [DOI] [PubMed] [Google Scholar]
- Qian X, Karpova T, Sheppard AM, McNally J, Lowy DR 2004. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. EMBO J 23: 1739–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AB, Roczniak-Ferguson A 2004. Emerging roles for p120-catenin in cell adhesion and cancer. Oncogene 23: 7947–7956 [DOI] [PubMed] [Google Scholar]
- Runko AP, Sagerstrom CG 2003. Nlz belongs to a family of zinc-finger-containing repressors and controls segmental gene expression in the zebrafish hindbrain. Dev Biol 262: 254–267 [DOI] [PubMed] [Google Scholar]
- Runko AP, Sagerstrom CG 2004. Isolation of nlz2 and characterization of essential domains in Nlz family proteins. J Biol Chem 279: 11917–11925 [DOI] [PubMed] [Google Scholar]
- Sarrio D, Perez-Mies B, Hardisson D, Moreno-Bueno G, Suarez A, Cano A, Martin-Perez J, Gamallo C, Palacios J 2004. Cytoplasmic localization of p120ctn and E-cadherin loss characterize lobular breast carcinoma from preinvasive to metastatic lesions. Oncogene 23: 3272–3283 [DOI] [PubMed] [Google Scholar]
- Talvinen K, Tuikkala J, Nykanen M, Nieminen A, Anttinen J, Nevalainen OS, Hurme S, Kuopio T, Kronqvist P 2010. Altered expression of p120catenin predicts poor outcome in invasive breast cancer. J Cancer Res Clin Oncol 136: 1377–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen H, Syed A, Nguyen BT, Lukacsovich T, Purcell J, Srivastava GP, Iron D, Gaudenz K, Nie Q, Wan FY, et al. 2007. Wingless directly represses DPP morphogen expression via an armadillo/TCF/Brinker complex. PLoS ONE 2: e142 doi: 10.1371/journal.pone.0000142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson P, Jones GE, Frame MC, Brunton VG 2001. Coordination of cell polarization and migration by the Rho family GTPases requires Src tyrosine kinase activity. Curr Biol 11: 1836–1846 [DOI] [PubMed] [Google Scholar]
- van Hengel J, van Roy F 2007. Diverse functions of p120ctn in tumors. Biochim Biophys Acta 1773: 78–88 [DOI] [PubMed] [Google Scholar]
- Wang Z, Sandiford S, Wu C, Li SS 2009. Numb regulates cell–cell adhesion and polarity in response to tyrosine kinase signalling. EMBO J 28: 2360–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welm BE, Dijkgraaf GJ, Bledau AS, Welm AL, Werb Z 2008. Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell 2: 90–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamana N, Arakawa Y, Nishino T, Kurokawa K, Tanji M, Itoh RE, Monypenny J, Ishizaki T, Bito H, Nozaki K, et al. 2006. The Rho–mDia1 pathway regulates cell polarity and focal adhesion turnover in migrating cells through mobilizing Apc and c-Src. Mol Cell Biol 26: 6844–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa M, Huveldt D, Kreinest P, Lohse CM, Cheville JC, Parker AS, Copland JA, Anastasiadis PZ 2008. A p120 catenin isoform switch affects Rho activity, induces tumor cell invasion, and predicts metastatic disease. J Biol Chem 283: 18344–18354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E 2008. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol 9: 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZQ, Streicher KL, Ray ME, Abrams J, Ethier SP 2006. Multiple interacting oncogenes on the 8p11-p12 amplicon in human breast cancer. Cancer Res 66: 11632–11643 [DOI] [PubMed] [Google Scholar]
- Yap AS, Niessen CM, Gumbiner BM 1998. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J Cell Biol 141: 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook JI, Li XY, Ota I, Fearon ER, Weiss SJ 2005. Wnt-dependent regulation of the E-cadherin repressor snail. J Biol Chem 280: 11740–11748 [DOI] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Fitch DH, Herman MA 2002. TLP-1 is an asymmetric cell fate determinant that responds to Wnt signals and controls male tail tip morphogenesis in C. elegans. Development 129: 1497–1508 [DOI] [PubMed] [Google Scholar]