Abstract
Mitochondrial dysfunction is a prominent feature of various neurodegenerative diseases as strict regulation of integrated mitochondrial functions is essential for neuronal signaling, plasticity, and transmitter release. Many lines of evidence suggest that mitochondrial dysfunction plays a central role in the pathogenesis of Parkinson's disease (PD). Several PD-associated genes interface with mitochondrial dynamics regulating the structure and function of the mitochondrial network. Mitochondrial dysfunction can induce neuron death through a plethora of mechanisms. Both mitochondrial dysfunction and neuroinflammation, a common denominator of PD, lead to an increased production of reactive oxygen species, which are detrimental to neurons. The transcription factor nuclear factor E2-related factor 2 (Nrf2, NFE2L2) is an emerging target to counteract mitochondrial dysfunction and its consequences in PD. Nrf2 activates the antioxidant response element (ARE) pathway, including a battery of cytoprotective genes such as antioxidants and anti-inflammatory genes and several transcription factors involved in mitochondrial biogenesis. Here, the current knowledge about the role of mitochondrial dysfunction in PD, Nrf2/ARE stress-response mechanisms, and the evidence for specific links between this pathway and PD are summarized. The neuroprotection of nigral dopaminergic neurons by the activation of Nrf2 through several inducers in PD is also emphasized as a promising therapeutic approach.
1. Introduction
Parkinson's disease (PD) is a prevalent and progressive neurodegenerative incurable movement disorder. The disease is characterized by the loss of dopaminergic neurons in the substantia nigra pars compacta (SNpc) and the presence of proteinaceous deposits within neuronal perikarya (Lewy bodies) and processes (Lewy neurites). These deposits are composed of α-synuclein, ubiquitin, neurofilaments, and molecular chaperones [1]. Nigral dopaminergic neuron death leads to a deficiency of the neurotransmitter dopamine in the striatum and consequent dysregulation of basal ganglia circuitries accounting for motor symptoms of bradykinesia, hypokinesia, progressive rigidity, resting tremor, and postural instability. Lewy body deposition is also associated with nonmotor features such as autonomic dysfunction, sleep disturbances, depression, and cognitive impairment [1, 2]. A major risk factor is aging, as in other neurodegenerative diseases, although people with the familial monogenic forms of the disease can present before 45 years of age [1]. Since the sporadic and monogenic forms of PD share important clinical, pathological, and biochemical features, notably the progressive demise of dopaminergic neurons in the substantia nigra (SN), inherited mutations underlying familial forms have provided insight into the molecular mechanisms of disease pathogenesis [1, 3]. Oxidative stress, neuroinflammation, mitochondrial dysfunction, aberrant protein aggregation, excitotoxicity, and alterations in the autophagic-lysosomal pathway are implicated in the development and progression of PD [4–6]. PD also seems to have a mitochondrial component, so events that would modulate normal mitochondrial functions may compromise neuronal survival.
Nuclear factor E2-related factor 2 (Nrf2, NFE2L2) is a master regulator that induces a battery of cytoprotective genes including antioxidative enzymes, anti-inflammatory mediators, the proteasome, and several transcription factors involved in mitochondrial biogenesis. Thus, it may be a promising target to counteract mitochondrial dysfunction and its consequences in PD. Following its nuclear translocation, Nrf2 binds the antioxidant response elements (AREs) in the promoter region of its target genes. Several lines of evidence including in vivo Nrf2-deficient mouse studies, postmortem studies of PD brains, and genetic association studies of patients indicate a link between Nrf2 dysregulation and PD pathogenesis. Endogenous responses to upregulate Nrf2 and mitochondrial biogenesis in PD may be insufficient to prevent the progression of neurodegeneration. Thus, further activation of the Nrf2/ARE system using exogenous inducers may be a plausible therapeutic approach for PD.
This paper provides a brief overview of mitochondrial dynamics, mitochondrial dysfunction in PD, and the Nrf2/ARE system. In addition, recent studies indicating a link between Nrf2 and PD are summarized. Finally, recent efforts employing inducers of Nrf2 activity to provide neuroprotection of nigral dopaminergic neurons in PD are emphasized.
2. The Functions, Biogenesis, and Dysfunction of Mitochondria
Mitochondria are membrane-enclosed organelles, which uniquely contain genetic material independent of nuclear DNA. These 1–10 μm long cellular structures contain their own genome, which encodes tRNAs, rRNAs and 13 mitochondrial proteins [7, 8], while other components of the mitochondrial respiratory chain are encoded by the nuclear genome [9]. Multiple copies of the circular DNA are present in each mitochondrion. In humans, the size of the mitochondrial genome is 16,569 bp and encodes for 2 ribosomal RNA molecules (16S and 12S rRNA) and 22 transfer RNA molecules [9, 10].
The structure of a mitochondrion includes outer membrane, intermembrane space, inner membrane, and matrix [8]. The topography of the inner membrane is complex and includes the electron transport system complexes, the adenosine triphosphate (ATP) synthetase complex, and transport proteins [11]. Mitochondria produce most of the cell's energy in the form of ATP through oxidative phosphorylation (OXPHOS). Depending on this process, there are five intramembrane complexes and two mobile electron carriers, called coenzyme Q and cytochrome c [12]. Cytochrome c, which is an essential component of the electron transport chain, is located in the outer face of the inner membrane (intermembrane space) of the mitochondrion [13]. Cristae are structures that are formed by the folding of the inner membrane and provide increased surface area for chemical reactions, which take place in mitochondria [14].
Mitochondrial biogenesis is a process that involves interaction between the genetic systems of the organelle and the nucleus. During this process, mitochondria are newly formed in the cell. Many different signals can activate mitochondrial biogenesis. The main regulators of mitochondrial biogenesis include the peroxisome proliferator-activated receptor gamma coactivator (PGC) family of transcriptional activators, which consists of PGC-1α, PGC-1β, and PGC-related coactivator (PRC) [15]. PGC-1α plays a role in the activation of nuclear respiratory factor 2 and together they co-activate nuclear respiratory factor 1. Consequently, nuclear respiratory factor 1 activates Tfam, which is important for mitochondrial DNA (mtDNA) transcription, translation, and repair. Thus, PGC-1 family coactivators act as mediators between the environment and the transcriptional machinery regulating the biogenesis of mitochondria [16].
Mitochondria also have roles in signaling, cellular differentiation, cell growth and cell death [17]. Dysfunction of the mitochondria is implicated in primary mitochondrial disorders, cardiac dysfunction, and the aging process. As one of the main sources of reactive oxygen species (ROS), these organelles can themselves be affected by oxidative damage, potentially membrane permeability transition, affecting the aging process and disease pathogenesis [18]. Oxidative damage to mitochondrial macromolecules can also transform into an apoptosis mechanism, which is the process of programmed cell death. Thus, many apoptotic responses occur in mitochondria which are altered in electron transport, the loss of membrane potential, the changes in oxidation-reduction potential, the release of caspase regulators, and the attendance of pro- and anti-apoptotic Bcl-2 family proteins. Therefore, it is plausible that the effects of mitochondrial disruptions can play a role in human aging processes and degenerative diseases such as PD [19]. Sometimes, mitochondria containing a mutated genome are removed from the cell by mitophagy, a term for autophagy of the mitochondria [20].
3. Mitochondrial Dysfunction in Parkinson's Disease
The combination of mitochondrial dysfunction and increased oxidative stress are assumed to assist in the pathogenesis of PD [21]. Many lines of evidence suggest that mitochondrial dysfunction plays a central role in the pathogenesis of PD. The first observation in the early 1980s was that an inhibitor of complex I of the electron transport chain can induce parkinsonism. Moreover, mitochondria are targeted for the actions of parkinsonian neurotoxins such as 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), and its metabolite 1-methyl-4-phenylpyridinium (MPP+), 6-hydroxydopamine (6-OHDA), rotenone, and paraquat [22, 23].
Deficiency in mitochondrial respiratory chain complex I and cristae disruption have been consistently described in PD [24]. It has been known that mitochondrial respiratory complex I (NADH-quinone oxidoreductase) activity declines in the SNpc of PD patients [25, 26]. Moreover, the first transgenic mouse model for complex I deficiency has recently been generated. However, in this model, ATP levels were at the normal level, and oxygen consumption was not affected in NADH:ubiquinone oxidoreductase iron-sulfur protein 4 (NDUFS4) knocked-out mice. Furthermore, ROS formation in the neuronal cultures derived from those mice did not increase as was expected. It is clear that more in vivo models are needed in order to explain the role of complex I deficiency in PD [27].
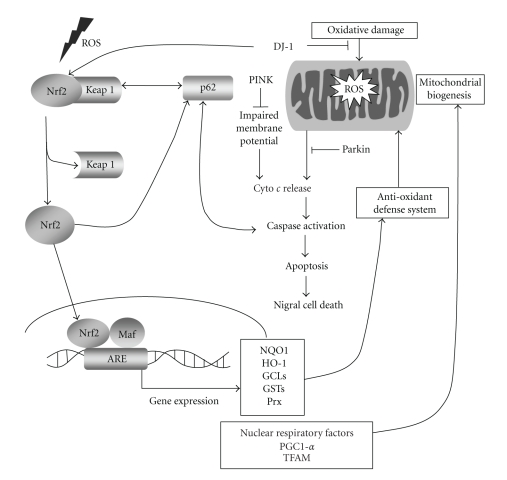
As in vivo evidence of mitochondrial dysfunction in PD, Hattingen et al. used a combination of anatomical magnetic resonance imaging and phosphorus and proton magnetic resonance spectroscopic imaging. They reported the presence of mitochondrial dysfunction of nigrostriatal dopaminergic neurons (see Figure 1). They also suggested that mitochondrial dysfunction phenomenon occurs in the early stages of PD pathogenesis [28].
Figure 1.
The role of the Nrf2 pathway in PD pathogenesis. In the case of oxidative damage, mitochondria produce increased amounts of ROS. Then, ROS activate Nrf2 bound to Keap1 in the cytoplasm, and it translocates into the nucleus to transactivate the transcription of ARE-bearing genes, which in turn activates the antioxidant defense system and mitochondrial biogenesis. In this pathway, PD-related genes are also involved. DJ-1 is found to inhibit oxidative damage in mitochondria. Another PD-related gene is PINK, which prevents impaired membrane potential of mitochondria and prevents apoptosis by counteracting cytochrome c release that leads to apoptosis of nigral cells. Lastly, Parkin was found to inhibit cytochrome c release that leads to caspase activation and apoptosis of nigral cells. Ubiquitin-interacting p62 also has a role in Nrf2 activation. It normally plays a role in transportation of ubiquitinated proteins to autophagosome. p62 was found to be interacting with Keap1 and transports it for autophagic degradation and provides indirect activation of Nrf2. p62 also has ARE in its promoter region which creates a positive feedback loop between Nrf2 and p62.
mtDNA has an increased susceptibility to mutations because of less efficient DNA-repair mechanisms and the absence of protective histones. There are several clinical studies indicating a link between PD and specific mtDNA point mutations [29]. There is also a link between mtDNA and neurodegeneration, which was verified by genetic mouse models. Mitochondrial transcription factor A (Tfam) is a gene encoding the Tfam protein, which plays a role in promoters within the D-loop region of mtDNA and provides regulation of the transcription and replication [30]. Both nuclear respiratory factor 1 and nuclear respiratory factor 2 can adjust the expression of the Tfam gene by attaching to consensus-binding sites. When a conditional Tfam knockout mouse in midbrain dopaminergic neurons was generated, decreased mtDNA expression, respiratory chain deficiency, and neuronal cell death leading to staggered L-dopa-responsive impairment of motor functions were observed [31].
Remarkably, recent studies indicate that several PD-associated genes, directly or indirectly, impinge on mitochondrial integrity, thereby providing a specific link to the mitochondrial dysfunction observed in sporadic PD [3, 32]. The protein products of PD genes, including alpha-synuclein, Parkin, PTEN-induced kinase 1 (PINK1), DJ-1, leucine-rich repeat kinase 2 (LRRK2) and HTR2A are localized to the mitochondria under certain conditions [33]. Functional studies in animal and cellular model systems have shown that PINK1 and Parkin play important roles in maintaining mitochondrial integrity and regulate mitochondrial morphology [3]. Loss-of-function mutations in the genes for these proteins are the primary cause of early-onset autosomal recessive forms of PD [1]. Parkin is an E3 ubiquitin-protein ligase, and impaired proteasome function has been described in sporadic PD. Parkin, therefore, promotes the degradation of dysfunctional mitochondria in cell culture conditions [34]. PINK1 represents the only kinase known to exhibit a canonical N-terminal mitochondrial localization signal [35]. These proteins are involved in mitophagy, the degradation process of terminally dysfunctional mitochondria in the lysosome [3, 32, 36]. Mutations in LRRK2 cause autosomal-dominant familial PD. This kinase modulates vulnerability to mitochondrial dysfunction induced by neurotoxins [37]. Furthermore, DJ-1 localizes to mitochondria during oxidative stress, where it exhibits peroxiredoxin-like activity [38]. Mutations in DJ-1 render animals and cultured cells more susceptible to oxidative stress and mitochondrial toxins implicated in sporadic PD, lending support to the hypothesis that some PD cases may be caused by gene-environmental factor interactions [39, 40]. A small proportion of alpha-synuclein is imported into mitochondria, where it accumulates in the brains of PD patients and may impair respiratory complex I activity [41]. Alpha-synuclein binds to mitochondria and leads to mitochondrial fragmentation [42].
4. The Nrf2/ARE Pathway
In pathology of neurodegenerative disorders, the generation of ROS may be harmful affecting proteins, lipids, and nucleic acids [45]. ROS itself regulates redox homeostasis activating a group of genes and signal transduction pathways [46]. The Nrf2 pathway is one of the pathways that respond to ROS by activating the transcription of phase II detoxification enzymes [47]. It was first identified by Moi et al. in 1994, as controlling the expression of β-globin gene [47]. It belongs to the cap and collar family of transcription factors having a distinct basic leucine-zipper motif [48]. Nrf2 is found in the cytosol bound to its inhibitor kelch-like ECH-associated protein (Keap1). When redox balance is tipped toward the oxidative side, Nrf2 translocates into the nucleus and activates the transcription of ARE-containing genes [49]. During this activation, Keap1, which is sensitive to electrophilic and oxidative stimuli, regulates Nrf2 modification, holding Nrf2 in the cytosol along with actin filaments [50]. Unless it is activated, Nrf2 is ubiquitinated by the E3-ubiquitin ligase-like domain of Keap1, followed by 26S proteasomal degradation [50]. Therefore, Keap1 regulates Nrf2 negatively, by promoting its sequestration and degradation.
Nrf2 is regulated by interactions between the conserved motifs DLG and ETGE within the Neh2 domain of Nrf2, the domain which regulates cellular stress responses, and the DGR region on Keap1. All of these domains provide correct positioning of Nrf2 for ubiquitination. When ROS accumulation increases in the cell, the binding to the DLG motif weakens; thus, ubiquitination is prevented [51–53]. In this way, degradation of Nrf2 decreases and stability of Nrf2 increases [43]. Extracellular signal-regulated kinase (ERK), c-Jun N-terminal protein kinase (JNK), and p38 mitogen-activated protein kinase (MAPK) pathways were shown to regulate Nrf2 transcriptional activity with an unknown mechanism. ERK and JNK appear to regulate positively the Nrf2 pathway after ARE-inducing substances, whereas p38 MAPK was reported to regulate Nrf2 both positively and negatively [43, 54]. ERK, JNK, and p38 MAPK act by phosphorylating N-terminal serine residues on Nrf2, which results as a response to electrophiles and oxidative stress [55]. Moreover, one of main signaling pathways, cytoprotective phosphotidylinositol-3 kinase (PI3K), which regulates maintenance of cellular homeostasis, was shown to regulate hemeoxygenase-1 (HO-1) under inflammatory stress [56]. Thus, it is proposed that the PI3K/Akt signaling pathway acts upstream of Nrf2 signaling. There are different upstream mechanisms, other than MAPK and PI3K/Akt, which are pancreatic endoplasmic reticulum kinase (PERK) and protein kinase C (PKC). In the case of redox imbalance, ROS can induce endoplasmic reticulum (ER) stress resulting in improper folding of proteins. There are different mechanisms to deal with this, one of which is PERK. Cullinan et al. found that Nrf2-target genes are activated in ER stress proposing PERK as the activator of Nrf2 [57]. Another Nrf2 mediator is PKC, which was first found by Huang et al. in 2000. As a result of inhibition experiments, they proposed that PKC is an upstream activator of Nrf2 signaling [58].
There are two hypotheses to explain the mechanism of Nrf2 translocation into the nucleus. According to the first hypothesis, there is nuclear localization signal in its basic region and two nuclear export signals in its leucine zipper and transactivation domain in order to provide Nrf2 nuclear translocation. In normal conditions, the nuclear localization signal is balanced by the nuclear export signal, and the sequestration of Nrf2 occurs in the cytosol. However, in stress conditions, the redox sensitive nuclear export signal found in the transactivation domain is disrupted. Thus, the nuclear localization signal drives translocation into the nucleus [59, 60]. However, this hypothesis fails to explain constitutive transcription of drug-metabolizing enzymes under normal conditions. For this reason, an alternative hypothesis was proposed. In this second hypothesis, Nrf2 is constitutively expressed and translocates into the nucleus. Its regulator and Keap1 enter into the nucleus by the CRM1/exportin pathway for removal and degradation of Nrf2, and after degradation it gets back into the cytosol. Consequently, under oxidative stress conditions, Nrf2 is expressed at the normal rate; however, its degradation rate decreases. In this way, its nuclear accumulation and transcriptional activity are enhanced [61, 62]. In addition to the above hypotheses, a novel ubiquitin-binding protein, p62 (sequestosome 1, SQSTM1), was recently shown to activate Nrf2. Studies suggest that p62 interacts with the Keap1-binding site of Nrf2, resulting in activation and triggering of downstream events of the Nrf2 pathway [63, 64].
In cells, there are mechanisms to deal with oxidative stress, including phase II detoxification enzymes, which have ARE in its promoter or enhancer regions. According to genetic analyses, the ARE sequence is found in many genes, which play important roles in gene regulation [65, 66]. Nrf2 also interacts with the AP-1 family proteins Jun, Fos, and Maf. Maf binds to the maf recognition element (MARE), which is very similar to the core sequence of ARE [67–70]. However, maf proteins are reported to regulate ARE-containing genes negatively [69]. Furthermore, some maf genes, MafF, and MafG modulate the expression of ARE-containing genes [48]. Overexpression studies on Jun revealed that Jun acts as a positive regulator of the transcription of ARE-regulated genes. Moreover, according to the studies which unravel the relationship between Fos and Jun, it was concluded that Fos acts as negative regulator, in an opposite fashion to Jun [71].
Genes transcribed after Nrf2 activation are called the “Nrf2 regulon.” This regulon performs several cellular functions, such as drug metabolism, ROS scavenging, glutathione homeostasis, efflux transport pathways, and activation of stress response proteins, which are vital for redox homeostasis [61, 72]. In the last decade, the role of Nrf2 in disease pathogenesis and toxic insults was shown to be protective, and it is confirmed to be a multiorgan protector [44]. The Nrf2 regulon genes can be classified into various classes based on their functions: glutathione homeostasis, drug metabolism, stress-response protein or iron metabolism and excretion/transporter. The functions of each gene group and genes are summarized in Table 1. In some studies, microarray analysis of the Nrf2 regulon revealed more genes than classical ARE-containing genes by comparing expression patterns of neurons and astrocytes, which are indicated as other genes in Table 1 [44]. In addition to the genes listed in Table 1, we performed in silico transcription factor binding analysis using the JASPAR database by searching for ARE sequence in the promoter [73]. Novel Nrf2 targets were obtained, such as nuclear respiratory factor-1, Tfam, and peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC1A). Nuclear respiratory factor-1 was experimentally validated to be a target gene for Nrf2 activation, which leads to mitochondrial biogenesis. In this way, apoptosis and necrosis can be prevented [74]. Peroxisome proliferator-activated receptor gamma (PPAR-γ) is a protein that has many beneficial effects including anti-inflammation in multiple tissues. Cho et al. first showed the regulation of PPAP-γ by Nrf2. They showed antioxidant effects of both Nrf2 and PPAR-γ, and they suggested that a PPAR-γ agonist can be used to combat oxidative damage [75].
Table 1.
Nrf2 target genes.
| Group | Gene symbol | Gene name | Function |
|---|---|---|---|
| GSH | γ-glutamyl-cysteinyl-glycine | Maintains redox homeostasis during oxidative stress | |
| GST | Glutathione-S-transferase | Cellular detoxification | |
| Gcl | Glutamate cysteine ligase | Catalyzes glutathione synthesis | |
| Glutathione homeostasis | Gcs | Glutamate cysteine synthetase | Catalyzes glutathione synthesis |
| GS | Glutathione synthetase | Catalyzes glutathione synthesis | |
| GPx | Glutathione Peroxidase | Catalyzes reduction of H2O2 or organic hyperoxides to water | |
| GR | Glutathione Reductase | Catalyzes reduction of oxidized GSSH to GSH | |
| NQO-1 | NAD(P)H quinone oxidoreductase-1 | Catalyzes two-electron reduction of quinones | |
| Drug metabolism | Ugt | UDP-glucuronosyltransferases | Catalyzes endogenous and exogenous substances with glucuronic acid |
| mEH | Microsomal epoxide hydrolase | Inactivates epoxides converting to vicinal dihydrodiol | |
| Stress response proteins/iron metabolism | Ferritin | Ferritin | Iron binding protein having role in iron oxidative stress |
| HO-1 | Heme oxygenase-1 | Catalyzes oxidative cleavage of Fe-protoporphyrin-IX | |
| Excretion/transporter | Mdr | Multi-drug resistance protein | Drug efflux pump for xenobiotic compounds |
| Mrp | Multidrug resistance associated protein | Multispecific organic anion transporter | |
| G6PDH | Glucose-6-phosphate dehydrogenasea | Glycolysis | |
| Other genes metabolism | Taldo | Transaldolasea | Pentose phosphate pathway enzyme |
| Tkt | Transketolasea | Channeling of excess glucose phosphates to glycolysis | |
| Pafah | PAF acetylhydrolasea | Catalyzes the degradation of platelet-activating factor to biologically inactive products | |
| Immune system | Ptgs2 | Prostaglandin-endoperoxide synthase 2a | Prostaglandin biosynthesis |
| Dig | Dithiolethione-inducible genea | Inhibition of chemically induced tumorigenesis | |
| Tac2 | Tachykinin 2a | Peptide neurotransmitter | |
| Calb1 | Calbindin-28Kb | Calcium binding protein | |
| Calcium homeostasis | Syt1 | Synaptotagmin-1b | Synaptic transmission |
| Hpca | Hippocalcinb | Calcium binding protein | |
| S100A1 | S100 calcium binding protein A1b | Calcium binding protein | |
| Ngfg | Nerve growth factor-γb | Growth factor for neuron survival | |
| Growth factor | FGF-13 | Fibroblast growth factor-13b | Nervous system development and function |
| FGF-14 | Fibroblast growth factor-14b | Nervous system development and function | |
| BDNF | Brain-derived neurotrophic factorb | Growth factor for neuron survival | |
| nGEF | Neuronal GEFb | Intracellular signaling networks | |
| Prkcb | Protein Kinase C-βb | Signal transduction | |
| Intracellular signaling | Gng3 | G-protein-γ3b | Signal transduction |
| Adm | Adrenomedullinb | Adrenal development | |
| Crh | Corticotropin-releasing hormoneb | Hormone released in response to stress | |
| Neurotransmission/channel | Clcn | Chloride channelb | Ion channel protein |
| Gabr1 | GABA-A receptor-1b | Neurotransmitter receptor | |
| Gabrg3 | GABA-A receptor, gamma 3b | Neurotransmitter receptor | |
| Gabbr1 | GABA-B receptor-1b | Neurotransmitter receptor | |
Nrf2 activates a battery of ARE-driven genes, also known as classic Nrf2-target genes, which are classified into 4 groups as glutathione homeostasis, drug metabolism, stress-response protein/iron metabolism, and excretion/transporter [43]. In addition to classic genes, microarray experiments revealed cell-type specific target genes. From (a) primary astrocyte and (b) primary neuronal cultures, novel targets were identified [44]. They also are stated as other genes.
5. The Nrf2/ARE Pathway and Parkinson's Disease
Several lines of evidence suggest the involvement of the Nrf2/ARE pathway in the pathogenesis of PD. The first evidence came from a postmortem study that examined the expression and localization of Nrf2 in susceptible neuron populations in AD and PD brain tissues [76]. Ramsey et al. showed that in neurodegenerative diseases, Nrf2 expression is altered in both neurons and astrocytes. As compared with age-matched normal controls, nuclear Nrf2 staining was decreased in hippocampal CA1 neurons and surrounding glia in AD brains, whereas nuclear localization of Nrf2 was induced in SN in PD brains, even though the response appeared insufficient to protect neurons from degeneration. Consistent with the findings of the study by Ramsay et al., increased glutathione was found in PD patients, probably reflecting a response to oxidative damage [77]. Another study by Spencer et al. found decreased glutathione levels [78]. Since Nrf2 regulates several enzymes involved in glutathione synthesis, increased Nrf2 in surviving PD nigral neurons could be interpreted as an appropriate neuronal response to oxidative stimuli, even though the response appears insufficient to protect neurons from neurodegeneration. Similarly, in the MPTP model of PD, ARE-dependent gene expression was decreased in striatum whereas it increased in SN [79]. Because subcellular trafficking is critical to the activity of the Nrf2/ARE pathway, Ramsey et al. concluded that disrupted or insufficient ARE responses likely occur downstream of Nrf2 nuclear localization in PD [76]. Additional downstream mechanisms may interfere with ARE transactivation despite nuclear stabilization of Nrf2. Further investigation is needed to understand why Nrf2 nuclear translocation is not sufficient to prevent oxidative stress or ongoing neurodegeneration in SN. Ramsey et al. did not report increased nuclear Nrf2 staining in surrounding glia in SN in postmortem brains from PD individuals and questioned the necessity of astroglial Nrf2 activation in neuroprotection. In reduced in vitro cell culture systems, increased neuronal Nrf2 activation protects neurons from the oxidative insult induced by parkinsonian neurotoxins such as paraquat, 6-OHDA, MPP+, and rotenone even in the absence of astroglia [80–88].
Primary cell culture systems used in these studies typically utilize either neuronal cell lines or dissociated cells. Nevertheless, it is likely that increased Nrf2 activity in both neurons and glia contributes to neuronal survival in disease states. The glia-enriched and mixed neuron-glia culture studies by Shih et al. suggest that in vitro neuroprotection against glutamate toxicity is not limited to increased neuronal Nrf2 activation, because activation of glial Nrf2 system also protects cultured neurons from insult [89]. A microarray analysis used to evaluate potential glial versus neuron-specific contributions to the neuroprotective effects of ARE activation and Nrf2 dependence showed that Nrf2 induction activates the expression of different genes in cultured astrocytes as compared with cultured cortical neurons. These results suggest that Nrf2-dependent genetic changes alter neuron-glia interactions resulting in neuroprotection [90]. The unique combination of cells found within organotypic nigrostriatal cocultures provides an ideal system by which we can examine this relationship between neurons and glia [91]. The neuroprotective effects of in vitro Nrf2 activation were also demonstrated in such organotypic nigrostriatal cocultures although the astroglial component was not studied [91]. All neurodegenerative diseases have different causes and mechanisms including progression, age of onset quality and severity of symptoms, survival after onset and cell population affected. Nrf2 was found to be regulated differently in neurons versus astrocytes [90]. Therefore, Ramsey et al. proposed that Nrf2 might regulate different gene products in various neuronal subpopulations (i.e., hippocampal neurons versus nigral neurons) [76]. Further studies are needed to explore how Nrf2 localization is regulated in a cell-type-specific manner, how it differs in various cellular populations and neuronal subpopulations, and how these differences may contribute to neuronal protection.
The link between the Nrf2/ARE pathway and PD was also studied using in vivo neurotoxin-based animal models of PD [84, 92]. Nrf2 knockout mice display increased vulnerability to 6-OHDA, and the induction of the Nrf2/ARE pathway by transplantation of astrocytes overexpressing Nrf2 can protect against 6-OHDA-induced damage in mouse brain [84]. Genetic deficiency of Nrf2 increases MPTP sensitivity in mice [79]. Furthermore, Nrf2 expression restricted to astrocytes is sufficient to protect against MPTP in transgenic mice with Nrf2 under control of the astrocyte-specific promoter for the glial fibrillary acidic protein (GFAP)-Nrf2. These results show that astroglial modulation of the Nrf2/ARE pathway also plays a pivotal role in neuroprotection in vivo. According to the results of a very recent study based on the MPTP model, Nrf2 also modulates toxin-mediated activation of microglia, another glial cell type in the CNS [93]. Dopaminergic neurodegeneration and microglial activation induced by chronic injection of MPTP were more severe in Nrf2 knockout mice than in wild-type mice. Characteristic of classical microglial activation, the levels of proinflammatory cytokines interleukin-6 (IL-6) and tumor necrosis factor-alpha (TNF-α) and the enzymes inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2) were increased, while anti-inflammatory markers attributable to alternative microglial activation were decreased [94]. These results demonstrate a crucial role of Nrf2 in the modulation of microglial dynamics [93]. There may be crosstalk between glial cell subpopulations characterized by the modulation of the activation status of microglia by astroglia [95]. The astrocytes, the most abundant cells in the brain, can secrete one or more factors capable of modulating microglial activation by regulating microglial Nrf2 activation. Treatment of microglia with astrocyte culture-conditioned media induces the nuclear translocation of Nrf2 and enhanced HO-1 promoter activity in an ARE-dependent manner and increases the expression and activity of HO-1 in microglia. Furthermore, treatment with astrocyte culture-conditioned media suppresses Interferon-gamma- (IFN-γ-)induced ROS production, leading to reduced iNOS expression and nitric oxide (NO) release [95]. However, as demonstrated by a very recent in vivo study, HO-1 may not be involved in the neuroprotection elicited by Nrf2 in experimental parkinsonism [92]. Deficiency of HO-1 does not protect or enhance sensitivity to neuronal death, whereas Nrf2-knockout mice showed exacerbated gliosis and dopaminergic nigrostriatal degeneration in the MPTP model of PD [96, 97].
Genetic evidence indicating a link between Nrf2/ARE pathway and PD came from a recent study in which a complete haplotype analysis of the NFE2L2 and Keap1 genes in relation to the risk of PD was performed in two independent case-control materials [98]. A protective NFE2L2 haplotype was found in both European case-control materials. The molecular consequence of this haplotype may be increased efficiency in the Keap1-Nrf2-ARE response to oxidative stress and thereby higher capacity to withstand endogenous or environmental risk factors for PD. Genetic variation in Keap1 did not show any associations. These results together with recent preclinical data provide another link between oxidative stress and the pathogenesis of PD and support NFE2L2 as a novel susceptibility gene for PD [98].
The interaction of Nrf2 with a parkinsonian gene, DJ-1 (PARK7), may provide another link between Nrf2 function and PD. A recently discovered function of DJ-1 is to stabilize Nrf2 by preventing its interaction with Keap1 and Nrf2's subsequent ubiquitination [99]. Furthermore, protein expression of Nrf2 significantly decreases in paraquat-treated DJ-1-deficient mice [100]. However, the findings of the study by Gan et al. using primary mouse embryonic fibroblasts were not confirmed in primary cortical neuronal and astrocyte cultures in vitro and ARE-driven human placental alkaline phosphatase (hPAP; ARE-hPAP) transgenic reporter mice in vivo [101]. Nrf2 activation, Nrf2-dependent gene induction, and Nrf2-mediated neuroprotection do not appear to be dependent on the presence of DJ-1 in the brain or in primary cultures derived from the brain. The most probable explanation for this discrepancy is the different cell types used in the two studies, suggesting that the relationship between DJ-1 and Nrf2 is cell-type specific. Neuroprotection by DJ-1 was confirmed in the second study [101].
Studies examining the Nrf2-activating effects of PD drugs support the link between the Nrf2/ARE pathway and PD. Apomorphine (Apo) is a drug used for clinical therapy of PD. It is a dopamine D(1)/D(2) receptor agonist and has scavenger and protective effects in ROS-induced cell death. Hara et al. reported that Apo enhances protection in 6-OHDA in vitro PD model in SH-SY5Y neuroblastoma cells. In that study, the involvement of the Nrf2 pathway in Apo-enhanced protection was also investigated. They observed nuclear translocation of Nrf2 into the nucleus and induced the expression of HO-1 gene in a dose-dependent fashion. On the other hand, cotreatment of Apo with antioxidant N-acetylcysteine suppressed the induction of HO-1 expression. Thus, Apo acts by producing intracellular ROS and activating the Nrf2 pathway to promote neuroprotective effects [81]. In addition to Apo, there is another cytoprotective drug, Deprenyl (Selegiline), that is, a promising candidate for neuroprotection, but its mode of action was unknown. However, Nakaso et al. reported a novel mechanism for the mode of action of Deprenyl, which involves PI3K and Nrf2 downstream oxidative-stress-related proteins. It increased the expression levels of HO-1, PrxI, TrxI, TrxRxI, gamma-GCS, and p62/A170, induced Nrf2 nuclear accumulation, and increased the strength of Nrf2 binding to ARE site. Nrf2 activation led to the induction of PI3K-controlled antioxidant molecules, and TrkB was identified as the upstream element of the PI3K/Nrf2 mechanism. Thus, protective effects of Deprenyl depend on PI3K-Nrf2 activation, which switches the antioxidant mechanism leading to cytoprotection [63]. The latest clinically used PD drug is bromocriptine, which is a dopamine agonist. Not only does it normally improve motor deficits by dopamine D2 receptor activation, but it also has neuroprotective and antioxidant activities. As reported by Lim et al., bromocriptine upregulates NAD(P)H quinone oxidoreductase-1 (NQO1) expression and increases its activity, thus leading to the protection of PC12 rat adrenal pheochromocytoma cells against oxidative damage. Bromocriptine also increased Nrf2 expression and nuclear translocation. It is known that bromocriptine leads to cytoprotection and antioxidant effects via the PI3K/Akt pathway, but independent from dopamine receptor activation. Also, dopamine receptor D2 antagonist does not affect the cytoprotective effect of bromocriptine and Nrf2-ARE activation by bromocriptine in dopamine D2 receptor expressing and nonexpressing cells. Consequently, NQO1 is a novel therapeutic target for PD, which can be upregulated by PI3K and Nrf2 activation [102].
Specific links between Nrf2 function and PD have been revealed. The activity of Nrf2 decreases with aging [103, 104], the major risk factor for the development and progression of PD [105]. Dysregulation of the Nrf2/ARE pathway is seen in PD. Nrf2 activators including known antiparkinsonian drugs (deprenyl and apomorphine [63, 81]) protect dopaminergic neurons against parkinsonian neurotoxins both in vitro and in vivo. Deficiency of Nrf2 aggravates experimental parkinsonism. Finally, apart from the stringent link between the Nrf2/ARE pathway and pathophysiological processes involved in PD such as oxidative stress, neuroinflammation, and ER stress, Nrf2 has the potential to interact with several molecules implicated in mitochondrial biogenesis (nuclear respiratory factors, PGC-1α, Tfam) and p62 which plays a role in mitophagy and the ubiquitin proteasome system (UPS) implicated in the pathogenesis of PD.
6. Nrf2/ARE Pathway Activation as a Novel Therapeutic Approach to Mitochondrial Dysfunction in Parkinson's Disease
As evidenced from postmortem tissue analyses and genetic analyses in humans and pathological studies in animal models, mitochondrial dysfunction plays a major role in PD, which leads to oxidative stress, DNA damage, and altered mitochondrial morphology and physiology. Therefore, therapeutic agents targeted to the mitochondria are thought to be promising tools for PD patients, such as agents targeting energy metabolism, mitochondria-targeted antioxidants, and more promising drugs that target the ARE/Nrf2/Keap1 pathway.
The Nrf2 pathway plays a role in redox homeostasis in cells. The main functions of Nrf2 were described in the previous part of this paper. The Nrf2 pathway is activated by ROS accumulation in cells; however, for therapuetic purposes, it is found to be activated by synthetic triterpenoids (TP), which are analogs of olenolic acid. They function as inhibitors of oxidative stress and cellular inflammatory processes and were shown to be protective in cancer models [106]. Moreover, the use of dietary compounds, synthetic chemicals, and xenobiotics decreases the incidence of diseases. For many decades, chemical substances from plants, called phytochemicals, have been shown to have chemopreventive activities, and most of them are suggested as Nrf2 inducers [107]. The potent chemopreventive compounds that induce Nrf2 can be listed as sulforaphane from cruciferous vegetables [108], curcumin [109], epigallocatechin-3-gallate from green tea [110], resveratrol from grape [111], caffeic acid phenethyl ester [109], wasabi [112], cafestol, kahweol [113], cinnamon based compounds [114], zerumbone [115], garlic organosulfur compounds [116], lycopene [117], carnosol [118], and avicins [119]. Novel compounds have been continuously discovered.
TPs are a group of Nrf2-activating compounds. Recently, novel Nrf2-activating, synthetic TP, CDDO methylamide (CDDO-MA), was tested by Yang et al. for neuroprotective effects in 3-nitropropionic acid (3-NP) rat and MPTP mouse models. CDDO-MA showed significant protection against both models and tert-butylhydroxyperoxide-induced ROS degeneration [120]. In addition to protective effects, CDDO-MA upregulated the expression of genes involved in mitochondria biogenesis, glutathione synthesis, and antioxidant mechanisms. Thus, this compound was thought to be useful for the treatment of neurodegenerative diseases.
Other neuroprotective agents that induce the expression of hemeoxygenase-1 are electrophilic neurite outgrowth-promoting prostaglandin (NEPP) compounds [121]. NEPP compounds are cyclopentenone prostaglandin derivatives and function as neurotrophic factors. According to a report by Satoh et al., NEPPs were preferentially taken up into neurons, where they bind to Keap1 in a thiol-dependent fashion. They showed that NEPPs are neuroprotective in a glutamate excitotoxicity model in vitro and in a stroke model in vivo. They suggest that NEPPs activate the Keap1/Nrf2 pathway to modulate the prevention of excitotoxicity; therefore, they can be used as therapeutics in stroke and neurodegenerative diseases [122].
Shih et al. showed that dietary administration of Nrf2 inducer (tert-Butylhydroxyquinone) reduced susceptibility to 3-NP and increased striatal and cortical gamma-glutamyl-cysteinyl- glycine (GSH) levels in Nrf2-expressing mice but not in Nrf2−/−. These results suggested that Nrf2 inducers to be dietary administration may be alternative drugs for the treatment of neurodegenerative disorders [123].
Neuroprotective agents such as erythropoietin (Epo), which is known to pass the blood brain barrier [124], also exert Nrf2-activating effects. Epo provides neuroprotection in MPTP- and 6-OHDA-induced toxin models of PD [125, 126]. Recently, Epo has been found to activate Nrf2 in SH-SY5Y cells [127]. Epo induced Nrf2 translocation to the nucleus and upregulated the expression of the HO-1 gene. Also, MAPK and PI3K inhibitors caused decreased translocation of Nrf2 caused by Epo. These results suggest that Epo activates the Nrf2 pathway and the PI3K and MAPK pathways as well. It is interesting that Epo, which is reported as neuroprotective in PD animal models, triggers neuronal Nrf2 translocation. Thus, the neuroprotective effects of Epo can be mediated by Nrf2.
7. Conclusions
Mitochondrial dysfunction plays a central role in the pathogenesis of PD. For this reason, new therapeutic strategies in PD targeting the restoration of mitochondrial dysfunction are being developed. The failure of antioxidant therapy strategies alone in clinical trials has encouraged efforts to find novel approaches strengthening endogenous antioxidant defense systems and inducing mitochondrial biogenesis and anti-inflammatory responses [123]. In this regard, Nrf2, a master regulator of the induction of a battery of genes affecting several cytoprotective systems, may be a promising target to counteract mitochondrial dysfunction and its consequences in PD. Thus, exogenous Nrf2 inducers should be tested for therapeutic potential in PD.
Acknowledgement
The authors thank Professor Anne Frary for critical reading of the manuscript for English editing.
Conflict of Interests
The authors declare no competing financial interests.
Abbrevation
- MPTP:
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- MPP+:
1-methyl-4-phenylpyridinium
- 3-NP:
3-nitropropionic acid
- 6-OHDA:
6-hydroxydopamine
- ATP:
Adenosine triphosphate
- ARE:
Antioxidant response element
- Apo:
Apomorphine
- hPAP; ARE-hPAP:
ARE-driven human placental alkaline phosphatase
- CDDO-MA:
CDDO methylamide
- JNK:
c-Jun N-terminal protein kinase
- COX-2:
Cyclooxygenase-2
- Epo:
Erythropoietin
- ERK:
Extracellular signal-regulated kinase
- GSH:
Gamma-glutamiyl-cysteinyl- glycine
- GFAP:
Glial fibrillary acidic protein
- HO-1:
Hemeoxygenase-1
- hPAP:
Human placental alkaline phosphatase
- iNOS:
Inducible nitric oxide synthase
- IFN-γ:
Interferon-gamma
- IL-6:
Interleukin-6
- Keap1:
Kelch-like ECH-associated protein
- LRRK2:
Leucine-rich repeat kinase 2
- MARE:
Maf recognition element
- mtDNA:
Mitochondrial DNA
- MAPK:
Mitogen-activated protein kinase
- Tfam:
Mitochondrial transcription factor A
- NDUFS4 gene:
NADH:ubiquinone oxidoreductase iron-sulfur protein 4
- NADPH:
Nicotinamide adenine dinucleotide phosphate
- NQO1:
NAD(P)H quinone oxidoreductase-1
- NEPP:
Neurite outgrowth-promoting prostaglandin
- NO:
Nitric oxide
- Nrf2:
Nuclear factor E2-related factor 2
- OXPHOS:
Oxidative phosphorylation
- PD:
Parkinson's disease
- PPAR-γ:
Peroxisome proliferator-activated receptor gamma
- PGC:
Peroxisome proliferator-activated receptor gamma coactivator
- PGC1A:
Peroxisome proliferator-activated receptor gamma coactivator 1 alpha
- PRC:
PGC-related coactivator
- PINK1:
PTEN-induced kinase 1
- ROS:
Reactive oxygen species
- SQSTM1:
Sequestosome 1
- SN:
Substantia nigra
- Snpc:
Substantia nigra pars compacta
- TP:
Triterpenoids
- TNF-α:
Tumor necrosis factor-alpha
- UPS:
Ubiquitin proteasome system.
References
- 1.Lees AJ, Hardy J, Revesz T. Parkinson’s disease. The Lancet. 2009;373(9680):2055–2066. doi: 10.1016/S0140-6736(09)60492-X. [DOI] [PubMed] [Google Scholar]
- 2.Braak H, Rüb U, Gai WP, Del Tredici K. Idiopathic Parkinson’s disease: possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. Journal of Neural Transmission. 2003;110(5):517–536. doi: 10.1007/s00702-002-0808-2. [DOI] [PubMed] [Google Scholar]
- 3.Deas E, Wood NW, Plun-Favreau H. Mitophagy and Parkinson's disease: the PINK1-parkin link. doi: 10.1016/j.bbamcr.2010.08.007. Biochimica et Biophysica Acta. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navarro A, Boveris A. Brain mitochondrial dysfunction and oxidative damage in Parkinson’s disease. Journal of Bioenergetics and Biomembranes. 2009;41(6):517–521. doi: 10.1007/s10863-009-9250-6. [DOI] [PubMed] [Google Scholar]
- 5.Witte ME, Geurts JJG, de Vries HE, van der Valk P, van Horssen J. Mitochondrial dysfunction: a potential link between neuroinflammation and neurodegeneration? Mitochondrion. 2010;10(5):411–418. doi: 10.1016/j.mito.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Di Filippo M, Chiasserini D, Tozzi A, Picconi B, Calabresi P. Mitochondria and the link between neuroinflammation and neurodegeneration. Journal of Alzheimer's Disease. 2010;20(supplement 2):S369–S379. doi: 10.3233/JAD-2010-100543. [DOI] [PubMed] [Google Scholar]
- 7.Henze K, Martin W. Essence of mitochondria. Nature. 2003;426(6963):127–128. doi: 10.1038/426127a. [DOI] [PubMed] [Google Scholar]
- 8.Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. Abington, UK: Garland Science, Taylor & Francis; 2008. Energy conversion: mitochondria and chloroplasts; pp. 815–827. [Google Scholar]
- 9.Anderson S, Bankier AT, Barrell BG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 10.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283(5407):1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 11.Herrmann JM, Neupert W. Protein transport into mitochondria. Current Opinion in Microbiology. 2000;3(2):210–214. doi: 10.1016/s1369-5274(00)00077-1. [DOI] [PubMed] [Google Scholar]
- 12.Wittig I, Schägger H. Supramolecular organization of ATP synthase and respiratory chain in mitochondrial membranes. Biochimica et Biophysica Acta. 2009;1787(6):672–680. doi: 10.1016/j.bbabio.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 13.Chipuk JE, Bouchier-Hayes L, Green DR. Mitochondrial outer membrane permeabilization during apoptosis: the innocent bystander scenario. Cell Death and Differentiation. 2006;13(8):1396–1402. doi: 10.1038/sj.cdd.4401963. [DOI] [PubMed] [Google Scholar]
- 14.Freya TG, Mannellab CA. The internal structure of mitochondria. Trends in Biochemical Sciences. 2000;25(7):319–324. doi: 10.1016/s0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- 15.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabolism. 2005;1(6):361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Scarpulla RC. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Annals of the New York Academy of Sciences. 2008;1147:321–334. doi: 10.1196/annals.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Current Biology. 2006;16(14):R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 18.Reddehase S, Grumbt B, Neupert W, Hell K. The disulfide relay system of mitochondria is required for the biogenesis of mitochondrial Ccs1 and Sod1. Journal of Molecular Biology. 2009;385(2):331–338. doi: 10.1016/j.jmb.2008.10.088. [DOI] [PubMed] [Google Scholar]
- 19.Wallace DC. Mitochondrial DNA mutations in disease and aging. Environmental and Molecular Mutagenesis. 2010;51(5):440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- 20.Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends in Cell Biology. 2007;17(9):422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Nicholls DG, Budd SL. Mitochondria and neuronal survival. Physiological Reviews. 2000;80(1):315–360. doi: 10.1152/physrev.2000.80.1.315. [DOI] [PubMed] [Google Scholar]
- 22.Banerjee R, Starkov AA, Beal MF, Thomas B. Mitochondrial dysfunction in the limelight of Parkinson’s disease pathogenesis. Biochimica et Biophysica Acta. 2009;1792(7):651–663. doi: 10.1016/j.bbadis.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winklhofer KF, Haass C. Mitochondrial dysfunction in Parkinson’s disease. Biochimica et Biophysica Acta. 2010;1802(1):29–44. doi: 10.1016/j.bbadis.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 24.Trimmer PA, Swerdlow RH, Parks JK, et al. Abnormal mitochondrial morphology in sporadic Parkinson’s and Alzheimer’s disease cybrid cell lines. Experimental Neurology. 2000;162(1):37–50. doi: 10.1006/exnr.2000.7333. [DOI] [PubMed] [Google Scholar]
- 25.Schapira AHV, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. Lancet. 1989;1(8649):p. 1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- 26.Schapira AHV, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD. Mitochondrial Complex I deficiency in Parkinson’s disease. Journal of Neurochemistry. 1990;54(3):823–827. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- 27.Choi WS, Kruse SE, Palmiter RD, Xia Z. Mitochondrial complex I inhibition is not required for dopaminergic neuron death induced by rotenone, MPP, or paraquat. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(39):15136–15141. doi: 10.1073/pnas.0807581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hattingen E, Magerkurth J, Pilatus U, et al. Phosphorus and proton magnetic resonance spectroscopy demonstrates mitochondrial dysfunction in early and advanced Parkinson’s disease. Brain. 2009;132(12):3285–3297. doi: 10.1093/brain/awp293. [DOI] [PubMed] [Google Scholar]
- 29.Reeve AK, Krishnan KJ, Turnbull D. Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Annals of the New York Academy of Sciences. 2008;1147:21–29. doi: 10.1196/annals.1427.016. [DOI] [PubMed] [Google Scholar]
- 30.Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(4):1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ekstrand MI, Terzioglu M, Galter D, et al. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(4):1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu CT. Tickled PINK1: mitochondrial homeostasis and autophagy in recessive Parkinsonism. Biochimica et Biophysica Acta. 2010;1802(1):20–28. doi: 10.1016/j.bbadis.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arduíno DM, Esteves AR, Oliveira CR, Cardoso SM. Mitochondrial metabolism modulation: a new therapeutic approach for Parkinson’s disease. CNS and Neurological Disorders. 2010;9(1):105–119. doi: 10.2174/187152710790966687. [DOI] [PubMed] [Google Scholar]
- 34.Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway: a mitochondrial quality control system? Journal of Bioenergetics and Biomembranes. 2009;41(6):499–503. doi: 10.1007/s10863-009-9253-3. [DOI] [PubMed] [Google Scholar]
- 35.Dagda RK, Zhu J, Chu CT. Mitochondrial kinases in Parkinson’s disease: converging insights from neurotoxin and genetic models. Mitochondrion. 2009;9(5):289–298. doi: 10.1016/j.mito.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuda N, Tanaka K. Uncovering the roles of PINK1 and parkin in mitophagy. Autophagy. 2010;6(7):952–954. doi: 10.4161/auto.6.7.13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saha S, Guillily MD, Ferree A, et al. LRRK2 modulates vulnerability to mitochondrial dysfunction in Caenorhabditis elegans. Journal of Neuroscience. 2009;29(29):9210–9218. doi: 10.1523/JNEUROSCI.2281-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cookson MR. DJ-1, PINK1, and their effects on mitochondrial pathways. Movement Disorders. 2010;25(supplement 1):S44–S48. doi: 10.1002/mds.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irrcher I, Aleyasin H, Seifert EL, et al. Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Human Molecular Genetics. 2010;19(19):3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- 40.Hao LY, Giasson BI, Bonini NM. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(21):9747–9752. doi: 10.1073/pnas.0911175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Büeler H. Impaired mitochondrial dynamics and function in the pathogenesis of Parkinson’s disease. Experimental Neurology. 2009;218(2):235–246. doi: 10.1016/j.expneurol.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 42.Kamp F, Exner N, Lutz AK, et al. Inhibition of mitochondrial fusion by α-synuclein is rescued by PINK1, Parkin and DJ-1. EMBO Journal. 2010;29(20):3571–3589. doi: 10.1038/emboj.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh S, Vrishni S, Singh BK, Rahman I, Kakkar P. Nrf2-ARE stress response mechanism: a control point in oxidative stress-mediated dysfunctions and chronic inflammatory diseases. Free Radical Research. 2010;44(11):1267–1288. doi: 10.3109/10715762.2010.507670. [DOI] [PubMed] [Google Scholar]
- 44.Lee JM, Li J, Johnson DA, et al. Nrf2, a multi-organ protector? FASEB Journal. 2005;19(9):1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- 45.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chemico-Biological Interactions. 2006;160(1):1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 46.Finkel T. Reactive oxygen species and signal transduction. IUBMB Life. 2001;52(1-2):3–6. doi: 10.1080/15216540252774694. [DOI] [PubMed] [Google Scholar]
- 47.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the β-globin locus control region. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(21):9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends in Molecular Medicine. 2004;10(11):549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Dhakshinamoorthy S, Jaiswal AK. Functional characterization and role of INrf2 in antioxidant response element-mediated expression and antioxidant induction of NAD(P)H: quinone oxidoreductase1 gene. Oncogene. 2001;20(29):3906–3917. doi: 10.1038/sj.onc.1204506. [DOI] [PubMed] [Google Scholar]
- 50.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annual Review of Pharmacology and Toxicology. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 51.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Molecular and Cellular Biology. 2006;26(8):2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Molecular and Cellular Biology. 2003;23(22):8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi A, Kang MI, Watai Y, et al. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Molecular and Cellular Biology. 2006;26(1):221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naidu S, Vijayan V, Santoso S, Kietzmann T, Immenschuh S. Inhibition and genetic deficiency of p38 MAPK up-regulates heme oxygenase-1 gene expression via Nrf2. Journal of Immunology. 2009;182(11):7048–7057. doi: 10.4049/jimmunol.0900006. [DOI] [PubMed] [Google Scholar]
- 55.Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. Journal of Biological Chemistry. 2002;277(45):42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 56.Pischke SE, Zhou Z, Song R, et al. Phosphatidylinositol 3-kinase/Akt pathway mediates heme oxygenase-1 regulation by lipopolysaccharide. Cellular and Molecular Biology. 2005;51(5):461–470. [PubMed] [Google Scholar]
- 57.Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. Journal of Biological Chemistry. 2004;279(19):20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- 58.Huang HC, Nguyen T, Pickett CB. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(23):12475–12480. doi: 10.1073/pnas.220418997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain AK, Bloom DA, Jaiswal AK. Nuclear import and export signals in control of Nrf2. Journal of Biological Chemistry. 2005;280(32):29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- 60.Li W, Yu SIW, Kong ANT. Nrf2 possesses a redox-sensitive nuclear exporting signal in the Neh5 transactivation domain. Journal of Biological Chemistry. 2006;281(37):27251–27263. doi: 10.1074/jbc.M602746200. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen T, Sherratt PJ, Nioi P, Yang CS, Pickett CB. Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. Journal of Biological Chemistry. 2005;280(37):32485–32492. doi: 10.1074/jbc.M503074200. [DOI] [PubMed] [Google Scholar]
- 62.Rubiolo JA, Mithieux G, Vega FV. Resveratrol protects primary rat hepatocytes against oxidative stress damage: activation of the Nrf2 transcription factor and augmented activities of antioxidant enzymes. European Journal of Pharmacology. 2008;591(1–3):66–72. doi: 10.1016/j.ejphar.2008.06.067. [DOI] [PubMed] [Google Scholar]
- 63.Nakaso K, Nakamura C, Sato H, Imamura K, Takeshima T, Nakashima K. Novel cytoprotective mechanism of anti-parkinsonian drug deprenyl: PI3K and Nrf2-derived induction of antioxidative proteins. Biochemical and Biophysical Research Communications. 2006;339(3):915–922. doi: 10.1016/j.bbrc.2005.11.095. [DOI] [PubMed] [Google Scholar]
- 64.Komatsu M, Kurokawa H, Waguri S, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature Cell Biology. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 65.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element: activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. Journal of Biological Chemistry. 1991;266(18):11632–11639. [PubMed] [Google Scholar]
- 66.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel NRF2-regulated antioxidant response element (ARE) in the mouse NAD(P)H: quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochemical Journal. 2003;374(2):337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Venugopal R, Jaiswal AK. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene. 1998;17(24):3145–3156. doi: 10.1038/sj.onc.1202237. [DOI] [PubMed] [Google Scholar]
- 68.Itoh K, Chiba T, Takahashi S, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochemical and Biophysical Research Communications. 1997;236(2):313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 69.Nguyen T, Huang HC, Pickett CB. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. Journal of Biological Chemistry. 2000;275(20):15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 70.Zhu M, Fahl WE. Functional characterization of transcription regulators that interact with the electrophile response element. Biochemical and Biophysical Research Communications. 2001;289(1):212–219. doi: 10.1006/bbrc.2001.5944. [DOI] [PubMed] [Google Scholar]
- 71.Curran T, Franza BR., Jr. Fos and jun: the AP-1 connection. Cell. 1988;55(3):395–397. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- 72.Reisman SA, Buckley DB, Tanaka Y, Klaassen CD. CDDO-Im protects from acetaminophen hepatotoxicity through induction of Nrf2-dependent genes. Toxicology and Applied Pharmacology. 2009;236(1):109–114. doi: 10.1016/j.taap.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nature Reviews Genetics. 2004;5(4):276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 74.Piantadosi CA, Carraway MS, Babiker A, Suliman HB. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circulation Research. 2008;103(11):1232–1240. doi: 10.1161/01.RES.0000338597.71702.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cho H-Y, Gladwell W, Wang X, et al. Nrf2-regulated PPARγ expression is critical to protection against acute lung injury in mice. American Journal of Respiratory and Critical Care Medicine. 2010;182(2):170–182. doi: 10.1164/rccm.200907-1047OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramsey CP, Glass CA, Montgomery MB, et al. Expression of Nrf2 in neurodegenerative diseases. Journal of Neuropathology and Experimental Neurology. 2007;66(1):75–85. doi: 10.1097/nen.0b013e31802d6da9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bogdanov M, Matson WR, Wang L, et al. Metabolomic profiling to develop blood biomarkers for Parkinson’s disease. Brain. 2008;131(2):389–396. doi: 10.1093/brain/awm304. [DOI] [PubMed] [Google Scholar]
- 78.Spencer JPE, Jenner P, Halliwell B. Superoxide-dependent depletion of reduced glutathione by L-DOPA and dopamine. Relevance to Parkinson’s disease. NeuroReport. 1995;6(11):1480–1484. doi: 10.1097/00001756-199507310-00004. [DOI] [PubMed] [Google Scholar]
- 79.Chen PC, Vargas MR, Pani AK, et al. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson’s disease: critical role for the astrocyte. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(8):2933–2938. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Niso-Santano M, González-Polo RA, Bravo-San Pedro JM, et al. Activation of apoptosis signal-regulating kinase 1 is a key factor in paraquat-induced cell death: modulation by the Nrf2/Trx axis. Free Radical Biology and Medicine. 2010;48(10):1370–1381. doi: 10.1016/j.freeradbiomed.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 81.Hara H, Ohta M, Adachi T. Apomorphine protects against 6-hydroxydopamine-induced neuronal cell death through activation of the Nrf2-ARE pathway. Journal of Neuroscience Research. 2006;84(4):860–866. doi: 10.1002/jnr.20974. [DOI] [PubMed] [Google Scholar]
- 82.Hwang YP, Jeong HG. The coffee diterpene kahweol induces heme oxygenase-1 via the PI3K and p38/Nrf2 pathway to protect human dopaminergic neurons from 6-hydroxydopamine-derived oxidative stress. FEBS Letters. 2008;582(17):2655–2662. doi: 10.1016/j.febslet.2008.06.045. [DOI] [PubMed] [Google Scholar]
- 83.Cao TT, Ma L, Kandpal G, Warren L, Hess JF, Seabrook GR. Increased nuclear factor-erythroid 2 p45-related factor 2 activity protects SH-SY5Y cells against oxidative damage. Journal of Neurochemistry. 2005;95(2):406–417. doi: 10.1111/j.1471-4159.2005.03377.x. [DOI] [PubMed] [Google Scholar]
- 84.Jakel RJ, Townsend JA, Kraft AD, Johnson JA. Nrf2-mediated protection against 6-hydroxydopamine. Brain Research. 2007;1144(1):192–201. doi: 10.1016/j.brainres.2007.01.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wruck CJ, Claussen M, Fuhrmann G, et al. Luteolin protects rat PC12 and C6 cells against MPP+ induced toxicity via an ERK dependent Keap1-Nrf2-ARE pathway. Journal of Neural Transmission.Supplementum. 2007;72:57–67. doi: 10.1007/978-3-211-73574-9_9. [DOI] [PubMed] [Google Scholar]
- 86.Lee JM, Shih AY, Murphy TH, Johnson JA. NF-E2-related factor-2 mediates neuroprotection against mitochondrial complex I inhibitors and increased concentrations of intracellular calcium in primary cortical neurons. Journal of Biological Chemistry. 2003;278(39):37948–37956. doi: 10.1074/jbc.M305204200. [DOI] [PubMed] [Google Scholar]
- 87.Satoh T, Harada N, Hosoya T, Tohyama K, Yamamoto M, Itoh K. Keap1/Nrf2 system regulates neuronal survival as revealed through study of keap1 gene-knockout mice. Biochemical and Biophysical Research Communications. 2009;380(2):298–302. doi: 10.1016/j.bbrc.2009.01.063. [DOI] [PubMed] [Google Scholar]
- 88.MacKenzie EL, Ray PD, Tsuji Y. Role and regulation of ferritin H in rotenone-mediated mitochondrial oxidative stress. Free Radical Biology and Medicine. 2008;44(9):1762–1771. doi: 10.1016/j.freeradbiomed.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shih AY, Johnson DA, Wong G, et al. Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress. Journal of Neuroscience. 2003;23(8):3394–3406. doi: 10.1523/JNEUROSCI.23-08-03394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kraft AD, Johnson DA, Johnson JA. Nuclear factor E2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult. Journal of Neuroscience. 2004;24(5):1101–1112. doi: 10.1523/JNEUROSCI.3817-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Siebert A, Desai V, Chandrasekaran K, Fiskum G, Jafri MS. Nrf2 activators provide neuroprotection against 6-hydroxydopamine toxicity in rat organotypic nigrostriatal cocultures. Journal of Neuroscience Research. 2009;87(7):1659–1669. doi: 10.1002/jnr.21975. [DOI] [PubMed] [Google Scholar]
- 92.Innamorato NG, Jazwa A, Rojo AI, et al. Different susceptibility to the parkinson's toxin MPTP in mice lacking the redox master regulator Nrf2 or its target gene heme oxygenase-1. PLoS One. 2010;5(7, article e11838) doi: 10.1371/journal.pone.0011838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rojo AI, Innamorato NG, Martín-Moreno AM, De Ceballos ML, Yamamoto M, Cuadrado A. Nrf2 regulates microglial dynamics and neuroinflammation in experimental Parkinson’s disease. Glia. 2010;58(5):588–598. doi: 10.1002/glia.20947. [DOI] [PubMed] [Google Scholar]
- 94.Colton CA. Heterogeneity of microglial activation in the innate immune response in the brain. Journal of Neuroimmune Pharmacology. 2009;4(4):399–418. doi: 10.1007/s11481-009-9164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Min KJ, Yang MS, Kim SU, Jou I, Joe EH. Astrocytes induce hemeoxygenase-1 expression in microglia: a feasible mechanism for preventing excessive brain inflammation. Journal of Neuroscience. 2006;26(6):1880–1887. doi: 10.1523/JNEUROSCI.3696-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marro S, Chiabrando D, Messana E, et al. Heme controls ferroportin1 (FPN1) transcription involving Bach1, Nrf2 and a MARE/ARE sequence motif at position -7007 of the FPN1 promoter. Haematologica. 2010;95(8):1261–1268. doi: 10.3324/haematol.2009.020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pietsch EC, Chan JY, Torti FM, Torti SV. Nrf2 mediates the induction of ferritin H in response to xenobiotics and cancer chemopreventive dithiolethiones. Journal of Biological Chemistry. 2003;278(4):2361–2369. doi: 10.1074/jbc.M210664200. [DOI] [PubMed] [Google Scholar]
- 98.von Otter M, Landgren S, Nilsson S, et al. Association of Nrf2-encoding NFE2L2 haplotypes with Parkinson’s disease. BMC Medical Genetics. 2010;11(1) doi: 10.1186/1471-2350-11-36. Article ID 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JPY. DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(41):15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang W, Chen L, Ding Y, Zhuang X, Kang UJ. Paraquat induces dopaminergic dysfunction and proteasome impairment in DJ-1-deficient mice. Human Molecular Genetics. 2007;16(23):2900–2910. doi: 10.1093/hmg/ddm249. [DOI] [PubMed] [Google Scholar]
- 101.Gan LI, Johnson DA, Johnson JA. Keap1-Nrf2 activation in the presence and absence of DJ-1. European Journal of Neuroscience. 2010;31(6):967–977. doi: 10.1111/j.1460-9568.2010.07138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lim JH, Kim KM, Kim SW, Hwang O, Choi HJ. Bromocriptine activates NQO1 via Nrf2-PI3K/Akt signaling: novel cytoprotective mechanism against oxidative damage. Pharmacological Research. 2008;57(5):325–331. doi: 10.1016/j.phrs.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 103.Duan W, Zhang R, Guo Y, et al. Nrf2 activity is lost in the spinal cord and its astrocytes of aged mice. In Vitro Cellular and Developmental Biology Animal. 2009;45(7):388–397. doi: 10.1007/s11626-009-9194-5. [DOI] [PubMed] [Google Scholar]
- 104.Sykiotis GP, Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Science Signaling. 2010;3(112):p. re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hindle JV. Ageing, neurodegeneration and Parkinson’s disease. Age and Ageing. 2010;39(2):156–161. doi: 10.1093/ageing/afp223. [DOI] [PubMed] [Google Scholar]
- 106.Liby KT, Yore MM, Sporn MB. Triterpenoids and rexinoids as multifunctional agents for the prevention and treatment of cancer. Nature Reviews Cancer. 2007;7(5):357–369. doi: 10.1038/nrc2129. [DOI] [PubMed] [Google Scholar]
- 107.Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacological Research. 2008;58(5-6):262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kensler TW, Curphey TJ, Maxiutenko Y, Roebuck BD. Chemoprotection by organosulfur inducers of phase 2 enzymes: dithiolethiones and dithiins. Drug Metabolism and Drug Interactions. 2000;17(1–4):3–22. doi: 10.1515/dmdi.2000.17.1-4.3. [DOI] [PubMed] [Google Scholar]
- 109.Balogun E, Hoque M, Gong P, et al. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochemical Journal. 2003;371(3):887–895. doi: 10.1042/BJ20021619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Na HK, Surh YJ. Modulation of Nrf2-mediated antioxidant and detoxifying enzyme induction by the green tea polyphenol EGCG. Food and Chemical Toxicology. 2008;46(4):1271–1278. doi: 10.1016/j.fct.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 111.Chen CY, Jang JH, Li MH, Surh YJ. Resveratrol upregulates heme oxygenase-1 expression via activation of NF-E2-related factor 2 in PC12 cells. Biochemical and Biophysical Research Communications. 2005;331(4):993–1000. doi: 10.1016/j.bbrc.2005.03.237. [DOI] [PubMed] [Google Scholar]
- 112.Morimitsu Y, Nakagawa Y, Hayashi K, et al. A sulforaphane analogue that potently activates the Nrf2-dependent detoxification pathway. Journal of Biological Chemistry. 2002;277(5):3456–3463. doi: 10.1074/jbc.M110244200. [DOI] [PubMed] [Google Scholar]
- 113.Higgins LG, Cavin C, Itoh K, Yamamoto M, Hayes JD. Induction of cancer chemopreventive enzymes by coffee is mediated by transcription factor Nrf2. Evidence that the coffee-specific diterpenes cafestol and kahweol confer protection against acrolein. Toxicology and Applied Pharmacology. 2008;226(3):328–337. doi: 10.1016/j.taap.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 114.Liao B-C, Hsieh C-W, Liu Y-C, Tzeng T-T, Sun Y-W, Wung B-S. Cinnamaldehyde inhibits the tumor necrosis factor-α-induced expression of cell adhesion molecules in endothelial cells by suppressing NF-κB activation: effects upon IκB and Nrf2. Toxicology and Applied Pharmacology. 2008;229(2):161–171. doi: 10.1016/j.taap.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 115.Nakamura Y, Yoshida C, Murakami A, Ohigashi H, Osawa T, Uchida K. Zerumbone, a tropical ginger sesquiterpene, activates phase II drug metabolizing enzymes. FEBS Letters. 2004;572(1–3):245–250. doi: 10.1016/j.febslet.2004.07.042. [DOI] [PubMed] [Google Scholar]
- 116.Chen C, Pung D, Leong V, et al. Induction of detoxifying enzymes by garlic organosulfur compounds through transcription factor Nrf2: effect of chemical structure and stress signals. Free Radical Biology and Medicine. 2004;37(10):1578–1590. doi: 10.1016/j.freeradbiomed.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 117.Ben-Dor A, Steiner M, Gheber L, et al. Carotenoids activate the antioxidant response element transcription system. Molecular Cancer Therapeutics. 2005;4(1):177–186. [PubMed] [Google Scholar]
- 118.Satoh T, Kosaka K, Itoh K, et al. Carnosic acid, a catechol-type electrophilic compound, protects neurons both in vitro and in vivo through activation of the Keap1/Nrf2 pathway via S-alkylation of targeted cysteines on Keap1. Journal of Neurochemistry. 2008;104(4):1116–1131. doi: 10.1111/j.1471-4159.2007.05039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Handas V, Kim S-O, Nishimura G, Hausladen A, Stamler JS, Gutterman JU. Avicinylation (thioesterification): a protein modification that can regulate the response to oxidative and nitrosative stress. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(29):10088–10093. doi: 10.1073/pnas.0504430102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang L, Calingasan NY, Thomas B, et al. Neuroprotective effects of the triterpenoid, CDDO methyl amide, a potent inducer of Nrf2-mediated transcription. PLoS One. 2009;4(6, article e5757) doi: 10.1371/journal.pone.0005757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Satoh T, Furuta K, Tomokiyo K, et al. Facilitatory roles of novel compounds designed from cyclopentenone prostaglandins on neurite outgrowth-promoting activities of nerve growth factor. Journal of Neurochemistry. 2000;75(3):1092–1102. doi: 10.1046/j.1471-4159.2000.0751092.x. [DOI] [PubMed] [Google Scholar]
- 122.Satoh T, Okamoto S-I, Cui J, et al. Activation of the Keap1/Nrf2 pathway for neuroprotection by electrophillic phase II inducers. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(3):768–773. doi: 10.1073/pnas.0505723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Beal MF. Therapeutic approaches to mitochondrial dysfunction in Parkinson’s disease. Parkinsonism and Related Disorders. 2009;15(supplement 3):S189–S194. doi: 10.1016/S1353-8020(09)70812-0. [DOI] [PubMed] [Google Scholar]
- 124.Brines ML, Ghezzi P, Keenan S, et al. Erythropoietin crosses the blood-brain barrier to protect against experimental brain injury. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(19):10526–10531. doi: 10.1073/pnas.97.19.10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Genc S, Kuralay F, Genc K, et al. Erythropoietin exerts neuroprotection in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-treated C57/BL mice via increasing nitric oxide production. Neuroscience Letters. 2001;298(2):139–141. doi: 10.1016/s0304-3940(00)01716-x. [DOI] [PubMed] [Google Scholar]
- 126.Signore AP, Weng Z, Hastings T, et al. Erythropoietin protects against 6-hydroxydopamine-induced dopaminergic cell death. Journal of Neurochemistry. 2006;96(2):428–443. doi: 10.1111/j.1471-4159.2005.03587.x. [DOI] [PubMed] [Google Scholar]
- 127.Genc K, Egrilmez MY, Genc S. Erythropoietin induces nuclear translocation of Nrf2 and heme oxygenase-1 expression in SH-SY5Y cells. Cell Biochemistry and Function. 2010;28(3):197–201. doi: 10.1002/cbf.1639. [DOI] [PubMed] [Google Scholar]