Abstract
Single-chain antibody fragments (scFv), consisting of two linked variable regions (VH and VL), are a versatile format for engineering and as potential antigen-specific therapeutics. Although the analogous format for T cell receptors (TCRs), consisting of two linked V regions (Vα and Vβ; referred to here as scTv), could provide similar opportunities, all wild-type scTv proteins examined to date are unstable. This obstacle has prevented scTv fragments from being widely used for engineering or therapeutics. To further explore whether some stable human scTv fragments could be expressed, we used a yeast system in which display of properly folded domains correlates with ability to express the folded scTv in soluble form. We discovered that, unexpectedly, scTv fragments that contained the human Vα2 region (IMGT: TRAV12 family) were displayed and properly associated with different Vβ regions. Furthermore, a single polymorphic residue (Serα49) in the framework region conferred additional thermal stability. These stabilized Vα2-containing scTv fragments could be expressed at high levels in Escherichia coli, and used to stain target cells that expressed the specific pep-HLA-A2 complexes. Thus, the scTv fragments can serve as a platform for engineering TCRs with diverse specificities, and possibly for therapeutic or diagnostic applications.
Keywords: directed evolution, peptide-MHC, single-chain, T cell receptors, yeast display
Introduction
T cell receptors (TCRs) mediate recognition of a foreign or self-peptide presented in the context of a self major histocompatibility (MHC) complex protein (reviewed in Rudolph et al., 2006). During development, T cells that express the highest affinity receptors are deleted in the thymus in a process called negative selection. Survival of the T cell, however, depends on a minimal affinity for self-MHC (positive selection), assuring that T cell recognition is MHC-restricted (reviewed in Starr et al., 2003). After development, if recognition of peptide:MHC (pepMHC) is of sufficient affinity, the T cell becomes activated and secretes cytokines or causes lysis of the target cell. The affinity of TCR:pepMHC interactions is relatively low, with KD values in the range of 1–500 μM (Davis et al., 1998; van der Merwe and Davis, 2003; Stone et al., 2009).
In recent years, in vitro engineering by yeast and phage display has yielded TCRs with >1000-fold improvements in affinity for class I MHC ligands (Holler et al., 2000, 2003; Chlewicki et al., 2005; Li et al., 2005; Dunn et al., 2006; Varela-Rohena et al., 2008) and class II MHC ligands (Weber et al., 2005). To date, there have been only a few MHC-restricted human TCRs engineered for improved affinity by phage display (reviewed in Richman and Kranz, 2007). These receptors were engineered in a full-length TCR format (Li et al., 2005; Dunn et al., 2006; Varela-Rohena et al., 2008), with the addition of two non-native cysteines in each of the TCR constant domains to facilitate TCR heterodimer formation through an interchain disulfide bond (Boulter et al., 2003). The addition of this disulfide bond allows the TCR to be expressed on the surface of yeast (Richman et al., 2009) or phage (Boulter et al., 2003).
Other affinity engineering efforts have focused on using a single-chain TCR (scTv) format, consisting of the variable domains of the TCR connected by a flexible linker (Vα–linker–Vβ or Vβ–linker–Vα; called scTv here to distinguish it from antibody scFv fragments, and from scTCRs that contain three domains, typically Vα, Vβ and Cβ; Chung et al., 1994; Plaksin et al., 1997; Card et al., 2004). Although analogous to antibody fragments in many aspects, unlike antibody scFv, the scTv requires mutations to allow for stable expression of the variable domains in the absence of the constant domains (Richman et al., 2009). Previously, scTv fragments have been isolated by yeast display through random mutagenesis and subsequent selection with clonotypic antibodies to select for mutations that stabilize the scTv and allow for yeast surface expression and/or thermal stability (Kieke et al., 1999; Shusta et al., 2000; Weber et al., 2005). These stable scTv scaffolds were subsequently used for engineering receptors of enhanced affinity and for expression as stable, soluble scTv (Shusta et al., 1999; Holler et al., 2000; Weber et al., 2005).
Like scFv fragments compared with the full antibody molecule, scTv fragments potentially afford significant advantages compared with the full-length TCR format for engineering, soluble protein expression and therapeutic potential. Higher surface levels of the scTv can be achieved relative to the full-length TCR in the yeast display system (Richman et al., 2009). This allows for more TCR molecules to be displayed on the surface of each yeast cell, and improved avidity effects in the detection of bound peptide-MHC multimers. In addition, the scTv is produced as a single polypeptide, which avoids the requirement for production of each TCR chain as a separate polypeptide. This can enable production of large quantities of scTv fragments in relatively short time in comparison to proteins expressed and refolded in the full-length format. Most importantly, the scTv has potential applications as therapeutic or diagnostic reagents in vivo similar to scFv fragments, e.g. as bispecific single-chain molecules (Bargou et al., 2008) or as immunotoxins (Wayne et al., 2010). The scTv has the added potential of improved tissue penetration because of the reduced size of the scTv relative to the full-length receptor. This is especially important in cases where the TCR is directed against a tumor antigen, where tumor stroma could be targeted to effectively destroy cancerous cells (Zhang et al., 2007, 2008; Thomas et al., 2009).
Previous efforts to engineer scTvs were dependent on having probes available to assess folding of the TCR Vα and Vβ domains. In the well-characterized 2C (Kieke et al., 1999) and 3L.2 (Weber et al., 2005) TCR systems, stabilized scTv fragments were identified by probing with clonotypic antibodies that recognized the conformation of the properly folded and associated Vα and Vβ domains. Because the wild-type affinity of these receptors for specific pepMHC is relatively low, pepMHC tetramers could not be used to detect properly folded, stabilized scTv fragments. It has been known for some time that expression of the extracellular regions of the TCR as soluble protein is often problematic (reviewed in Rudolph et al., 2006), and that the scTv format is typically unstable (Richman et al., 2009). The explanations for scTv instability are probably related to the long-standing observations that particular VH families are more stable as scFv than other VH families. Thus, the human VH3 family confers enhanced stability, but stability is influenced by framework residues and complementarity determining regions (CDRs) (Honegger et al., 2009). Efforts to understand the structural basis of these effects are important because various scFv fragments as bispecific proteins are currently in various phases of clinical testing (reviewed in Thakur and Lum, 2010), and the development of a single, highly stable human scFv platform for different antigen specificities would be desirable (Miller et al., 2010). To this end, scFv libraries with randomized residues in all six CDR loops have been produced to obtain antibodies with high affinity for various antigens (Rothe et al., 2008).
In the current study, we explored whether human scTv fragments might also be developed using a single platform, allowing the natural antigens of the TCR, peptides bound to HLA products, to be targeted. We used in vitro engineered, high-affinity TCRs in a single-chain format to isolate the first human stabilized scTv fragments (Vβ–linker–Vα), and we determined various features of the scTv that allowed them to be expressed as stable proteins, both on the surface of yeast and in soluble form from Escherichia coli. High-affinity TCRs were used as templates for random mutagenesis (error-prone PCR), and stabilized scTv fragments were isolated by selection with soluble peptide:HLA-A2 tetramers. We describe the isolation of variants of two stabilized, human scTv fragments, the A6 scTv that is specific for a peptide derived from the human T cell lymphotrophic virus Tax protein (peptide: Tax11–19, LLFGYPVYV), and the 868 scTv that is specific for a peptide derived from the human immunodeficiency virus Gag protein (peptide: SL977–85, SLYNTVATL). Both of these TCRs used the same exceptionally stable Vα domain: Vα2 (IMGT: TRAV12 family), whereas other human scTv fragments that were not able to be displayed, contained other Vα regions. We also demonstrated that a single polymorphic residue at position 49 of the Vα2 controlled susceptibility to thermal denaturation and influenced the stability of the corresponding Vβ domain of the scTv. Furthermore, the two scTv fragments could be expressed in soluble form from E.coli and were capable of specifically recognizing their cognate peptides bound to HLA-A2 on antigen-presenting cells.
Materials and methods
Antibodies, peptide:HLA.A2 tetramers and flow cytometry
Antibodies used to detect yeast surface expression included: anti-human Vβ5.2, clone 1C1 (FisherThermo Scientific), anti-HA eptiope tag clone HA.11 (Covance), anti-human Cβ clone 8A3 (Endogen/Pierce), anti-human Cα clone 3A8 (Endogen/Pierce), goat anti-mouse IgG F(ab′)2 AlexaFluor 488 secondary antibody (Invitrogen), goat-anti-mouse IgG F(ab′)2 AlexaFluor 647 secondary antibody (Invitrogen) and streptavidin-phycoerythrin (SA:PE, BD Pharmingen). Peptides that bind to HLA.A2 [Tax11–19: LLFGYPVYV, NYESO-Val157–165: SLLMWITNV, SL977–85 (HIV-Gag): SLYNTVATL and WT-1126–134: RMFPNAPYL] were synthesized by standard F-moc (N-(9-fluorenyl)methoxycarbonyl) chemistry at the Macromolecular Core Facility at Penn State University College of Medicine (Hershey, PA, USA). HLA.A2 heavy chain was produced as inclusion bodies and refolded in vitro with HLA.A2 binding peptides and human β-2 microglobulin as described (Garboczi et al., 1996a,b). The HLA.A2 heavy chain contains a biotinylation substrate sequence which allows for in vitro biotinylation (Avidity, BirA enzyme) and subsequently for the formation of streptavidin:phycoerythrin peptide/MHC tetramer. Tetramer and antibody staining of yeast cells was performed on ice for 45 min using ∼1 × 106 cells. Cells were subsequently washed with 500 μl PBS/BSA (0.5%) and analyzed by flow cytometry with an Accuri C6 flow cytometer.
Cloning and expression of single-chain and full-length TCR genes in yeast display vectors
scTv fragments and VβCβ for full-length TCR constructs were expressed from yeast display plasmid pCT302 (Vβ–L–Vα scTvs) (Boder and Wittrup, 2000) or p315 (VβCβ)p315 generously provided by K. Dane Wittrup, MIT), which contain a galactose-inducible AGA2 fusion and allow for growth in Trp or Leu media, respectively. The full sequence of the AGA-2 fusions with scTv genes A6 and 868 (see below) is shown in Supplementary data, Fig. S1. VαCα for full-length TCR constructs was expressed from pCT302-secreted, a galactose-inducible system that allows for growth in Trp media. Gene induction involved growth of transformed EBY100 yeast cells to stationary phase in selection media followed by transfer to galactose-containing media.
The single-chain genes for TCRs were synthesized by Genscript (Piscataway, NJ, USA) with mutations originally isolated for improved affinity by phage display of full-length TCR with a non-native disulfide bond in the C regions (Boulter et al., 2003). The TCRs are specific for various peptides in the context of HLA.A2: the A6 mutant clone 134 (KD = 2.5 nM) specific for Tax from human T cell lymphotrophic virus (Li et al., 2005), the 1G4 mutant clone C49C50 (KD = 1.3 nM) specific for NYESO (Sami et al., 2007) and an 868 mutant clone (KD of at least 10 nM) specific for human immunodeficiency virus Gag peptide SL9 (Varela-Rohena et al., 2008). For the 868 mutant, the most prevalent clone listed from CDR2β (YVRGEE, affinity mutations underlined) and CDR3α (CAVRGAHDYALN) was generated (Varela-Rohena et al., 2008) (sequences of clones and published affinities are provided in Supplementary data, Table S1 and Fig. S2). The scTv genes (Fig. 1) consisted of the variable domains attached by the linker region GSADDAKKDAAKKDGKS (as described in Soo Hoo et al., 1992; Weber et al., 2005). The scTv genes were introduced into the yeast display vector pCT302 using NheI and XhoI restriction sites. The DNA and protein sequences of the stabilized clones A6-X15 and 868-Z11 are provided in Supplementary data, Fig. S1.
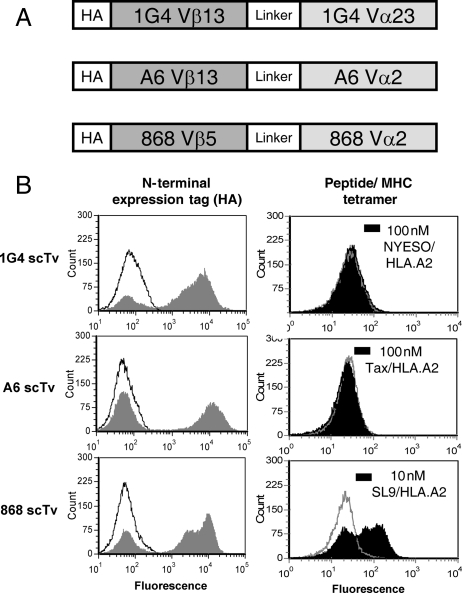
Fig. 1.
Yeast surface display of scTv variable fragments. (A) Schematic of scTv fusions for the human TCRs 1G4, A6 and 868, which recognize peptides NYESO, Tax and HIV-Gag (SL9) in the context of HLA.A2 (Li et al., 2005; Dunn et al., 2006; Varela-Rohena et al., 2008). (B) Yeast surface expression of scTv fusions was monitored for expression of N-terminal expression tag [hemagluttanin (HA), solid gray] with anti-HA antibody and goat anti-mouse IgG alexa 488 secondary antibody, or secondary antibody only as a control (black line). The negative peak in each histogram is due to yeast that have lost plasmid, and serves as an internal control for each induced yeast sample. To assess display of properly folded scTv fusions, cells were incubated with pep:HLA.A2 streptavidin:phycoerythrin tetramers. Null tetramer staining histograms are shown (gray line) as a control, along with the cognate tetramers, as indicated (black, solid).
For construction of full-length TCR constructs, V-region genes were introduced into human full-length VαCα and VβCβ as described (Boulter et al., 2003; Richman et al., 2009). In brief, Vβ genes of 868 and A6 were introduced into the p315 plasmid as a fusion to AGA-2 by ligation into NheI and BglII restriction sites and a BglII site was introduced into the Cβ gene. Vα genes were introduced into NheI and BamHI sites, and a BamHI site was introduced into the Cα gene, and both were cloned into a pCT302 secretion plasmid. The full-length TCR constructs contained a non-native Cys in each constant domain (α: Thr48Cys, β: Ser57Cys) to facilitate the formation of a disulfide bond (Boulter et al., 2003). C region cysteine residues that normally participate in an interchain disulfide bond in the stalk region were replaced with a stop codon and the Cβ cysteine at position 71 was mutated to a serine (Cβ: Cys71Ser).
Construction, display and selection of mutated scTv yeast display libraries
Error-prone PCR was used to generate random mutations, as described previously (Richman et al., 2009). PCR products were electroporated along with NheI and XhoI digested vector into yeast strain EBY100 to generate libraries by homologous recombination. Random mutational libraries of the A6, 868 and 1G4 scTv genes were induced in galactose-containing media (SG-CAA) for 48 h, washed with 500 μl 0.5% PBS/BSA and stained with corresponding pep:MHC SA:PE tetramers (100 nM). Cells were washed (5 ml, 0.5% PBS/BSA), and the most fluorescent cells were selected using either a MoFlo (Cytomation) or a FACS Aria (BD Bioscience) high-speed sorter. In some cases, libraries were subjected to additional rounds of thermal stability sorting whereby yeast cells were incubated at higher temperatures (A6: 42°C for 45 min, 868: 40°C for 30 min) prior to staining.
Cloning of single-site or multiple-site scTv mutants
The PheVα49Ser mutation was introduced using Quikchange Lightning Kit (Stratagene, Agilent Technologies) with forward primer: 5′ GGTAAATCTCCAGAATTGATCATGTCCATCTACTCTAATGGTGACAAAGAAG 3′ and reverse complement primer: 5′ CTTCTTTGTCACCATTAGAGTAGATGGACATGATCAATTCTGGAGATTTACC 3′. The 868 scTv with CDR2β (Sequence: YYEEEE) and CDR3α (Sequence: CAVRTNSGYALNFG) wild-type residues was synthesized by Genscript. The high-affinity mutations for each CDR were introduced into this template by splicing by overlap extension PCR (Warrens et al., 1997).
Expression in E.coli, folding and biotinylation of soluble scTv fragments
The 868-Z11 and A6-X15 scTv mutants were introduced into the pET28a expression vector using NcoI and EcorI (868-Z11, forward primer: 5′TATACCATGGGCAGCAGCCATCATCATCATCATCACAGCAGCGGCCTGGTGCCGCGCGGCAGCGAAGCTGGTGTTACTCAATCTCC 3′ 868-Z11, reverse primer: AAATGAATTCTTAAATATGTGGAGTAACCCAAAAAGAAGTACC). Plasmids were transformed into the BL21 cell line, and after culture and gene induction, cells were passed through a microfluidizer (Microfluidics Corporation, Newton, MA, USA) and inclusion bodies were isolated and protein was purified as described (Garcia et al., 2001). scTv proteins were folded from inclusion bodies and purified with Ni agarose resin (Qiagen, Valencia, CA) followed by gel filtration (Superdex 200). Folded proteins were biotinylated using N-hydroxysuccinimide (NHS) biotin ester (EZ-Link Sulfo-NHS-LC-Biotin Kit, Pierce/Thermo Scientific). Biotinylation was verified by gel-shift with streptavidin by SDS–PAGE.
Binding of scTv proteins measured by surface plasmon resonance
The binding of purified refolded scTv proteins to cognate peptide-HLA.A2 was monitored with surface plasmon resonance (SPR) using a Biacore 3000 instrument. Complexes of the Tax11–19 and SL977–85 peptides with HLA-A*02:01 (called HLA.A2 hereafter) were generated by refolding from bacterially expressed heavy chain and β2m inclusion bodies as previously described (Davis-Harrison et al., 2005). Due to the high affinities of the scTv proteins, a kinetic titration assay was utilized, in which increasing concentrations of analyte were sequentially injected over the surface without the requirement for disruptive regeneration injections (Karlsson et al., 2006). Experiments were performed with 868-Z11 and A6-X15 amine coupled to a standard CM5 sensor chip. Peptide/MHC analyte was sequentially injected at various concentrations. The amount of immobilized scTv was kept below 500 RU and the flow rate was set to the maximum of 100 μl/min to minimize mass transport effects. Data were analyzed using Biaevaluation 4.1 as described (Karlsson et al., 2006). Solution conditions were 10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P-20, 25°C.
Binding of scTv proteins to peptide-loaded antigen-presenting cells
To examine binding of soluble scTv protein to peptide presented by HLA.A2 on cells, T2 cells (HLA-A*02:01+) were incubated for 2 h with various concentrations of peptides. Cells were then washed twice with 0.5% PBS/BSA, and incubated at room temperature for 30 min with biotinylated scTv at various concentrations. Cells were washed twice with 0.5% PBS/BSA, followed by incubation with SA:PE for 30 min at room temperature. Cells were washed twice and analyzed using an Accuri C6 Flow Cytometer.
Results
Yeast display of human scTvs
To determine whether human TCRs could be expressed in single-chain formats, we cloned three human TCRs (Fig. 1) that had high affinity for their respective pep-HLA.A2 ligands, thus allowing the pepMHC to be used as high-affinity probes for properly folded and associated VαVβ binding sites. The TCRs were engineered previously for higher affinity, using a full-length TCR format and phage display (Li et al., 2005; Sami et al., 2007; Varela-Rohena et al., 2008). The variable domains contained various mutations in the CDRs that conferred high affinity, and they were fused by a linker peptide (L) in the orientation Vβ–L–Vα (Supplementary data, Fig. S1). The single-chain constructs were cloned at the C-terminus of the yeast cell wall protein Aga2, followed by a hemagglutinin (HA) peptide tag for detection of displayed proteins.
Yeast cells containing the 1G4, A6 and 868 scTv fusions were induced, and the cells were stained with an anti-HA antibody (Fig. 1B). All three constructs showed high levels of expression of the HA tag, as evidenced by a positive population (the negative population has been found in all yeast displayed systems, and is due to a fraction of yeast cells that lose the plasmid during induction). To detect properly folded VαVβ domains, the respective peptide/A2 tetramers were used for staining the same induced yeast cells. As expected, based on previous studies with various scTv fragments (Richman et al., 2009), neither the 1G4 nor A6 scTv were positive with their respective cognate pep/HLA.A2 ligands (Fig. 1B), consistent with the absence of properly folded scTv proteins on the surface of yeast. Staining of the 1G4 and A6 cells with an antibody to Vβ13 was also negative (unpublished data), confirming that these scTv proteins were not expressed on the yeast surface. Surprisingly, the 868 scTv was positive for staining with its cognate ligand, SL9/HLA.A2, even at the concentration of 10 nM (Fig. 1B). This represented the first time that an scTv fragment could be detected as a properly folded protein, without the need for random mutation and selection (Kieke et al., 1999; Weber et al., 2005; Richman et al., 2009).
Engineering of stabilized surface displayed scTv fragments by random mutagenesis
Although the features of the 868 V regions that allowed display were unknown, this TCR uses a Vβ (Vβ5) that differs from 1G4 (Vβ13) and A6 (Vβ13), while it uses a Vα that is similar to A6 (Vα2). This may imply that the Vβ5 region of 868 confers stability on the protein. To gain insight into the mechanisms that enable scTv stability and surface display, we performed mutagenesis and selections for displayed proteins. First, to determine if the 1G4 and A6 scTv could be stabilized by specific V-region mutations, the scTv genes were subjected to rounds of random mutagenesis and libraries were sorted with pep/HLA.A2 tetramers to isolate scTv mutants with improved surface expression. Despite multiple efforts, through various rounds of mutagenesis and sorting with NYESO/HLA.A2 tetramers, yeast displayed 1G4 scTv could not be isolated (data not shown).
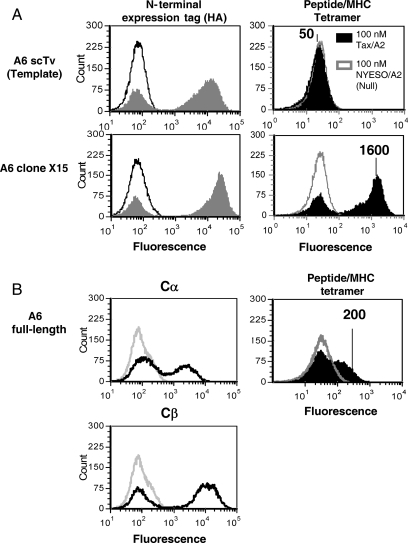
In contrast, stabilized mutants of the A6 scTv were isolated after mutagenesis and sorting with the Tax/HLA.A2 tetramers (100 nM) (data not shown). Previous experiments with the mouse TCRs 2C and 3.L2 demonstrated that selection of scTv fragments with increased resistance to thermal denaturation yielded mutants with significantly higher levels of display that correlated with soluble secretion efficiency (Shusta et al., 2000; Weber et al., 2005). Consequently, the A6 scTv library was sorted with Tax/HLA.A2 tetramer after incubation at a higher temperature (42°C) and several mutants, including a clone called A6-X15 with improved yeast surface levels, were isolated (Fig. 2). The scTv mutant A6-X15 (mean fluorescence intensity MFI = 1600, sequence provided in Supplementary data, Fig. S1) was expressed at higher surface levels than the A6 template scTv (MFI = 50) or the full-length TCR (MFI = 200) (Fig. 2), with stabilized C region cysteines, as monitored by Tax/HLA.A2 tetramer. In the full-length A6 TCR, each of the Cα and Cβ antibodies detected higher surface levels than tetramer, suggesting that the fully folded and assembled dimer represents only a subset of the total α and β chains on the surface. Notably, introduction of the Vβ mutations from A6-X15 into the 1G4 scTv, which shares Vβ usage with A6, did not allow for surface expression of the 1G4 TCR (data not shown).
Fig. 2.
Isolation of stabilized A6 scTv mutants and yeast display of with full-length A6 TCR. (A) The high-affinity A6 TCR was cloned as an scTv, and a library of random mutants was sorted with Tax/HLA.A2 tetramer. Clone A6-X15, isolated after two rounds of mutagenesis and selection with Tax/A2 tetramer, was stained with anti-HA antibody (left panels, solid gray histogram), secondary antibody only (left panels, black line histograms), 100 nM Tax/A2 tetramer (right panels, solid black histogram) or 100 nM null peptide NYESO/A2 tetramer (right panels, gray line histogram). (B) The full-length A6 TCR with β chain expressed as a fusion to AGA-2 and α chain secreted was used to compare full-length and single-chain formats. Surface expression of α and β chains was monitored with anti-Cα or anti-Cβ antibody (left panels, black line histograms) or with goat anti-mouse IgG secondary antibody only as a control (left panels, gray line histograms). Cells were stained with 100 nM Tax/A2 tetramer (right panel, solid black histogram) or 100 nM null peptide NYESO/A2 tetramer (right panel, gray line histogram). Mean fluorescence units of positive peaks are shown.
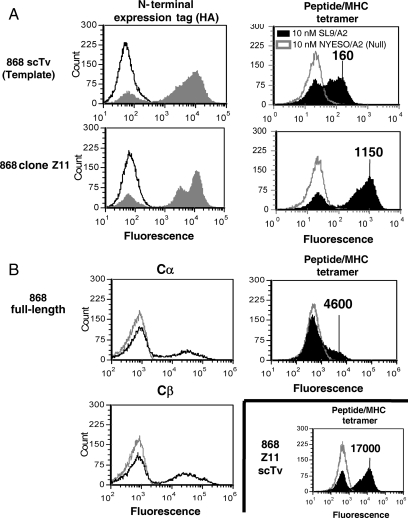
Based on our previous results (Shusta et al., 2000; Weber et al., 2005), we also sought to identify mutants of the 868 scTv that would exhibit increased stability. The 868 scTv gene was randomly mutagenized by error-prone PCR, and the library was subjected to several rounds of sorting with SL9/HLA.A2 tetramer, including a round after incubation at 40°C. Various temperature stable clones were isolated, including a mutant called 868-Z11 that bound to SL9/HLA.A2 tetramer at concentrations as low as 1 nM as detected by flow cytometry (Fig. 3 and data not shown, sequence provided in Supplementary data, Fig. S1). Clone 868-Z11 exhibited improved surface levels that were almost 10-fold improved relative to the 868 scTv template (Fig. 3). In addition, like the A6 TCR, the scTv forms of the 868 TCR were also detected at higher levels than the full-length format that contained C regions with the stabilizing cysteines (Fig. 3). This demonstrated that the single-chain format allows for much higher surface levels of productively associated Vα:Vβ pairs relative to the full-length display format. This is consistent with a previous finding using the mouse 2C system, comparing the temperature-stabilized scTv and full-length TCR formats (Richman et al., 2009).
Fig. 3.
Isolation of stabilized 868 scTv mutants and yeast display of scTv and full-length 868 TCR. (A) The high-affinity 868 TCR was cloned as an scTv, and a library of random mutants was sorted with HIV Gag SL9/HLA.A2 (SL9/A2). Clone 868-Z11, isolated after one round of mutagenesis and selection with SL9/A2 tetramer, was stained with anti-HA antibody (left panels, solid gray histogram), secondary antibody only (left panels, black line histograms), 10 nM SL9/A2 tetramer (right panels, solid black histogram) or 10 nM null peptide NYESO/A2 tetramer (right panels, gray line histogram). (B) The full-length 868 TCR with β chain expressed as a fusion to AGA-2 and α chain secreted was used to compare full-length and single-chain formats. Surface expression of α and β chains was monitored with anti-Cα or anti-Cβ antibody (left panels, black line histograms) or with goat anti-mouse IgG secondary antibody only as a control (left panels, gray line histograms). Cells were stained with 10 nM SL9/A2 tetramer (right panels, solid black histogram) or 10 nM null peptide NYESO/A2 tetramer (right panels, gray line histogram). Note that the 868-Z11 scTv staining with tetramer is shown again, as in this experiment different flow cytometry settings were used compared to (A), resulting in the indicated mean fluorescence units for comparison with full-length 868 TCR.
To examine what mutations in the A6 and 868 scTv conferred enhanced yeast surface levels and temperature stability, various clones, including A6-X15 and 868-Z11, were sequenced (Fig. 4). The A6-X15 clone showed only mutations in the Vβ13 region, with no mutations in the Vα2. Several of these Vβ mutations (A52V, N63D, N66K, Q106L) were also shared with the stabilized clone A6-X3 (Fig. 4A). Thus, it appeared that in A6, the wild-type Vα2 region, but not the Vβ13 region, was sufficiently stable to allow its expression, proper folding and assembly with the mutated Vβ on the yeast surface.
Fig. 4.
Sequences of selected A6 and 868 scTv mutants showing highest surface levels following thermal denaturation. Mutants of the A6 scTv (A) or the 868 scTv (B) were isolated after mutagenesis of the scTv genes, followed by surface expression, high-temperature exposure and selection with tetramers. Sequences are coded as follows: yellow, high-affinity CDR mutations for A6 (Li et al., 2005) and for 868 (Varela-Rohena et al., 2008); red, conserved immunoglobulin fold residues, black-boxed residues, mutations identified in selected mutants. Note: for ease of comparing with published structures, residues of each variable domain of the scTv are numbered sequentially, and thus do not correspond to IMGT nomenclature.
In contrast to the A6 scTv mutants, the stabilized 868 scTv clones did not show any shared mutations in the Vβ region, but they contained a single shared mutation in the Vα2 region, F49αS (Fig. 4B). (Note that the numbering follows that used in the pdb file of A6; based on IMGT sequences, this residue is numbered 55.) Interestingly, the Ser49 residue is also found in the Vα2 of the A6 TCR, and represents a polymorphism at this position (Supplementary data, Fig. S2).
Polymorphic residue 49 of Vα2 influences Vα and Vβ domain stability
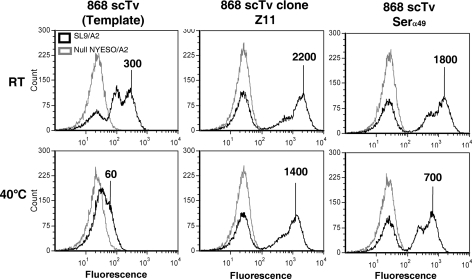
As indicated, the sequences of the five most stable 868 scTv clones (out of 20) isolated after temperature stability sorting shared a single Vα mutation, PheVα49Ser, and this was the only Vα mutation present in the most stable 868 clone, 868-Z11 (Fig. 4B). To assess whether this mutation alone, and not the Vβ mutations, was responsible for the enhanced stability of the 868-Z11 mutant, the PheVα49Ser mutation was introduced into the 868 scTv. Staining of this mutant after incubation at 40°C showed that this single mutation provided the protein with improved resistance to thermal denaturation, resulting in over a 10-fold increase in the surface level (Fig. 5).
Fig. 5.
Residue 49 of the Vα2 region influences thermal stability of the 868 scTv. The serine polymorphism/mutation at position 49 was introduced into the 868 scTv template. Yeast cells were incubated at room temperature (top row) or at 40°C for 30 min and then stained with 10 nM SL9/A2 SA:PE tetramer (black line histograms) or null peptide Tax/A2 SA:PE tetramer (gray line histograms). Mean fluorescence units of positive peaks are shown.
Relative effects of CDR and Serα49 mutations on yeast display of scTv
Our previous work with the mouse TCRs 2C and 3.L2 has shown that some CDR mutations can also confer increased stability to the scTv format (Kieke et al., 1999; Weber et al., 2005). It is possible that the 868 CDR mutations that were selected for higher affinity binding to SL9/HLA.A2 also contributed to stability of the scTv format. Accordingly, the wild-type CDR residues were cloned into the 868 scTv format (WT), with either a Pheα49 or Serα49 residue. To approach this question, we could not use the tetramer for detection, due to the low affinity of the wild-type TCR. Thus, an antibody to the TCR Vβ5 domain was used as a surrogate to assess surface levels (Fig. 6). The WT 868 scTv with the Serα49 residue showed significantly improved surface levels, relative to the WT 868 scTv with the Pheα49 residue, which was barely detectable with the anti-Vβ5 antibody. Although the Serα49 residue yielded enhanced surface levels with the high-affinity mutations (MUT) in the CDRs, in this case even the scTv with the Pheα49 residue was easily detected with the anti-Vβ5 antibody (Fig. 6). Thus, the CDR mutations that yielded higher affinity also contributed to enhanced stability. We have observed a reciprocal selection scenario, in which stabilizing mutations in the CDR1α of an scTv also yielded higher affinity (Weber et al., 2005).
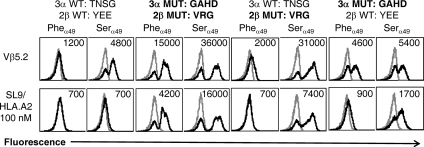
Fig. 6.
CDR mutations and the PheVα49Ser mutation each improve the surface display of the 868 scTv. Various 868 scTv proteins were expressed in the yeast display system with wild-type CDR residues or with high-affinity CDR mutations (Fig. 4B), with either Phe or Ser at position α49. Top row, cells stained with anti-Vβ5.2 antibody and goat anti-mouse (black line histograms) or secondary antibody only (gray-lined histograms). Bottom row, surface expression was also monitored with SL9/A2 tetramers (black line histograms) or null peptide Tax/A2 tetramers (gray-lined histograms). Mean fluorescence units of positive peaks are shown (or in cases where only a negative peak is present, the mean of this peak).
To further investigate whether the high-affinity mutations in the CDR2β and/or the CDR3α were responsible for the enhanced surface stability, the wild-type CDR2β and CDR3α sequences were individually introduced into the template high-affinity 868 scTv, with the Pheα49 or Serα49 residue, and yeast surface levels were monitored by staining with anti-Vβ5 antibody. The high-affinity mutations in CDR3α conferred improved Vβ surface levels as monitored with the anti-Vβ5 antibody, in comparison to the WT scTv with Pheα49 (Fig. 6). In contrast, the high-affinity mutations in CDR2β did not increase substantially the Vβ surface levels in the presence of Pheα49 but appeared to act synergistically in the presence of Serα49. Thus, it appears that both CDR mutations and the Serα49 mutation provided enhanced stability to the scTv format of this TCR. The maximal scTv surface levels observed for binding of SL9/HLA.A2 tetramers to proteins that contained the high-affinity mutations (Fig. 6) also substantiated these findings.
Expression of soluble human scTv proteins in E.coli, and peptide:HLA.A2 binding
Our previous studies with mouse TCRs 2C and 3.L2 have shown that the generation of yeast surface displayed mutants of the scTv proteins allowed the mutants to be expressed in E.coli in both secreted form and refolded from inclusion bodies (Weber et al., 2005; Jones et al., 2006 , 2008). To determine if this were possible with the human scTv mutants, the A6-X15 and 868-Z11 scTv genes were cloned into an E.coli expression vector and the induced E.coli cells expressed the expected size proteins (data not shown). The scTv proteins were refolded from inclusion bodies with yields of >1 mg/l of culture, yielding purified scTv proteins of the expected 30 kDa (Fig. 7A).
Fig. 7.
Expression, purification and SPR binding studies of soluble scTv proteins A6-X15 and 868-Z11. A6 clone X15 and 868 clone Z11 were expressed in the E.coli pET28 expression system. Proteins were refolded from inclusion bodies, and purified by Ni-column and size exclusion chromatography. (A) SDS–PAGE of purified A6-X15 and 868-Z11 scTv proteins, and indicated molecular weight markers (M). (B) SPR traces of Tax/A2 binding immobilized A6-X15. Concentrations of injected pepMHC analyte are indicated below each injection. Black line represents a fit to a 1:1 binding interaction using a kinetic titration scheme as described in the Materials and methods section. Fitted parameters (KD, kon, koff) are shown in the inset. (C) SPR traces of SL9/A2 binding immobilized 868-Z11.
Two different binding studies were conducted. First, SPR was performed with purified peptide/HLA.A2 binding immobilized scTv to determine solution binding affinities and kinetics. A kinetic titration experiment was performed, obviating the need for potentially destabilizing regeneration steps necessary when high-affinity interactions are studied with SPR (Karlsson et al., 2006). Analysis of the data for the A6-X15-Tax/HLA.A2 and 868-Z11-SL9/HLA.A2 interactions yielded nanomolar and picomolar binding affinities for the A6-X15 and 868-Z11 interactions, respectively (Fig. 7B and C). The relative values correlated with the affinities seen with the full-length proteins and confirmed the single-chain constructs retained specificity and high affinity for their cognate ligand. Any differences with the previously published affinities (Li et al., 2005; Varela-Rohena et al., 2008; Piepenbrink et al., 2009) may reflect difficulties quantifying high-affinity binding via SPR, or possibly an influence of the mutations generated via yeast display, or the linker, on binding affinity. These possibilities are currently being investigated.
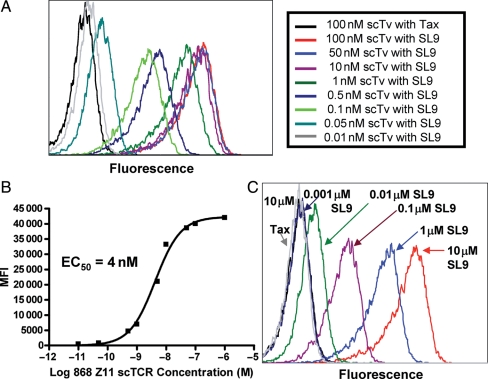
In a second approach, we determined whether cell surface pep/HLA.A2 complexes could be detected with soluble scTv protein. The 868 scTv protein was biotinylated using a biotin-succinimidyl cross-linking agent that reacts with free amines (the 868 scTCR linker contains multiple lysines, whereas the scTv contains no lysines in the CDR loops). The HLA.A2-positive human cell line T2 was loaded with SL9 peptide or Tax (868-null) peptide at 10 μM and cells were subsequently incubated with various concentrations of biotinylated-868-Z11 scTv followed by streptavidin:PE at room temperature (Fig. 8A). Soluble 868-Z11 scTv detected SL9 peptide loaded cells at scTv concentrations below 1nM, whereas cells loaded with the null peptide Tax were not positive even at the highest concentration of 868-Z11 scTv (Fig. 8A). The concentration that yielded 50% maximal staining, an estimate of the KD of the reaction, was ∼4 nM scTv (Fig. 8B). This approach, using flow cytometry, has provided affinity estimates that are in good agreement with values derived using SPR and soluble proteins, in the KD range of ∼1–200 nM (Feldhaus et al., 2003; Holler et al., 2003).
Fig. 8.
Recognition of peptide-loaded antigen-presenting cells with soluble 868-Z11 scTv. (A) Titration of various concentrations of the biotinylated 868 scTv on antigen-presenting cell line T2 (HLA.A2+) pre-loaded with SL9 (10 μM) peptide, or null peptide Tax (10 μM). Cells were stained at room temperature for 30 min with scTv, washed, and stained with SA:PE for 30 min. Analysis was performed after two additional washes. (B) Mean fluorescence intensity (MFI) values of scTv titration shown in (A) were plotted versus scTv concentration, to determine the concentration that yielded 50% maximal binding, EC50, for 868 Z11 scTv. (C) The SL9 peptide was titrated on T2 cells, and subsequently stained with a saturating concentration (100 nM) of biotinylated 868-Z11 scTv.
To examine the peptide sensitivity of binding by the scTv, the SL9 peptide was titrated on T2 cells and the antigen-presenting cells were stained with biotinylated 868-Z11 scTv at 100 nM (Fig. 8C). Without any optimization of staining signal, the scTv detected the peptide at concentrations as low as 10 nM. It remains to be seen how this concentration corresponds to levels associated with endogenous processing and presentation of the SL9 peptide. Nevertheless, the scTv fragments appear to be quite stable, as even after storage at 4°C for over 1 year, the protein was able to bind target cells loaded with low nanomolar peptide concentrations (data not shown). Thus, they should be useful in addressing the challenges that remain with targeting of the natural HIV epitope, or other pepMHC, which are presented at relatively low levels compared with most current antibody-targeted antigens (Richman and Kranz, 2007).
Discussion
It has been known for many years that the expression and crystallization of the extracellular region of αβ TCRs have posed significant problems, when compared with either antibodies or MHC proteins (reviewed in Rudolph et al., 2006). The explanation for this difficulty lies, at least in part, in the behavior of TCR variable regions. This also accounts for problems that have plagued efforts to use scTv V-region fragments in the same way that has been done with antibody single-chain Fv fragments (Richman et al., 2009). Here, we show that it is possible to at least in part overcome these inherent problems by using a specific human V-region family, Vα2 (IMGT: TRAV12 family). This Vα region appears to be over-represented among human T cells directed against a number of HLA.A2-associated antigenic peptides, perhaps most notably peptides from MART1/MelanA (ELAGIGILTV) and Epstein Barr Virus protein BMLF1 (GLCTLVAML) (Mantovani et al., 2002; Trautmann et al., 2002; Dietrich et al., 2003). It is possible that the inherent stability of Vα2, observed in the context of scTv proteins described here, might contribute to its preferential use in some T cell responses, and in the original isolation of the HTLV-1 Tax and HIV-1 Gag-specific TCRs, A6 and 868 (Utz et al., 1996; Varela-Rohena et al., 2008).
In previous studies, we have attempted to provide a possible molecular rationale for V-region mutations that enabled the yeast surface display of mouse scTv (Kieke et al., 1999; Shusta et al., 2000; Weber et al., 2005; Richman et al., 2009) and individual Vβ regions (Buonpane et al., 2005). The underlying premise was based on our observations that surface display was directly correlated with thermal stability of the scTv (Shusta et al., 1999; Orr et al., 2003; Weber et al., 2005). In some cases, a possible explanation involved enhanced hydrophilicity at positions where a residue was buried at the would be V:C interface, or mutations at the Vα:Vβ interface that improved domain association. However, a significant number of mutations, such as those present within or at the base of CDR loops, have less obvious explanations. For example, it is unknown how the CDR3α or CDR2β mutations in the 868 scTv that conferred high affinity also yielded enhanced surface display and stability, in the unbound state. In addition, A6 scTv mutants with improved stability each contained a mutation within CDR2β, AlaVβ52Val. We have argued that some of the CDR mutations could confer local stability on the CDR loops, based on modeling predictions (Holler et al., 2003). Similar results with stabilizing CDR mutations in scFv fragments have also been observed (Honegger et al., 2009; Miller et al., 2010).
Additional mutations in the framework 3 region or hypervariable 4 region of the Vβ domains may also have contributed to the stability of the A6 and 868 scTv fragments. In the case of A6, the AsnVβ62Asp and AsnVβ66Lys mutations within framework 3 were conserved among isolated clones. Although the contributions of each individual mutation have not been determined, their conservation among 14 out of 20 scTv clones of the A6 TCR (data not shown) suggests that they play a role in surface display and stability.
The mechanism by which residue 49 of the Vα domain confers stability is also not clear. It is near the CDR3β (e.g. it is 2.8 Å from Glu105β in A6) loop, such that it is possible that its primary role is in stabilization of the Vα:Vβ interface (Fig. 9), a location that has been considered important in the design of stabilized scFv fragments (Miller et al., 2010). This area of the Vβ (i.e. near Glu105β, which can be encoded by either N-region addition, the D gene, or the N-terminus of the Jβ gene) could in turn influence the stabilizing potential of the Vα and Ser49α. The residue adjacent to Glu105β, Gln106β, was mutated to Leu in each of the surface displayed scTv of A6, suggesting that this region can be further optimized for enhanced stability in the context of the Ser49α residue. Furthermore, CDR3α and CDR3β stabilizing mutations (L104αP and T105βA) have been identified previously in the mouse 2C scTv (Kieke et al., 1999). Thus, it is apparent that various CDR and framework regions can have complex and interdependent impacts on the biophysical properties of the scTv molecules, not unlike antibody scFv fragments.
Fig. 9.

Position of the Serα49 residue in the structure of the A6 TCR. V regions of the A6:Tax/A2 complex (Garboczi et al., 1996a,b) are shown, highlighting the position of the Serα49 residue which is 2.8 Å from CDR3β residue Gluβ105. The Tax peptide (magenta) is shown for reference. PDB file 1AO7 was used in PyMol for graphics.
Studies with soluble full-length TCRs (VαCα/VβCβ) have shown that an interchain disulfide bond formed by a pair of introduced cysteines in the Cα and Cβ regions can confer additional stability on the molecule, enabling expression as a soluble protein (Boulter et al., 2003). This form of the TCR has been used for binding studies, and as a platform for affinity engineering (Li et al., 2005; Dunn et al., 2006; Varela-Rohena et al., 2008). The design of stable scTv fragments that bind to the specific pepMHC ligands, as shown here, provides additional opportunities to use the scTv as a platform for various applications, as has been done widely with antibody scFv fragments (reviewed in Holliger and Hudson, 2005). These include the use of display technologies for affinity engineering (Boder and Wittrup, 2000; Hoogenboom, 2005) and more facile production of biophysically well-behaved single-chain proteins in E.coli (Worn and Pluckthun, 2001; Ewert et al., 2003). The scTv form also provides a platform for use in chimeric, single-chain antigen receptors (reviewed in Sadelain et al., 2009) or in soluble form as bispecific fusions that could include, anti-CD3 antibodies, toxins or cytokines (Bargou et al., 2008; Miller et al., 2010). As we have discussed in a previous review (Richman and Kranz, 2007), the most significant challenge with the use of soluble TCRs, whether single-chain or full-length forms, involves the relatively low surface densities associated with processed and presented antigens.
Based on the diverse sequences of the HLA.A2-restricted peptides that have elicited Vα2+ T cells (e.g. MART1, ELAGIGILTV; EBV, GLCTLVAML; Tax, LLFGYPVYV; Gag, SLYNTVATL), we anticipate that it should be possible to generate Vα2+ scTv proteins against a wide array of diverse antigens. Two structures are available for Vα2+ TCRs in complex with pep-HLA.2, the A6 TCR:Tax/A2 complex (Garboczi et al., 1996a,b) and the MEL5 TCR/MART-1/A2 complex (Cole et al., 2009). The two TCRs dock in almost identical orientations on their pep-A2 ligands, and they share CDR1α interactions with both the α2 helix and peptide positions 1 and 4. Peptide specificity appears to be achieved in large part through interactions with CDR3β, providing support to the notion that the scTv proteins described here, which use distinct Vβ regions, can maintain their antigen specificity through the involvement of other CDRs.
Supplementary data
Supplementary data are available at PEDS online.
Funding
This work was supported by the National Institutes of Health (R01 GM55767 and P01 CA097296 to D.M.K., R01 GM067079 to B.M.B.) and a grant from the McDonnell Foundation (to D.M.K.). K.H.P. and F.K.I. were supported by the Notre Dame CBBI training program, funded by NIH grant T32 GM075762.
Acknowledgements
We thank the University of Illinois Flow Cytometry Facility for technical assistance and Jennifer Stone, Sarah Richman, Lindsay Jones and Natalie Bowerman for useful discussions.
References
- Bargou R., Leo E., Zugmaier G., et al. Science. 2008;321:974–977. doi: 10.1126/science.1158545. [DOI] [PubMed] [Google Scholar]
- Boder E.T., Wittrup K.D. Methods Enzymol. 2000;328:430–444. doi: 10.1016/s0076-6879(00)28410-3. [DOI] [PubMed] [Google Scholar]
- Boulter J.M., Glick M., Todorov P.T., Baston E., Sami M., Rizkallah P., Jakobsen B.K. Protein Eng. 2003;16:707–711. doi: 10.1093/protein/gzg087. [DOI] [PubMed] [Google Scholar]
- Buonpane R.A., Moza B., Sundberg E.J., Kranz D.M. J. Mol. Biol. 2005;353:308–321. doi: 10.1016/j.jmb.2005.08.041. [DOI] [PubMed] [Google Scholar]
- Card K.F., Price-Schiavi S.A., Liu B., et al. Cancer Immunol. Immunother. 2004;53:345–357. doi: 10.1007/s00262-003-0450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlewicki L.K., Holler P.D., Monti B.C., Clutter M.A., Kranz D.M. J. Mol. Biol. 2005;346:223–239. doi: 10.1016/j.jmb.2004.11.057. [DOI] [PubMed] [Google Scholar]
- Chung S., Wucherpfenning K.A., Friedman S.M., Hafler D.A., Strominger J.L. Proc. Natl Acad. Sci. USA. 1994;91:12654–12658. doi: 10.1073/pnas.91.26.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D.K., Yuan F., Rizkallah P.J., Miles J.J., Gostick E., Price D.A., Gao G.F., Jakobsen B.K., Sewell A.K. J. Biol. Chem. 2009;284:27281–27289. doi: 10.1074/jbc.M109.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.M., Boniface J.J., Reich Z., Lyons D., Hampl J., Arden B., Chien Y. Annu. Rev. Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- Davis-Harrison R.L., Armstrong K.M., Baker B.M. J. Mol. Biol. 2005;346:533–550. doi: 10.1016/j.jmb.2004.11.063. [DOI] [PubMed] [Google Scholar]
- Dietrich P.Y., Le Gal F.A., Dutoit V., et al. J. Immunol. 2003;170:5103–5109. doi: 10.4049/jimmunol.170.10.5103. [DOI] [PubMed] [Google Scholar]
- Dunn S.M., Rizkallah P.J., Baston E., et al. Protein Sci. 2006;15:710–721. doi: 10.1110/ps.051936406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewert S., Huber T., Honegger A., Pluckthun A. J. Mol. Biol. 2003;325:531–553. doi: 10.1016/s0022-2836(02)01237-8. [DOI] [PubMed] [Google Scholar]
- Feldhaus M., Siegel R., Opresko L., et al. Nat. Biotechnol. 2003;21:163–170. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- Garboczi D.N., Ghosh P., Utz U., Fan Q.R., Biddison W.E., Wiley D.C. Nature. 1996a;384:134–141. [PubMed] [Google Scholar]
- Garboczi D.N., Utz U., Ghosh P., Seth A., Kim J., VanTienhoven E.A., Biddison W.E., Wiley D.C. J. Immunol. 1996b;157:5403–5410. [PubMed] [Google Scholar]
- Garcia K.C., Radu C.G., Ho J., Ober R.J., Ward E.S. Proc. Natl Acad. Sci. USA. 2001;98:6818–6823. doi: 10.1073/pnas.111161198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler P.D., Holman P.O., Shusta E.V., O'Herrin S., Wittrup K.D., Kranz D.M. Proc. Natl Acad. Sci. USA. 2000;97:5387–5392. doi: 10.1073/pnas.080078297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler P.D., Chlewicki L.K., Kranz D.M. Nat. Immunol. 2003;4:55–62. doi: 10.1038/ni863. [DOI] [PubMed] [Google Scholar]
- Holliger P., Hudson P.J. Nat. Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- Honegger A., Malebranche A.D., Rothlisberger D., Pluckthun A. Protein Eng. Des. Sel. 2009;22:121–134. doi: 10.1093/protein/gzn077. [DOI] [PubMed] [Google Scholar]
- Hoogenboom H.R. Nat. Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- Jones L.L., Brophy S.E., Bankovich A.J., Colf L.A., Hanick N.A., Garcia K.C., Kranz D.M. J. Biol. Chem. 2006;281:25734–25744. doi: 10.1074/jbc.M604343200. [DOI] [PubMed] [Google Scholar]
- Jones L.L., Colf L.A., Bankovich A.J., Stone J.D., Gao Y.G., Chan C.M., Huang R.H., Garcia K.C., Kranz D.M. Biochemistry. 2008;47:12398–12408. doi: 10.1021/bi801349g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson R., Katsamba P.S., Nordin H., Pol E., Myszka D.G. Anal. Biochem. 2006;349:136–147. doi: 10.1016/j.ab.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Kieke M.C., Shusta E.V., Boder E.T., Teyton L., Wittrup K.D., Kranz D.M. Proc. Natl Acad. Sci. USA. 1999;96:5651–5656. doi: 10.1073/pnas.96.10.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Moysey R., Molloy P.E., et al. Nat. Biotechnol. 2005;23:349–354. doi: 10.1038/nbt1070. [DOI] [PubMed] [Google Scholar]
- Mantovani S., Palermo B., Garbelli S., Campanelli R., Robustelli Della Cuna G., Gennari R., Benvenuto F., Lantelme E., Giachino C. J. Immunol. 2002;169:6253–6260. doi: 10.4049/jimmunol.169.11.6253. [DOI] [PubMed] [Google Scholar]
- Miller B.R., Demarest S.J., Lugovskoy A., et al. Protein Eng. Des. Sel. 2010;23:549–557. doi: 10.1093/protein/gzq028. [DOI] [PubMed] [Google Scholar]
- Orr B.A., Carr L.M., Wittrup K.D., Roy E.J., Kranz D.M. Biotechnol. Prog. 2003;19:631–638. doi: 10.1021/bp0200797. [DOI] [PubMed] [Google Scholar]
- Piepenbrink K.H., Borbulevych O.Y., Sommese R.F., Clemens J., Armstrong K.M., Desmond C., Do P., Baker B.M. Biochem. J. 2009;423:353–361. doi: 10.1042/BJ20090732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaksin D., Polakova K., McPhie P., Margulies D.H. J. Immunol. 1997;158:2218–2227. [PubMed] [Google Scholar]
- Richman S.A., Kranz D.M. Biomol. Eng. 2007;24:361–373. doi: 10.1016/j.bioeng.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Richman S.A., Aggen D.H., Dossett M.L., Donermeyer D.L., Allen P.M., Greenberg P.D., Kranz D.M. Mol. Immunol. 2009;46:902–916. doi: 10.1016/j.molimm.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C., Urlinger S., Lohning C., et al. J. Mol. Biol. 2008;376:1182–1200. doi: 10.1016/j.jmb.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Rudolph M.G., Stanfield R.L., Wilson I.A. Annu. Rev. Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- Sadelain M., Brentjens R., Riviere I. Curr. Opin. Immunol. 2009;21:215–223. doi: 10.1016/j.coi.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sami M., Rizkallah P.J., Dunn S., et al. Protein Eng. Des. Sel. 2007;20:397–403. doi: 10.1093/protein/gzm033. [DOI] [PubMed] [Google Scholar]
- Shusta E.V., Kieke M.C., Parke E., Kranz D.M., Wittrup K.D. J. Mol. Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- Shusta E.V., Holler P.D., Kieke M.C., Kranz D.M., Wittrup K.D. Nat. Biotechnol. 2000;18:754–759. doi: 10.1038/77325. [DOI] [PubMed] [Google Scholar]
- Soo Hoo W.F., Lacy M.J., Denzin L.K., Voss E.W.J., Hardman K.D., Kranz D.M. Proc. Natl Acad. Sci. USA. 1992;89:4759–4763. doi: 10.1073/pnas.89.10.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.K., Jameson S.C., Hogquist K.A. Annu. Rev. Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- Stone J.D., Chervin A.S., Kranz D.M. Immunology. 2009;126:165–176. doi: 10.1111/j.1365-2567.2008.03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A., Lum L.G. Curr. Opin. Mol. Ther. 2010;12:340–349. [PMC free article] [PubMed] [Google Scholar]
- Thomas D.L., Kim M., Bowerman N.A., Narayanan S., Kranz D.M., Schreiber H., Roy E.J. J. Immunol. 2009;183:1828–1837. doi: 10.4049/jimmunol.0802322. [DOI] [PubMed] [Google Scholar]
- Trautmann L., Labarriere N., Jotereau F., Karanikas V., Gervois N., Connerotte T., Coulie P., Bonneville M. Eur. J. Immunol. 2002;32:3181–3190. doi: 10.1002/1521-4141(200211)32:11<3181::AID-IMMU3181>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Utz U., Banks D., Jacobson S., Biddison W.E. J. Virol. 1996;70:843–851. doi: 10.1128/jvi.70.2.843-851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe P.A., Davis S.J. Annu. Rev. Immunol. 2003;21:659–684. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- Varela-Rohena A., Molloy P.E., Dunn S.M., et al. Nat. Med. 2008;14:1390–1395. doi: 10.1038/nm.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrens A.N., Jones M.D., Lechler R.I. Gene. 1997;186:29–35. doi: 10.1016/s0378-1119(96)00674-9. [DOI] [PubMed] [Google Scholar]
- Wayne A.S., Kreitman R.J., Findley H.W., Lew G., Delbrook C., Steinberg S.M., Stetler-Stevenson M., Fitzgerald D.J., Pastan I. Clin. Cancer Res. 2010;16:1894–1903. doi: 10.1158/1078-0432.CCR-09-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K.S., Donermeyer D.L., Allen P.M., Kranz D.M. Proc. Natl Acad. Sci. USA. 2005;102:19033–19038. doi: 10.1073/pnas.0507554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worn A., Pluckthun A. J. Mol. Biol. 2001;305:989–1010. doi: 10.1006/jmbi.2000.4265. [DOI] [PubMed] [Google Scholar]
- Zhang B., Bowerman N.A., Salama J.K., et al. J. Exp. Med. 2007;204:49–55. doi: 10.1084/jem.20062056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zhang Y., Bowerman N.A., Schietinger A., Fu Y.X., Kranz D.M., Rowley D.A., Schreiber H. Cancer Res. 2008;68:1563–1571. doi: 10.1158/0008-5472.CAN-07-5324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.