Abstract
Monoclonal antibodies are a commercially successful class of drug molecules and there are now a growing number of antibodies coupled to toxic payloads, which demonstrate clinical efficacy. Determining the precise epitope of therapeutic antibodies is beneficial in understanding the structure–activity relationship of the drug, but in many cases is not done due to the structural complexity of, in particular, conformational protein epitopes. Using the immunotoxin CAT-8015 as a test case, this study demonstrates that a new methodology, hybrid β-lactamase display, can be employed to elucidate a complex epitope on CD22. Following insertion of random CD22 gene fragments into a permissive site within β-lactamase, proteins expressed in Escherichia coli were first screened for correct folding by resistance to ampicillin and then selected by phage display for affinity to CAT-8015. The optimal protein region recognised by CAT-8015 could then be used as a tool for fine epitope mapping, using alanine-scanning analysis, demonstrating that this technology is well suited to the rapid characterisation of antibody epitopes.
Keywords: beta-lactamase display, CD22, epitope mapping, immunotoxin
Introduction
The elucidation of an antibody epitope is a key step in understanding the relationship between structure and function at the antigen–antibody interface. This can enable the future isolation of higher affinity, more potent, antibody analogues and also has the potential to enable design of smaller antibody mimetics, peptides and even small molecules, which can mimic the antibody-binding mechanism and interact with the same epitope region. Perhaps more importantly, epitope studies can shed light on how the structure of the target antigen determines its natural biological function.
While an X-ray co-crystal structure of the antigen–antibody complex remains the gold standard of epitope determination, it is not a widely available technology and is not always feasible due to the difficulty of obtaining high concentrations of good-quality proteins and also in finding the correct conditions to produce diffracting crystals. In the absence of a crystal complex, hydrogen/deuterium exchange (e.g. DXMS, H/D-Ex, etc.) is an attractive option. Unfortunately, these techniques require large quantities of soluble, homogenous, highly purified and stable antigens and may run the risk that the purified recombinant antigen does not closely resemble the native protein.
Of the many other epitope mapping techniques which exist, all are faced with the same problem which is the fine mapping of conformational epitopes and the attribution of the key amino acids on the 3D structure of the antigen when one is available. For instance, epitope mapping on a peptide microarray allows the study of thousands of specific-binding sequences in a single experiment (Reineke et al., 2001). This technique is very efficient for mapping of linear epitopes and the CLIPS™ technology developed by Pepscan, which stabilises peptides in biologically relevant structures, can give some informative results about the characterisation of conformational epitopes (Timmerman et al., 2007). Unfortunately, these approaches are complex with regard to the vast combinations of peptides to be analysed before finding out the appropriate conformation.
The screening of phage libraries displaying random peptides has had limited impact on the mapping of conformational epitopes because the selected peptides are regarded as mimotopes of the genuine epitope of the antibody's interacting antigen. Typically, an algorithm is used to align a set of query peptides from the phage display process onto the solved structure of the antigen. Following a clustering step, which combines the most significant matches, only one predicted epitope is inferred in the better cases (Mayrose et al., 2007). If no crystal structure is available, the result of this approach is typically a consensus mimotope sequence, which is difficult to locate over a primary amino acid sequence.
Methods based on mutagenesis of the target antigen, in particular combinatorial alanine scanning (Weiss et al., 2000), are attractive due to their relative speed and the precise analysis of individual side-chain contributions to the binding energy. However, such methods do require significant mutagenesis efforts when used to probe the entire surface area of a large target antigen. To reduce the mutagenesis burden, one may employ a domain deletion approach to focus the epitope search (Lacal and Aaronson, 1986). Such methods can be successful but it is worth noting that domain deletion experiments measure a negative outcome, i.e. the loss of binding, which could be explained by other factors, such as effects on protein stability, loss of tertiary or quaternary structure or the introduction of an aggregation liability.
In this context, the expression of random antigen fragments into a highly-permissive and solvent-exposed loop of the Bacillus licheniformis β-lactamase BlaP and the screening of these hybrid β-lactamases using phage display offers several benefits. First, it has been demonstrated that the use of BlaP as a protein carrier makes easier the expression of protein domains known to be insoluble or difficult to express and that the protein carrier confers enough conformational flexibility to the inserts for refolding and interacting with their native ligands (Chevigne et al., 2007; Vandevenne et al., 2007, 2008; Ruth et al., 2008). Next, the random insertion of fragments of the target gene into the blaP gene allows a large number of protein fragments with different sizes to be expressed from the same polypeptide region. As a result, this region can adopt a large number of different folds. This feature allows the generation of epitope libraries which do not correspond to mimotopes because they are directly related to the nucleotide sequence of the antigen. By alteration of the fragment sizes, the library can be made more representative of linear or conformational epitopes. In addition, this method can be viewed as an original methodology to expose cryptic epitopes when these are not accessible in the native antigen. In this procedure, the β-lactam resistance conferred to Escherichia coli infected with phages expressing soluble and functional hybrid β-lactamases is pivotal because it allows a positive selection of phages during the phage library construction and provides an easy method to measure the enrichment of the phage library during successive rounds of biopanning. Moreover, the fact that the epitope is selected as a bifunctional hybrid protein, which associates an epitope with a specific and efficient enzymatic activity, allows rapid characterisation of the antigen–antibody interaction.
In this case study, we applied the hybrid β-lactamase display method to identify the epitope of the anti-CD22 immunotoxin CAT-8015. Therapeutically, CD22 is of interest because it is a specific marker present on the cell surface of malignant B cells and is rapidly internalised upon binding, making it an appealing target for an antibody drug conjugate or immunotoxin approach (Clark, 1993; Du et al., 2008). CAT-8015 is an immunotoxin which combines a CD22-specific antibody variable fragment (Fv) with a Pseudomonas exotoxin A (PE38) payload and has demonstrated noteworthy clinical activity in chronic lymphocytic leukaemia (CLL), hairy cell leukaemia (HCL) (Alderson et al., 2009) and paediatric acute lymphoblastic leukaemia (Mussai et al., 2010). It is one of the several anti-CD22 antibodies currently in clinical trials, including epratuzumab (Dorner et al., 2006) and inotuzumab ozogomicin (Wong and Dang, 2010).
CD22 itself is a cell surface glycoprotein with multiple, immunoglobulin (Ig)-like domains within its extracellular portion. The N-terminal Ig-like V-type domain, which mediates sialic acid binding (May et al., 1998), is followed by varying numbers of Ig-like C2-type domains (Crocker and Varki, 2001a,b). No structural data exist for CD22 and relatively little structural information is available for related family members, due to their high levels of glycosylation. However, some general information exists on the epitope of anti-CD22 antibodies, including the predecessors of CAT-8015, epratuzumab and inotuzumab. This information has arisen from antibody competition-binding studies and the binding of antibodies to domain deletion mutants of CD22 and suggests that CAT-8015 and epratuzumab both bind to the C-like domain 2 while inotuzumab binds to the V-like domain (Li et al., 1989; Stein et al., 1993; Engel et al., 1995; DiJoseph et al., 2005). However, such studies do not give the precise location of the antibody epitopes.
In the present study we were interested to achieve a more comprehensive epitope mapping analysis of the antibody CAT-8015 to identify the CD22 residues involved in the interaction. Random fragments of the human CD22 gene were cloned into the blaP gene and the bifunctional hybrid β-lactamases were affinity selected on the antibody CAT-8015 by phage display. This approach enabled the identification of a CD22 sub-region conferring high affinity for CAT-8015, which could then be used in an alanine-scanning study to characterise the key amino acids involved in binding. These results support the assumption that the extracellular domains of CD22 could interact together and participate to form the CAT-8015 epitope.
Materials and methods
Phage display methodology
Preparation of CD22 gene fragments
The gene fragment encoding the extracellular domain (Asp20 to Arg687) of the human CD22 protein (CD22, accession number NM_001771; Wilson et al., 1991) was chemically synthesised to avoid the use of rare codons; the synthetic gene was optimised according to the E.coli codon usage. We also avoided introducing regions with very high (>80%) or very low (<30%) GC content, internal TATA boxes, ribosomal entry sites, repeat sequences and RNA secondary structures. The absence of RNA secondary structure is of crucial importance because this parameter governs the intensity and the specificity of the signals generated with the epitope array. The gene coding for the CD22 extracellular domain (CD22ed) was PCR amplified using Taq and Pfu DNA polymerase from the synthetic gene, obviating the need for large-scale purification and digestion of plasmid DNA. The cloning of DNA fragments not related to the cd22ed is then not possible. The DNAse Shotgun® cleavage kit (Novagen) was used on the purified PCR products to produce random gene fragments ranging from 50- to 1000-bp in length. Four digestions were done in a total volume of 60 µl in the presence of 0.048 Unit of DNAse and 12.5 µg of PCR products. The digestion reactions were stopped after 1, 2, 3 or 4 min with 10 mM EDTA. Five microlitres of each reaction was loaded on agarose gel to control the range of the DNA fragments. The rest of the digestions were pooled and successively purified, repaired using Pfu polymerase and finally purified once more. The end-repaired gene fragments were dephosphorylated using calf intestine phosphatase to avoid multiple cloning of cd22ed fragments into the same vector. All the purifications were done with the QIAquick Nucleotide Removal Kit (Qiagen).
Construction of the fd-Tet BlaP/CD22ed gene-fragments library
The blaP gene was modified by introducing a SmaI restriction site in order to clone blunt-ended heterologous gene fragments. The recombinant blaP-SmaI gene was fused with the phage pIII gene to generate fd-Tet BlaP/SmaI (Chevigne et al., 2007). The CD22ed gene fragments were ligated into the SmaI-digested fd-Tet BlaP/SmaI. The ligation products was dialysed against water and used to transform electrocompetent E.coli TG1 (Stratagene). Electrotransformed cells were regenerated in the SOC medium for 1 h at 37°C and plated on LB agar medium supplemented with 7.5 µg/ml tetracycline and 5 µg/ml ampicillin.
Phage preparation
The transformants were scraped and suspended in the LB liquid medium supplemented with 7.5 µg/ml tetracycline and 5 µg/ml ampicillin. Phages were amplified at 28°C for 16 h and recovered from the culture supernatant by two polyethylene glycol precipitations. After the first precipitation, a filtration through a 0.45-µm filter was performed to remove membrane fragments. Phages were suspended in TBS (50 mM Tris, 150 mM NaCl at pH 7.6) and the phage concentration was estimated by measuring the absorbance at 265 nm.
Checking the expression of hybrid β-lactamases
The presence of a cloned CD22ed gene fragment into the blaP gene was checked by PCR from individual colonies with primers BlapSmaseq (5′-CAGAGTTAAATGAAGTGAATCCGGGTGAAACTCAGGAT-3′) and BlapEnd (5′-GTGGTGCCCTTTCCCGTTCATGTTTAAGGC-3′). PCR fragments >400 bp confirmed the presence of an insert into blaP.
Bio-panning procedure
The affinity selection of the phage library was achieved against the CAT-8015-related monoclonal antibody (mAb 8015). The rabbit monoclonal antibody anti-CD22 provided by Epitomics (mAb EP498Y), which recognises the 20 N-terminus amino acids of the CD22 (DSSKWVFEHPETLYAWEGAC), was chosen as a control for linear epitope selection. Five hundred nanograms of mAb 8015 and 200 ng of mAb EP498Y diluted in carbonate buffer (50 mM NaHCO3, pH 9.6) were coated onto Maxisorp® microplate strips (Nalgen Nunc International) for 16 h at 4°C. After washing with Tween/TBS (Tris Buffered Saline with 0.05 % Tween 20), the wells were blocked with BSA 1% in TTBS for 1 h at 37°C and washed again with TTBS. Then 1012 phage particles were added per well and incubated overnight at 4°C. Unbound phages were removed by successive washings with TTBS and bound phages were eluted in 25-mM glycine–HCl buffer (pH 2.2) for 5 min. The eluate was transferred to a tube and neutralised with 2 M Tris–HCl (pH 8). The eluted phages were amplified by infecting log phase E.coli TG1.
Phage ELISA
The analysis of the binding properties of phage was performed as described for the bio-panning procedure except that elution step was replaced by the addition of a colorimetric substrate. The immobilised β-lactamase activity carried by phage was measured by following the hydrolysis of 100 µM nitrocefin in 50 mM phosphate buffer pH 7.5 at 482 nm.
Epitope-array methodology
Construction of oligonucleotide microarray
The CD22ed oligonucleotide microarray was designed using 72 non-overlapping oligonucleotides that cover the whole sequence coding for the extracellular domain. Each oligonucleotide was spotted in duplicate. The microarrays were printed by Eurogentec using the VersArray® ChipWriter Pro System (Bio-Rad). Positive control oligonucleotides, complementary to BlaP sequences upstream and downstream of the insertion site, were included as well as mismatched controls (90, 80 and 60% of mismatch) to monitor washing steps and hybridisation specificity.
Probe preparation
Fluorescent DNA probes were prepared from initial and enriched phage libraries by PCR amplification using Taq polymerase with primers BlapSmaseq and BlapEnd. The probes were labelled with Cy3-dCTP (GE Healthcare).
Hybridisation and washing
Fluorescent probe mixtures were diluted in the hybridisation buffer provided by Eurogentec and added with calf thymus DNA (Sigma). Fluorescent probe mixtures were denatured at 95°C for 5 min, and applied onto the chip under a cover glass. Chips were hybridised at 42°C for 16–18 h. The hybridised chips were then washed at room temperature successively with the washing buffer 1 (30-mM NaCl, 3-mM sodium citrate, 0.1% SDS, pH 7) and the washing buffer 2 (30-mM NaCl, 3-mM Sodium citrate, pH 7). The chips were dried at room temperature before scanning.
Detection and analysis
The chips were scanned with a GenePix 4000A fluorescence reader (Axon) to detect emission from the Cy3 label at 650 nm, following laser excitation at 550 nm. The acquired images were analysed by GenePix Pro analysis software (Axon).
Modelling of CD22
The C-type domains 2–3 of CD22 were modelled separately. The MODELER program in Accelrys Discovery Studio software package was used for modelling these domains. The intervening sequences between the domains were also included in the modelling. The model of carcinoembryonic antigen (CEA) which consists of seven extracellular Ig-like domains similar to that predicted for CD22 was used as the structural template for assembling the modelled domains. Alternative orientations of the C-type domains 2 and 3 were also modelled based on the structures of either human Fcγ receptor (2fcb) or leukocyte Ig-like receptor A5 (2d3v). Models were subject to energy minimisation with CHARMm forcefield in the Accelrys Discovery Studio package.
Analysis of CD22 sub-domain binding
Expression and purification of the CD22 sub-domain V234–G435 into BlaP
Gene-fragments coding for the native and the 36 single-alanine mutants of the CD22 sub-domain (V234–G435) were chemically synthesised and cloned into the permissive SmaI insertion site of BlaP carried by the pNY vector (Chevigne et al., 2007). pNY allows E.coli full constitutive expression of BlaP. The nucleotidic sequence of the hybrid genes were verified by dideoxy sequencing. To achieve production of the BlaP CD22 hybrid proteins, E.coli BL21 transformed with the pNY derivative plasmids was grown in Terrific Broth supplemented with 75 µg/ml spectinomycin and 10 µg/ml ampicillin at 37°C. Cells from an overnight culture (500 ml) were harvested by centrifugation (9000 g for 15 min).
The periplasmic proteins were harvested by a cold shock osmotic procedure using a successive treatment of cells with TES (20% sucrose, 30-Tris–HCl, 5-EDTA, pH 8) and 5-MgSO4. The periplasmic proteins were purified on the automated Profinia Imac purification system (Bio-rad) using the preprogrammed method. The eluted proteins were conditionned in PBS (50-mM phosphate, 150-mM NaCl, pH 7.5).
Comparative-binding assays
Two distinct procedures were used to compare the binding activity of the BlaP CD22 hybrid proteins to mAb 8015. In the first procedure, 100 ng of monoclonal antibody diluted in carbonate buffer were coated for 16 h at 4°C onto Maxisorp® microplate strips (Nalge Nunc International). After washing with PBST (PBS with 0.05% tween 20), wells were blocked with 2% BSA in PBST for 1 h at 37°C. Native and mutated CD22 sub-domains displayed within the HIS-tagged BlaP β-lactamase were incubated upon the immobilised antibody for 2 h at RT. After washing with PBS, anti-polyhistidine HRP-conjugate (Roche) was diluted in PBS (1/2500) and incubated in the wells for 1 h at 37°C. Unbound antibodies were removed by washes with PBS. ABTS [2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid)] (0.2 mg/ml final) in 50 mM citric acid (pH 4) was used as substrate to measure captured peroxidase activity at 405 nm.
In the second procedure, ELISA was carried out on HIS-Select® High Sensitivity Nickel Coated Microplates (Sigma). Five hundred nanograms of BlaP CD22 hybrid proteins diluted in PBS were coated for 16 h at 4°C. After washing with PBST (PBS with 0.05% Tween 20), wells were blocked with 2% BSA in PBST for 1 h at 37°C. Bound hybrid protein was detected with 100 ng of mAb 8015 diluted in PBS for 1 h at 37°C. Unbound mAb 8015 were removed by several washes with PBS. Anti-human HRP conjugate (Promega) was diluted in PBS (1/2500) and incubated in the wells for 1 h at 37°C. Unbound antibodies were removed by washes with PBS and ABTS was used as substrate to measure captured peroxidase activity.
In order to take into account the non-specific binding, data were adjusted by subtracting the non-specific binding value of an irrelevant antibody from the mean binding score, following the formula: binding of BlaP CD22/C2C3 mutant = (mean binding score of BlaP CD22/C2C3 mutant to mAb 8015 – mean binding score of BlaP CD22/C2C3 mutant to irrelevant mAb).
Then data were normalised to the average-binding score of the wild-type strain in order to have a reference level at 100 for the wild-type strain: normalised binding of BlaP CD22/C2C3 mutant = binding of BlaP CD22/C2C3 mutant/binding of BlaP CD22/C2C3 WT × 100.
Data were analysed by the software package R (Chambers, 2008) using a two-way ANOVA considering strain and experiment as factors. Pairwise differences between strains were then obtained with the Tukey's method (Honest significant difference). This method tests the difference between factor levels, while adjusting P-values in order to control the family-wise error rate that inflates in the case of multiple comparisons.
Dissociation constant measurements
The dissociation constant (Kd) of the BlaP CD22–mAb 8015 complex was determined according to the previous ELISA (Friguet et al., 1985). The BlaP CD22 concentrations that yielded a linear signal with 1 µg of coated mAb 8015 were determined by following the immobilised β-lactamase activity as described above. In the assays, the percentage of BlaP CD22 bound to the immobilised mAb 8015 was below 2.5% of the total amount of hybrid protein added in the well to avoid modification of the equilibrium in solution. To determine the Kd, various concentrations of free mAb 8015 were preincubated with BlaP CD22. After the equilibrium was reached (1 h at room temperature), the concentration of free BlaP CD22 in each sample was determined with the help of mAb 8015-coated microplates as described above. The β-lactamase activities were plotted versus the concentration of free mAb 8015 mixed to BlaP CD22. The titration curve obeys the following equation:
where [E]0 and [L]0 are the initial concentrations of BlaP CD22 and the total concentration of mAb 8015 in solution, respectively; Kd is the dissociation constant; A0 and A are the activities measured on coated mAb 8015 before or after preincubation with mAb 8015, respectively. The Kd was deduced by fitting the experimental points to the equation given above.
Results
Isolation of minimal CD22 sub-regions conferring high affinity to mAb 8015
Although previous studies using antibody competition assays suggested that RFB4, the parent antibody of CAT-8015, bound to the C-like domain 2 region, the entire extracellular domain of CD22 (Supplementary data, Fig. 1) was chosen as a starting point to construct the CD22 gene-fragment phage library. Following gene fragmentation and cloning, 6 × 105 transformants were obtained and a PCR analysis indicated that more than 95% of the clones were positive for the presence of a CD22 gene fragment in blaP and that the size distribution of the inserts was from 50 bp up to 1000 bp. Epitope-array analysis confirmed that the entire CD22 gene was well represented in the β-lactamase positive infectious phage library except for two small regions encoding the short amino acid sequences WKPHGAW and PETIGRR (data not shown). The lower representation of these regions could be due to a lower permissivity into the carrier protein, to an unexpected mis-hybridation or even more to an insufficient spotting.
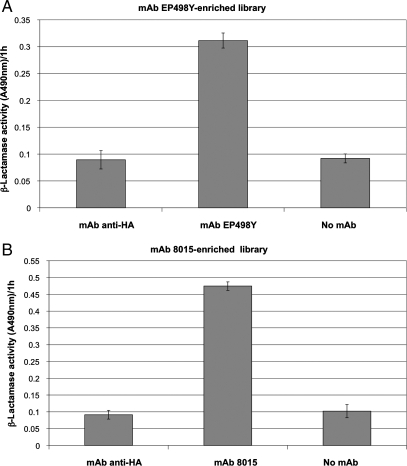
The phage library was affinity enriched on two distinct monoclonal antibodies, mAb 8015 and mAb EP498Y. The mAb EP498Y was used as a control to monitor the efficiency of the bio-panning procedure to identify a linear epitope. After three rounds of bio-panning, the binding properties of both enriched phage libraries were analysed by measuring the immobilised β-lactamase activity on the coated mAbs. Figure 1 shows that both libraries have been specifically enriched with hybrid β-lactamases displaying a specific binding activity against their related mAb. The epitope-arrays (Fig. 2) show that the CD22 gene fragments from libraries enriched upon mAb EP498Y and mAb 8015 have diverged during the successive rounds of bio-panning.
Fig. 1.
Phage ELISA performed with the phage libraries obtained after three rounds of panning on mAb 8015 and mAb EP498Y. (A) Phage ELISA performed with the mAb EP498Y-enriched library. Two hundred nanograms of mAb EP498Y was coated in the assays. (B) Phage ELISA performed with the mAb 8015-enriched library. Two hundred nanograms of mAb 8015 was coated in the assays. Nitrocefin (100 µM) was used as substrate. The mAb anti-HA (200 ng per well) was used as a control for non-specific-binding activity.
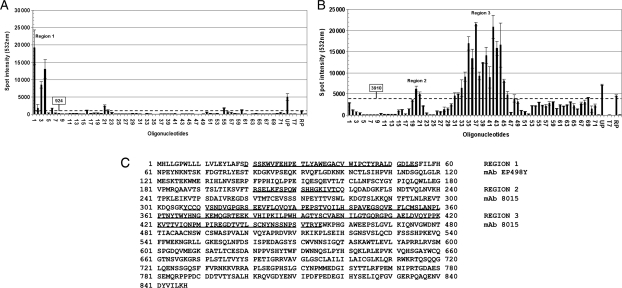
Fig. 2.
Epitope-arrays performed with the phage libraries obtained after three rounds of panning with mAb EP498Y (A) and mAb 8015 (B). The arrays, consisting of 72 oligonucleotides spanning the nucleotide sequence of the extracellular portion of CD22, were probed with fluorescently labelled PCR products from the round three phage selection outputs. Hybridisation controls UP and RP are positive controls for labelling and library amplification, which are complementary to BlaP sequences upstream and downstream the insertion site. T7 and surrounders bars are mismatched UP controls (90, 80 and 60% of mismatch) used to monitor washing steps and hybridisation specificity. (C) Representation of regions 1, 2 and 3 on the primary amino acid sequence of human CD22.
According to the results, the protein region mapped with mAb EP498Y (region 1) corresponds to the N-terminus of CD22 reported from the mAb provider and does not overlap with the two protein regions identified with mAb 8015. With mAb EP498Y, the highest signal was obtained on oligonucleotide 1 suggesting that the peptide sequence DSSKWVFEHP is of crucial importance for the binding of the antibody (region 1 of Fig. 2A).
Of the two regions highlighted in the epitope-array as being important for mAb 8015 binding (Fig. 2B), region 3 has higher overall fluorescence intensity, suggesting that the contribution of region 3 is more important for the binding activity. This region also includes the C-type domain 2 previously defined as relevant for the binding to RFB4.
Selection of a relevant hybrid β-lactamase for studying mAb 8015-binding properties
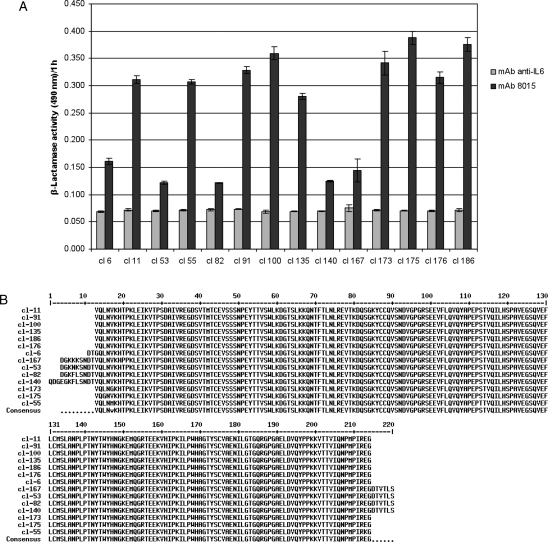
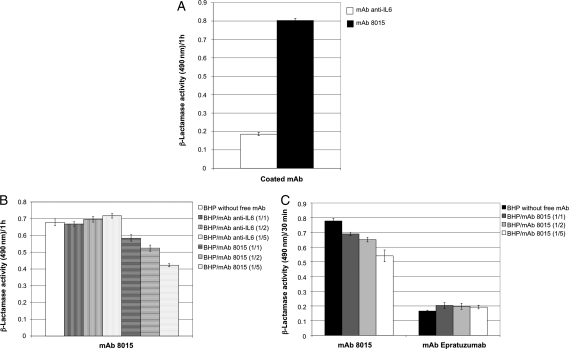
To identify the most appropriate hybrid β-lactamase for conducting the characteriation of mAb 8015, we performed PCR and phage ELISA analysis on single clones (Fig. 3A). Phage ELISA was used to confirm that the selected clones express a hybrid protein that displays a CD22 fragment able to interact strongly and specifically with mAb 8015. From these experiments, 14 candidates were selected and sequenced. The multiple sequence alignment (Fig. 3B) suggested that the minimal region conferring high affinity for mAb 8015 encompasses a region of 202 residues in length (V234–G435) that includes the C-terminal amino acids of the C-type domain 1, the whole of C-type domains 2 and 3 and the N-terminal amino acids of the C-type domain 4. Further truncation of this fragment resulted in significant reduction in binding to mAb 8015 (data not shown). The hybrid β-lactamase displaying this fragment (BlaP CD22/C2C3) was overexpressed in E.coli and purified by affinity chromatography. The graphs presented in Fig. 4 show that this hybrid protein binds specifically to mAb 8015 (Fig. 4A) with a Kd calculated to 26.2 ± 1.4 nM, that free mAb 8015 competes with the coated mAb 8015 for binding to BlaP CD22/C2C3 (Fig. 4B) and that the CD22 antibody epratuzumab does not share the same epitope (Fig. 4C).
Fig. 3.
Isolation of a hybrid protein, comprising beta-lactamase with a CD22 fragment insertion, for conducting the mutational analysis. (A) Phage ELISA of different hybrid proteins performed on 200 ng of coated mAb 8015 with supernatants of isolated phage culture. Nitrocefin (100 µM) was used as reporter substrate. (B) Multiple sequence alignment of the 14 candidates selected in (A).
Fig. 4.
Binding and competition assays performed with the hybrid β-lactamase containing CD22 residues V234–G435, referred to as the bifunctional hybrid protein (BHP). (A) Binding assay performed on 200 ng of coated mAb anti-IL6 or mAb 8015 with 130 ng of BHP. (B) Competition assays performed on 200 ng of coated mAb 8015 with 1.3 µg of BHP supplemented with 1.3 µg (1/1), 2.6 µg (1/2) or 7.5 µg (1/5) of free mAb anti-IL6 or free mAb 8015. (C) Competition assays performed on 100 ng of coated mAb 8015 and mAb epratuzumab with 1.3 µg of BHP supplemented with 1.3 µg (1/1), 2.6 µg (1/2) or 7.5 µg (1/5) of free mAb 8015. Nitrocefin (100 µM) was used as reporter substrate.
Alanine-scanning analysis
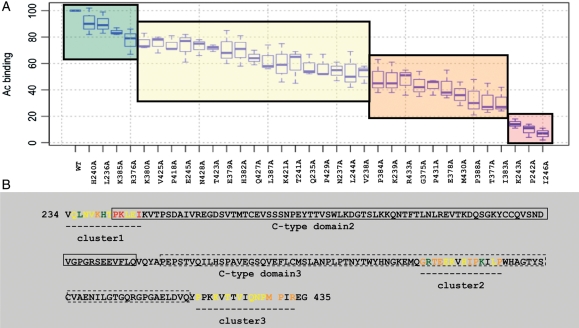
To investigate the interaction network of the complex between CD22 and mAb8015, we used molecular modelling to first identify residues in the CD22 fragment (V234–G435) displayed within the BlaP CD22/C2C3 construct that would be predicted to have some degree of solvent accessibility. From this study, 36 residues were chosen to perform single-point mutation to alanine. As shown in Fig. 5, these mutations form three clusters: cluster 1 (Q235-I246) is located in the junction between the C-type domains 1 and 2, cluster 2 (G375-P388) is in the C-type domain 3 and cluster 3 (P418-R433) encompasses both the junction between the C-type domains 3 and 4, and the N-terminal part of the C-type domain 4. Among 36 mutated hybrid proteins, only two were not expressed due to recurrent sequence deletion located after the substituted residue. The 34 active mutants were overexpressed in E.coli and purified by affinity chromatography.
Fig. 5.
Summary of the binding of CD22 fragments (V234–G435), each containing single-alanine substitutions, to CAT-8015 IgG1, normalised to the binding of the wild-type CD22 fragment. (A) Statistical analysis of the residual binding data according to the ANOVA method allowed grouping into four distinct groups: coloured red, orange, yellow and green, based on the loss of binding signal as a result of alanine substitution (red, 0–20% residual binding; orange, 20–50% residual binding; yellow, 50–80% residual binding; green, 80–100% residual binding). Results are presented as box-and-whisker plots, with the bold line representing the median value and boundaries of the box representing the 25th and 75th percentile. The bars represent the data extremes. (B) Representation of the same residues, with the same colouring scheme, on the amino acid sequence of CD22 fragment (V234–G435).
The binding activity of each mutant to mAb 8015 was measured by ELISA and compared with the binding of the wild-type BlaP CD22/C2C3 protein. Statistical analysis of the data showed that the experiments were highly correlated and that the residual affinities measured with both procedures did not differ significantly from each other (data not shown). Figure 5 shows the mutation effects ordered by a decreasing binding. From this statistical analysis, it appears that the BlaP CD22/C2C3 mutants corresponding to L236A, H240A, R376A and K385A are not significantly different from the wild-type at a family-wise error of 5% while the BlaP CD22/C2C3 mutants corresponding to P242A, K243A and I246A cause the most drastic effect on binding to mAb 8015. The other mutants can be classified in two intermediate groups depending on whether the mutation has minor or important impact on the binding. Considering the 13 mutations that have a percentage of residual binding below 50%, 4/13 correspond to proline (P242, P384, P388 and P431), 4/13 are charged residues (K239, K243, E378 and R433), 4/13 are non-polar residues (I246, G375, I383 and M430) and only one residue is a polar residue with a neutral side chain (T377).
Modelling of CD22
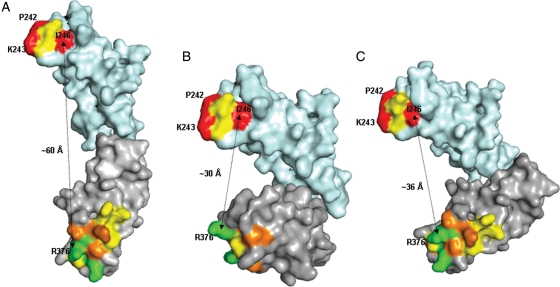
The C-type domains 2 and 3 of CD22, which contained the mAb 8015 epitope clusters 1 and 2 were modelled separately based on homologies to the C2-type domain 3 of human CEA model (1e07, Boehm and Perkins, 2000) and C2-type domain of human TAG-1 (2om5, Mortl et al., 2007), respectively. The separate models were then assembled together in different orientations based on the solved structures of other multi-domain Ig-like proteins, carcinoembryonic antigen (Fig. 6A), human Fcγ-receptor (Fig. 6B) and leukocyte Ig-like receptor A5 (Fig. 6C). Ramachandran plot statistics showed the final models were of acceptable quality (89.6% of residues in allowed region; 9.5% in generously allowed region; 0.9% in the disallowed region). The domain arrangements based on human Fcγ-receptor and leukocyte Ig-like receptor A5 resulted in a shorter distance (∼30–36 Å) between the two epitope clusters than the linear arrangement based on carcinoembryonic antigen, where the distance between epitope clusters was ∼60 Å.
Fig. 6.
Alternative arrangements of C2 (cyan) and C3 (grey) domains of CD22 obtained by superposition of the models of these domains on (A), a model of carcinoembryonic antigen (domains 3 and 4; 1e07.pdb), (B) crystal structure of human Fcγ-receptor (2fcb.pdb) and (C) crystal structure of leukocyte Ig-like receptor A5 (2d3v.pdb). Residues mutated to alanine are coloured according to the percentage of wild-type binding to CAT-8015, upon alanine substitution (red, 0–20% residual binding; orange, 20–50% residual binding; yellow, 50–80% residual binding; green, 80–100% residual binding). Approximate distance between C-α atoms of I246 in the C2 domain and R376 in the C3 domain are shown as an indication of the proximity of the epitope clusters (cluster 1 at the top and cluster 2 at the bottom).
Discussion
CAT-8015 is an immunotoxin of significant clinical interest, which has shown activity in both CLL and HCL patients. This drug exerts its effect via binding to CD22 and consequent, rapid internalisation into target cancer cells to deliver the toxic component PE38. It has previously been shown by antibody competition binding studies that antibody RFB4, the parent of CAT-8015, is likely to bind an epitope in the C-type domain 2 of CD22. However, as antibody competition assays can provide imprecise epitope locations, this study was performed using the hybrid β-lactamase technology. This new approach was applied in order to isolate from a β-lactamase displaying CD22 fragment library the minimal sub-region of CD22 that is able to be bound efficiently by mAb 8015, to produce it as a soluble hybrid protein carrying an enzymatic reporting activity and to use this ‘antigen-like’ material for characterising the CD22/mAb 8015 interaction. The aim of this work was to understand whether the epitope of CAT-8015 had unique properties, which may confer its ability to rapidly internalise and deliver toxins into target cells.
The mAb 8015 was obtained by affinity maturation of mAb RFB4 resulting in a high-affinity monoclonal antibody with a dissociation constant (Kd) below the nanomolar range (Ho et al., 2005). Such affinity gains are generally associated with mutations that make the interaction between the antibody paratope and the antigen epitope more energetically stable or create new interaction sites. As a consequence, the mean surface area between an antibody variable domain and its antigen is 600–800 Å2 allowing the establishment of a large network of interactions.
The complexity of the interaction between mAb 8015 and CD22 was confirmed by our results since the minimal sub-region of CD22 conferring high affinity for mAb 8015 encompasses a 202 residue protein fragment (V234–G435) including the whole of C-type domains 2 and 3. Selection of smaller fragments failed, suggesting that the most important structural elements required for the binding to mAb 8015 are present in the CD22 sub-domain V234–G435. The large size of this fragment suggests that the mAb 8015-related epitope is discontinuous and that both the C-type domains 2 and 3, and possibly their junction regions, play a role in the formation of this epitope. Interestingly, the present study has highlighted a greater level of epitope complexity than previously reported. It was suggested from previous antibody competition and CD22 domain deletion studies that both mAb 8015 and epratuzumab bound to the C-like domain 2 whereas here we show that the mAb 8015 epitope encompasses regions of both C-like domains 2 and 3 and further that epratuzumab is unable to bind to the same CD22 fragment.
To investigate the binding properties of mAb 8015, we overexpressed and purified the CD22 sub-domain V234–G435 as a soluble hybrid protein. The implementation of this bifunctional protein in immunoassays enabled us to confirm the specificity of the interaction with mAb 8015 by using the β-lactamase enzyme as a reporting activity. It is worth acknowledging that the 26-nM affinity observed with the CD22 sub-domain was lower than that for the entire CD22 extracellular region, previously reported as 0.8 nM (Ho et al., 2005). This suggests that some element of the native epitope could be missing in the CD22 minimal epitope fragment inserted within the BlaP. For instance, this could be the case if we consider that region 2 depicted on Fig. 3B, which resides in C-type domain 1 and is not included in the sub-domain V234–G435, also plays a minor role in the binding upon mAb 8015. Another explanation could be due to steric hindrance, whereby the β-lactamase protein carrier obstructs the epitope, or a subtle structural modification that affects the reconstituted epitope. A more mundane explanation could be that the method used previously to determine affinity, which was a saturation binding analysis on cells (Ho et al., 2005), was substantially different from the method employed here, which used recombinant antigen.
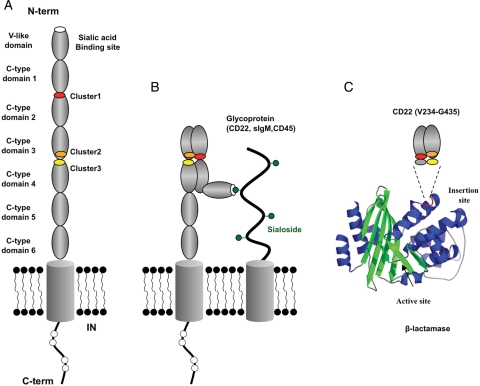
To refine the mapping of the mAb 8015-related epitope, we submitted the hybrid protein to an alanine-scanning study. From this experiment, we concluded that residues in three distinct epitope clusters have an important impact on the mAb binding. Considering only the point mutations giving a percentage of residual binding below 50% of wild type, we observed that the key residues in clusters 1, 2 and 3 are (K239/P242/K243/I246), (G375/T377/E378/I383/P384/P388) and (M430/P431/R432), respectively. Most of the key residues map to clusters 1 and 2, located in the C-type domains 2 and 3, respectively, and the modelling of different orientations of these two domains strongly suggested that a U-shaped arrangement of these domains would bring the clusters into close enough proximity to form an antibody epitope. The distance between clusters of 30–36 Å would be broadly in line with a total surface interaction area of between 600 and 800 Å2, typical for discontinuous antibody epitopes. Although cluster 3 could not be included in the model due to the lack of a suitable template structure in that region, we hypothesise that the U-shaped arrangement would also bring cluster 3 into close enough proximity to be incorporated into the mAb 8015 epitope (Fig. 7). Encouragingly, there is some precedent for other Ig-like proteins, such as the neural cell adhesion molecule axonin-1/TAG-1 (Freigang et al., 2000) and hemolin (Su et al., 1998), adopting U-shaped forms by incorporating tight turns between adjacent Ig-like domains. Furthermore, previous work has shown that the β-lactamase BlaP is a rigid protein but very permissive to polypeptide, protein fragment or entire protein insertion. This feature enables the β-lactamase to keep its structural organisation, which is essential for enzymatic activity. As the β-lactamase activity of the hybrid protein BlaP CD22/C2C3 is relatively well conserved (data not shown), we assume that the N- and C-terminal parts of CD22/C2C3 are quite close to each other into BlaP supporting the concept that C-type domains 2 and 3 can adopt a U-shaped configuration (Fig. 7C).
Fig. 7.
Hypothetical linear and U-shaped arrangements of CD22. The hypothetical linear arrangement results in the epitope clusters being far apart (A), whereas the hypothetical U-shaped arrangement formed by the C-type domains 1–4 brings the epitope clusters into closer proximity for binding to CAT-8015 and potentially enables cis interactions with sialic acid groups on other cell surface glycoproteins (B). Also shown is a schematic representation of the CD22 fragment (V234–G435) in the context of β-lactamase (C).
The hypothesis that some Ig-like domains of CD22 could interact together to give a more compact structural organisation is consistent with the fact that CD22 is known to interact in cis with sialic acid groups of other B cell glycoproteins (Razi and Varki, 1998). Using in situ photoaffinity cross-linking of glycan ligands to CD22 Han et al. (2005) observed that CD22 also recognises glycans of neighbouring CD22 molecules as cis ligands, forming homomultimeric complexes. It is thought that these cis interactions of CD22 on the B cell surface play an important role in masking the sialic acid binding V-like domain and preventing CD22 from interacting in trans with other carbohydrate groups. The hypothetical structure (Fig. 7B) suggested by this study would be consistent with such cis interactions.
In conclusion, the present study has explored the molecular epitope of the investigational therapeutic immunotoxin CAT-8015 and shown it to be a complicated structural arrangement, involving at least three discontinuous regions of the CD22 polypeptide. Whether or not the same approach will work for other antigens with contrasting protein structures requires further epitope mapping studies. However, the hybrid β-lactamase technology has now been used successfully to resolve epitopes of antigens, which do not resemble the Ig-like domain structure of CD22 (data not shown), suggesting that the method could be broadly applicable to different antigen structures. In this example, the technology allowed the identification of the minimal epitope fragment of CD22, despite its structural complexity, due to β-lactamase enabling the correct folding of the embedded antigen. Alanine scanning of this CD22 sub-domain reagent allowed further definition of the key-binding residues demonstrating that this technology is suited to the characterisation of complex epitopes.
Supplementary data
Supplementary data are available at PEDS online.
Funding
This work was funded by MedImmune.
Acknowledgements
The authors would like to thank Changshou Gao and Ronald Herbst for their initial discussions and for the provision of reagents. We would also like to thank Lutz Jermutus, Ira Pastan and Mitchell Ho for their helpful comments on the manuscript.
References
- Alderson R.F., Kreitman R.J., Chen T., Yeung P., Herbst R., Fox J.A., Pastan I. Clin Cancer Res. 2009;15:832–839. doi: 10.1158/1078-0432.CCR-08-1456. doi:10.1158/1078-0432.CCR-08-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M.K., Perkins S.J. FEBS Lett. 2000;475:11–16. doi: 10.1016/s0014-5793(00)01612-4. doi:10.1016/S0014-5793(00)01612-4. [DOI] [PubMed] [Google Scholar]
- Chambers J.M. Software for Data Analysis: Programming with R. New York: Springer; 2008. [Google Scholar]
- Chevigne A., Yilmaz N., Gaspard G., Giannotta F., Francois J.M., Frere J.M., Galleni M., Filee P. J. Immunol. Methods. 2007;320:81–93. doi: 10.1016/j.jim.2006.12.009. doi:10.1016/j.jim.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Clark E.A. J. Immunol. 1993;150:4715–4718. [PubMed] [Google Scholar]
- Crocker P.R., Varki A. Immunology. 2001a;103:137–145. doi: 10.1046/j.0019-2805.2001.01241.x. doi:10.1046/j.0019-2805.2001.01241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker P.R., Varki A. Trends Immunol. 2001b;22:337–342. doi: 10.1016/s1471-4906(01)01930-5. doi:10.1016/S1471-4906(01)01930-5. [DOI] [PubMed] [Google Scholar]
- DiJoseph J.F., Popplewell A., Tickle S., et al. Cancer Immunol. Immunother. 2005;54:11–24. doi: 10.1007/s00262-004-0572-2. doi:10.1007/s00262-004-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner T., Kaufmann J., Wegener W.A., Teoh N., Goldenberg D.M., Burmester G.R. Arthritis Res. Ther. 2006;8:R74. doi: 10.1186/ar1942. doi:10.1186/ar1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Beers R., Fitzgerald D.J., Pastan I. Cancer Res. 2008;68:6300–6305. doi: 10.1158/0008-5472.CAN-08-0461. doi:10.1158/0008-5472.CAN-08-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P., Wagner N., Miller A.S., Tedder T.F. J. Exp. Med. 1995;181:1581–1586. doi: 10.1084/jem.181.4.1581. doi:10.1084/jem.181.4.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freigang J., Proba K., Leder L., Diederichs K., Sonderegger P., Welte W. Cell. 2000;101:425–433. doi: 10.1016/s0092-8674(00)80852-1. doi:10.1016/S0092-8674(00)80852-1. [DOI] [PubMed] [Google Scholar]
- Friguet B., Chaffotte A.F., Djavadi-Ohaniance L., Goldberg M.E. J. Immunol. Methods. 1985;77:305–319. doi: 10.1016/0022-1759(85)90044-4. doi:10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- Han S., Collins B.E., Bengtson P., Paulson J.C. Nat. Chem. Biol. 2005;1:93–97. doi: 10.1038/nchembio713. doi:10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- Ho M., Kreitman R.J., Onda M., Pastan I. J. Biol. Chem. 2005;280:607–617. doi: 10.1074/jbc.M409783200. [DOI] [PubMed] [Google Scholar]
- Lacal J.C., Aaronson S.A. Mol. Cell. Biol. 1986;6:1002–1009. doi: 10.1128/mcb.6.4.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.L., Shen G.L., Ghetie M.A., et al. Cell. Immunol. 1989;118:85–99. doi: 10.1016/0008-8749(89)90359-6. doi:10.1016/0008-8749(89)90359-6. [DOI] [PubMed] [Google Scholar]
- May A.P., Robinson R.C., Vinson M., Crocker P.R., Jones E.Y. Mol. Cell. 1998;1:719–728. doi: 10.1016/s1097-2765(00)80071-4. doi:10.1016/S1097-2765(00)80071-4. [DOI] [PubMed] [Google Scholar]
- Mayrose I., Shlomi T., Rubinstein N.D., Gershoni J.M., Ruppin E., Sharan R., Pupko T. Nucleic Acids Res. 2007;35:69–78. doi: 10.1093/nar/gkl975. doi:10.1093/nar/gkl975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortl M., Sonderegger P., Diederichs K., Welte W. Protein Sci. 2007;16:2174–2183. doi: 10.1110/ps.072802707. doi:10.1110/ps.072802707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussai F., Campana D., Bhojwani D., Stetler-Stevenson M., Steinberg S.M., Wayne A.S., Pastan I. Br. J. Haematol. 2010;150:352–358. doi: 10.1111/j.1365-2141.2010.08251.x. doi:10.1111/j.1365-2141.2010.08251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi N., Varki A. Proc. Natl. Acad. Sci. USA. 1998;95:7469–7474. doi: 10.1073/pnas.95.13.7469. doi:10.1073/pnas.95.13.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke U., Volkmer-Engert R., Schneider-Mergener J. Curr. Opin. Biotechnol. 2001;12:59–64. doi: 10.1016/s0958-1669(00)00178-6. doi:10.1016/S0958-1669(00)00178-6. [DOI] [PubMed] [Google Scholar]
- Ruth N., Quinting B., Mainil J., Hallet B., Frere J.M., Huygen K., Galleni M. FEBS J. 2008;275:5150–5160. doi: 10.1111/j.1742-4658.2008.06646.x. doi:10.1111/j.1742-4658.2008.06646.x. [DOI] [PubMed] [Google Scholar]
- Stein R., Belisle E., Hansen H.J., Goldenberg D.M. Cancer Immunol. Immunother. 1993;37:293–298. doi: 10.1007/BF01518451. doi:10.1007/BF01518451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X.D., Gastinel L.N., Vaughn D.E., Faye I., Poon P., Bjorkman P.J. Science (New York, NY. 1998;281:991–995. doi: 10.1126/science.281.5379.991. [DOI] [PubMed] [Google Scholar]
- Timmerman P., Puijk W.C., Meloen R.H. J. Mol. Recognit. 2007;20:283–299. doi: 10.1002/jmr.846. doi:10.1002/jmr.846. [DOI] [PubMed] [Google Scholar]
- Vandevenne M., Filee P., Scarafone N. Protein Sci. 2007;16:2260–2271. doi: 10.1110/ps.072912407. doi:10.1110/ps.072912407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandevenne M., Gaspard G., Yilmaz N., Giannotta F., Frere J.M., Galleni M., Filee P. Protein Eng. Des. Sel. 2008;21:443–451. doi: 10.1093/protein/gzn021. doi:10.1093/protein/gzn021. [DOI] [PubMed] [Google Scholar]
- Weiss G.A., Watanabe C.K., Zhong A., Goddard A., Sidhu S.S. Proc. Natl. Acad. Sci. USA. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. doi:10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G.L., Fox C.H., Fauci A.S., Kehrl J.H. J. Exp. Med. 1991;173:137–146. doi: 10.1084/jem.173.1.137. doi:10.1084/jem.173.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.