Abstract
Human interleukin-15 (hIL-15) and its receptor α (hIL-15Rα) are co-expressed in antigen presenting cells allowing trans-presentation of the cytokine to immune effector cells. We exploited the high-affinity interactions between hIL-15 and the extracellular hIL-15Rα sushi domain (hIL-15RαSu) to create a functional scaffold for the design of multispecific fusion protein complexes. Using single-chain T cell receptors (scTCRs) as recognition domains linked to the IL-15:IL-15Rα scaffold, we generated both bivalent and bispecific complexes. In these fusions, the scTCR domains retain the antigen-binding activity and the hIL-15 domain exhibits receptor binding and biological activity. As expected, bivalent scTCR fusions exhibited improved antigen binding due to increased avidity, whereas fusions comprising two different scTCR domains were capable of binding two cognate peptide/MHC complexes. Bispecific molecules containing scTCR and scCD8αβ domains also exhibit enhanced binding to peptide/MHC complexes, demonstrating that the IL-15:IL-15Rα scaffold displays flexibility necessary to support multi-domain interactions with a given target. Surprisingly, functional heterodimeric molecules could be formed by co-expressing the TCR α and β chains separately as fusions to the hIL-15 and hIL-15RαSu domains. Together, these properties indicate that the hIL-15 and hIL-15RαSu domains can be used as versatile, functional scaffold for generating novel targeted immune molecules.
Keywords: bispecific fusion protein, CD8, interleukin-15, protein scaffold, T cell receptor
Introduction
Previous studies have shown that novel dimeric targeting proteins can be created for the purposes of either augmenting effective affinity through formation of homodimers or broadening the spectrum of recognition through formation of multiple specific heterodimers. A variety of protein interaction domains have been employed to generate such molecules. Initially, leucine zipper domains were used as association partners for dimerization of fusion proteins. In this approach, hydrophobic interaction of leucine zipper domains is mediated by regularly spaced leucines in parallel α-helices, while the dimerization partner is determined primarily by charged residues immediately outside of the hydrophobic core, that form salt bridges (Kouzarides and Ziff, 1988). This interaction is exemplified by the Fos and Jun family of proteins, which preferentially form heterodimers without significantly affecting the binding activity of fused recognition domain. This approach provides a versatile scaffold to create multimeric complexes (Rieker and Hu, 2000). However, there are limitations to this approach for development of therapeutic proteins. Most prominently, Fos and Jun are intracellular proteins that accumulate almost exclusively within the nucleus. Thus, soluble and secreted Fos and Jun fusions are usually produced using baculovirus or insect cell expression systems (Stern and Wiley, 1992; Sloan et al., 1995), a relatively low yielding and not easily scalable manufacturing process. In an attempt to create functional bispecific molecules, antibody (Ab) domains linked to Fos and Jun were produced in bacterial or mammalian cells, but subunit homodimerization was observed (Kostelny et al., 1992; de Kruif and Logtenberg, 1996), which complicated the purification process and reduced the overall yield.
In addition to leucine zipper motifis, helix-turn-helix self-dimerizing peptides, tri- and tetrameric subdomains of collagen and p53 have been used as scaffolds by which to create multivalent molecules (Holliger and Hudson, 2005; Kubetzko et al., 2006; Cuesta et al., 2009; Hayden-Ledbetter et al., 2009). These interaction domains primarily serve as molecular scaffolds and lack other functional activities per se. Moreover, fusion proteins containing these domains often require further optimization to promote stable multimer formation and specialized production cell lines and purification methods that are tedious or impose regulatory hurdles for therapeutic development. Many of these scaffolds are derivatives of either non-human protein domains or non-native components of plasma that may exhibit poor pharmacokinetic properties and pose the risk of immunogenic responses that could limit their therapeutic potential.
IgG domains, particularly the Fc fragment, have been used successfully as dimeric scaffolds for a number of therapeutic molecules including approved biologic drugs. For example, dimerization of the soluble human tumor necrosis factor-α (TNF-α) receptor linked to the human IgG1 Fc domain results in up to 1000 times more potent TNF-α antagonist activity than the monomeric receptor and provides the fusion with a 5-fold longer serum half-life than the monomeric form (Mohler et al., 1993). In addition to its dimerization activity, the Fc fragment also provides cytotoxic effector functions through the complement activation and interaction with Fcγ receptors displayed on natural killer (NK) cells, neutrophils, phagocytes and dendritic cells (Nimmerjahn and Ravetch, 2008). In the context of anti-cancer therapeutic Abs and other Ab domain-Fc fusion proteins, these activities likely play an important role in efficacy observed in animal tumor models and in cancer patients (Weiner, 2007). Thus, IgG domains have been used as a scaffold to form bispecific Abs to improve the quality and quantity of products generated by the traditional hybridoma fusion technology (Shen et al., 2006; Lu and Zhu, 2009). Although these methods bypass the shortcomings of leucine zipper and the synthetic scaffolds, there continue to be difficulties in producing bispecific Abs in mammalian cells at levels sufficient to support clinical development and use. Additionally, the Fc-mediated effector functions of such fusions may not be adequate or appropriate in a number of therapeutic applications. There has been considerable interest in improving and expanding on the effector activity of the Fc domain and developing other means of recruiting cytolytic immune responses, including T cell activity, to the disease site via targeted therapeutic molecules (Weiner, 2007; Baeuerle et al., 2009).
In an effort to develop a new, human-derived immunostimulatory multimeric scaffold, we focused on the use of human IL-15 (hIL-15) and IL-15 receptor domains. Human IL-15 is a member of the small four α-helix bundle family of cytokines that associates with the hIL-15 receptor α-chain (hIL-15Rα) with a high binding affinity (equilibrium dissociation constant (KD) ∼10−11 M) (Mortier et al., 2006). The resulting complex is then trans-presented to the human IL-2/15 receptor β/common γ chain (hIL-15RβγC) complexes displayed on the surface of T cells and NK cells. This cytokine/receptor interaction results in expansion and activation of effector T cells and NK cells, which play an important role in eradicating virally infected and malignant cells (Waldmann, 2006). Normally, hIL-15 and hIL-15Rα are co-produced in dendritic cells to form complexes intracellularly that are subsequently secreted and displayed as heterodimeric molecules on cell surfaces (Bergamaschi et al., 2008). Thus, the characteristics of hIL-15 and hIL-15Rα interactions suggest that these inter-chain-binding domains could serve as a novel, human-derived immunostimulatory scaffold to make soluble dimeric molecules capable of target-specific binding. We have previously reported the use of fusion proteins comprising soluble T cell receptor (TCR) proteins as disease antigen-specific recognition domains (Mosquera et al., 2005; Wen et al., 2008). In this report, we extend these studies to describe the generation and characterization of a number of fusion proteins comprising TCR- and CD8-binding domains as building blocks to demonstrate the feasibility of using hIL-15:hIL-15Rα scaffold to create both soluble bivalent dimers with increased functional binding affinity toward target antigens and heterodimers for multiple site-specific protein complexes. We also show that these fusion proteins retain potent hIL-15 activity capable of stimulating immune effector cell responses.
Materials and methods
Cell culture
The HLA-A*0201-restricted, p53 [amino acids (aa) 149–157] peptide-specific cytotoxic T lymphocytes (CTLs) were obtained from Dr L. Sherman (The Scripps Research Institute, La Jolla, CA; Theobald et al., 1995). The H-2Kb-restricted, ovalbumin (OVA) (aa257–264) peptide-specific CTLs, derived from OT1 transgenic mice, were kindly provided by Dr L. Lefrancois (University of Connecticut, Farmington, CT). The 293GP cell line was provided by Dr R. Morgan (National Cancer Institute, Bethesda, MD). Chinese hamster ovary (CHO), CTLs, T2, EL4 and 293GP cells were cultured in complete Iscove's modified Dulbecco's medium (IMDM) [IMDM plus 10% fetal bovine serum (HyClone, Logan, UT)]. 32Dβ cells (Zhu et al., 2009) were cultured in IMDM complete medium plus 10 ng/ml rhIL-15 (kindly provided by Dr J. Yovandich, NCI-Frederick, MD).
Materials
The p53 (aa149–157: STPPPGTRV), p53 (aa264–272: LLGRNSFEV), OVA (aa257–264: SIINFEKL), vesicular stomatitis virus (VSV; nucleoprotein aa52–59: RGYVYQGL) peptides were purchased from Peptide 2.0 Inc. (Chantilly, VA). Bacterial expression vectors for producing human β2 microglobulin (β2m) and soluble HLA-A*0201 molecules were provided by Dr J. Altman (Emory University, Atlanta, GA). All oligonucleotide primers were purchased from Sigma Aldrich Corp. (Woodlands, TX). The anti-human TCR Cβ (BF1) 8A3.31 and W4F.5B monoclonal Abs (mAbs) and anti-mouse TCR H57-597 mAb were purified via protein A sepharose from the supernatants of hybridoma cells purchased from ATCC (Manassas, VA). Assay reagents include anti-mouse CD8α and β mAbs (Biolegend, San Diego, CA), anti-human IL-15 mAb (R&D Systems, Minneapolis, MN), streptavidin-horse radish peroxidase (SA-HRP) and streptavidin-phycoerythrin (SA-PE; Jackson ImmunoResearch, West Grove, PA). The pNEF38 and pDEF38 vectors used in this study were described previously (Deer and Allison, 2004). The pMSGV-1 vector (Zhao et al., 2007) with the puromycin-resistant gene (Zhu et al., 2009) was further modified by introducing the CMV promoter to enhance gene expression.
Construction of scTCR fusion expression vectors
Generation of a soluble single-chain (sc) three-domain TCR construct, c264scTCR, has been described previously (Belmont et al., 2006). This protein recognizes the human p53 peptide (aa264–272) presented in the context of HLA-A*0201. Fusions between the c264scTCR domain and human IL-15 (hIL-15), hIL-15 N72D and D8N muteins or the sushi domain of human IL-15Rα (aa1–66 of hIL-15Rα; hIL-15RαSu) linked via a mutated human IgG1 hinge region were generated as described previously (Zhu et al., 2009). The hIL-15RαSu gene construct was further modified by the addition of sequence encoding a flexible linker and birA tag (GGLNDIFEAQKIEWHE) at the 3′end of the construct by PCR. The details of the linker sequences between the protein domains of the fusion proteins are provided in Supplementary data, Table S1.
Similar scTCR fusion constructs were generated from TCR α and β chain genes cloned from either H-2Kb-restricted, OVA (aa257–264) peptide-specific or HLA-A*0201 restricted, p53 (aa149–157) peptide-specific CTLs using SMART RACE cDNA Amplification Kit (Clonetech, Mountain View, CA). The resulting constructs encode an OVA peptide-specific scTCR, referred to as OT1scTCR, comprising murine TCR Vα linked to murine TCR Vβ–Cβ domains via a flexible peptide linker or a p53 (aa149–157) peptide-specific scTCR, referred to as c149scTCR, comprising a murine TCR Vα–linker–murine TCR Vβ–human TCR Cβ fusion. These scTCR genes were linked to the hIL-15 mutein and hIL-15RαSu/birA genes in a similar approach used for the c264scTCR fusions except the c149scTCR/hIL-15N72D construct did not contain the human IgG1 hinge region, which could be deleted without affecting IL-15 activity. The OT1scTCR gene was also fused to the birA tag sequence for generation of the OT1scTCR/birA fusion. To construct an OT1 TCRα/β heterodimer, the OT1 TCR Vα–Cα and Vβ–Cβ-coding regions were linked to the hIL-15 and hIL-15RαSu/birA genes, respectively, to create the OT1TCRα/hIL-15 and OT1TCRβ/hIL-15RαSu/birA genes. The 264TCRα/hIL-15D8N and 264TCRβ/hIL-15RαSu/birA genes were constructed using a similar approach. For the scCD8 fusions, the murine CD8α and CD8β genes were amplified using the SuperScript One-Step RT-PCR method (Invitrogen, Carlsbad, CA) from total RNA extracted from T cells obtained from C57BL/6 mice. A flexible linker sequence was inserted between the CD8α and CD8β chain genes and the scCD8 construct was fused to the hIL-15RαSu/birA gene. The resulting c264scTCR/hIL-15RαSu/birA, OT1scTCR/hIL-15RαSu/birA, OT1scTCR/hIL-15D8N, OT1TCRβ/hIL-15RαSu/birA and OT1scTCR/birA fusion genes were expressed in the neoR-based pNEF38 vector, whereas the OT1TCRα/hIL-15 fusion was expressed in the DHFR-based pDEF38 vector. The c149scTCR/hIL-15N72D, 264TCRα/hIL-15D8N, 264TCRβ/hIL-15RαSu/birA and scCD8/hIL-15RαSu/birA fusion genes were expressed in the modified pMSGV retroviral vector.
Fusion protein production and purification
Expression vectors containing the various fusion constructs were introduced into CHO cells by either transfection (pNEF38 and pDEF38 vectors) or transduction (pMSGV retroviral vectors). The retroviral particles carrying the pMSGV fusion constructs were generated by transfecting 293GP packaging cells as previously described (Yang et al., 2008). The scTCR fusion proteins were purified from recombinant CHO cell-culture supernatants by immunoaffinity chromatography, using the anti-human TCR Cβ mAb (BF1) 8A3.31 for c264scTCR fusion proteins and the anti-mouse TCR Cβ mAb H57-597 for OT1scTCR fusion proteins as described previously (Card et al., 2004; Belmont et al., 2006). The purified fusion proteins with birA tag were biotinylated with biotin ligase (Avidity, Denver, CO) in the presence of excess biotin, according to the manufacturer's instructions. The biotinylated fusion proteins were multimerized with SA-HRP and SA-PE for ELISA and flow cytometry, respectively.
The purified fusion proteins were analyzed by reducing SDS polyacrylamide gel electrophoresis (SDS-PAGE; 12% Bis Tris gel) followed by staining with SimplyBlue Safe Stain (Invitrogen). Formation of hIL-15:hIL-15RαSu dimers was characterized by size-exclusion chromatography (SEC). The purified protein samples (200 μl) were applied to Superdex 200HR 10/30 column (GE Healthcare, Piscataway, NJ) at concentration of 0.33 mg/ml in PBS. The samples were eluted at a flow rate 0.7 ml/min.
Preparation of peptide/MHC class I (pMHCI) tetramers
The murine H-2Kb gene was cloned from total RNA extracted from C57BL/6 mouse lymphocytes as described above. The extracellular region was ligated into the HLA-A*0201 heavy chain expression vector replacing the HLA-A*0201-coding sequence (Garboczi et al., 1992). The β2m, HLA-A*0201 and H-2Kb expression vectors were individually transformed into E.coli. The recombinant proteins were induced and expressed in insoluble inclusion bodies as described previously (Garboczi et al., 1992). The active and soluble proteins in complex with the peptides were obtained by the re-folding method described at http://www.microbiology.emory.edu/altman/jdaWebSite_v3/ptetPrepOverview.shtml. The p53 (aa264–272) and (aa149–157) peptide/HLA-A*0201 reagents are referred to as A2/p53.264–272 and A2/p53.149–157, respectively, and the OVA (aa257–264) peptide/H-2Kb is referred to as Kb/OVA.257–264.
ELISA
Immunoplates were coated with (BF1) 8A3.31 mAb for capturing c264scTCR fusion proteins or with H57–597 mAb for capturing OT1scTCR fusion proteins. After washing, the proteins were detected using various probes as detailed in the Results section. ABTS (2,2′-azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt) substrate was then added and absorbance was measured at 405 nm using a 96-well plate reader.
Flow cytometry
For characterization of the c264scTCR fusion protein complexes, T2 cells were pulsed with p53 (aa264–272) peptide at 37°C for 2 h in the presence of peptide-loading enhancer (PLE, Altor BioScience Corp., Miramar, FL). For the OT1scTCR fusion protein complexes, murine lymphoma EL4 cells were pulsed with OVA peptide at 100 μg/ml and PLE at 37°C for 6 h. The various birA fusion proteins (complexed with SA-PE) were added and incubated at 4°C for 1 h. The samples were washed two times and analyzed on an FACScan flow cytometer using CellQuest software (BD Biosciences, San Jose, CA). To assess IL-15 domain binding activity, 32Dβ cells were incubated with 320 nM of the c264scTCR fusion protein complexes for 30 min at 4°C. The binding of the proteins was in turn detected with biotinylated (BF1) 8A3.31 mAb for 15 min and SA-PE (5 μg/ml each) for 15 min. The stained cells were analyzed by flow cytometry as described above.
Cell proliferation assays
Cell proliferation was measured as previously described (Zhu et al., 2009). Briefly, 32Dβ cells (1 × 104 cells/well) were incubated with increasing concentrations of scTCR/hIL-15 or scTCR/hIL-15 muteins in the presence or absence of an equal molar concentration of scTCR/hIL-15RαSu for 48 h at 37°C. Cell proliferation reagent WST-1 (Roche Applied Science, Indianapolis, IN) was added during the last 4 h of cell growth according to the manufacturer's procedures. Conversion of WST-1 to the colored formazan dye by metabolically active cells was determined through absorbance measurements at 440 nm. The EC50 was determined with the dose–response curve generated from the experimental data by non-linear regression variable slope curve fitting with Prizm4 software (GraphPad Software, La Jolla, CA).
Surface plasmon resonance
The affinity constants of the OT1scTCR fusion proteins to their cognate pMHCI were determined using surface plasmon resonance (SPR) methodology on a BIAcore 2000 instrument (GE Healthcare). Biotinylated pMHCI complexes were immobilized onto the streptavidin-coated surface of an SA5 sensor chip (GE Healthcare) by injecting protein at 2 µg/ml in HBS buffer (10 mM HEPES, 150 mM NaCl, 3.4 mM EDTA, 0.005% P20 surfactant, pH 7.4) at a flow rate of 10 µl/min. This resulted in 1000–1200 RU of immobilized pMHCI complexes. The purified OT1scTCR fusion proteins were diluted to 1, 0.5 and 0.25 µM in HBS. Each concentration was injected once (50 µl) at a flow rate of 10 µl/min over a freshly immobilized pMHCI surface as well as over a control streptavidin surface blocked with biotin (baseline) and the binding curves were registered. The dissociation constant (KD) and the association (kon) and dissociation (koff) rates were calculated from the corrected binding curves (baseline subtracted) using the BIAevaluation 4.1.1 software (GE Healthcare).
Results
Creation of biologically active bivalent dimers using the hIL-15:hIL-15Rα scaffold
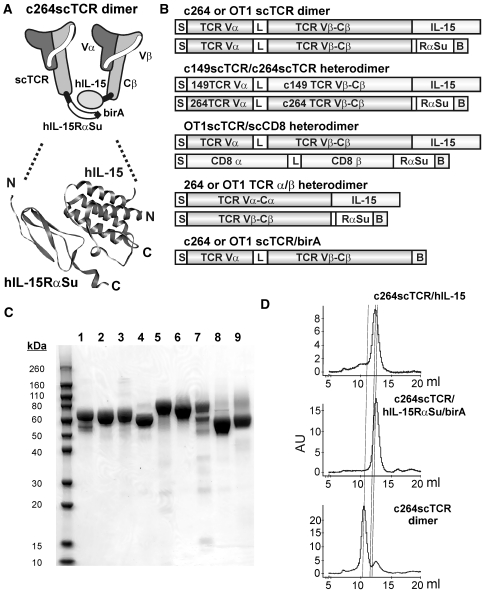
We have previously shown that a biologically active, bifunctional fusion protein, designated as c264scTCR/hIL-15, could be created by fusing the N-terminus of hIL-15 to a scTCR (c264scTCR; Zhu et al., 2009; Fig. 1A and B). In this construct, c264scTCR was used as a functional recognition domain based on its specific-binding activity to the p53 (aa264–272) peptide/HLA-A*0201 complex. We constructed a similar fusion protein with c264scTCR and the sushi-binding domain (aa1–66) of human IL-15Rα, which has been shown to contain the structural elements responsible for hIL-15 binding (Wei et al., 2001). This fusion protein was genetically linked at the C-terminus to a birA peptide tag to allow for biotinylation and subsequent multimerization in the presence of streptavidin (Zhu et al., 2006). This fusion protein is designated as c264scTCR/hIL-15RαSu/birA (Fig. 1A and B) and its expression and purification from CHO cells were similar to that of c264scTCR/hIL-15. These hIL-15RαSu/birA and hIL-15 fusion proteins (and others described below) could be readily produced and purified from cell-culture supernatants at the 2–8 mg/liter level without much efforts of cell line screening for high producers or process optimization. Additionally, the analysis by reducing SDS-PAGE indicated that the purified preparations predominantly consisted of a protein band that migrates at a molecular weight (MW) of 10–15 kDa greater than that calculated from the protein sequence, presumably due to glycosylation expected for these mammalian cell-produced proteins (Fig. 1C). Based on the high specific-binding activity between the hIL-15 and hIL-15RαSu domains, we anticipated that the fusion proteins could form a heterodimeric complex. In addition, examination of the crystal structure of the human IL-15:IL-15Rα complex indicated that the N-termini of the two proteins are at opposite ends of the complex ∼50 Å apart (Chirifu et al., 2007) (Fig. 1A). Hence, fusion of the scTCR domains to these regions is not expected to block complex formation. Initial evidence of binding between the c264scTCR/hIL-15 and c264scTCR/hIL-15RαSu/birA fusion proteins was observed in ELISAs using the plate-bound c264scTCR/hIL-15RαSu/birA to capture hIL-15 and c264scTCR/hIL-15 proteins (Zhu et al., 2009).
Fig. 1.
(A) Schematic diagram of the c264scTCR/hIL-15:c264scTCR/hIL-15RαSu/birA complex (c264scTCR dimer). The model of the dimeric hIL-15:hIL-15RαSu domains is based on the published crystal structure of the human IL-15:IL-15Rα complex (Chirifu et al., 2007; PDB 2Z3Q). (B) Schematic diagram of the hIL-15:hIL-15Rα fusion protein constructs described in this study: S, signal peptide; L, peptide linker; B, linker-birA tag sequence. Linker sequences between the protein domains of the fusion proteins are provided in Supplementary data, Table S1. (C) SDS-PAGE analysis of purified hIL-15 and hIL-15Rα fusion proteins under reducing conditions. Lanes: (1) c264scTCR/hIL-15, (2) c264scTCR/hIL-15N72D, (3) c264scTCR/hIL-15D8N, (4) c264scTCR/hIL-15RαSu/birA, (5) OT1scTCR/hIL-15D8N, (6) OT1scTCR/hIL-15RαSu/birA, (7) OT1scTCR/hIL-15D8N:scCD8αβ/hIL-15RαSu/birA complex, (8) OT1TCRα/hIL-15:OT1TCRβ/hIL-15RαSu/birA complex, and (9) OT1scTCR/birA. (D) SEC analysis of c264scTCR fusion proteins. Panels show comparative analysis of c264scTCR/hIL-15 (top), c264scTCR/hIL-5RαSu/birA (middle) and c264scTCR/hIL-15:c264scTCR/hIL-15RαSu/birA complex (c264scTCR dimer) (bottom) with dashed lines indicating relative protein peaks.
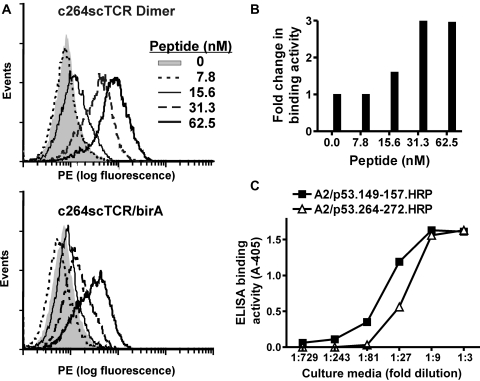
To further characterize the dimeric c264scTCR fusion protein complexes (referred to as c264scTCR dimer), equal molar amounts of purified c264scTCR/hIL-15 and c264scTCR/hIL-15RαSu/birA fusion proteins were mixed and allowed to associate at room temperature for more than 10 min. The complexes and the individual protein fusions were evaluated by SEC. As shown in Fig. 1D, the mixture of the two fusion proteins resulted in a major peak with an MW corresponding to dimeric form of the major species found in the individual fusion protein preparations. Thus, the appearance of the larger MW species in the c264scTCR dimer preparations is evidence that the heterodimeric complex has been generated. The c264scTCR dimer was compared with monomeric c264scTCR/birA protein for their ability to bind the TCR-specific antigen, p53 (aa264–272)/HLA-A*0201. In each case, the proteins were biotinylated with biotin ligase followed by complexing with SA-PE to generate multimeric flow cytometry staining reagents as previously described (Zhu et al., 2006). When used to stain HLA-A*0201-positive T2 cells pulsed with varying concentrations of p53 (aa264–272) peptide, both reagents exhibited antigen-specific binding that increased in a peptide concentration-dependent manner (Fig. 2A). However, the staining reagents comprising the c264scTCR dimer stained up to three times better than the monomer-derived c264scTCR/birA counterparts (Fig. 2B). These data suggest that dimerization through IL-15:IL-15Rα interaction preserves the functional activity of the scTCRs and increases the effective affinity of scTCR fusion complex to its cognate HLA/peptide through increased avidity. Similar results were observed when biotinylation via the birA tag was directed to the C-terminus of the scTCR/hIL-15 of the complex (data not shown). This demonstrates that the C termini of both the IL-15 and IL-15Rα domains are accessible to conjugation to molecular probes of significant size (MW of streptavidin is ∼60 kDa) without interfering with either the dimerization or antigen-binding domains of the fusion protein complex.
Fig. 2.
Characterization of c264scTCR dimer and c264scTCR/c149scTCR heterodimer binding activity. (A) T2 cells were pulsed with 0–62.5 nM of p53 (aa264–272) peptide as described in the section Materials and methods. The cells were stained with equivalent amounts (80 nM) of PE-conjugated multimers of the c264scTCR dimer or c264scTCR/birA. (B) The relative increase in cell staining comparing c264scTCR dimer with c264scTCR/birA reagents was determined at different peptide concentrations. Fold increase = (Geo mean of T2 cells stained by c264scTCR dimer)/(Geo Mean of T2 cells stained by c264scTCR/birA). (C) The p53 peptide/HLA-A*0201 binding activity of c264scTCR/c149scTCR heterodimer was determined by ELISA. Anti-hIL-15 mAb was used as a capturing reagent. A2/p53.264–272.HRP (open triangles) or A2/p53.149–157.HRP (closed squares) tetramers were used as the probes. The data represent the means ± SD of triplicate determinations. The results are representative of at least two independent assays.
These studies were extended to examine the possibility of generating bispecific molecules. A second scTCR (c149scTCR) was created, which recognizes an HLA-A*0201-restricted epitope of the human p53 protein spanning the amino acid residues of 149–157 (Theobald et al., 1995). The sequence encoding this scTCR fused to hIL-15 domain, designated as c149scTCR/hIL-15, was co-expressed in CHO cells with the c264scTCR/hIL-15αSu/birA fusion (Fig. 1B). The fusion complex generated in the supernatant of the recombinant CHO cell culture was immobilized using an anti-IL-15 Ab and probed either by HRP-labeled p53 (aa264–272) or p53 (aa149–157) peptide/HLA-A*0201 tetramers. As shown in Fig. 2C, the anti-IL-15 Ab captured fusion protein complex was able to bind both the peptide-loaded HLA tetramers. The result demonstrates that the individual scTCR molecules retain functional activity when fused to the hIL-15:hIL-15RαSu scaffold and the spatial arrangement of hIL-15:hIL-15RαSu complex does not significantly interfere with the packing of the scTCR domains, which have individual MWs of ∼40 kDa.
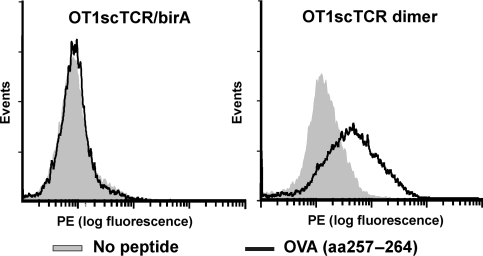
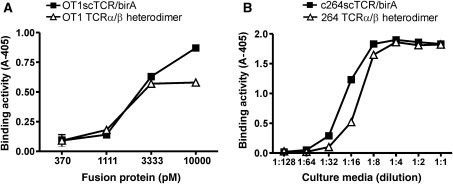
To demonstrate the broad utility of the hIL-15:hIL-15RαSu scaffold for protein dimerization, we created a second dimeric scTCR fusion complex by pairing two OT1scTCRs, one fused to the N-terminus of hIL-15 and another to the N-terminus of hIL-15RαSu/birA protein (Fig. 1B). OT1 is a well-characterized TCR recognizing an epitope of OVA protein spanning the amino acid residues 257–264 in the context of murine H-2Kb (Hogquist et al., 1994). OT1scTCR gene was generated and fused to either the hIL-15D8N or hIL-15RαSu/birA domain for recombinant CHO cell expression (Fig. 1C, lanes 5 and 6). The purified OT1scTCR fusion proteins were then mixed to form the OT1scTCR dimer protein complex. The individual fusions and protein complex were found to have pMHCI-binding activity in ELISA using anti-mouse TCR Cβ H57 Ab as a capture reagent and HRP-labeled, OVA (aa257–264) peptide-loaded H-2Kb tetramer as a probe (Supplementary data, Fig. S1). To distinguish the difference in binding activity between the OT1scTCR dimer and OT1scTCR/birA monomer, we conducted flow cytometry analysis similar to that described above for the c264scTCR dimers but with H-2Kb-positive EL4 cells loaded with OVA (aa257–264) peptide. As shown in Fig. 3, SA-PE tetramers comprising the OT1scTCR dimer indeed stained significantly better than those comprising monomeric OT1scTCR/birA fusions. We also performed surface plasmon resonance assays to assess the binding affinity of the OT1scTCR monomer and dimer against the biotinylated OVA (aa257–264) peptide-loaded H2-Kb/birA complexes immobilized on a streptavidin sensor chip. The apparent binding affinity (KD) of the OT1scTCR dimer to OVA peptide/H-2Kb complexes was estimated to be about 30 μM, whereas no binding was observed for the monomeric OT1scTCR/birA fusion protein (Table 1). These data confirm that dimerization through hIL-15:hIL-15Rα interaction preserves the biological activity of the scTCRs and increases the effective affinity of the scTCR molecule to its cognate pMHCI complexes through increased avidity.
Fig. 3.
Characterization of OT1scTCR dimer binding activity. EL4 cells were loaded with OVA (aa257–264) peptide and stained with OT1scTCR/birA-SA-PE (left) and OT1scTCR dimer-SA-PE (right) at 200 nM.
Table I.
OT1scTCR fusion protein binding kinetics and affinity values determined by SPR
| Analyte | Immobilized ligand | kon (M−1 s−1) | koff (s−1) | KD (μM) |
|---|---|---|---|---|
| OT1scTCR/birA (monomer) | OVA-(257–264)/H-2Kb | No binding | ||
| OT1scTCR dimer | OVA-(257–264)/H-2Kb | 8.9 × 102 | 0.029 | 32 |
| OT1scTCR dimer | VSV/H-2Kb | No binding | ||
| OT1scTCR/scCD8 heterodimer | OVA-(257–264)/H-2Kb | 1.0 × 104 | 0.027 | 2.6 |
| OT1scTCR/scCD8 heterodimer | VSV/H-2Kb | No binding | ||
Creation of a biologically active heterodimer
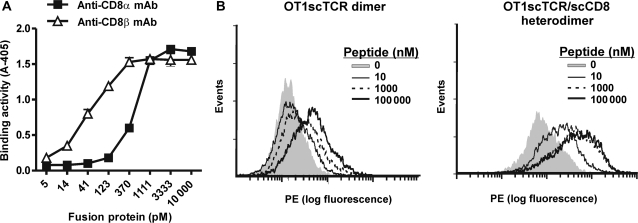
Since the CD8 molecule has been previously demonstrated to play a pivotal role in the interaction between OT1 TCR and its cognate OVA peptide/H2-Kb complex (Schott and Ploegh, 2002), the hIL-15:hIL-15RαSu scaffold provides an opportunity to assess whether the CD8 molecule enhances OT1 TCR binding affinity to OVA peptide/H-2Kb expressed on the cell surface and under cell-free and adhesion molecule-free conditions. To achieve this, we first created a murine CD8 molecule in sc format (scCD8) by fusing the extracellular domains of the α and β chains of the murine CD8 using a flexible linker. This fusion gene was fused to the hIL-15RαSu/birA construct in a retroviral expression vector. Recombinant retrovirus was then used to infect a CHO cell line expressing the OT1scTCR/hIL-15D8N fusion protein in order to create an scTCR/scCD8 heterodimeric complex (Fig. 1B). The fusion protein complex was purified from the supernatant of the cultured recombinant CHO cells using the anti-TCR Ab-based affinity chromatography as described above. SDS-PAGE analysis of the purified protein complex revealed protein bands of the appropriate MW for the OT1scTCR/hIL-15D8N (calculated MW 56 kDa) and scCD8/hIL-15RαSu/birA fusions (calculated MW 48 kDa; Fig. 1C, lane 7). The protein complex was subjected to ELISA using anti-TCR Ab as the capture reagent and either the biotinylated anti-mCD8α or anti-mCD8β mAbs as probes. As shown in Fig. 4A, the anti-TCR Ab-immobilized fusion complex contains both the CD8α and CD8β and, thus, indicates formation of an OT1scTCR/scCD8 heterodimer. We used flow cytometry analysis to compare the binding activity of the OT1scTCR/scCD8 heterodimer with the OT1scTCR dimer to varying amounts of OVA peptide/H-2Kb complexes displayed on the cell surface. As shown in Fig. 4B, SA-PE staining reagents comprising the OT1scTCR/scCD8 heterodimer could readily detect OVA peptide/H-2Kb complexes on EL4 cells loaded with as little as 10 ng/ml OVA peptide, whereas little or no staining was observed at this peptide concentration when comparable reagents comprising the OT1scTCR dimer were used. Higher background OT1scTCR/scCD8 heterodimer staining was observed on EL4 cells that were not pulsed with peptide, suggesting peptide-independent interactions were occurring between the CD8 domain and MHC molecules on the cell surface. Similar effects have been reported for pMHCI tetramers binding to CD8 molecules expressed on T cells (Neveu et al., 2006).
Fig. 4.
OTscTCR/scCD8 heterodimer exhibits enhanced pMHCI binding activity. (A) Murine CD8 expression of OT1scTCR/scCD8 heterodimer was determined by ELISA. Anti-mTCR H57-597 mAb was used as a capturing reagent. The biotinylated anti-murine CD8α (closed squares) or CD8β (open triangles) mAb was used as a probe followed by SA-HRP. The data represent the means ± SD of triplicate determinations. The results are representative of at least two independent assays. (B) EL4 cells were loaded with OVA (aa257–264) peptide at the indicated concentration and stained with OT1scTCR dimer-SA-PE (left) and OT1scTCR/scCD8 heterodimer-SA-PE (right) at 200 nM.
The results for peptide-specific interactions of the OT1scTCR/scCD8 heterodimer were further confirmed by surface plasmon resonance analysis. The binding affinity (KD) of the OT1scTCR/scCD8 heterodimer to OVA peptide/H-2Kb complexes was estimated to be 2.6 μM, which is significantly higher than the ∼30 μM observed for the OT1scTCR dimer (Table 1, Supplementary data, Fig. S2). Neither fusion protein showed any binding to control VSV peptide/H-2Kb complexes. The difference in specific pMHCI-binding activity is surprising given that the bivalent nature of the OT1scTCR dimer is expected to provide increased functional affinity in this assay format. Additionally, similar SPR binding studies conducted with soluble TCR, CD8 α/β and pMHCI proteins as independent components showed only weak interactions (KD 30–100 μM) between CD8 protein and peptide/H-2Kb complexes and no apparent cooperative effects of CD8 on TCR:peptide/H-2Kb interactions (Kern et al., 1999; Arcaro et al., 2001). Taken together, these data indicate that the addition of the CD8 α/β domain to the OT1scTCR fusion has a greater impact on pMHCI binding than creation of the bivalent OT1scTCR molecule.
Creation of functional two-chain heterodimers
As indicated above, the N-termini of the hIL-15 and hIL-15Rα domains are at distal ends of the complex raising questions as to whether this scaffold is suitable for fusions to polypeptides of a multi-chain protein. To determine whether a soluble, biologically active, heterodimeric TCR α/β could be constructed using the hIL-15 and hIL-15RαSu scaffold, the C-terminal ends of the extracellular OT1 TCR Vα–Cα and Vβ–Cβ domains were linked to the N-termini of hIL-15 and hIL-15RαSu/birA chains, respectively. Based on the published α/β TCR crystal structures, the TCR Cα and Cβ C-terminal amino acids of the properly folded OT1 TCR α/β molecule are expected to be ∼18 Å apart (Garboczi et al., 1996). The OT1 TCRα/hIL-15 and OT1 TCRβ/hIL-15RαSu/birA fusion genes were cloned into two separate expression vectors and co-transfected into CHO cells to generate a heterodimeric TCR α/β complex (Fig. 1B). The secreted fusion protein complex was purified using anti-TCR Cβ mAb affinity chromatography as described above. When analyzed by SDS-PAGE under reducing condition, the purified protein bands migrated at 50–60 kDa, consistent with the calculated monomeric MW (40 kDa) of each of the two fusion molecules (Fig. 1C, lane 8). The purified protein was further characterized in the functional ELISA (anti-TCR Cβ mAb capture: OVA peptide/H2-Kb tetramer probe). As shown in Fig. 5A, the purified protein was found to have equivalent pMHCI-binding activity as OT1 TCR in the sc format. Similar results were observed for hIL-15:hIL-15RαSu/birA fusions to the Vα–Cα and Vβ–Cβ chains of the p53-specific 264 TCR (Fig. 5B). Previous attempts to produce soluble α/β TCR heterodimers in mammalian cells have been largely unsuccessful (Traunecker et al., 1989; Lin et al., 1990). Thus, our results suggest that the TCR α and β chains were appropriately folded through the association of the hIL-15 and hIL-15RαSu/birA domains within the transfected cells. Intriguingly, the fusion to N-termini of the hIL-15:hIL-15RαSu scaffold is able to provide the spatial arrangement sufficient for functionally independent binding domains as observed with the c264scTCR/c149scTCR heterodimeric complex while retaining flexibility to permit folding of closely paired chains such as the α and β chains of OT1 TCR and 264 TCR.
Fig. 5.
Fusion proteins containing TCR α/β heterodimers retain pMHCI binding activity. (A) Binding activity of OT1scTCR/birA (closed squares) and OT1 TCRα/β heterodimer (open triangles) to OVA (aa257–264)/H-2Kb complex was determined by ELISA. Anti-mTCR H57-597 mAb was used as a capturing reagent. Kb/OVA.257-264.HRP tetramer was used as a probe. (B) Binding activity of 264scTCR/birA (closed squares) and 264 TCRα/β heterodimer (open triangles) to p53 (aa264–272)/HLA-A*0201 complex was determined by ELISA. Anti-TCR mAb was used as a capturing reagent. A2/p53.264–272.HRP tetramer was used as a probe. The data represent the means ± SD of triplicate determinations. The results are representative of at least two independent assays.
Biological activity of the hIL-15 domain for the hIL-15:hIL-15RαSu fusion complexes
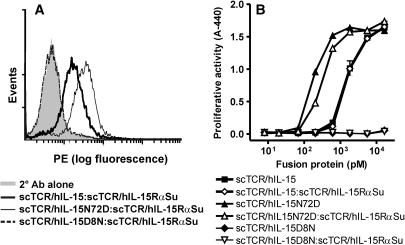
The IL-15 receptor (IL-15RβγC) binding capability of the hIL-15:hIL-15Rα domain of the c264scTCR dimer was evaluated by flow cytometry analysis using 32Dβ cells, which carry the hIL-15Rβ and the murine γC (mγC) chains. These studies were carried out using c264scTCR dimers containing the wild-type hIL-15 domain, as well as dimers with hIL-15 mutein domains previously shown to enhance (N72D) or reduce (D8N) binding to the hIL-15Rβ chain without affecting formation of the hIL-15:hIL-15RαSu complex (Zhu et al., 2009). Following incubation with the c264scTCR dimers, the 32Dβ cells were stained with anti-TCR mAb to detect cell-bound fusion protein dimers. As shown in Fig. 6A, the 32Dβ cells were stained positively by the c264scTCR dimers containing hIL-15 wild-type or hIL-15N72D domains but not with those containing the hIL-15D8N domain, indicating that the IL-15:hIL-15RαSu portion of the complex retains the expected IL-15RβγC binding activity.
Fig. 6.
IL-15 binding and functional activity of fusion protiens. (A) 32Dβ cells were incubated with 320 nM of the c264scTCR dimers comprising IL-15 wild type or IL-15N72D or IL-15D8N mutein domains. The binding of the fusion proteins was in turn detected with anti-human TCR Cβ Ab. (B) The ability of the c264scTCR dimers comprising IL-15 wild-type or mutein domains to support proliferation of 32Dβ cells was determined as described in the section Materials and methods. The data represent the means ± range of duplicate determinations.
The hIL-15 biological activity of the fusion protein dimers were also examined in cell proliferation assays using 32Dβ cells. As shown in Fig. 6B, the hIL-15 wild-type domain in the scTCR/hIL-15 fusion or scTCR/hIL-15:scTCR/hIL-15RαSu fusion complex was able to support the growth of 32Dβ cells in a concentration-dependent manner, exhibiting half-maximal stimulation (EC50) at ∼300 pM. The hIL-15N72D or D8N domains either increased or eliminated the biological activity of the fusion proteins, respectively, regardless of whether they were present in the monomeric or dimeric fusions. These results are consistent with the functional activity observed for non-fusion IL-15 cytokine carrying the N72D or D8N mutations (Zhu et al., 2009). Thus, formation of the fusion protein complex containing two independent TCR domains does not significantly change the biological activity of the IL-15 domain. In contrast, there was at least a 3-fold loss of IL-15 activity for the OT1 TCRα/β heterodimer complex (data not shown), suggesting that formation of the heterodimeric TCR structure inhibits, to some extent, the ability of the hIL-15 domain to interact with hIL-15RβmγC. Additionally, these results indicate that the hIL-15 domain can be readily manipulated to allow enhanced or reduced receptor binding and functional activity, thus providing additional flexibility for the use of the hIL-15:hIL-15RαSu scaffold in different applications.
Discussion
In these studies, we demonstrate the potential uses of a hIL-15:hIL-15RαSu-based scaffold to create novel, dimeric targeting molecules. The dimeric fusion protein complexes retained immunostimulatory effects and binding activity of their hIL-15 domains and target-specific recognition domains, respectively. These results indicate that the addition of hIL-15 and hIL-15Rα did not significantly alter the spatial arrangement of the fusion domains and provided an adequate degree of conformational flexibility without eliminating cytokine activity. Thus, this scaffold could be used to form multivalent fusion complexes, such as the c264scTCR dimer, to increase the overall binding affinity of molecules or bispecific molecules, such as the c264scTCR/c149scTCR heterodimer. In all cases, the soluble fusion proteins were produced at relatively high levels in recombinant CHO cell culture (up to 8 mg per liter purified from cell-culture supernatant without extensive cell line screening or process optimization) and could be readily purified from the cell-culture supernatants. We also demonstrated that the utility of the hIL-15:hIL-15RαSu-based scaffold could be expanded to create soluble, biologically active, two-chain molecules, such as α/β TCR heterodimers, by fusing the extracellular domains of the two polypeptide chains to the N-termini of hIL-15 and hIL-15RαSu. This format resulted in a moderate decrease in hIL-15 activity, possibly due to steric hindrance between the interfolded TCR α/β chains (fused at distal ends of the hIL-15:hIL-15RαSu complex) and the hIL-15RβγC-binding site located in the middle of the complex. We are currently assessing the activity of additional heterodimeric complexes and different linker formats to further optimize the biological activity of these fusions.
We have previously shown in xenograft tumor models that tumor antigen-specific scTCR-IgG dimers exhibit potent antitumor activity via IgG effector activity (Mosquera et al., 2005). The results presented here indicate that the hIL-15:hIL-15RαSu scaffold could be exploited much like the IgG scaffold to generate multivalent or multispecific targeted therapeutics. With its potent activity for stimulating proliferation and activation of effector NK and CD8+ memory T cells, the hIL-15 domain expands the scope of immunotherapeutic mechanisms beyond antibody-dependent cellular cytotoxicity and complement activation associated with IgG-based approaches. Based on its immunostimulatory activity, IL-15 has been recently viewed as the most promising anti-cancer candidate among 12 immunotherapy drugs (Cheever, 2008). Moreover, disease-targeting approaches, such as those using Ab or TCR recognition domains, could further enhance therapeutic potential of IL-15 while limiting systemic toxicities. Indeed, it has been shown that a fusion protein composed of IL-15 linked to an anti-fibronectin scAb exhibits targeted, CD8 T cell-mediated antitumor activity against both subcutaneous and metastatic tumors in immunocompetent murine models (Kaspar et al., 2007).
Employing approaches similar to those previously used to manipulate the IgG Fc domain activity, we have also found that the IL-15 domain can be mutated to increase or decrease its functional activity. We have shown that hIL-15:hIL-15RαSu fusion molecule containing an N72D mutation in the IL-15 domain exhibit a 3–4-fold increase in biological activity, whereas IL-15 D8N mutation exhibit little or no activity (Zhu et al., 2009). While IL-15 superagonist-based fusion proteins could serve as targeted immunotherapeutics for cancer and infectious diseases, an IL-15 antagonist capable of inhibiting IL-15 responsive cells at the disease site may have therapeutic potential in treating allograft rejection and inflammatory autoimmune diseases, particularly if memory CD8 T cells play a role in disease pathology (Waldmann, 2006). A non-targeted IL-15 mutant/Fcγ2a antagonist protein has already been shown to be effective at inhibiting islet and cardiac allograft rejection and preventing development and progression of arthritis in experimental animal models (Ferrari-Lacraz et al., 2004; Zheng et al., 2006). We are investigating similar approaches with IL-15 antagonist domains in the context of the hIL-15:hIL-15RαSu fusion proteins. In addition, under certain circumstances, it may be desirable to have a functionally inert scaffold for generation of multimeric molecules. For example, we have found that scTCR/hIL-15:scTCR/hIL-15RαSu fusions containing an IL-15 D8N mutation, which eliminates interactions with IL-15Rβγ, provide better TCR antigen-specific staining of cells displaying IL-15 receptor complex.
Additionally, we demonstrate that the hIL-15:hIL-15RαSu scaffold could be used to create heterodimer comprising CD8 α/β and TCR domains capable of binding the same pMHCI complex but at a spatially distinct sites. Previous studies using soluble pMHCI reagents have determined that CD8 stabilizes and enhances TCR:pMHCI interactions at the cell surface through effects on both the off-rate and the on-rate (Arcaro et al., 2001; Gakamsky et al., 2005; Laugel et al., 2007). However, several binding studies using soluble purified CD8 α/β, TCR and pMHCI proteins have shown that TCR:pMHCI interactions are not affected by the presence or absence of CD8, suggesting no cooperative binding effects (Arcaro et al., 2001; Cole et al., 2008). The results of our cell-based and SPR binding studies with the OT1scTCR/scCD8 heterodimer are in contrast with these earlier reports in showing that TCR and CD8 domains displayed on the same soluble molecule exhibited much better pMHC binding activity than was observed with molecules carrying monovalent or divalent TCR domains. This effect is reflected in a faster on-rate of the pMHCI:OT1scTCR/scCD8 heterodimer complex, consistent with the observations for pMHCI binding to CD8 and TCR molecules on T cells (Gakamsky et al., 2005). Thus, the OT1scTCR/scCD8 heterodimer mimics binding of the OT1 TCR on T cells, which exhibits a strong dependence of CD8 coreceptor activity for pMHC interactions. These results indicate that the scTCR/scCD8 heterodimer and variants of this molecule could serve as very useful tools for further dissecting molecular interactions between the tertiary TCR:pMHCI:CD8 complex in a cell-free system.
The results of our SPR experiments on the OT1scTCR fusions differ from those reported by Alam et al. where the binding affinity of monovalent OT1 TCRα/β heterodimer to immobilized OVA peptide/H-2Kb complex was shown to be ∼6 μM (Alam et al., 1999). In our studies, we were unable to detect OVA peptide/H-2Kb binding of the OT1scTCR/birA monomer and the OT1scTCR dimer exhibited an apparent KD of 30 μM. It is possible that the OT1 TCR exhibited decreased binding activity when formatted as a sc Vα–linker–Vβ–Cβ molecule. However, we observed equivalent activity when comparing OT1scTCR/birA and a two-chain construct. Moreover, previous studies have shown that OVA peptide/H-2Kb tetramers with Kb mutations that abrogate CD8 binding exhibit little or no specific binding activity to OT1 TCR-bearing cells whereas tetramers without these mutations brightly stain OT1 TCR cells. These results are consistent with the very low binding affinity between OT1 TCR and its cognate pMHCI and the CD8 dependence effects on these interactions observed in our studies.
Although we have focused on TCRs and CD8 molecules as target-recognition domains for demonstration purposes in this study, it is conceivable that the hIL-15:hIL-15RαSu scaffold could be used to construct other novel binding molecules with protein domains derived from Abs, adhesion molecules or other receptors. It may also be possible to create protein domain fusions to the C-termini of the hIL-15 and hIL-15RαSu which, based on the crystal structure, are accessible for modification. The resulting molecules could contain up to four different target-recognition capabilities. With the appropriate fusion partners, these types of molecules could promote the conjugation of immune effectors cells and target cells and achieve effective killing of target cells. As noted above, the IL-15 domain of the complex could further augment these processes by providing immunostimulatory activity to support effector cell proliferation and cytotoxicity. Currently, we have constructed a variety of multifunctional molecules based on this concept and are in the process of characterizing them as anti-cancer and anti-viral immunotherapeutic agents.
Previously, the poor expression level in standard mammalian cell system limited the development of recombinant hIL-15 as a therapeutic (Ward et al., 2009). In this study, we found that expression of scTCR/hIL-15:scTCR/hIL-15RαSu complexes is at levels capable of supporting clinical development and potentially product commercialization. In addition, it has been shown that the IL-15Rα chain enhances the in vivo activity of hIL-15, possibly by improving the pharmacokinetics of the cytokine (Rubinstein et al., 2006; Stoklasek et al., 2006; Dubois et al., 2008; Epardaud et al., 2008). These two characteristics of hIL-15:hIL-15RαSu complexes, in combination with its multivalent nature and/or multispecific targeting design, may provide an opportunity to capture the full potential of hIL-15 as an immunotherapeutic agent against cancer and viral infections.
Supplementary data
Supplementary data are available at PEDS online.
Conflict of interest statement
Bai Liu, Xiaoyun Zhu, Lijing You, Lin Kong, Kai-ping Han, Hyung-il Lee, Pierre-Andre Chavaillaz, Peter R. Rhode, Hing C. Wong were employees and equity holders of Altor BioScience Corp. during the course of this study.
Funding
This work was supported in part by funding from the National Institutes of Health [R43 CA139810 to P.R.R.].
Acknowledgement
We would like to thank Dr Dean Taylor for his thoughtful review of this manuscript.
References
- Alam S.M., Davies G.M., Lin C.M., Zal T., Nasholds W., Jameson S.C., Hogquist K.A., Gascoigne N.R., Travers P.J. Immunity. 1999;10:227–237. doi: 10.1016/s1074-7613(00)80023-0. [DOI] [PubMed] [Google Scholar]
- Arcaro A., Gregoire C., Bakker T.R., et al. J. Exp. Med. 2001;194:1485–1495. doi: 10.1084/jem.194.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle P.A., Kufer P., Bargou R. Curr. Opin. Mol. Ther. 2009;11:22–30. [PubMed] [Google Scholar]
- Belmont H.J., Price-Schiavi S., Liu B., et al. Clin. Immunol. 2006;121:29–39. doi: 10.1016/j.clim.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Bergamaschi C., Rosati M., Jalah R., Valentin A., Kulkarni V., Alicea C., Zhang G.M., Patel V., Felber B.K., Pavlakis G.N. J. Biol. Chem. 2008;283:4189–4199. doi: 10.1074/jbc.M705725200. [DOI] [PubMed] [Google Scholar]
- Card K.F., Price-Schiavi S., Liu B., et al. Cancer Immunol. Immunother. 2004;53:345–357. doi: 10.1007/s00262-003-0450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever M.A. Immunol. Rev. 2008;222:357–368. doi: 10.1111/j.1600-065X.2008.00604.x. [DOI] [PubMed] [Google Scholar]
- Chirifu M., Hayashi C., Nakamura T., Toma S., Shuto T., Kai H., Yamagata Y., Davis S.J., Ikemizu S. Nat. Immunol. 2007;8:1001–1007. doi: 10.1038/ni1492. [DOI] [PubMed] [Google Scholar]
- Cole D.K., Dunn S.M., Sami M., Boulter J.M., Jakobsen B.K., Sewell A.K. Mol. Immunol. 2008;45:2700–2709. doi: 10.1016/j.molimm.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Cuesta A.M., Sanchez-Martin D., Sanz L., Bonet J., Compte M., Kremer L., Blanco F.J., Oliva B., Alvarez-Vallina L. PLoS One. 2009;4:e5381. doi: 10.1371/journal.pone.0005381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kruif J., Logtenberg T. J. Biol. Chem. 1996;271:7630–7634. doi: 10.1074/jbc.271.13.7630. [DOI] [PubMed] [Google Scholar]
- Deer J.R., Allison D.S. Biotechnol. Prog. 2004;20:880–889. doi: 10.1021/bp034383r. [DOI] [PubMed] [Google Scholar]
- Dubois S., Patel H.J., Zhang M., Waldmann T.A., Muller J.R. J. Immunol. 2008;180:2099–2106. doi: 10.4049/jimmunol.180.4.2099. [DOI] [PubMed] [Google Scholar]
- Epardaud M., Elpek K.G., Rubinstein M.P., Yonekura A.R., Bellemare-Pelletier A., Bronson R., Hamerman J.A., Goldrath A.W., Turley S.J. Cancer Res. 2008;68:2972–2983. doi: 10.1158/0008-5472.CAN-08-0045. [DOI] [PubMed] [Google Scholar]
- Ferrari-Lacraz S., Zanelli E., Neuberg M., et al. J. Immunol. 2004;173:5818–5826. doi: 10.4049/jimmunol.173.9.5818. [DOI] [PubMed] [Google Scholar]
- Gakamsky D.M., Luescher I.F., Pramanik A., Kopito R.B., Lemonnier F., Vogel H., Rigler R., Pecht I. Biophys. J. 2005;89:2121–2133. doi: 10.1529/biophysj.105.061671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garboczi D.N., Hung D.T., Wiley D.C. PNAS. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garboczi D.N., Ghosh P., Utz U., Fan Q.R., Biddison W.E., Wiley D.C. Nature. 1996;384:134–141. [PubMed] [Google Scholar]
- Hayden-Ledbetter M.S., Cerveny C.G., Espling E., et al. Clin. Cancer Res. 2009;15:2739–2746. doi: 10.1158/1078-0432.CCR-08-1694. [DOI] [PubMed] [Google Scholar]
- Hogquist K.A., Jameson S.C., Heath W.R., Howard J.L., Bevan M.J., Carbone F.R. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Holliger P., Hudson P.J. Nat. Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- Kaspar M., Trachsel E., Neri D. Cancer Res. 2007;67:4940–4948. doi: 10.1158/0008-5472.CAN-07-0283. [DOI] [PubMed] [Google Scholar]
- Kern P., Hussey R.E., Spoerl R., Reinherz E.L., Chang H.-C. J. Biol. Chem. 1999;274:27237–27243. doi: 10.1074/jbc.274.38.27237. [DOI] [PubMed] [Google Scholar]
- Kostelny S., Cole M., Tso J. J. Immunol. 1992;148:1547–1553. [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. Nature. 1988;336:646–651. doi: 10.1038/336646a0. [DOI] [PubMed] [Google Scholar]
- Kubetzko S., Balic E., Waibel R., Zangemeister-Wittke U., Pluckthun A. J. Biol. Chem. 2006;281:35186–35201. doi: 10.1074/jbc.M604127200. [DOI] [PubMed] [Google Scholar]
- Laugel B., van den Berg H.A., Gostick E., Cole D.K., Wooldridge L., Boulter J., Milicic A., Price D.A., Sewell A.K. J. Biol. Chem. 2007;282:23799–23810. doi: 10.1074/jbc.M700976200. [DOI] [PubMed] [Google Scholar]
- Lin A., Devaux B., Green A., Sagerstrom C., Elliott J., Davis M. Science. 1990;249:677–679. doi: 10.1126/science.1696397. [DOI] [PubMed] [Google Scholar]
- Lu D., Zhu Z. Methods Mol. Biol. 2009;525:377–404. doi: 10.1007/978-1-59745-554-1_20. xiv. [DOI] [PubMed] [Google Scholar]
- Mohler K., Torrance D., Smith C., Goodwin R., Stremler K., Fung V., Madani H., Widmer M. J. Immunol. 1993;151:1548–1561. [PubMed] [Google Scholar]
- Mortier E., Quemener A., Vusio P., Lorenzen I., Boublik Y., Grotzinger J., Plet A., Jacques Y. J. Biol. Chem. 2006;281:1612–1619. doi: 10.1074/jbc.M508624200. [DOI] [PubMed] [Google Scholar]
- Mosquera L.A., Card K.F., Price-Schiavi S.A., et al. J. Immunol. 2005;174:4381–4388. doi: 10.4049/jimmunol.174.7.4381. [DOI] [PubMed] [Google Scholar]
- Neveu B., Echasserieau K., Hill T., Kuus-Reichel K., Houssaint E., Bonneville M., Saulquin X. Int. Immunol. 2006;18:1139–1145. doi: 10.1093/intimm/dxl048. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F., Ravetch J.V. Nat. Rev. Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- Rieker J.D., Hu J.C. Methods Enzymol. 2000;328:282–296. doi: 10.1016/s0076-6879(00)28403-6. [DOI] [PubMed] [Google Scholar]
- Rubinstein M.P., Kovar M., Purton J.F., Cho J.H., Boyman O., Surh C.D., Sprent J. Proc. Natl. Acad Sci. USA. 2006;103:9166–9171. doi: 10.1073/pnas.0600240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott E., Ploegh H.L. Eur. J. Immunol. 2002;32:3425–3434. doi: 10.1002/1521-4141(200212)32:12<3425::AID-IMMU3425>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Shen J., Vil M.D., Jimenez X., Iacolina M., Zhang H., Zhu Z. J. Biol. Chem. 2006;281:10706–10714. doi: 10.1074/jbc.M513415200. [DOI] [PubMed] [Google Scholar]
- Sloan V.S., Cameron P., Porter G., Gammon M., Amaya M., Mellins E., Zaller D.M. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- Stern L.J., Wiley D.C. Cell. 1992;68:465–477. doi: 10.1016/0092-8674(92)90184-e. [DOI] [PubMed] [Google Scholar]
- Stoklasek T.A., Schluns K.S., Lefrancois L. J. Immunol. 2006;177:6072–6080. doi: 10.4049/jimmunol.177.9.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald M., Biggs J., Dittmer D., Levine A.J., Sherman L.A. Proc. Natl. Acad Sci. USA. 1995;92:11993–11997. doi: 10.1073/pnas.92.26.11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traunecker A., Dolder B., Oliveri F., Karjalainen K. Immunol. Today. 1989;10:29–32. doi: 10.1016/0167-5699(89)90062-5. [DOI] [PubMed] [Google Scholar]
- Waldmann T.A. Nat. Rev. Immunol. 2006;6:595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- Ward A., Anderson M., Craggs R.I., Maltby J., Grahames C., Davies R.A., Finch D., Pattison D., Oakes H., Mallinder P.R. Protein Expr. Purif. 2009;68:42–48. doi: 10.1016/j.pep.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Wei X.-Q., Orchardson M., Gracie J.A., Leung B.P., Gao B.-M., Guan H., Niedbala W., Paterson G.K., McInnes I.B., Liew F.Y. J. Immunol. 2001;167:277–282. doi: 10.4049/jimmunol.167.1.277. [DOI] [PubMed] [Google Scholar]
- Weiner L.M. Nat. Rev. Cancer. 2007;7:701–706. doi: 10.1038/nrc2209. [DOI] [PubMed] [Google Scholar]
- Wen J., Zhu X., Liu B., You L., Kong L., Lee H.I., Han K.P., Wong J.L., Rhode P.R., Wong H.C. Cancer Immunol. Immunother. 2008;57:1781–1794. doi: 10.1007/s00262-008-0504-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Rosenberg S.A., Morgan R.A. J. Immunother. 2008;31:830–839. doi: 10.1097/CJI.0b013e31818817c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Bennett A.D., Zheng Z., et al. J. Immunol. 2007;179:5845–5854. doi: 10.4049/jimmunol.179.9.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X.X., Gao W., Donskoy E., Neuberg M., Ruediger M., Strom T.B., Moll T. Transplantation. 2006;81:109–116. doi: 10.1097/01.tp.0000188139.11931.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Belmont H.J., Price-Schiavi S., Liu B., Lee H.I., Fernandez M., Wong R.L., Builes J., Rhode P.R., Wong H.C. J. Immunol. 2006;176:3223–3232. doi: 10.4049/jimmunol.176.5.3223. [DOI] [PubMed] [Google Scholar]
- Zhu X., Marcus W.D., Xu W., Lee H.I., Han K., Egan J.O., Yovandich J.L., Rhode P.R., Wong H.C. J. Immunol. 2009;183:3598–3607. doi: 10.4049/jimmunol.0901244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.