Abstract
Adolescent idiopathic scoliosis (AIS) is an unexplained and common spinal deformity seen in otherwise healthy children. Its pathophysiology is poorly understood despite intensive investigation. Although genetic underpinnings are clear, replicated susceptibility loci that could provide insight into etiology have not been forthcoming. To address these issues, we performed genome-wide association studies (GWAS) of ∼327 000 single nucleotide polymorphisms (SNPs) in 419 AIS families. We found strongest evidence of association with chromosome 3p26.3 SNPs in the proximity of the CHL1 gene (P < 8 × 10−8 for rs1400180). We genotyped additional chromosome 3p26.3 SNPs and tested replication in two follow-up case–control cohorts, obtaining strongest results when all three cohorts were combined (rs10510181 odds ratio = 1.49, 95% confidence interval = 1.29–1.73, P = 2.58 × 10−8), but these were not confirmed in a separate GWAS. CHL1 is of interest, as it encodes an axon guidance protein related to Robo3. Mutations in the Robo3 protein cause horizontal gaze palsy with progressive scoliosis (HGPPS), a rare disease marked by severe scoliosis. Other top associations in our GWAS were with SNPs in the DSCAM gene encoding an axon guidance protein in the same structural class with Chl1 and Robo3. We additionally found AIS associations with loci in CNTNAP2, supporting a previous study linking this gene with AIS. Cntnap2 is also of functional interest, as it interacts directly with L1 and Robo class proteins and participates in axon pathfinding. Our results suggest the relevance of axon guidance pathways in AIS susceptibility, although these findings require further study, particularly given the apparent genetic heterogeneity in this disease.
INTRODUCTION
Adolescent idiopathic scoliosis (AIS) is the most common pediatric spinal deformity, affecting ∼3% of school age children worldwide. Through unknown mechanisms, the vertebrae become misaligned such that the spine or a segment of the spine deviates from the midline laterally in the coronal plane, is rotated axially toward the side of deviation, and in the sagittal plane may deviate anteriorly (a lordotic deviation) or posteriorly (kyphotic deviation). AIS occurs in otherwise healthy children who bear no obvious deficiencies in the components of the spinal column itself, and the onset is highly correlated with the adolescent growth spurt (1). School screenings are recommended due to the morbidity and rapid onset of disease (2). Intervention is required in cases where the spinal deformity continues to worsen, so-called progressive disease. A first approach is bracing with an orthotic device that is designed to limit progression by holding the torso erect (3). However, in extreme cases (∼1% of patients), bracing will not halt the progression of the deformity, and patients risk significant deformity with possible cardiopulmonary compromise. Treatment in these cases typically requires surgical intervention involving spinal instrumentation with fusion of selected vertebrae.
The etiology of AIS is poorly understood as is implied by the name (4). Indeed, although at least superficially it is a disease of the spinal column, the underlying pathophysiology is not clear. Lesions of the bony composition of the vertebrae, the vertebral endplates, the paraspinous muscles or the neurologic system each have been proposed at various times to explain disease pathogenesis. However, observational studies have had limited power in discerning whether these features are secondary to AIS or causative (5). Progress in this regard has been hampered by the lack of an obvious AIS animal model. Many vertebrate strains (i.e. mouse and zebrafish) have been described with deformities of the spine or tail (6–9). Although it is anticipated that an appropriate vertebrate model will be invaluable for elucidating disease pathogenesis, at present it is not clear which if any of these animals recapitulates the patho-etiology of human AIS.
Genetic contributions to AIS have been described for decades (10). About one quarter of AIS patients report a positive family history of disease, and twin studies have consistently supported shared genetic factors in disease (reviewed in 4,5). Although extended families are described, the genetic architecture of AIS appears to be generally complex. Accordingly, genome-wide linkage studies of multiplex families have identified many chromosomal regions of interest, but replication is reported for only one linkage peak, on chromosome 9q31.2–34.2 (11,12). Likewise, replicated causal genes or variants are also lacking. We previously performed targeted linkage studies in a cohort of 53 AIS multiplex families, and in follow-up studies we found association with haplotypes in the CHD7 gene encoded on chromosome 8q. Rare mutations in CHD7 are responsible for the CHARGE (coloboma of the eye, heart defects, atresia of the choanae, retardation of growth and/or development, genital and/or urinary abnormalities, and ear abnormalities and deafness) syndrome of multiple anomalies that can include scoliosis (13). From this study, we concluded that other genes responsible for rare Mendelian syndromes involving scoliosis might contribute to AIS, and that genome-wide studies of common variation in AIS cohorts would be fruitful in identifying susceptibility loci (14).
In the present study, we searched more comprehensively for common AIS risk loci by performing genome-wide association studies (GWAS) in 419 Texas families. We identified top loci of interest using family-based tests of association that are robust to population stratification. We then tested replication of our most significant findings in additional independent cohorts of cases and controls ascertained in Texas and elsewhere in the USA. To our knowledge, this is the first reported GWAS for AIS. We provide genome-wide data and our top 100 single nucleotide polymorphism (SNP) associations so that others may replicate our findings. Our results provide candidate loci and genes worthy of further study, and particularly underscore genes involved in axon guidance pathways in AIS susceptibility.
RESULTS
Probands, other affected family members and parents were ascertained primarily in orthopedic clinics in Dallas, Texas and St Louis, Missouri (see Materials and Methods and Supplementary Material, Table S1 for a description of study populations). Samples from Texas AIS family trios (probands and parental controls) were genotyped using the Illumina HumanCNV370-quad platform that interrogates over 370 000 human polymorphisms. After applying stringent quality control, we performed tests of transmission disequilibrium (TDT) for 326 498 SNPs using the PLINK analysis program (15) in two ways (see Materials and Methods for description of statistical methods). In the first analysis, genotypes for all 419 families (n = 1122) of all self-reported ethnicities were used, as the TDT statistic is robust to population stratification (16). The second analysis was restricted to the subset of self-reported non-Hispanic white families, our largest ethnic group. To define the latter group, we corrected possible stratification by performing identity-by-state (IBS) analysis of unrelated probands using PLINK. Plotting the first three dimensions of a multidimensional scaling analysis of pairwise IBS distances identified one outlier family that was removed from further analyses (Supplementary Material, Fig. S1). A plot of resulting −2log(e) P-values against expected results under the null hypothesis (quantile–quantile plot, Fig. 1) suggested a modest excess of associations without evidence of stratification (λGC = 1.0025) within the non-Hispanic white cohort. TDT results for the two data sets are shown in the form of Manhattan plots in Figures 2 and 3. We also examined cryptic relatedness that could produce overly inflated results. Using pairwise inheritance-by-descent (IBD) estimation, we did not detect closely related pairs, with πhat values <0.15 for all samples (see Materials and Methods).
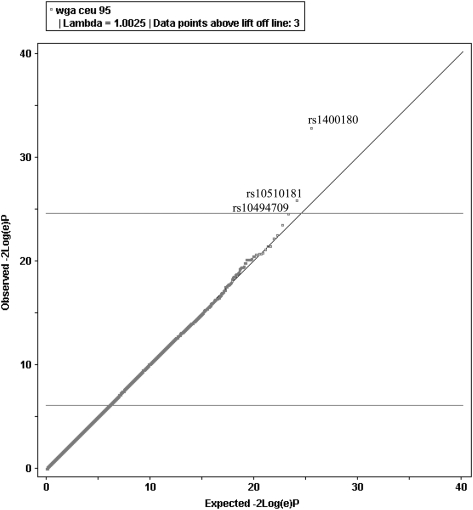
Figure 1.
Quantile–quantile (Q–Q) plot of actual versus expected −2log(e)P under the null hypothesis for a genome-wide data in non-Hispanic white probands. This plot was constructed using WGAViewer software (49). The plot suggests departure from the null (λGC = 1.0025), with three SNPs (labeled in black) exceeding lift-off.
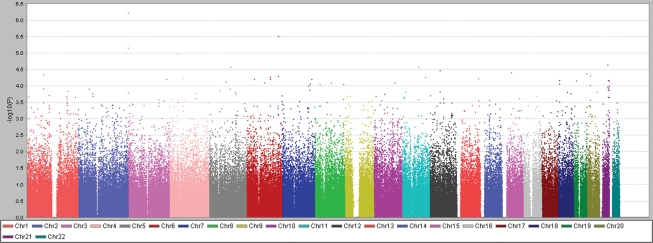
Figure 2.
Scatterplot (plotted using Haploview) of SNP −log10 P-values (Y-axis) obtained by TDT analyses in 419 Texas AIS family trios versus autosomal position (X-axis). P-values for SNPs on same chromosomes are represented in same color.
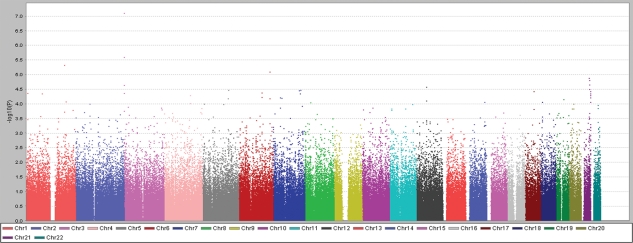
Figure 3.
Scatterplot (plotted using Haploview) of SNP −log10 P-values (Y-axis) obtained by TDT analyses in 339 (340 families—one outlier) non-Hispanic white AIS trios versus autosomal position (X-axis). P-values for SNPs on same chromosomes are represented in same color.
We estimated that our discovery cohort would provide (dependent on allele frequencies) ∼90% power to detect disease associations with effect sizes [odds ratios (ORs)] ≥ 2.0 at a significance level P = 5.0 × 10−8 (Supplementary Material, Fig. S2a), and 70% power to detect loci with an effect size of 1.8, but only 10% power to detect weaker effect sizes of 1.5 or less at P = 5.0 × 10−8 (17). However, this cohort was potentially enriched for genetic risk factors, as 21% of the cases were familial. Only three SNPs met or exceeded a significance threshold P ≤ 1 × 10−5; however, genomic clustering suggested non-random association. Specifically, we noted that in the total data set, our most significant result was obtained for the SNP rs1400180 [OR = 1.92, 95% confidence interval (CI) = 1.48–2.49; P = 6.2 × 10−7], with nearby SNP rs10510181 among the top-ranked SNPs (OR = 1.88, 95% CI = 1.42–2.49; P = 7.1 × 10−6). These two SNPs are within 21 kb of each other at distal chromosome 3p and were ranked highest in the non-Hispanic white data set (Fig. 3 and Supplementary Material, Table S2). The evidence for association for the chromosome 3 SNPs rs1400180 (OR = 2.13; P = 7.9 × 10−8) and rs10510181 (OR = 2.03; P = 2.6 × 10−6) increased in the non-Hispanic white cohort despite the fact that this subset contained 80 fewer families than the total (results for non-white families are given in Supplementary Material, Table S2).
We also imputed genotypes at untyped loci to potentially increase genome-wide coverage, given the relatively low density of the CNV370-quad platform. We imputed 2 271 581 genotypes in the non-Hispanic white families and tested these SNPs for association using the TDT statistic in PLINK as before. This analysis produced additional signals of interest (in terms of clustering and P-values), in particular for loci on chromosomes 1, 6 and 21 (Supplementary Material, Fig. S3). However, SNPs clustering in the region of rs1400180 and rs10510181 remained the most significant by imputation (Supplementary Material, Tables S3 and S4 and Fig. S4), and we prioritized this region for further study (Fig. 4).
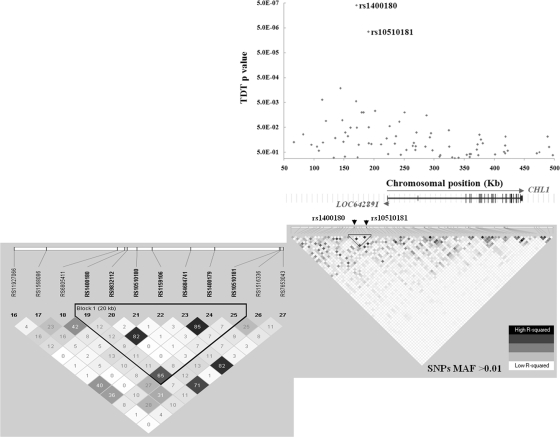
Figure 4.
TDT results and LD structure for SNPs in the chromosome 3p26.3 (50 000–500 000 bp) region. Results are shown relative to physical locations as denoted in assembly GRCh37, build 37.1. Shown are TDT P-values plotted versus chromosomal location for SNPs present on the Illumina HumanCNV370-quad chip from the region in 419 discovery set families. The respective locations of the two genes CHL1 (long arrow) and hypothetical gene LOC642891 (short arrowhead) are also shown. Below are the pairwise r2 values for SNPs in the region represented graphically, where darker squares indicate greater correlation between SNPs. Only SNPs with MAF > 0.01 are shown. The inverted triangle highlights the region including the top two SNPs rs1400180 and rs10510181. The call out box is the same region magnified to better show the respective r2 values.
We subsequently tested SNPs rs1400180 and rs10510181 for evidence of allelic association with AIS in additional independent cohorts. In the first replication study, we genotyped samples from 375 Texas cases of non-Hispanic white ethnicity as defined by self-report as well as by IBS analyses using genotypes from 384 ancestry-informative markers (as described above and Materials and Methods, Supplementary Material, Fig. S5). Close relationship within or between this cohort and the discovery cohort was unlikely per extensive review of pedigree and demographic information. For both SNPs, we observed that the same allele that was overtransmitted in families was over-represented in cases when compared with controls (i.e. the same direction of effect). Strongest results were obtained for rs10510181, where the frequency in cases was 0.37 when compared with 0.32 in controls. In addition, a logistic regression analysis incorporating gender and age at onset as covariates yielded essentially the same results (Table 1). In the second replication study, we genotyped samples from 187 cases ascertained in US clinics outside of Texas and 222 controls, and again observed the same direction of effect for both SNPs and strongest results for SNP rs10510181 (Table 1). Combining the results of the two replication studies (562 cases, 666 controls) yielded OR = 1.36, 95% CI = 1.14–1.61, P = 0.0005 for rs10510181.
Table 1.
AIS risk association results for chromosome 3p26.3 loci
| Discovery |
Rep 1 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All families (419) | White, non-Hispanic families (339) | Dallas (375:444) | |||||||||||
| Marker | Location | Risk allele | T:U | P-value | ORallelic (95% CI) | T:U | P-value | ORallelic (95% CI) | Risk allele Ca:Co | P-value | ORallelic (95% CI) | P-value* | OR* (95% CI) |
| rs1400180 | 170,968 | C | 165:86 | 6.15 × 10−7 | 1.92 (1.48–2.49) | 151:71 | 7.91 × 10−8 | 2.13 (1.60–2.82) | 0.46:0.41 | 0.067 | 1.21 (0.98–1.47) | 0.045 | 1.22 (1.0–1.49) |
| rs9754850 | 190,290 | T | ND | ND | ND | 120:80 | 0.0047 | 1.5 (1.13–1.99) | 0.49:0.43 | 0.022 | 1.26 (1.03–1.54) | 0.013 | 1.3 (1.06–1.6) |
| rs9754552 | 190,411 | A | ND | ND | ND | 120:80 | 0.0047 | 1.5 (1.13–1.99) | 0.49:0.44 | 0.031 | 1.25 (1.02–1.52) | 0.018 | 1.29 (1.04–1.59) |
| rs10510181 | 191,047 | A | 141:75 | 7.10 × 10−6 | 1.88 (1.42–2.49) | 128:63 | 2.56 × 10−6 | 2.03 (1.50–2.75) | 0.37:0.32 | 0.018 | 1.29 (1.04–1.59) | 0.013 | 1.3 (1.06–1.62) |
| Rep 2 | Rep 3 | Combined | |||||||||||
| St Louis/USA (187:222) | CHOP (137:2126) | Discovery white non-Hispanic + Rep1 + Rep2 | Discovery white non-Hispanic + Rep1 + Rep2 + Rep3 | ||||||||||
| Marker |
Location |
Risk allele |
Risk allele Ca:Co |
P-value |
ORallelic (95% CI) |
Risk allele Ca:Co |
P-value |
PFisher |
ORallelic (95% CI) |
PFisher |
ORMeta (95% CI) |
||
| rs1400180 | 170,968 | C | 0.43:0.41 | 0.56 | 1.09 (0.82–1.45) | 0.40:0.44 | 0.25 | 6.35 × 10−8 | 1.37 (1.19–1.58) | 1.33 × 10−6 | 1.30 (1.14–1.48) | ||
| rs9754850 | 190,290 | T | 0.51:0.44 | 0.044 | 1.35 (1.01–1.80) | 0.45:0.44 | 0.82 | 1.50 × 10−4 | 1.34 (1.16–1.54) | 1.56 × 10−3 | 1.29 (1.13–1.47) | ||
| rs9754552 | 190,411 | A | 0.51:0.44 | 0.049 | 1.34 (1.00–1.79) | ND | ND | 2.28 × 10−4 | 1.33 (1.15–1.53) | ND | ND | ||
| rs10510181 | 191,047 | A | 0.38:0.30 | 0.021 | 1.42 (1.05–1.92) | 0.31:0.33 | 0.44 | 2.58 × 10−8 | 1.49 (1.29–1.73) | 8.22 × 10−7 | 1.37 (1.20–1.58) | ||
Location, base pair position of SNP (Build GRCh37); T:U = transmitted:untransmitted count; Rep, replication cohort; Ca:Co, case:control risk allele frequencies; P-value, P-value assuming a central chi-square distribution with one degree of freedom and a statistic value equal to TDT-CHISQ; P*-value, P-value adjusted for gender, age and four dimension values c1, c2, c3 and c4 from MDS by logistic regression; OR, odds ratio; OR*, odds ratio adjusted for gender, age and four dimension values c1, c2, c3 and c4 from MDS by logistic regression; 95% CI, 95% confidence interval for ORX; PFisher, Fisher-combined P-values.
Two overlapping genes, CHL1 and LOC642891, are nearest the region of association that we observed and are predicted to be transcribed in opposite orientation. CHL1 encodes Close Homologue of L1, a member of the family of immunoglobulin-class L1 neural cell adhesion molecules. Chl1 functions in axonal guidance and neuronal migration (18,19); however, whether LOC642891 encodes a functional protein is unknown. We confirmed transcription of both genes in fetal and adult brain (Supplementary Material, Fig. S6 and S7). The CHL1 gene spans >212 kb and its putative promoter sequences are ∼45 kb distal to the associated haplotype. The predicted LOC642891 gene spans ∼1.5 kb, where its 5′-end lies within CHL1 intron 1 and its 3′-end lies within the putative CHL1 promoter (Fig. 4). Forty-nine imputed SNPs from this region (50–500 kb at chromosome 3p26.3) including the CHL1 and LOC642891 genes produced P-values < 9 × 10−4 by TDT analysis (Supplementary Material, Table S4) of discovery data. To validate these findings, we selected four imputed SNPs (rs965084, rs1400182, rs9754850 and rs9754552) near rs1400180 and rs10510181 for genotyping in the discovery and Rep1 cohorts (Supplementary Material, Tables S4 and S5). SNPs rs9754850 and rs9754552 yielded evidence for association (P < 0.05) with the same direction of effect as in the discovery phase, but SNPs rs965084 and rs1400182 did not. SNPs rs9754850 and rs9754552 are within 800 bp of rs10510181 and are moderately correlated with this SNP: r2 (rs9754850: rs10510181) = 0.56, 0.57, and r2 (rs9754552: rs10510181) = 0.56, 0.58, respectively, in case–control and discovery cohorts. We obtained similar results for rs9754850 and rs9754552 in the Rep2 cohort (Table 1).
In a third replication study (Rep 3), we compared our data to a separate GWAS of 137 AIS cases and 2126 controls ascertained at Children's Hospital of Philadelphia (CHOP). Allele frequencies for SNPs rs1400180, rs10510181 and rs9754850 did not differ in cases compared with controls in that study (Table 1). To assess potential bias in our locally ascertained controls, we examined SNP risk allele frequencies in available data sets. We found that risk allele frequencies for the four SNPs rs1400180, rs9754850, rs9754552 and rs10510181 were similar between our controls and four control data sets (total n = 3917 individuals), but not HapMap CEU, a difference we attribute to the relative few chromosomes represented in the HapMap data set (n = 60) (Supplementary Material, Table S6).
Taken together, these results suggest that a genomic region correlated with SNP rs10510181, proximal to the CHL1 and LOC642891 genes, is associated with increased AIS risk. The lack of replication in the CHOP GWAS may reflect issues of heterogeneity and power to detect modest effect sizes.
Many prior observations have indirectly linked scoliosis and neuropathology (4). Clear evidence that improper axonal targeting specifically can evoke scoliosis is evident in the rare autosomal recessive disease horizontal gaze palsy with progressive scoliosis (HGPPS, MIM #607313) that is remarkable for absent horizontal eye movements and severe progressive scoliosis. This disease is caused by homozygous or compound heterozygous mutations in the ROBO3 gene encoding a transmembrane receptor that controls commissural axon guidance (20). Brain imaging and neurophysiologic studies of HGPPS patients have revealed hindbrain anomalies and improper motor and sensory axonal projections (21). We have noted with interest that Chl1 and Robo3 proteins belong to the same molecular (immunoglobulin transmembrane receptor) and functional (axon guidance and neurite outgrowth) classes. Thus, CHL1 is a plausible candidate gene for AIS susceptibility. We selected 90 families having multiple members affected with AIS for analysis of the CHL1 gene, with the rationale that such families could be more likely to harbor highly penetrant alleles (22). Analysis of four markers in the region produced suggestive evidence for linkage (HLOD = 1.93, P = 0.001 at rs1400180) (Supplementary Material, Table S7). We re-sequenced coding exons and flanking intronic regions of the CHL1 gene in 10 unrelated probands from AIS families with positive evidence of linkage to the region (LOD ≥ 1.0). We observed two coding changes that predict non-synonymous amino acid changes (rs2272522 and rs62230378) and are found in exons 3 and 17, respectively, of the CHL1 gene. We noted with interest that SNP rs2272522 was previously associated with susceptibility to schizophrenia in separate studies of Japanese and Han Chinese populations (23,24). This SNP was actually present on the CNV370-quad beadchip and was informative in our population, but did not produce evidence for association with AIS. Further investigation of rs62230378 also did not yield evidence that this SNP was associated with AIS. We also observed nine non-coding variants, none of which predicted alterations of known functional elements, such as transcription factor binding sites or consensus splice sites (Materials and Methods, Supplementary Material, Table S8).
We observed additional associations in our discovery data with SNPs in axon guidance genes. Among the top results were several SNPs clustering in the DSCAM gene located on chromosome 21 within the Down syndrome critical region (OR = 0.56, 95% CI = 0.42–0.73; P = 2.26 × 10−5 for rs2222973) (see top 100 SNP associations, Supplementary Material, Table S9, and Figs 2 and 3). DSCAM encodes Down syndrome cell adhesion molecule, an immunoglobulin-class neural cell adhesion molecule in the same molecular class with Chl1 and Robo3. Dscam likewise functions in axon guidance, including commissural axon pathfinding, as observed in both vertebrate and invertebrate model systems (25,26). We also noted rs11770843 in the CNTNAP2 gene within our top-associated SNPs (OR = 1.75, 95% CI = 1.32–2.30; P = 6.20 × 10−5) and some evidence for association with nearby loci, although SNP coverage in this region was poor (Supplementary Material, Table S10). This could be a random effect, given the size of the CNTNAP2 gene (2.3 Mb) and the number tests that we performed (297 SNPs from the CNTNAP2 gene were genotyped). However, the evidence for association with rs11770843 remained significant after correction for the 297 tests. CNTNAP2 is of interest, as this gene was previously linked with AIS (27). The extent of overlap with our data is unclear, but we conclude that further study of CNTNAP2 in AIS cohorts is warranted. CNTNAP2 encodes contactin-associated protein 2, also called neurexin IV, that binds to the immunoglobulin-class neural cell adhesion molecule contactin-2 in cis and to the L1 family of neural cell adhesion molecules (possibly including Chl1) in trans (28). In Drosophila, neurexin IV was recently shown to interact with Robo (29). These data provide additional evidence for variation in CNTNAP2 in AIS susceptibility. These and other top findings from our GWAS warrant further exploration in additional AIS cohorts.
DISCUSSION
AIS has been clinically recognized for centuries, yet the underlying molecular etiology and indeed the pathophysiology of the disease have remained elusive. We report here the first GWAS of AIS, which we conducted with the goal of identifying promising candidate loci for further analysis. We also anticipated that such loci could enlighten potential disease mechanisms. Our analyses yielded top results for a region of chromosome 3p26.3 associated with AIS risk in the proximity of the axon guidance gene CHL1. CHL1 is a plausible candidate gene, given the prior evidence that compound heterozygous mutations in the structurally and functionally related gene ROBO3 cause an extreme scoliosis phenotype, horizontal gaze palsy with progressive scoliosis (20). We did not detect obvious causal variation within CHL1 itself, but further re-sequencing in larger cohorts is needed to test this possibility. It is interesting to note that previous association studies have identified CHL1 as a candidate gene for schizophrenia (23,24). The SNP specifically associated in these studies was not associated with AIS in our study, suggesting the possibility of distinct CHL1-mediated pathways in the two disorders. While we propose CHL1 as a candidate gene in AIS susceptibility, we note that our experiments have shown LOC642891 expression in the brain, and we cannot exclude a role for this gene in disease risk. The physical overlap of CHL1 and LOC642891 on chromosome 3p26.3 also suggests the interesting possibility of correlated expression (30).
Our results suggest that variants in other immunoglobulin-class neural cell adhesion molecules and their interacting partners may contribute to AIS susceptibility. Specifically, candidate genes identified in this study underscore mechanisms of commissural axon guidance, i.e. axon crossing at the midline of the corticospinal tract. Under normal conditions, longitudinal axons are known to respond to attractive and repulsive cues at specified locations (31). However, when such molecular cues are disrupted, as in the case of ROBO3 mutations in HGPPS, axons may display aberrant growth and directionality. Clearly further study of the candidate genes and loci proposed in this study are needed to confirm causal roles. In this regard, we recently replicated association with SNPs in DSCAM in a second cohort of Texas AIS families (rs2222973 combined OR = 0.59, 95% CI = 0.48–0.74; P = 1.46 × 10−6). The role of Dscam in axon guidance and neurite outgrowth is well described, and on commissural axons it acts as a receptor for the attractive guidance molecule netrin (25,26). Dscam knockdowns in zebrafish embryos exhibit severe shortening of the anterior/posterior axis and, interestingly, partial knockdowns produce embryos with crooked tails (32). Our data also provide additional evidence that variation in the CNTNAP2 gene encoding neurexin IV contributes to AIS susceptibility. Neurexin IV is clearly involved in axon guidance pathways via its interactions with contactin and L1 neural cell adhesion molecules, and with Robo in a pathway of repulsive midline axon guidance (29).
The possibility of neuropathology underlying AIS has stemmed from several observations. Scoliosis is frequent in neurologic/neuromuscular diseases, including Duchenne's muscular dystrophy, spinal muscular atrophy etc. Spinal cord anomalies such as syrinx, a fluid-filled cavity in the spinal cord (or brainstem), is often detected in conjunction with scoliosis (4,5). Several investigations of AIS patients have suggested co-existing deficits in oculo-vestibular (visual/hearing) and proprioceptive function. As an example, one study measuring otolith vestibulo-ocular response in AIS found significant left–right asymmetry in horizontal eye movements in AIS patients compared with matched healthy control children; moreover, the average of horizontal, but not vertical, eye movements was significantly different (greater) in AIS patients than in controls (33–37). These data might suggest a disease continuum, with HGPPS at the extreme end of AIS clinical manifestations. How scoliosis develops in this disease is not clear, although secondary abrogation of muscle tone and locomotion has been hypothesized (20).
We also examined loci in our data set corresponding to previously published linkage peaks. Within the 9q31.2–34.2 interval, we observed modest associations clustering in four regions, with strongest results for the following SNPs (genes): rs4979321 (ZNF618; P = 0.0009), rs891725 (AMBP; P = 0.0003), rs1969944 (PALM2; P = 0.0005) and rs4836643 (intergenic; P = 0.0003). We also searched genomic intervals encoding the ROBO3 and CHD7 genes but did not observe significant associations, suggesting that common variation in these genes does not contribute significantly to overall disease risk in AIS populations. Complete re-sequencing of both genes in large AIS cohorts may be fruitful to test the possibility that they harbor rare disease-causing variation.
One conclusion from our GWAS is that no single locus contributes predominantly to AIS risk. This was not surprising, as such signals should have been detected in prior studies. Our data also suggested that genetic heterogeneity is a significant factor in AIS, illustrated in our replication studies of chromosome 3p candidate loci. Further studies in larger cohorts are needed to identify and confirm additional AIS associations with common variants. Our results suggest genomic regions where follow-up genotyping as well as deep re-sequencing to detect causal alleles in AIS cohorts may be fruitful.
MATERIALS AND METHODS
Study subjects
All affected subjects considered for inclusion in the study met criteria for a positive diagnosis of idiopathic scoliosis: lateral deviation from the midline greater than 10° as measured by the Cobb angle method from standing spinal radiographs, axial rotation toward the side of the deviation and exclusion of relevant co-existing diagnoses. Control study subjects were parents (for TDT analyses) or unrelated individuals without history of scoliosis (for case/control analyses). Clinical details are summarized in Supplementary Material, Table S1.
Ascertainment of Dallas, Texas study subjects
All participating research subjects were recruited under a protocol approved by the University of Texas Southwestern Medical Center Institutional Review Board. Affected probands included in discovery and initial replication studies were ascertained in orthopedic clinics at Texas Scottish Rite Hospital for Children (TSRHC) as previously described (14). Parents, and other affected family members where possible, were routinely ascertained as well. A pediatric orthopedic surgeon confirmed a positive diagnosis of AIS by exclusion of co-existing diagnoses and positive findings from standing spinal radiographs. Blood samples were obtained by venipuncture. In some cases, saliva samples were self-collected using the Oragene DNA kit (DNA Genotek, Inc.). Four hundred nineteen probands and parents of all self-reported ethnicities were included in the discovery phase as detailed below. This cohort, as reported for the proband, included 81% white non-Hispanic, 9% black non-Hispanic, 6% Hispanic/Latino, 1% Asian/Pacific Islander and 3% other. Three hundred seventy-six affected probands were similarly ascertained in orthopedic clinics at TSRHC and included in the first replication (Rep1 cohort).
Ascertainment of St Louis, Missouri study subjects
All study subjects were recruited under protocols approved by either Washington University or the St Louis Shriners Hospital for Children Institutional Review Board. Affected probands were ascertained by a single pediatric orthopedic surgeon (M.B.D.) at St Louis Shriners Hospital for Children. Non-hispanic white St Louis study subjects (n = 174) were included in the Rep2 cohort.
USA study subjects
Additional probands were ascertained by pediatric orthopedic surgeons at the time of treatment at Shriners Hospital for Children, Lexington, KY, USA (T. Milbrandt, V. Talwalkar, H. J. Iwinski); Hasbro Childrens' Hospital, Providence, RI, USA (C. P. Eberson); University of Massachusetts Memorial Medical Center, Worcester, MA, USA (A. Lapinsky); Childrens Hospital of Wisconsin, Milwaukee, WI, USA (J. C. Tassone, X. C. Liu) and Akron Children's Hospital, Akron, OH, USA (W. Schrader). All study subjects were recruited according to the protocols approved by local corresponding ethics boards or the University of Texas Southwestern Medical Center Institutional Review Board. USA study subjects (n = 13) were included in the Rep2 cohort (Supplementary Material, Table S1).
Controls
Control individuals were ascertained from within the local Texas population or non-orthopedic clinics at TSRHC. A diagnosis of scoliosis was excluded by questionnaire. Six hundred sixty-six of these were of self-reported white, non-Hispanic ethnicity. These individuals were randomly assigned as either Rep1 (444 individuals) or Rep2 (222 individuals) controls in proportion to the number of cases in each group.
We also obtained genotypes for three publicly available control data sets: (i) National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) IBDGC Crohn's Disease Genome-Wide Association Study; (ii) National Institute of Neurological Disorders and Stroke (NINDS) Genome Wide Association in Familial Parkinson Disease; and (iii) European American controls from the New York Health project Intragen Population Genetics database (38). For the purposes of this study, we refer to these as the ‘NIDDK', ‘NINDS' and ‘Intragen' controls. NIDDK controls consisted of 895 individuals of non-Jewish, European ancestry that were genotyped with the Illumina HumanHap300 Beadchip and were without history of inflammatory bowel disease. The NINDS controls consisted of 943 Caucasian, non-Hispanic individuals that were genotyped with the Illumina HumanCNV370 Beadchip and were negative for neurological disease by self-report. The data retrieved from IntraGen Population Genetics Database (IntraGenDB) consisted of 478 individuals between the ages of 30 and 60, of non-Jewish white ancestry, genotyped with the Illumina HumanHap300 genotyping platform.
Discovery genotyping and quality assurance
We genotyped 1128 samples from 422 families (probands and parents) in the discovery phase. The majority of DNA samples were derived from blood; 66 (5.4%) of DNA samples were derived from saliva. Each DNA sample was quantified in triplicate with the Quanti-iT PicoGreen dsDNA reagent (Invitrogen) and assessed for quality by gel electrophoresis prior to genotyping. Each DNA sample (250–375 ng) was subsequently genotyped on Illumina Human CNV370-Quad arrays (Illumina). Genotypes with quality metrics (‘GenCall scores') > 0.15 were called. For the remaining SNPs, the average success rate for each SNP was 99.20%. To test reproducibility of SNP calls, we genotyped 42 samples (3.54% of the total) in duplicate on separate arrays. The average concordance for the 42 samples was >99.99%. We then filtered individual samples and SNPs that failed the following quality control measures. Three families displayed mis-inheritances and their samples were excluded from analysis. For the remaining 419 families (1122 samples), the percent of SNPs called successfully was >95% in all but 9 samples (<1%). These nine samples were removed from further analysis, leaving 1113 samples with an average SNP success rate >0.998. We note that the SNP success rate was 100% for both rs1400180 and rs10510181 after quality control. Individual SNPs were further pruned for low call rates (<0.95), deviation from Hardy–Weinberg equilibrium (P < 0.00001), and minor allele frequency (MAF) < 0.01. In total, 326 498 autosomal SNPs were retained for further analyses.
Statistical analyses in the discovery cohort
We used the SNP genotypes in 1113 samples that remained after quality control (as described above) in statistical analyses. We performed statistical analyses using software within the BC/SNPmax database platform (Biocomputing Platforms Ltd.).
IBS and multidimensional scaling
We searched for evidence of population stratification in self-reported non-Hispanic white probands of our discovery set using IBS analysis as implemented in PLINK (15). For this purpose, we included autosomal SNPs present on the Illumina Human 370-quad chip and in HapMap release 23a for which we downloaded genotype data for 210 unrelated HapMap samples (60 CEU, 45 HCB, 45 JPT and 60 YRI) (www.hapmap.org) (39). Of these, one of each SNP pair with pairwise r2 > 0.5 was pruned, as were SNPs with call rate <99%, MAF < 0.01 and a possible disease association (P < 0.05, observed in 419 Texas AIS family trios) that left 8297 SNPs. Dimensions based on IBS distances in non-Hispanic white probands from the discovery set and HapMap samples were calculated, and multidimensional scaling (MDS) plots were made using the first three dimensions. Outliers (1 out of the 340 families) were identified from the discovery set and removed from further analyses (Supplementary Material, Fig. S1).
IBD analyses and πhat estimation
We calculated pairwise IBD coefficients using PLINK to assess cryptic relatedness between probands. SNPs were selected using the same criteria as for IBS analyses. We required πhat < 0.15 for each sample pair to exclude cryptic relationship (on the order of second cousins in an outbred population). All proband pairs passed the πhat < 0.15 threshold, with πhat = 0.072 being the highest value observed.
TDT analyses in the discovery set
TDT results, displayed as Manhattan plots in Figures 2 and 3, were computed using the methods implemented in the PLINK software (15). We also computed TDT results using the TDTae statistic that is robust to genotyping errors and missing genotypes (40). These results are given in Supplementary Material, Table S8. Pedigrees were subsequently assigned to one of two groups, ‘non-Hispanic white' (n = 339 after removal of outlier family), or ‘non-white' (n = 79), and TDT was computed for each in the same manner as above. We also performed a genome-wide TDT analysis in the non-Hispanic black trios, our largest non-white group. The resultant P-values for each of the two markers on chromosome 3 are presented in Supplementary Material, Table S2. None of the neighboring markers (within 500 kb) in these two groups showed TDT P-values less than 0.01 (data not shown).
Power analyses
Power analyses were performed for the discovery cohort as described in the text. We also calculated power provided by our replication cohorts for P = 0.05 (41) (Supplementary Material, Fig. S2a and b).
Imputation
To predict individual genotypes at un-typed loci, we used statistics contained in the software application MaCH 1.0 (42), applying HAPMAP reference haplotype release #22 data for population CEU. Genome-wide genotypes were imputed for the 339 non-Hispanic white families and analyzed using the TDT in PLINK as before (Supplementary Material, Tables S3 and S4 and Figs S3 and S4).
Statistical analyses in the replication cohorts
In our first replication study, we evaluated the possibility of biases due to population stratification using multidimensional scaling analysis of IBS distances as previously described. For this, we designed a panel of 384 ancestry-informative markers selected from Illumina databases and published literature (43). We selected autosomal SNPs >100 kb apart and present on the Illumina CNV370 beadchip (a full list of SNPs is available upon request). As a positive control, we performed IBS analyses in the discovery set utilizing these 384 SNP genotypes (obtained in discovery) and obtained essentially the same results as with the 8297 described above (data not shown). We genotyped these SNPs in the Rep1 cohort and controls using the Illumina GoldenGate platform. The average success rate for each SNP was 99.4%; five SNPs with call rates <95% were excluded from analyses, leaving 379 SNPs. We included three duplicate samples and observed 100% concordant SNP calls. Subsequent IBS/MDS analysis identified one outlier individual in this data set that was removed, leaving 375 AIS individuals for further analyses (Supplementary Material, Fig. S5).
Individual genotyping
We performed follow-up genotyping for SNPs rs1400180, rs965084, rs1400182, rs9754850, rs9754552 and rs10501081 using Taqman genotyping as previously described (14). To check agreement across platforms, we included ∼30% of discovery samples in Taqman genotyping for rs1400180 and rs10510181 and found 100% concordance for the two methods. All genotyped SNPs displayed agreement with Hardy–Weinberg equilibrium (P > 0.05) with the exception of rs1400180 (P = 0.04) which showed marginal deviation from HWE.
Case–control statistics
We genotyped six SNPs: the original rs1400180 and rs10510181, and rs965084, rs1400182, rs9754850 and rs9754552, in the first replication study. The number of cases and controls included in this study (‘Rep1') was 375 and 444, respectively. We performed allelic tests of association in cases and controls. As above, the statistic was computed on all data using the method implemented in the PLINK software (15). We genotyped four of the SNPs rs1400180, rs10510181, rs9754850, and rs9754552 in the second replication study. The number of cases and controls included in this study (‘Rep2') was 187 and 222, respectively. We performed allelic tests of association as in the Rep1 study.
Combined statistics
We estimated combined ORs and CIs for TDT and case–control results using the method implemented in the Catmap software program (44) We also estimated the combined P-values using Fisher's method as implemented in the PVALUES software program (45). Results were as reported in the text.
LD structure
Pairwise r2 values were computed using Haploview software Version 4.1 (46). Only SNPs with MAF > 0.01 were considered in this analysis.
RT–PCR
RNA was isolated from frozen human lymphocytes using the RNeasy mini kit (Qiagen). Adult and fetal human brain poly A+ RNA (Clontech Laboratories) was procured. cDNA was prepared using the high-capacity cDNA Archive reagent (Applied Biosystems). Approximately 500 ng of each RNA was converted to cDNA using guidelines provided in the high-capacity cDNA archive reagent protocol (Applied Biosystems) in a 20 μl reaction. Primers were designed using Primer 3 software (47). To amplify CHL1 cDNA (derived from NM_006614.2), the forward primer was designed to span the splice junction of exons 6 and 7, and the reverse primer was designed within exon 7. For the hypothetical cDNA LOC642891 (derived from XM_931148.3), the forward primer was designed to span the junction of exons 2 and 3, and the reverse primer was designed within exon 5. This design theoretically avoids amplifying unspliced RNA or genomic DNA. Primer sequences and amplified region are given in Supplementary Material, Figure S6. Approximately 100 ng of each cDNA was amplified using 1 U of GoTaq DNA Polymerase (Promega) in a 12.5 μl reaction containing: 0.5 pmoles/μl of forward and reverse primer, 0.2 mm dNTPs, and 1.5 m MgCl2. Thirty-five cycles of PCR amplification were carried out at 94°–55°–72°, 30 s each cycle (Supplementary Material, Fig. S7).
Linkage analysis
Each family selected for analysis was represented in our original discovery cohort and was selected to be informative in a linkage analysis but without regard to genotype at any particular locus. Maximum heterogeneity LOD scores (HLODs) were computed over dominant and recessive models with a phenocopy rate of 0.5 and a disease allele frequency of 0.001 to approximate the frequency of familial AIS. Results were adjusted for inflation by subtracting 0.3 as described in Hodge et al. (48). The TDTae was computed to estimate evidence for association in the presence of linkage as previously described (40).
Genomic re-sequencing
Coding exons and splice junctions of CHL1 (Gene ID: 10752; Ensembl ID:ENSG00000134121; reference sequence mRNA NM_006614.2) were extracted from available databases. Genomic re-sequencing and analysis was performed as previously described (14) and by alignment with DNA Baser Sequence Assembler v2 (HeracleSoftware). Primer sequences and PCR conditions are available upon request. The SNP (rs2272522) that predicts a leucine-to-phenylalanine change was included in our original GWAS but did not show evidence of association with AIS in the total data set (OR = 0.92, 95% CI = 0.68–1.25; P = 0.59) nor in trios with family history of AIS (OR = 0.74, 95% CI = 0.42–1.32; P = 0.31). A second variant, SNP rs62230378 that predicts a glycine-to-alanine change, was observed in two unrelated affected individuals from families (91 families) with multiple members affected by AIS. Upon further sequencing, this variant was detected in 1/20 unrelated white, non-Hispanic controls and 1/29 unrelated white, non-Hispanic AIS cases, suggesting near-equal frequencies (3–5%) in cases and controls of this ethnic group.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This project was funded by NIH grant R01 HD052973, the Crystal Charity Ball, the Scoliosis Research Society, the Cain Foundation and the TSRHC Research Fund (to C.A.W.).
ACKNOWLEDGEMENTS
We thank the patients and families for their participation in these studies. We also particularly thank referring surgeons and associates: J. Birch, C. E. Johnston, L. A. Karol, K. E. Rathjen, B. S. Richards, D. J. Sucato, T. Milbrandt, V. Talwalkar, H. J. Iwinski, Jr., T. Cloar, C. P. Eberson, D. Adalio, K. Clark, A. Lapinsky, R. Merolli, J. C. Tassone, X. C. Liu, C. Blanchard, W. Schrader, M. Morsher; C. Horton, R. Wilson, and B. Crider in the McDermott Center core facility for help with genotyping and sequencing, B. Zhang in the UTSW microarray core facility for help with genotyping, R. Debaradinis for help with constructing questionnaires, J. Howell for administrative support and A. Bowcock for reviewing the manuscript. Two public data sets were made available through the Database of Genotypes and Phenotypes (dbGaP). The IBDGC Crohn's Disease Genome-Wide Association Study reported here was conducted by IBDGC Crohn's Disease Genome-Wide Association Study Investigators and supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). This manuscript was not prepared in collaboration with Investigators of the IBDGC Crohn's Disease Genome-Wide Association Study and does not necessarily reflect the opinions or views of the IBDGC Crohn's Disease Genome-Wide Association Study of the NIDDK. The Genome Wide Association Study in Familial Parkinson Disease reported here was conducted as part of PROGENI and GenePD and supported by the National Institute of Neurological Disorders and Stroke (NINDS).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Herring J.A., editor. Tachdjian's Pediatric Orthopaedics. Vol. 1. Philedelphia: W.B. Saunders Company; 2002. [Google Scholar]
- 2.Richards B.S., Vitale M.G. Screening for idiopathic scoliosis in adolescents. An information statement. J. Bone Joint Surg. Am. 2008;90:195–198. doi: 10.2106/JBJS.G.01276. doi:10.2106/JBJS.G.01276. [DOI] [PubMed] [Google Scholar]
- 3.Katz D.E., Herring J.A., Browne R.H., Kelly D.M., Birch J.G. Brace wear control of curve progression in adolescent idiopathic scoliosis. J. Bone Joint Surg. Am. 2010;92:1343–1352. doi: 10.2106/JBJS.I.01142. doi:10.2106/JBJS.I.01142. [DOI] [PubMed] [Google Scholar]
- 4.Wise C.A., Gao X., Shoemaker S., Gordon D., Herring J.A. Understanding genetic factors in idiopathic scoliosis, a complex disease of childhood. Curr. Genomics. 2008;9:51–59. doi: 10.2174/138920208783884874. doi:10.2174/138920208783884874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wise C.A., Sharma S. In: The Genetics and Development of Scoliosis. Kusumi K., Dunwoodie S.L., editors. Springer; 2009. pp. 167–190. [Google Scholar]
- 6.Giampietro P.F., Raggio C.L., Blank R.D. Synteny-defined candidate genes for congenital and idiopathic scoliosis. Am. J. Med. Genet. 1999;83:164–177. doi:10.1002/(SICI)1096-8628(19990319)83:3<164::AID-AJMG5>3.0.CO;2-D. [PubMed] [Google Scholar]
- 7.Semba K., Araki K., Li Z., Matsumoto K., Suzuki M., Nakagata N., Takagi K., Takeya M., Yoshinobu K., Araki M., et al. A novel murine gene, Sickle tail, linked to the Danforth's short tail locus, is required for normal development of the intervertebral disc. Genetics. 2006;172:445–456. doi: 10.1534/genetics.105.048934. doi:10.1534/genetics.105.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulkarni S., Nagarajan P., Wall J., Donovan D.J., Donell R.L., Ligon A.H., Venkatachalam S., Quade B.J. Disruption of chromodomain helicase DNA binding protein 2 (CHD2) causes scoliosis. Am. J. Med. Genet. A. 2008;146A:1117–1127. doi: 10.1002/ajmg.a.32178. doi:10.1002/ajmg.a.32178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan-Brown J., Schottenfeld J., Okabe N., Hostetter C.L., Serluca F.C., Thiberge S.Y., Burdine R.D. Zebrafish mutations affecting cilia motility share similar cystic phenotypes and suggest a mechanism of cyst formation that differs from pkd2 morphants. Dev. Biol. 2008;314:261–275. doi: 10.1016/j.ydbio.2007.11.025. doi:10.1016/j.ydbio.2007.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wynne-Davies R. Genetic aspects of idiopathic scoliosis. Dev. Med. Child Neurol. 1973;15:809–811. doi: 10.1111/j.1469-8749.1973.tb04919.x. doi:10.1111/j.1469-8749.1973.tb04919.x. [DOI] [PubMed] [Google Scholar]
- 11.Miller N.H., Justice C.M., Marosy B., Doheny K.F., Pugh E., Zhang J., Dietz H.C., 3rd, Wilson A.F. Identification of candidate regions for familial idiopathic scoliosis. Spine. 2005;30:1181–1187. doi: 10.1097/01.brs.0000162282.46160.0a. doi:10.1097/01.brs.0000162282.46160.0a. [DOI] [PubMed] [Google Scholar]
- 12.Ocaka L., Zhao C., Reed J.A., Ebenezer N.D., Brice G., Morley T., Mehta M., O'Dowd J., Weber J.L., Hardcastle A.J., et al. Assignment of two loci for autosomal dominant adolescent idiopathic scoliosis to chromosomes 9q31.2-q34.2 and 17q25.3-qtel. J. Med. Genet. 2008;45:87–92. doi: 10.1136/jmg.2007.051896. doi:10.1136/jmg.2007.051896. [DOI] [PubMed] [Google Scholar]
- 13.Doyle C., Blake K. Scoliosis in CHARGE: a prospective survey and two case reports. Am. J. Med. Genet. A. 2005;133A:340–343. doi: 10.1002/ajmg.a.30564. doi:10.1002/ajmg.a.30564. [DOI] [PubMed] [Google Scholar]
- 14.Gao X., Gordon D., Zhang D., Browne R., Helms C., Gillum J., Weber S., Devroy S., Swaney S., Dobbs M., et al. CHD7 gene polymorphisms are associated with susceptibility to idiopathic scoliosis. Am. J. Hum. Genet. 2007;80:957–965. doi: 10.1086/513571. doi:10.1086/513571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spielman R.S., McGinnis R.E., Ewens W.J. The transmission/disequilibrium test detects cosegregation and linkage. Am. J. Hum. Genet. 1994;54:559–560. author reply 560-553) [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C., Yang G., Buyske S., Matise T., Finch S.J., Gordon D. Transmission disequilibrium test power and sample size in the presence of locus heterogeneity. Stat. Appl. Genet. Mol. Biol. 2009;8 doi: 10.2202/1544-6115.1501. Article 44. [DOI] [PubMed] [Google Scholar]
- 18.Holm J., Hillenbrand R., Steuber V., Bartsch U., Moos M., Lubbert H., Montag D., Schachner M. Structural features of a close homologue of L1 (CHL1) in the mouse: a new member of the L1 family of neural recognition molecules. Eur. J. Neurosci. 1996;8:1613–1629. doi: 10.1111/j.1460-9568.1996.tb01306.x. doi:10.1111/j.1460-9568.1996.tb01306.x. [DOI] [PubMed] [Google Scholar]
- 19.Wright A.G., Demyanenko G.P., Powell A., Schachner M., Enriquez-Barreto L., Tran T.S., Polleux F., Maness P.F. Close homolog of L1 and neuropilin 1 mediate guidance of thalamocortical axons at the ventral telencephalon. J. Neurosci. 2007;27:13667–13679. doi: 10.1523/JNEUROSCI.2888-07.2007. doi:10.1523/JNEUROSCI.2888-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jen J.C., Chan W.M., Bosley T.M., Wan J., Carr J.R., Rub U., Shattuck D., Salamon G., Kudo L.C., Ou J., et al. Mutations in a human ROBO gene disrupt hindbrain axon pathway crossing and morphogenesis. Science. 2004;304:1509–1513. doi: 10.1126/science.1096437. doi:10.1126/science.1096437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sicotte N.L., Salamon G., Shattuck D.W., Hageman N., Rub U., Salamon N., Drain A.E., Demer J.L., Engle E.C., Alger J.R., et al. Diffusion tensor MRI shows abnormal brainstem crossing fibers associated with ROBO3 mutations. Neurology. 2006;67:519–521. doi: 10.1212/01.wnl.0000227960.38262.0c. doi:10.1212/01.wnl.0000227960.38262.0c. [DOI] [PubMed] [Google Scholar]
- 22.Goldstein D.B. Common genetic variation and human traits. N. Engl. J. Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. doi:10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 23.Sakurai K., Migita O., Toru M., Arinami T. An association between a missense polymorphism in the close homologue of L1 (CHL1, CALL) gene and schizophrenia. Mol. Psychiatry. 2002;7:412–415. doi: 10.1038/sj.mp.4000973. doi:10.1038/sj.mp.4000973. [DOI] [PubMed] [Google Scholar]
- 24.Chen Q.Y., Chen Q., Feng G.Y., Lindpaintner K., Chen Y., Sun X., Chen Z., Gao Z., Tang J., He L. Case-control association study of the close homologue of L1 (CHL1) gene and schizophrenia in the Chinese population. Schizophr. Res. 2005;73:269–274. doi: 10.1016/j.schres.2004.06.001. doi:10.1016/j.schres.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Andrews G.L., Tanglao S., Farmer W.T., Morin S., Brotman S., Berberoglu M.A., Price H., Fernandez G.C., Mastick G.S., Charron F., et al. Dscam guides embryonic axons by Netrin-dependent and -independent functions. Development. 2008;135:3839–3848. doi: 10.1242/dev.023739. doi:10.1242/dev.023739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu G., Li W., Wang L., Kar A., Guan K.L., Rao Y., Wu J.Y. DSCAM functions as a netrin receptor in commissural axon pathfinding. Proc. Natl Acad. Sci. USA. 2009;106:2951–2956. doi: 10.1073/pnas.0811083106. doi:10.1073/pnas.0811083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson L.M., Kenneth W. 2008 Patent WO/2008/033813.
- 28.Schmid R.S., Maness P.F. L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Curr. Opin. Neurobiol. 2008;18:245–250. doi: 10.1016/j.conb.2008.07.015. doi:10.1016/j.conb.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerjee S., Blauth K., Peters K., Rogers S.L., Fanning A.S., Bhat M.A. Drosophila neurexin IV interacts with Roundabout and is required for repulsive midline axon guidance. J. Neurosci. 2010;30:5653–5667. doi: 10.1523/JNEUROSCI.6187-09.2010. doi:10.1523/JNEUROSCI.6187-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buratowski S. Transcription. Gene expression—where to start? Science. 2008;322:1804–1805. doi: 10.1126/science.1168805. doi:10.1126/science.1168805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maness P.F., Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat. Neurosci. 2007;10:19–26. doi: 10.1038/nn1827. doi:10.1038/nn1827. [DOI] [PubMed] [Google Scholar]
- 32.Yimlamai D., Konnikova L., Moss L.G., Jay D.G. The zebrafish down syndrome cell adhesion molecule is involved in cell movement during embryogenesis. Dev. Biol. 2005;279:44–57. doi: 10.1016/j.ydbio.2004.12.001. doi:10.1016/j.ydbio.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Lonstein J.E. Adolescent idiopathic scoliosis. Lancet. 1994;344:1407–1412. doi: 10.1016/s0140-6736(94)90572-x. doi:10.1016/S0140-6736(94)90572-X. [DOI] [PubMed] [Google Scholar]
- 34.Wiener-Vacher S.R., Mazda K. Asymmetric otolith vestibulo-ocular responses in children with idiopathic scoliosis. J. Pediatr. 1998;132:1028–1032. doi: 10.1016/s0022-3476(98)70403-2. doi:10.1016/S0022-3476(98)70403-2. [DOI] [PubMed] [Google Scholar]
- 35.Rousie D., Hache J.C., Pellerin P., Deroubaix J.P., Van Tichelen P., Berthoz A. Oculomotor, postural, and perceptual asymmetries associated with a common cause. Craniofacial asymmetries and asymmetries in vestibular organ anatomy. Ann. NY Acad. Sci. 1999;871:439–446. doi: 10.1111/j.1749-6632.1999.tb09213.x. doi:10.1111/j.1749-6632.1999.tb09213.x. [DOI] [PubMed] [Google Scholar]
- 36.Guo X., Chau W.W., Hui-Chan C.W., Cheung C.S., Tsang W.W., Cheng J.C. Balance control in adolescents with idiopathic scoliosis and disturbed somatosensory function. Spine. 2006;31:E437–E440. doi: 10.1097/01.brs.0000222048.47010.bf. doi:10.1097/01.brs.0000222048.47010.bf. [DOI] [PubMed] [Google Scholar]
- 37.Mallau S., Bollini G., Jouve J.L., Assaiante C. Locomotor skills and balance strategies in adolescents idiopathic scoliosis. Spine. 2007;32:E14–E22. doi: 10.1097/01.brs.0000251069.58498.eb. doi:10.1097/01.brs.0000251069.58498.eb. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell M.K., Gregersen P.K., Johnson S., Parsons R., Vlahov D. The New York Cancer Project: rationale, organization, design, and baseline characteristics. J. Urban Health. 2004;81:301–310. doi: 10.1093/jurban/jth116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. doi:10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordon D., Heath S.C., Liu X., Ott J. A transmission/disequilibrium test that allows for genotyping errors in the analysis of single-nucleotide polymorphism data. Am. J. Hum. Genet. 2001;69:371–380. doi: 10.1086/321981. doi:10.1086/321981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordon D., Haynes C., Blumenfeld J., Finch S.J. PAWE-3D: visualizing power for association with error in case-control genetic studies of complex traits. Bioinformatics. 2005;21:3935–3937. doi: 10.1093/bioinformatics/bti643. doi:10.1093/bioinformatics/bti643. [DOI] [PubMed] [Google Scholar]
- 42.Li Y., Abecasis G. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. 2006;S79:2290. [Google Scholar]
- 43.Seldin M.F., Price A.L. Application of ancestry informative markers to association studies in European Americans. PLoS Genet. 2008;4:e5. doi: 10.1371/journal.pgen.0040005. doi:10.1371/journal.pgen.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicodemus K.K. Catmap: case-control and TDT meta-analysis package. BMC Bioinformatics. 2008;9:130. doi: 10.1186/1471-2105-9-130. doi:10.1186/1471-2105-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ott J. Analysis of Human Genetic Linkage. Baltimore: Johns Hopkins; 1999. [Google Scholar]
- 46.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. doi:10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 47.Rozen S., Skaletsky H. Series. Primer3 on the WWW for General Users and for Biologist Programmers. Totowa: Humana Press; 2000. [DOI] [PubMed] [Google Scholar]
- 48.Hodge S.E., Abreu P.C., Greenberg D.A. Magnitude of type I error when single-locus linkage analysis is maximized over models: a simulation study. Am. J. Hum. Genet. 1997;60:217–227. [PMC free article] [PubMed] [Google Scholar]
- 49.Ge D., Zhang K., Need A.C., Martin O., Fellay J., Urban T.J., Telenti A., Goldstein D.B. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–643. doi: 10.1101/gr.071571.107. doi:10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.