Abstract
Hyper- and hypomethylation at the IGF2-H19 imprinting control region (ICR) result in reciprocal changes in IGF2-H19 expression and the two contrasting growth disorders, Beckwith–Wiedemann syndrome (BWS) and Silver–Russell syndrome (SRS). DNA methylation of the ICR controls the reciprocal imprinting of IGF2 and H19 by preventing the binding of the insulator protein, CTCF. We here show that local changes in histone modifications and CTCF–cohesin binding at the ICR in BWS and SRS together with DNA methylation correlate with the higher order chromatin structure at the locus. In lymphoblastoid cells from control individuals, we found the repressive histone H3K9me3 and H4K20me3 marks associated with the methylated paternal ICR allele and the bivalent H3K4me2/H3K27me3 mark together with H3K9ac and CTCF–cohesin associated with the non-methylated maternal allele. In patient-derived cell lines, the mat/pat asymmetric distribution of these epigenetic marks was lost with H3K9me3 and H4K20me3 becoming biallelic in the BWS and H3K4me2, H3K27me3 and H3K9ac together with CTCF–cohesin becoming biallelic in the SRS. We further show that in BWS and SRS cells, there is opposing chromatin looping conformation mediated by CTCF–cohesin binding sites surrounding the locus. In normal cells, lack of CTCF–cohesin binding at the paternal ICR is associated with monoallelic interaction between two CTCF sites flanking the locus. CTCF–cohesin binding at the maternal ICR blocks this interaction by associating with the CTCF site downstream of the enhancers. The two alternative chromatin conformations are differently favoured in BWS and SRS likely predisposing the locus to the activation of IGF2 or H19, respectively.
INTRODUCTION
Chromosome 11p15.5 contains two independently regulated clusters of imprinted genes. An imprinting control region (ICR) in each cluster controls monoallelic expression of the surrounding genes. The ICRs correspond to sequences that are methylated on one parental allele, also known as differentially methylated regions (DMRs). The telomeric cluster harbours the IGF2 and H19 genes and the centromeric cluster contains, among others, the CDKN1C and KCNQ1OT1 genes. Opposite methylation defects at the ICR controlling IGF2-H19 lead to the fetal over growth syndrome, Beckwith–Wiedemann syndrome (BWS, OMIM130650) and the growth retardation syndrome, Silver–Russell syndrome (SRS, OMIM180680). BWS is also caused by the loss of ICR methylation and deregulation in the centromeric cluster or paternal uniparental disomy (pUPD), which results in aberrant methylation at the ICRs of both clusters (reviewed in 1,2).
The ICR at IGF2 and H19 is normally methylated on the paternal allele. Methylation is excluded from the maternal allele by CTCF binding. The presence of CTCF confers an insulator function upon the ICR, which blocks access of the IGF2 promoters to the enhancers downstream of the H19 gene (3–5). This function is lost on the methylated paternal chromosome and this enables IGF2 expression by allowing the promoters to access the enhancers. In mice, we and others have shown that the higher order chromatin structure at the locus differs between maternal and paternal alleles and that on the maternal allele CTCF mediates a looping structure that sequesters the enhancers away from the Igf2 promoters (6–9).
Cohesin has recently been shown to colocalize with CTCF at CTCF-binding sites genome wide (10–13). RNAi-mediated cohesin and CTCF knockdown experiments indicate that CTCF is required for cohesin loading, but that CTCF binding is independent of cohesin. Since cohesin is known to hold two sister chromatids together, it was postulated that cohesin may also function to hold different chromatid regions together in cis to facilitate the formation of chromatin loops. We have recently shown that CTCF and cohesin mediate intrachromosomal looping interactions at the human IGF2-H19 locus and that the insulator function of the ICR is determined by its interaction with other CTCF sites at this locus (14). Cohesin is required to stabilize such loops. Cohesin also associates with other transcription factor complexes independent of CTCF and may facilitate looping interactions between promoters and enhancers in these instances (15,16).
It has been shown in the mouse that the chromatin associated with the Igf2-H19 ICR carries allele-specific histone modifications (17). On one allele, H3K9me3 and H4K20me3 are associated with DNA methylation at the ICR, while H3K4me2/3 and H3/H4 acetylation are found on the chromosome carrying the unmethylated ICR. A functional role of histone modifications in imprinting control is indicated by the observation that the KDM1B-directed removal of H3K4 methylation is a prerequisite for establishment of DNA methylation imprints at maternally methylated ICRs (18).
The effects of abnormal methylation changes at the ICR on the underlying chromatin and long-range associations with neighbouring CTCF sites are not known. We used a panel of BWS and SRS cell lines with aberrant methylation at the IGF2-H19 ICR to address this question. Our data indicate that DNA methylation changes at the ICR are accompanied by allele-specific chromatin changes. Methylation and histone modifications at the ICR further affect chromatin conformation between several CTCF–cohesin binding sites at the locus, to generate contrasting structures in BWS and SRS cells.
RESULTS
Contrasting histone marks at the IGF2-H19 ICR in BWS and SRS cells
Individuals affected by BWS and SRS display DNA methylation abnormalities at 11p15.5 ICRs in DNA extracted from peripheral blood leukocytes (PBLs). Epstein Barr Virus (EBV)-transformed lymphoblastoid cell lines (EBV-LCLs) have been shown to maintain DNA methylation at the ICRs over several passages, indicating that epigenetic methylation imprints are maintained in these cells (19,20).
We established EBV-LCL from eight BWS patients [five with IGF2-H19 ICR hypermethylation (ICR-HM) and three with 11p15.5 pUPD] and four SRS patients with IGF2-H19 ICR hypomethylation. One of the hypermethylated BWS lines carried a 1.4 kb deletion resulting in the HM of the remaining ICR sequence (21). DNA methylation at the ICR in these cell lines was very similar to the originating patient's PBLs (Supplementary Material, Fig. S1). It has been shown that the gain of methylation at the maternal ICR results in biallelic IGF2 activation and H19 silencing, while the loss of ICR methylation is associated with IGF2 silencing and biallelic H19 activation (22,23). Although the expression of the IGF2 and H19 genes is generally low in EBV-LCLs, we observed that the SRS cell lines had H19 RNA levels 2–3-fold higher than the BWS and control cell lines (Supplementary Material, Fig. S2A). By cDNA sequencing, we also demonstrated biallelic H19 expression in three SRS cell lines which were informative for transcribed SNPs and biallelic IGF2 expression in one informative BWS cell line (Supplementary Material, Fig. S2B and C). These results are consistent with the gain of the insulator function on the paternal chromosome in the hypomethylated SRS cell lines and the loss of the insulator function on the maternal chromosome in the BWS cell lines.
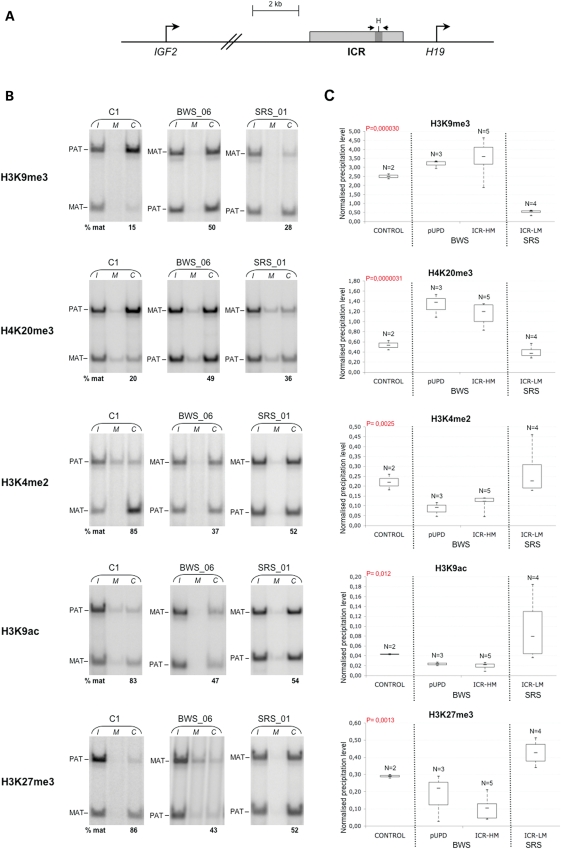
The BWS and SRS LCLs were further characterized for chromatin modifications at the ICR. We performed both allele-specific and quantitative analyses of DNA recovered after chromatin immunoprecipitation (ChIP) using H3K9me3, H4K20me3, H3K27me3, H3K4me2 and H3K9ac antibodies. The region analysed was located around the sixth CTCF-binding site of the human IGF2-H19 ICR (hg19 chr11: 2021143–2021271). A SNP (rs10732516) was used to distinguish the maternal and paternal alleles in non-UPD heterozygote samples. With the allele-specific ChIP assay, we first observed that in control EBV-LCLs, the repressive modifications H3K9me3 and H4K20me3 were enriched on the DNA-methylated paternal ICR allele, while the permissive modifications H3K4me2 and H3K9ac were enriched on the non-DNA-methylated maternal allele (Fig. 1B and Supplementary Material, Table S1). H3K27me3 was also found preferentially associated with the non-methylated maternal allele. We observed that in two hypermethylated heterozygote BWS cell lines (BWS_03 and BWS_06) and four hypomethylated heterozygote SRS cell lines (SRS_01, SRS_05, SRS_06 and SRS_08), the mat/pat asymmetric distribution of the five epigenetic marks examined was abolished or attenuated (Fig. 1B and Supplementary Material, Table S1). To better characterize the changes in histone marks at the ICR, we also measured their overall levels in all our cell lines using qPCR. Consistent with the demonstrated allele specificity of the histone marks, we observed that in pUPD BWS cells the paternal-specific H3K9me3 and H4K20me3 were increased while the maternal-specific H3K4me2, H3K9ac and H3K27me3 were decreased (Fig. 1C). A similar pattern was observed in the five BWS lines with ICR-HM, while opposite results with increased H3K4me2, H3K9ac and H3K27me3 and decreased H3K9me3 and H4K20me3 were found in the hypomethylated SRS cells (ICR-LM).
Figure 1.
Contrasting histone modifications at the ICR in BWS and SRS cell lines. (A)Diagram illustrating the region of the IGF2-H19 locus analysed. The ICR is indicated by a grey box and the IGF2 and H19 transcription start sites by bent arrows. The ICR region analysed by ChIP corresponding to the sixth CTCF target site is depicted in dark grey. The location of the PCR primers and the polymorphic HhaI site is indicated. (B). Allele-specific ChIP analysis of H3K9me3, H4K20me3, H3K4me2, H3K9ac and H3K27me3 in control (C), hypermethylated BWS and hypomethylated SRS EBV-LCLs. The DNAs extracted from input (I), non-specific IgG control (M) and specific antibody-bound (C) fractions were PCR amplified and digested with the polymorphic HhaI enzyme to distinguish the parental ICR alleles. As result of HhaI digestion, the 128 bp PCR product was divided in two co-migrating 64 bp fragments. The relative contribution of the paternal and maternal alleles in the immunoprecipitated chromatin after normalization to input is indicated below the gels. The results obtained on only one cell line of each group are shown. Complete results are reported in Supplementary Material, Table S1. (C). Real-time qPCR analysis of the histone marks in control, BWS and SRS EBV-LCLs. The percentage of input chromatin that was precipitated in the antibody-bound fraction at the ICR was normalized to the value obtained at a positive control region. Box plots show max., third quartile, median, first quartile and min. values for each group of samples. The BWS includes a group of three samples with paternal 11p15.5 UPD (pUPD) and a group of five samples with ICR-HM. The SRS samples include a single group of five samples with ICR hypomethylation (ICR-LM). Note that the control, BWS and SRS samples show significant differences in the histone modifications analysed at the ICR. ANOVA P-values are indicated.
These results show that the allele-specific histone modification status of the ICR is altered along with DNA methylation in BWS and SRS cells. In BWS, the gain of DNA methylation is associated with the gain of H3K9me3 and H4K20me3 and the loss of H3K4me2, H3K9ac and H3K27me3 on the maternal allele. In SRS, the loss of DNA methylation is associated with the loss of H3K9me3 and H4K20me3 and the gain of H3K4me2, H3K9ac and H3K27me3 on the paternal allele. The results also show that a bivalent H3K4me2/H3K27me3 mark together with H3K9ac is present on the unmethylated maternal allele in control cells. These marks are reduced in the BWS cells, and increased becoming biallelic in the SRS cells.
CTCF and cohesin binding in BWS and SRS cells
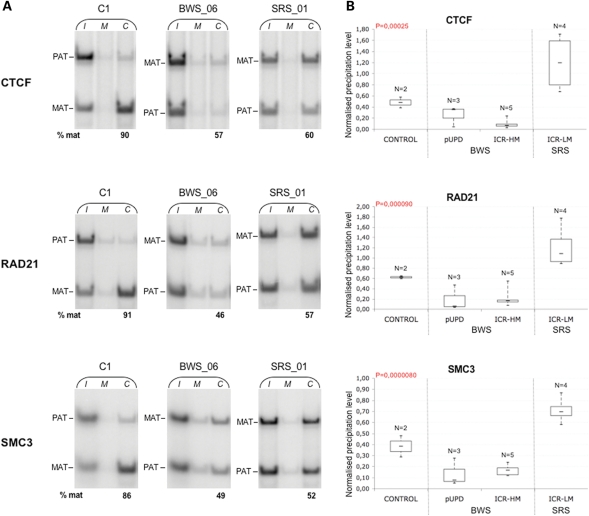
Having established that opposite changes in histone marks occur at the IGF2-H19 ICR in BWS and SRS cells, we looked at the binding of CTCF and cohesin. Allele-specific ChIP showed that CTCF and the cohesin subunits RAD21 and SMC3 were preferentially associated with the non-methylated maternal allele in control cells (Fig. 2A and Supplementary Material, Table S2). However, this allele-specific distribution was lacking or attenuated in the heterozygote hypermethylated BWS and hypomethylated SRS cells. The qPCR analysis demonstrated that the binding of CTCF and cohesin subunits to the ICR was decreased in all hypermethylated BWS cells as well as pUPD BWS, but increased in hypomethylated SRS cells (Fig. 2B), indicating that CTCF and associated cohesin were lost from both parental alleles in BWS and acquired on both alleles in SRS.
Figure 2.
Differential CTCF and cohesin binding at the ICR in BWS and SRS cell lines. (A). Allele-specific ChIP analysis of CTCF, RAD21 and SMC3 binding in control (C1), hypermethylated BWS and hypomethylated SRS EBV-LCLs. The relative contribution of the maternal and the paternal ICR alleles in the immunoprecipitated chromatin was determined as in Figure 1 (A). The results obtained on only one cell line of each group are shown. Complete results are reported in Supplementary Material, Table S2. (B) Real-time qPCR analysis of CTCF, RAD21 and SMC3 binding. The percentage of input chromatin that was precipitated with CTCF, RAD21 and SMC3 antibodies at the ICR was determined and is indicated as in Figure 1B. Note that the control, BWS and SRS samples show significant differences in the CTCF and cohesin binding at the ICR. ANOVA P-values are indicated.
We then tested CTCF and cohesin binding at the CTCF sites upstream and downstream of the IGF2-H19 locus. These regions have previously been shown to be unmethylated and to bind CTCF and cohesin on both alleles (14). Supplementary Material, Figure S3 indicates that CTCF and cohesin were both present at the CTCF site upstream of IGF2 (CTCF AD), at the centrally conserved domain (CCD) and the CTCF site downstream of the H19 endoderm and mesoderm enhancers (CTCF DS), in the normal as well as all the BWS and SRS cell lines, without significant binding differences. These results indicate that binding of CTCF–cohesin to CTCF AD, CCD and CTCF DS is not affected by the DNA methylation and chromatin changes occurring at the IGF2-H19 ICR in BWS and SRS.
CTCF-mediated chromatin conformation at the IGF2-H19 locus in normal and pUPD cell lines
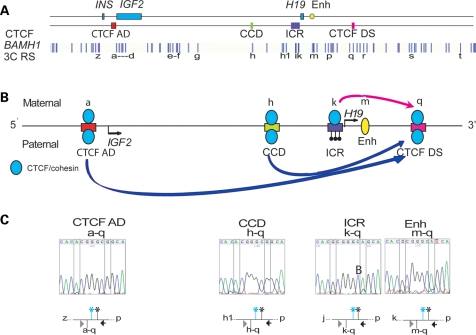
We previously observed long-range interactions between the CTCF sites in the IGF2-H19 region with the ICR interacting monoallelically with the CTCF DS in an adult epithelial breast cell line (14). We now examined the looping profile of a normal neonate fibroblast cell line (HS27), which had two informative SNPs in the CTCF DS site which enabled us to assign parental-specific interactions to the CTCF DS loops by sequencing the 3C products. Using 3C anchors at the ICR, the enhancer and the two CTCF sites flanking the locus (CTCF AD and CTCF DS), we found looping profiles similar to those we previously reported (Supplementary Material, Fig. S4). Consistent with previous results (14), we found that the CTCF DS associates monoallelically with the ICR (Fig. 3). Unexpectedly, however, the CTCF DS was found to interact monoallelically also with other sites at the locus. Specifically, CTCF DS site associates with the CTCF AD and CCD sites on one chromosome and with the ICR on the reciprocal chromosome (Fig. 3C). Thus, the ICR affects the allele-specific interactions of the surrounding CTCF sites at the locus. Since the ICR is methylated on the paternal allele, it is likely that the CTCF DS association with the ICR is on the maternal allele, and the CTCF DS association with the CTCF AD site is on the paternal allele.
Figure 3.
Allele specificity of looping associations with the CTCF site downstream of the enhancers. (A)Locus with the position of BamHI sites, and CTCF-binding sites relative to enhancers and genes. (B) Schematic of monoallelic interactions with the CTCF DS site. (C) Heterozygous SNPs in the CTCF DS enabled the analysis of the allele specificity in the HS27 cell line. The CTCF DS interacts with the CTCF AD and CCD on the ‘AC’ allele and with the ICR and enhancers on the ‘GA’ allele indicating that the CTCF DS interactions are monoallelic.
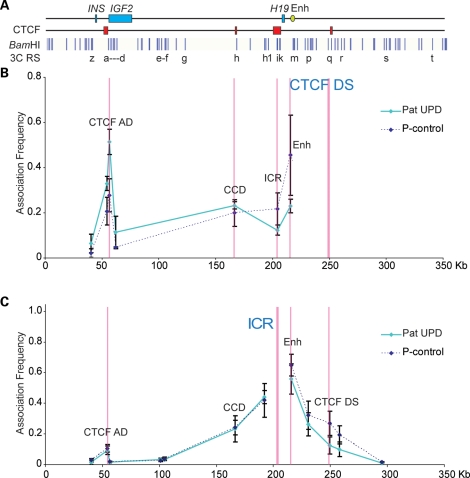
We therefore used a pUPD BWS EBV-LCL cell line to confirm that the associations between the CTCF AD and the CTCF DS are on the paternal allele as predicted. Using the CTCF DS and the ICR as anchors, the 3C profiles of the EBV-LCL control cell line were shown to be remarkably similar to those observed in the HS27 fibroblast and adult breast epithelial cell lines (data not shown). Figure 4B and C shows the topology of the interactions in the pUPD cells. Compared with the control cell line, the association between the CTCF DS and the CCD or the CTCF AD is stronger, whereas the CTCF DS association with the ICR and the enhancer is weaker (Fig. 4B). These results confirm that the CTCF DS associates with the ICR on the maternal chromosome and with the CTCF AD and the CCD on the paternal chromosome. This was also shown reciprocally, when we placed our 3C anchor at the ICR region, where very little CTCF was bound (Fig. 4C, see also Fig. 2B). Similar to the control cell lines, a weak association was evident between the ICR and the CTCF AD, which could indicate a non-CTCF-mediated interaction between these regions or represent an indirect effect of the CTCF DS–AD interaction. These results corroborate and further clarify our previous interpretation of allele-specific long-range chromatin interactions at the locus. Since both the endoderm and mesoderm enhancers are close to the CTCF DS, it is possible that the CTCF DS directs the enhancers to the IGF2 or H19 promoters through its association with different CTCF sites on the paternal and maternal chromosomes. The mesoderm enhancers are 5 kb downstream of the endoderm enhancers and it is therefore possible that the enhancers recruit tissue-specific transcription factors to mediate transcription, once they have been brought into proximity of the promoters through the CTCF DS–AS interaction.
Figure 4.
Looping profile of EBV-LCL cell line derived from a Beckwith patient with pUPD. (A) Locus as in Figure 3. (B) Looping profile of pUPD compared with EBV control when the CTCF DS is used as an anchor, showing strong interactions with elements 5′ of the ICR such as the CCD and the CTCF AD. No interaction is seen with the enhancer or ICR. (C). Using the ICR anchor, no difference is seen in the two cell lines for the ICR–CTCF AD interaction, whereas the ICR–CTCF DS interaction is absent in the pUPD cell line. The paternally methylated ICR does not interact with the CTCF DS, and the CTCF DS interacts with the CTCF AD on the paternal allele.
CTCF-mediated chromatin conformation in BWS and SRS cell lines with ICR DNA methylation defects
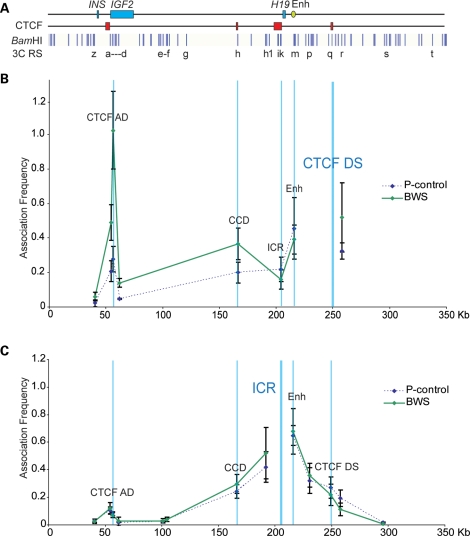
The BWS and SRS EBV-LCL allowed us to investigate the relationship between DNA methylation and histone marks and higher order chromatin conformation at the IGF2-H19 locus. We characterized three BWS cell lines with ICR-HM and two SRS cell lines with ICR hypomethylation for chromatin looping conformation. Figure 5B shows the average profiles of the BWS cases, with the CTCF DS as the 3C anchor compared with the baseline normal cell line. Similar to results obtained with the pUPD cells, we found that interactions with the CCD and the CTCF AD sites were increased while that with the ICR was decreased compared with the control cell lines. Using the ICR as an anchor, associations of similar intensity to the control were detected between the ICR and the CTCF AD, and the weaker association was detected between the ICR and CTCF DS region (Fig. 5C).
Figure 5.
Looping profile of cell lines from BWS patients with the gain of methylation at the ICR. (A) Locus as in Figure 3. (B) Looping profile at the locus using CTCF DS as anchor shows that BWS cell lines have strong interactions between the CTCF DS and the CCD compared with control. (C) Looping profiles at locus using ICR as anchor, showing weaker interactions between the ICR and the CTCF DS (n = 3 BWS cell lines representing three biological replicates, each sample being processed thrice as technical replicates).
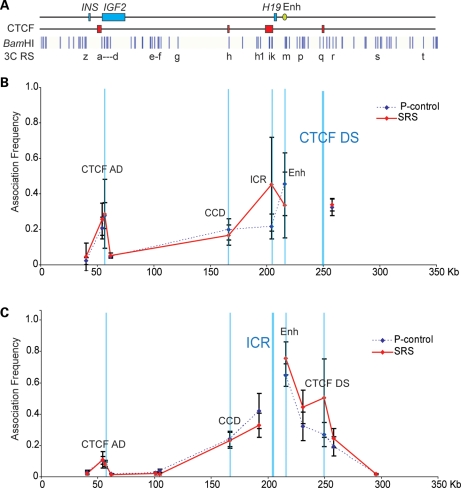
In contrast to the BWS, the SRS cell lines showed a strong association at the ICR when the CTCF DS was used as an anchor and a reciprocal strong association at the CTCF DS when the ICR was used as an anchor (Fig. 6B and C). However, with both anchors, peaks detected at the CTCF AD were similar to those of control cell lines. Overall, these results demonstrate that higher order chromatin conformation of the IGF2-H19 locus correlates with DNA methylation and histone mark deposition at the IGF2-H19 ICR. As a result of opposite CTCF–cohesin binding and histone modifications, different long-range interactions are established in BWS and SRS and are likely to influence IGF2-H19 expression in a reciprocal manner.
Figure 6.
Looping profile of cell lines from SRS patients with the loss of methylation at the ICR. (A) Locus as in Figure 3. (B) Looping profile at the locus using CTCF DS as anchor shows that SRS cell lines have strong interactions between the CTCF DS and the ICR compared with control. (C) Looping profiles at the locus using ICR as anchor, showing strong interactions between the ICR and the CTCF DS (n= 2 SRS cell lines representing three biological replicates, each sample being processed thrice as technical replicates).
DISCUSSION
We have previously shown that chromatin looping interactions between CTCF sites at the human IGF2-H19 locus are stabilized by cohesin in an adult breast epithelial cell line (14). Our current analysis utilizing BWS and SRS LCLs derived from patients with pUPD and methylation defects at the ICR has enabled us to assign specific patterns of histone modifications and chromatin looping profiles to maternal or paternal alleles and to determine the influence of the ICR on chromatin conformation at the locus.
In the first instance, we were able to demonstrate that in humans, as in the mouse, histone modifications mark the IGF2-H19 ICR in a parent of origin-specific manner, with the permissive H3K4me2 and H3K9ac on the non-DNA-methylated maternal allele and the repressive H3K9me3 and H4K20me3 on the DNA-methylated paternal allele. Unexpectedly, the repressive H3K27me3 was preferentially associated with the non-DNA-methylated allele, thus creating a ‘bivalent domain' at the maternal ICR. Enrichment of H3K27me3 at the non-DNA-methylated allele has been previously reported for other ICRs, and this histone mark is believed to be associated with the developmental regulation of linked transcripts rather than with imprinting control (24–26). However, data obtained by the ENCODE Project and available from the UCSC hg18 Genome Browser show that H3K27me3 is present at the IGF2-H19 ICR in several cell lines including some in which H19 is expressed (e.g. H1ES), thus indicating that the bivalent mark does not correlate with cell specificity or expression of the nearby H19 gene (Supplementary Material, Fig. S5). It is relevant to note that similarly to other bivalent domains, the IGF2-H19 ICR is frequently hypermethylated in cancer cells (27).
In the BWS and SRS cells, the allele-specific histone marks follow the DNA methylation changes of the IGF2-H19 ICR. The epigenetic asymmetry observed in control cells was lost when the ICR was hyper- or hypomethylated with reciprocal consequences on the occurrence of the histone marks. In parallel with DNA methylation and histone modifications, CTCF binding at the ICR also changed in BWS and SRS cells and this in turn affected cohesin recruitment. CTCF binding is known to be inhibited by DNA methylation in vitro (3). However, the local heterochromatinization of the paternal ICR allele brought about by H3K9me3 and H4K20me3 may also have a role in the control of the allele-specific interaction with non-histone proteins and/or contribute to the asymmetric higher order chromatin structure of the locus.
It is known that DNA methylation at the ICR determines methylation at somatic DMRs within the IGF2 locus presumably through long-range interactions. So we examined whether CTCF binding at the ICR affects CTCF binding and recruitment of cohesin to surrounding CTCF sites. Our data show that CTCF binding and recruitment of cohesin at sites other than the ICR is independent of the methylation state at the ICR. The CTCF sites at the IGF2-H19 locus other than the ICR are unmethylated and bind CTCF and cohesin on both alleles.
In our control lymphoblastoid and fibroblast cell lines, the chromatin conformation profiles for loops mediated by associations between CTCF-binding sites at the locus were found to be similar to those found previously in breast epithelial cells (14). SNP analysis in the fibroblast cell line indicates that the biallelic CTCF site downstream of the enhancers interacts allele specifically with the ICR and other biallelic CTCF sites. In a pUPD BWS cell line, the interactions between the CTCF DS and the ICR are notably low compared with the flanking enhancer and the CCD, attesting that the ICR–CTCF DS association is on the maternal chromosome which is normally unmethylated at the ICR. In contrast, we found that in pUPD cells, the CTCF DS associates strongly with the CTCF AD, indicating that this interaction is specific for the paternal allele. These novel findings of allele-specific interactions between CTCF sites outside the imprinted region suggest that we ought to look at promoter competition and insulator function slightly differently. One interpretation of our data would be that enhancers do not track, but that instead they are brought into the proximity of the promoters through nearby CTCF interactions. This interpretation would support a primary looping scaffold model where transcription factors could dock onto promoters and enhancers which are already in close proximity. This would explain why there is no difference in looping profiles between IGF2 expressing and non-expressing cell lines and why the enhancer–promoter interactions are weaker than the flanking CTCF-mediated interactions (14). CTCF binding at the AD, CCD and DS sites is conserved in the mouse locus (D. Schmidt, unpublished data), and it is likely that if these sites were included in 3C analysis in mice, the same loops would be detected as found in humans.
The 3C profiles of BWS cell lines derived from patients in which the ICR was hypermethylated had identical profiles to that of the pUPD cell line, suggesting biallelic CTCF AD–DS associations and the loss of ICR–DS interaction. Conversely, the SRS cell lines with hypomethylation at the ICR showed stronger interactions between the ICR and the CTCF DS. Interestingly, SRS cells did not lose the CTCF DS–AD interactions suggesting that the ICR–DS loop is the limiting step for IGF2 expression control. A fraction of BWS and SRS patients do not show any molecular defect at the 11p15.5 ICRs and coding sequences. It will be interesting to see if mutations or abnormal methylation of the CTCF DS or AD sites causes defective IGF2-H19 expression in these individuals.
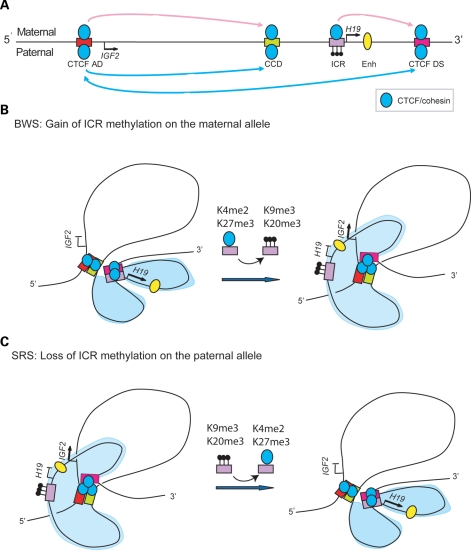
In summary, our data fit the modified model depicted in Figure 7, where CTCF bound sites, together with cohesin associate to form alternative looping interactions on the maternal and paternal chromosomes. CTCF bound to the ICR blocks interactions between the CTCF DS and CTCF AD, by inserting a new loop that separates the enhancers away from the IGF2 promoters. Thus, the allele-specific chromatin organization of the ICR determined by its asymmetric histone marks and DNA methylation appears to be the critical determinant of the long-range interactions at the locus. CTCF binding seems to be stable in all cell lines examined by genome-wide ChIP, which would be consistent with a constitutive CTCF–cohesin scaffold at this locus in all tissues. Upon this scaffold, the enhancers and promoters are brought into proximity and are poised for activation subject to recruitment of the appropriate transcription factors. Localized changes in the chromatin structure at the ICR differently affect the parent of origin-specific looping conformation and the expression potential of IGF2 and H19 in BWS and SRS, representing the first example of how alteration of transcriptional genomic neighbourhood can contribute to human disease (28).
Figure 7.
Summary of human allele-specific loops and changes in BWS and SRS patients. (A) Locus showing CTCF and cohesin complexes as blue circles, the CTCF-binding sites as bars and the enhancer as an oval. Position of the IGF2 and H19 genes are shown with arrows. The CTCF AD and CCD were previously shown to interact on both alleles. The unmethylated ICR interacts with the CTCF DS on the maternal allele. When the ICR is methylated, the CTCF DS interacts with the CTCF AD site on the paternal allele (B). Proposed model for the maternal allele showing that the ICR and CTCF DS interact and sequester the enhancers into proximity of the H19 promoter—gain of methylation and heterochromatic conformation at the ICR in BWS patients results in the CTCF DS site interacting with the CTCF AD and CCD. The CTCF DS essentially drags the enhancer into the vicinity of the IGF2 promoters. (C) Proposed model for the paternal allele where CTCF hub is formed between the CTCF sites at the locus. Methylation at the ICR excludes it from CTCF binding and keeps it outside of the CTCF hub. In SRS patients, the loss of heterochromatic marks enables the ICR to interact with the CTCF DS.
MATERIALS AND METHODS
Cell lines
LCLs were established by EBV transformation of PBLs of individuals affected by BWS and SRS and two normal controls. Cells were grown in the RPMI 1640 medium supplemented with 15% fetal bovine serum, l-glutamine and antibiotics at standard concentration.
DNA methylation analysis
ICR DNA methylation was determined by pyrosequencing and bisulphite treatment coupled with restriction enzyme digestion (COBRA) as previously described (14,29). For COBRA, after DNA modification with sodium bisulphite, the CTS6 region (surrounding the sixth CTCF-binding site of the ICR) was PCR amplified, the product digested with BstUI and the restriction fragments separated on a non-denaturing 6% polyacrylamide gel.
Expression analysis
RNAs were extracted from cultured EBV-LCL by use of Trizol (Invitrogen). For the detection of the H19 and IGF2 transcripts, 2 μg of total RNA was used for the cDNA synthesis with the QuantiTect Reverse Transcription Kit (Qiagen), according to the protocol of the manufacturer. Quantitative expression was determined by real-time PCR using iQ SYBR Green Supermix (BioRad). Reactions were run on a CFX96 Real-Time System + C1000 Thermal Cycler (Biorad). Two independent cDNA preparations from each RNA sample were analysed in triplicate. For the allele-specific analysis of H19 and IGF2, polymorphisms were typed by PCR amplification of DNA and cDNA, followed by sequencing. A nested PCR was required for the cDNA amplification of both transcripts. All primer sequences and PCR conditions are available upon request.
Chromatin immunoprecipitation (ChIP)
Chromatin from about 80 million cells was prepared as described previously (30) and 100 μg was used for each immunoprecipitation reaction with Protein A Agarose/Salmon Sperm DNA (Millipore, 16-157) and specific antibody. H3K9me3 (07-442), H3K4me2 (07-030), H3K4me3 (07-473), H3K27me3 (07-449), H3K9ac (07-352), H4ac (06-598) and CTCF (07-729) antibodies were provided by Millipore. Antibodies against H4K20me3 (AB9053), RAD21 (AB992) and SMC3 (AB9263) were obtained from ABCAM. For each ChIP, a fraction of the input chromatin (1%) was also processed for DNA purification and a mock immunoprecipitation with a neutral, unrelated IgG antiserum was carried out in parallel.
Allele-specific analysis of immunoprecipitated chromatin
In the input, mock and antibody-bound fractions of cell lines heterozygote for the HhaI RFLP rs10732516, parental alleles were distinguished by enzymatic digestion of an HOT-STOP 32P-dCTP-labeled PCR product. The DNA region across the polymorphic site was amplified using the following primer pair: For 5′-TCCTTCGGTCTCACCGCCTG-3′, Rev 5′-TTCCACGGGCGAACCCCAGT-3′. Products were resolved on 8% polyacrylamide gels and the relative band intensities were quantified by using ImageQuant (Molecular Dynamics).
qPCR analysis of immunoprecipitated chromatin
Levels of immunoprecipitated chromatin at specific loci were determined by quantitative real-time PCR amplification, carried out with the C1000 Thermal Cycler CFX96 Real-Time System, using the iQ SYBR Green Supermix (Biorad, 170-8882). Each PCR was run in triplicate and protein binding was quantified as a percentage of total input material, calculated as described by Wierzbicki (protocol available at: http://www.mcdb.lsa.umich.edu/labs/wierzbicki/protocols.php). The same primer pair was used for both allele-specific and qPCR analyses. In order to compare ChIP levels of different cell lines, precipitation levels obtained at the analysed region for each antibody were normalized to the level obtained on a positive control region. An heterochromatic satellite region on chromosome 4 was used as positive control for the H3K9me3 and H4K20me3 binding: For 5′-CTGCACTACCTGAAGAGGAC-3′, Rev 5′-GATGGTTCAACACTCTTACA-3′; the GAPD promoter was the positive control for the H3K4me2 and H3K9ac binding: For 5′-CAATTCCCCATCTCAGTCGT-3′, Rev 5′-GCAGCAGGACACTAGGGAGT-3′; the MYO-D promoter was the positive control for the H3K27me3 binding: For 5′-ACGGCTCTCTCTGCTCCTTT-3′, Rev 5′-GAGTGCTCTTCGGGTTTCAG-3′; an intergenic chromosome 6 region (region 25), described by Wendt et al. (13), was used for CTCF and cohesin binding: For 5′-CAGCTCTGTGTCCTGTCTTATCC-3′, Rev 5′-CAGCTATAATTGATGAAGAGGCG-3′. Normalized values of enrichment were grouped according to molecular defect at 11p15.5 and graphed as box plot.
Quantitative chromatin conformation capture (q3C)
q3C was performed as described previously (31–34) with the following modifications. Subconfluent cells from a 15 cm2 plate were fixed in 1% formaldehyde for 10 min at 37°C. After washing out the formaldehyde with ice-cold PBS, cells were collected and suspended in 1 ml lysis buffer (50 mm Tris–HCl pH 8, 1% SDS, 10 mm EDTA + 1/500 Protease Inhibitor Cocktail (PI) (P8340, Sigma)]. The cells were passed five times through a syringe needle and incubated on ice for 10 min. Nuclei were collected by centrifugation at 3500 rpm for 5 min. 1.5 × 106 nuclei were resuspended in 300 μl of 1X Buffer 3 [R0136M, New England Biolabs (NEB) with added protease inhibitors (P8340, Sigma) and 1.8% Triton X-100 (T8787, Sigma)]. Nuclei were digested overnight with 1000U of BamHI (R0136M, NEB) in a 300 μl reaction volume.
Ligation was carried out at 2.5 ng/μl final concentration of digested chromatin in a 1.5 ml reaction volume of T4 ligase buffer (B0202S, NEB) containing 3200 U of T4 ligase (M0202L, NEB). A further overnight digestion step with 1000 U of EcoRI (R0101M, NEB), which cuts outside the hybrid ligation products, was incorporated prior to reversal of crosslinks to shorten the size of the 3C template and increase the PCR efficiency. After this step, the samples were ethanol precipitated. Quality control checks for BamHI digestion efficiency was assessed by qPCR across each restriction site. The percentage of digestion was determined by comparing template amplification of digested (not relegated) and undigested fractions after normalizing to copy number of genomic DNA. If digestion efficiency was below 70%, the chromatin was discarded.
3C primers design and qPCR
3C qPCR primers flanking BamHI restriction sites were as previously described (14). PCR efficiency of each primer combination was assessed on a 3C PCR standard. Ten thousand copies of 3C template for each primer combination (measured by qPCR amplification of a region excluding BamHI sites) were then used in a qPCR reaction for the 3C analysis. A 384-well real-time machine (7900HT Fast Real time PCR system, Applied Biosystems) was used for the PCR amplification as previously described (14). Quantification of 3C interactions was based on standard curves for each primer set.
3C standard template preparation
A stock of PCR standard template was prepared similar to that described previously (34) by amplification of 36 genomic regions across BamHI RSs in the IGF2-H19 locus on commercially obtained genomic DNA (Becton Dickinson). These amplicons were column purified (DNA Clean α Concentrator™ 25 Kit, Zymo Research) and quantified using a Picogreen assay. Equimolar amounts of amplicons were mixed, BamHI digested, re-ligated, purified by ethanol precipitation and dissolved in H2O.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by a Cancer Research UK Senior Cancer Research Fellowship (A.M.); Association for International Cancer Research Project grant (06-0589) (A.M.), MIUR PRIN 2007 (A.R.), Telethon- Italia grant N. GGP07086 (A.R.), Associazione Italiana Ricerca sul Cancro (A.R.) and the support of the University of Cambridge, Cancer Research UK and Hutchison Whampoa Limited. A.S. was recipient of a fellowship from Fondazione Italiana Ricerca sul Cancro. Funding to pay the Open Access publication charges for this article was provided by Cancer Research UK.
ACKNOWLEDGEMENTS
We thank V. Orlando's group for their advice on ChIP protocols.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Weksberg R., Shuman C., Beckwith J.B. Beckwith-Wiedemann syndrome. Eur. J. Hum. Genet. 2010;18:8–14. doi: 10.1038/ejhg.2009.106. doi:10.1038/ejhg.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eggermann T. Silver-Russell and Beckwith-Wiedemann syndromes: opposite (epi)mutations in 11p15 result in opposite clinical pictures. Horm. Res. 2009;71(Suppl. 2):30–35. doi: 10.1159/000192433. doi:10.1159/000192433. [DOI] [PubMed] [Google Scholar]
- 3.Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. doi:10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 4.Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. doi:10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 5.Szabo P., Tang S.H., Rentsendorj A., Pfeifer G.P., Mann J.R. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. doi:10.1016/S0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- 6.Kurukuti S., Tiwari V.K., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z., Lobanenkov V., Reik W., Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl Acad. Sci. USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. doi:10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murrell A., Heeson S., Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. doi:10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 8.Engel N., Raval A.K., Thorvaldsen J.L., Bartolomei S.M. Three-dimensional conformation at the H19/Igf2 locus supports a model of enhancer tracking. Hum. Mol. Genet. 2008;17:3021–3029. doi: 10.1093/hmg/ddn200. doi:10.1093/hmg/ddn200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu X., Vu T.H., Lu Q., Ling J.Q., Li T., Hou A., Wang S.K., Chen H.L., Hu J.F., Hoffman A.R. A complex deoxyribonucleic acid looping configuration associated with the silencing of the maternal Igf2 allele. Mol. Endocrinol. 2008;22:1476–1488. doi: 10.1210/me.2007-0474. doi:10.1210/me.2007-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H.C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. doi:10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Rubio E.D., Reiss D.J., Welcsh P.L., Disteche C.M., Filippova G.N., Baliga N.S., Aebersold R., Ranish J.A., Krumm A. CTCF physically links cohesin to chromatin. Proc. Natl Acad. Sci. USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. doi:10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stedman W., Kang H., Lin S., Kissil J.L., Bartolomei M.S., Lieberman P.M. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. doi:10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. doi:10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 14.Nativio R., Wendt K.S., Ito Y., Huddleston J.E., Uribe-Lewis S., Woodfine K., Krueger C., Reik W., Peters J.M., Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. doi:10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt D., Schwalie P.C., Ross-Innes C.S., Hurtado A., Brown G.D., Carroll J.S., Flicek P., Odom D.T. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. doi:10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S., et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. doi:10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kacem S., Feil R. Chromatin mechanisms in genomic imprinting. Mamm. Genome. 2009;20:544–556. doi: 10.1007/s00335-009-9223-4. doi:10.1007/s00335-009-9223-4. [DOI] [PubMed] [Google Scholar]
- 18.Ciccone D.N., Su H., Hevi S., Gay F., Lei H., Bajko J., Xu G., Li E., Chen T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–418. doi: 10.1038/nature08315. doi:10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- 19.Higashimoto K., Urano T., Sugiura K., Yatsuki H., Joh K., Zhao W., Iwakawa M., Ohashi H., Oshimura M., Niikawa N., et al. Loss of CpG methylation is strongly correlated with loss of histone H3 lysine 9 methylation at DMR-LIT1 in patients with Beckwith-Wiedemann syndrome. Am. J. Hum. Genet. 2003;73:948–956. doi: 10.1086/378595. doi:10.1086/378595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grafodatskaya D., Choufani S., Ferreira J.C., Butcher D.T., Lou Y., Zhao C., Scherer S.W., Weksberg R. EBV transformation and cell culturing destabilizes DNA methylation in human lymphoblastoid cell lines. Genomics. 2010;95:73–83. doi: 10.1016/j.ygeno.2009.12.001. doi:10.1016/j.ygeno.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Cerrato F., Sparago A., Farina L., Ferrero G.B., Silengo M.C., Riccio A. Reply to ‘Microdeletion and IGF2 loss of imprinting in a cascade causing Beckwith-Wiedemann syndrome with Wilms’ tumor'. Nat. Genet. 2005;37:786–787. doi: 10.1038/ng0805-785. doi:10.1038/ng0805-786. [DOI] [PubMed] [Google Scholar]
- 22.Gicquel C., Rossignol S., Cabrol S., Houang M., Steunou V., Barbu V., Danton F., Thibaud N., Le Merrer M., Burglen L., et al. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat. Genet. 2005;37:1003–1007. doi: 10.1038/ng1629. doi:10.1038/ng1629. [DOI] [PubMed] [Google Scholar]
- 23.Sparago A., Cerrato F., Vernucci M., Ferrero G.B., Silengo M.C., Riccio A. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat. Genet. 2004;36:958–960. doi: 10.1038/ng1410. doi:10.1038/ng1410. [DOI] [PubMed] [Google Scholar]
- 24.Lindroth A.M., Park Y.J., McLean C.M., Dokshin G.A., Persson J.M., Herman H., Pasini D., Miro X., Donohoe M.E., Lee J.T., et al. Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus. PLoS Genet. 2008;4:e1000145. doi: 10.1371/journal.pgen.1000145. doi:10.1371/journal.pgen.1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McEwen K.R., Ferguson-Smith A.C. Distinguishing epigenetic marks of developmental and imprinting regulation. Epigenetics Chromatin. 2010;3:2. doi: 10.1186/1756-8935-3-2. doi:10.1186/1756-8935-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanz L.A., Chamberlain S., Sabourin J.C., Henckel A., Magnuson T., Hugnot J.P., Feil R., Arnaud P. A mono-allelic bivalent chromatin domain controls tissue-specific imprinting at Grb10. EMBO J. 2008;27:2523–2532. doi: 10.1038/emboj.2008.142. doi:10.1038/emboj.2008.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monk D. Deciphering the cancer imprintome. Brief Funct. Genomics. 2010;9:329–339. doi: 10.1093/bfgp/elq013. [DOI] [PubMed] [Google Scholar]
- 28.Elizondo L.I., Jafar-Nejad P., Clewing J.M., Boerkoel C.F. Gene clusters, molecular evolution and disease: a speculation. Curr. Genomics. 2009;10:64–75. doi: 10.2174/138920209787581271. doi:10.2174/138920209787581271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sparago A., Russo S., Cerrato F., Ferraiuolo S., Castorina P., Selicorni A., Schwienbacher C., Negrini M., Ferrero G.B., Silengo M.C., et al. Mechanisms causing imprinting defects in familial Beckwith-Wiedemann syndrome with Wilms’ tumour. Hum. Mol. Genet. 2007;16:254–264. doi: 10.1093/hmg/ddl448. doi:10.1093/hmg/ddl448. [DOI] [PubMed] [Google Scholar]
- 30.Breiling A., O'Neill L.P., D'Eliseo D., Turner B.M., Orlando V. Epigenome changes in active and inactive polycomb-group-controlled regions. EMBO Rep. 2004;5:976–982. doi: 10.1038/sj.embor.7400260. doi:10.1038/sj.embor.7400260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat. Methods. 2006;3:17–21. doi: 10.1038/nmeth823. doi:10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 32.Hagege H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. doi:10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 33.Miele A., Gheldof N., Tabuchi T.M., Dostie J., Dekker J. Mapping chromatin interactions by chromosome conformation capture. Curr. Protoc. Mol. Biol. 2006 doi: 10.1002/0471142727.mb2111s74. Chapter 21, Unit 21 11. [DOI] [PubMed] [Google Scholar]
- 34.Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. doi:10.1016/S1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.