Abstract
An expanded polyglutamine tract (>37 glutamines) in the N-terminal region of huntingtin (htt) causes htt to accumulate in the nucleus, leading to transcriptional dysregulation in Huntington disease (HD). In HD knock-in mice that express full-length mutant htt at the endogenous level, mutant htt preferentially accumulates in the nuclei of striatal neurons, which are affected most profoundly in HD. The mechanism underlying this preferential nuclear accumulation of mutant htt in striatal neurons remains unknown. Here, we report that serine 16 (S16) in htt is important for the generation of small N-terminal fragments that are able to accumulate in the nucleus and form aggregates. Phosphorylation of N-terminal S16 in htt promotes the nuclear accumulation of small N-terminal fragments and reduces the interaction of N-terminal htt with the nuclear pore complex protein Tpr. Mouse brain striatal tissues show increased S16 phosphorylation and a decreased association between mutant N-terminal htt and Tpr. These findings provide mechanistic insight into the nuclear accumulation of mutant htt and the selective neuropathology of HD, revealing potential therapeutic targets for treating this disease.
INTRODUCTION
Huntington disease (HD) is a late-onset neurodegenerative disorder caused by a polyglutamine (polyQ) expansion (>37 glutamines) in the N-terminal region of huntingtin (htt). Full-length htt, a 350 kDa protein, is predominantly localized in the cytoplasm; however, mutant htt with an expanded polyQ tract can accumulate in the nucleus and forms nuclear inclusions (1,2), the major histopathological hallmark of HD that is also seen in other polyQ diseases (3). The aberrant nuclear localization of mutant htt precedes neuropathology (4) and leads to transcriptional dysregulation via interactions with a number of transcription factors (5–7). Importantly, in HD knock-in (KI) mice that express full-length mutant htt under the control of the endogenous mouse htt promoter, mutant htt preferentially accumulates in the nuclei of neurons in the striatum (8–10), a brain region that is most profoundly affected in HD (11,12).
It is evident that mutant htt is localized in the nucleus and cytoplasm and affects a variety of cellular functions. Although the nuclear localization of mutant htt plays a critical role in gene transcriptional dysregulation, how mutant htt accumulates in neuronal nuclei remains a mystery. Transgenic mouse models of HD clearly show that N-terminal htt fragments can accumulate in neuronal nuclei in the brain and cause severe neurological symptoms (13,14). Consistently, various N-terminal htt fragments can be generated via proteolysis by a number of proteases, including caspases, calpain and matrix metalloproteinase (15–17). Recent studies showed that mimicking phosphorylation of both serine 13 and 16 in the N-terminal region of htt influences the nuclear localization and toxicity of mutant htt (18,19). These studies raised several important questions about HD pathogenesis. First, because full-length htt is found in the nucleus (20–22), but only N-terminal mutant htt is able to form nuclear inclusions (1,2), we need to know whether phosphorylation regulates the nuclear localization of full-length or N-terminal htt, a prerequisite for the toxic effect of mutant htt in the nucleus. Secondly, although double mutations of serine 13 and 16 indicate the effects of their phosphorylation on htt (18,19), knowing which serine residue is more important for the nuclear localization of mutant htt could point to a more specific therapeutic target. Also, because full-length htt plays vital roles in a variety of cellular functions, although N-terminal mutant htt causes a toxic gain of function, it is important to know whether post-translational modifications of different forms of htt lead to different consequences.
We found that phosphorylation of serine 16 (S16) promotes the nuclear localization and aggregation of N-terminal mutant htt. Mouse striatal tissues have increased S16 phosphorylation, which can reduce the association of N-terminal mutant htt with Tpr, a nuclear pore complex protein that is involved in the nuclear export of proteins (23–26). Our findings establish that S16 phosphorylation plays a critical role in the selective neuropathology of HD, suggesting a specific therapeutic target for its treatment.
RESULTS
Nuclear accumulation of N-terminal htt fragments is regulated by phosphorylation
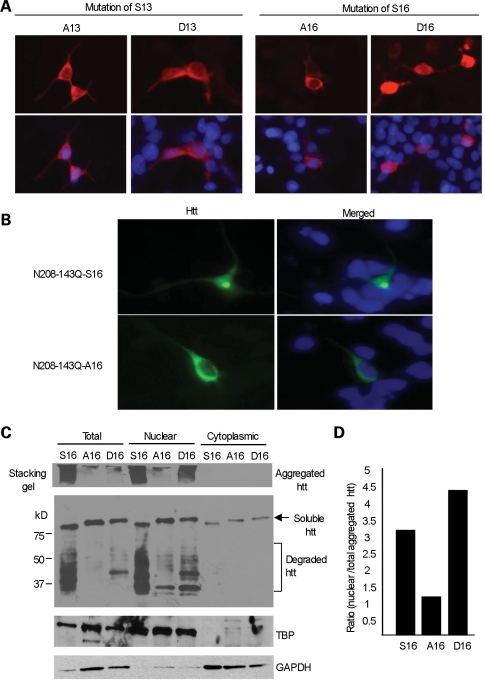
Investigation of various animal (27–33) and cellular (34–38) models has provided convincing evidence that small N-terminal fragments of mutant htt can accumulate in the nuclei of cells. Moreover, the nuclear accumulation of mutant htt is associated with neurological phenotypes in transgenic animals (37,38). Although phosphorylation of both S13 and S16 in N-terminal htt is known to increase the nuclear localization of htt in cultured cells (18), the question of which serine residue is most important for this nuclear localization remains unanswered. Using a mutagenesis approach to express transfected N-terminal htt (1–208 amino acids with 23Q) in HEK293 cells, we found that phosphorylation of S16 was more important than that of S13 in the mediation of htt nuclear localization, because replacing S16, rather than S13, with the phosphomimetic residue aspartic acid (D16) increased the nuclear localization of N-terminal htt in cultured HEK293 cells (Fig. 1A). To examine the subcellular localization of mutant htt, we transfected N-terminal mutant htt with S16 (N208-143Q-S16) in cultured primary rat brain striatal neurons. Increased nuclear localization of N208-143Q-S16 was also seen in transfected primary rat brain striatal neurons compared with N208-143Q-A16 (Fig. 1B). To more quantitatively assess the subcellular localization of transfected htt, we performed subcellular fractionation of htt-transfected HEK293 cells. Western blotting revealed that N208-143Q containing S16 substituted with the non-phosphorylatable residue, alanine (A16), displayed a decrease in the nuclear distribution of mutant htt (aggregated and degraded htt fragments) compared with htt containing S16 or the phosphomimetic residue aspartic acid (D16) (Fig. 1C). The ability of S16 phosphorylation to increase the nuclear distribution of htt is also supported by the ratios of nuclear aggregated htt to total aggregated htt in the lysates of those cells that expressed S16-, A16- or D16-htt (Fig. 1D).
Figure 1.
S16 of htt promotes the nuclear aggregation of N-terminal mutant htt. (A) HEK293 cells were transfected with N-terminal htt (N208-23Q), with S13 substituted by alanine (A13) or aspartic acid (D13), or with S16 substituted by alanine (A16) or aspartic acid (D16). Mimicking phosphorylation on S16 (D16) led to more nuclear distribution of htt than on S13 (D13). (B) Transfection of N208-143Q-S16 or -A16 in cultured rat striatal neurons showing that A16 substitution reduces the nuclear accumulation and aggregation of N-terminal mutant htt. (C) Subcellular fractionation of HEK293 cells transfected with mutant N-terminal htt (N208-143Q) containing S16, A16 or D16. EM48 western blotting reveals aggregated htt in the stacking gel, as well as soluble htt (arrow) and its degraded products (bracket) in the resolving gel. The blot was also probed with the antibody to the nuclear protein TATA-box-binding protein (TBP) and the cytoplasmic protein GAPDH. (D) The relative level of aggregated htt in the nucleus was determined using densitometry to calculate the ratio of nuclear aggregated htt to total aggregated htt.
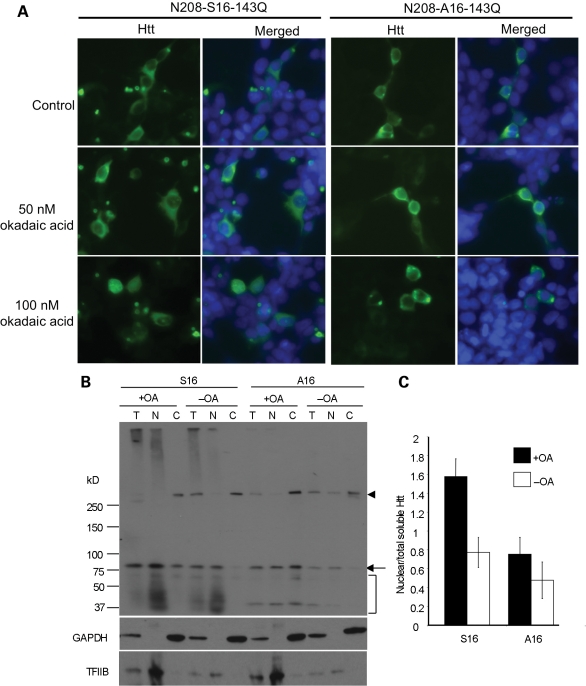
Because mutagenesis could induce protein conformational changes independent of phosphorylation, we also treated cultured cells with okadaic acid (OA), a phosphatase inhibitor that can induce cellular phosphorylation without affecting viability when it is used at 100 nm for 4 h. This treatment increased the nuclear accumulation of the aggregated and degraded forms of N208-143Q with S16, but not with A16 (Fig. 2). Immunocytochemical analysis confirmed the increased nuclear localization of transfected N208-143Q with S16 (Fig. 2A). We also noted that endogenous full-length human htt (arrowhead in Fig. 2B) in HEK293 cells showed no change in its nuclear distribution after OA treatment suggesting that only the nuclear localization of N-terminal mutant htt is regulated by S16 phosphorylation.
Figure 2.
OA-induced phosphorylation increased the levels of nuclear N208-143Q containing S16. (A) EM48 immunostaining of transfected HEK293 cells expressing N208-143Q-S16 or N208-143Q-A16 after treatment with OA (50 and 100 nm for 15 min). The control is cells without OA treatment. (B) Representative western blot (n = 3) of nuclear and cytosolic fractions of transfected HEK293 cells treated with 100 nm OA for 4 h. Substitution of S16 by alanine (A16) eliminated this increased nuclear level. The arrowhead indicates full-length normal htt in HEK293 cells. The arrow indicates soluble transfected htt. The bracket indicates degraded htt fragments. (C) Densitometry analysis of the ratio of nuclear soluble htt to htt in total lysates.
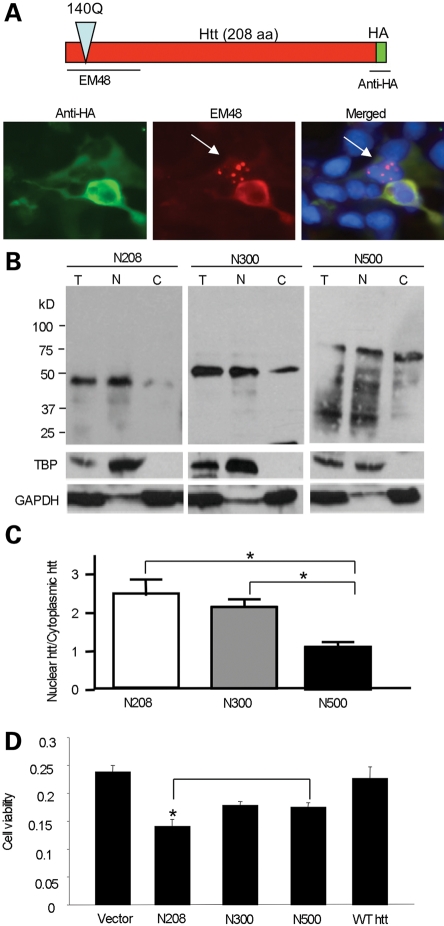
Because the A16 mutation reduces htt nuclear localization, as well as the amount of degraded products of N208-143Q and the aggregated htt (Fig. 1B), the smaller degraded products are likely to accumulate in the nucleus and form nuclear aggregates, which can also be promoted by S16 phosphorylation. To test this idea, we expressed N208-143Q, which was tagged with the HA epitope at its C-terminus, in HEK293 cells. As evidenced by the loss of anti-HA staining, the degraded htt products formed by the removal of the C-terminal HA epitope were able to accumulate in the nucleus (Fig. 3A). This finding indicates that proteolytic cleavage is an important step in generating smaller fragments that can more readily accumulate and form aggregates in the nucleus.
Figure 3.
Shorter N-terminal htt fragments are more prone to nuclear accumulation than longer N-terminal fragments. (A) Expression of N208-143Q with the HA epitope at its C-terminus in HEK293 cells. Double immunofluorescent staining reveals that proteolytic N-terminal fragments without the HA epitope form nuclear aggregates (arrow) that were only labeled by EM48. (B) N-terminal htt (N208, N300 or N500) of varying lengths (208, 300 or 500 amino acids) with 140-143Q was transfected into HEK293 cells. Subcellular fractionation followed by western blotting with antibodies to htt (EM48), the nuclear protein TBP and the cytoplasmic protein GAPDH. (C) The ratio of nuclear to cytoplasmic htt was calculated via densitometry of western blots (n = 3) to compare the amounts of nuclear accumulation of each N-terminal fragment (N208, N300, N500). (D) HEK293 cells were transfected with N208, N300 or N500 containing 140Q for 48 h. A wild-type N-terminal htt with 23Q (N208-23Q) served as a control. Cell lysates were used for an MTS assay to compare cell viability expressing htt fragments of different sizes. *P < 0.05.
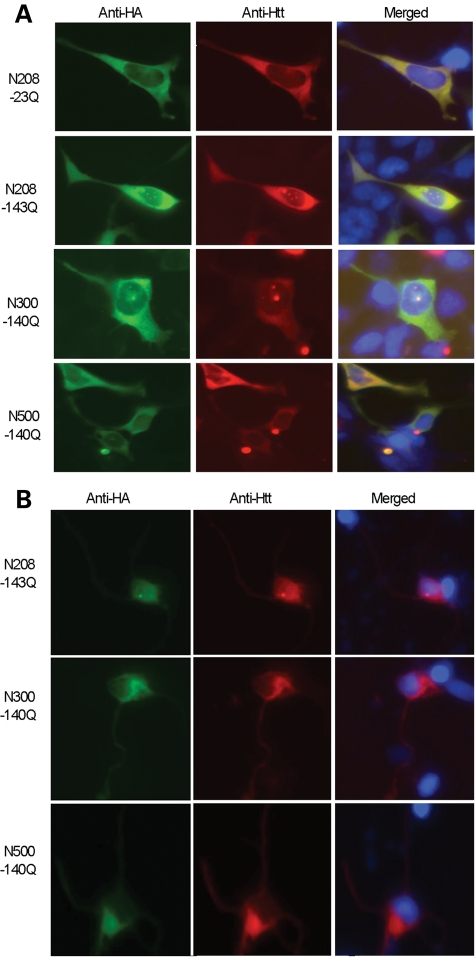
The nuclear localization of N-terminal mutant htt may occur via passive diffusion and may depend on the size of the N-terminal fragments. Subcellular fractionation confirmed that smaller N-terminal htt fragments show more nuclear localization than larger N-terminal htt fragments (Fig. 3B and C). This is also supported by immunofluorescent staining of transfected HEK293 cells and primary rat cortical neurons (Fig. 4). Furthermore, N208-143Q reduced the viability of cultured HEK293 cells compared with larger N-terminal mutant htt fragments (Fig. 3D). The increased nuclear accumulation of N208-104Q compared with larger fragments is consistent with the idea that nuclear localization of mutant htt can increase its cytotoxicity (34–38).
Figure 4.
Small N-terminal mutant htt fragments show increased nuclear accumulation in transfected cells. EM48 immunostaining of HEK293 cells (A) or primary rat striatal neurons (B) transfected with N-terminal mutant htt of different lengths (208, 300 and 500 amino acids) containing 140-143Q. Transfected htt was tagged with the HA epitope at its C-terminus. The nuclei were stained with Hoechst dye.
Increased S16 phosphorylation in mouse striatal tissues
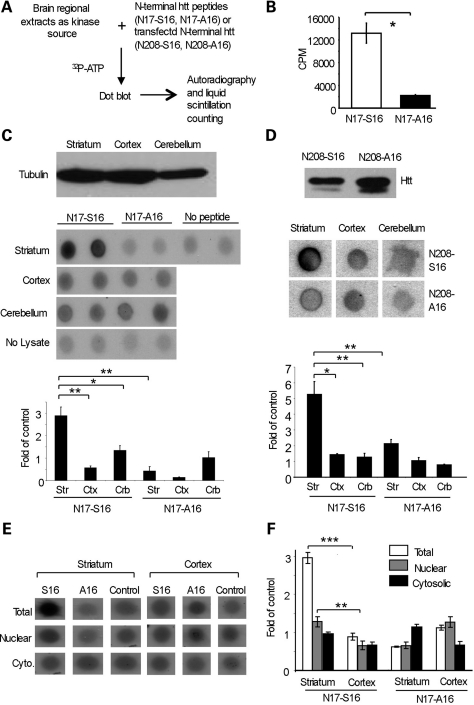
Because striatal neurons show preferential accumulation of mutant htt and because S16 phosphorylation can promote the nuclear localization of mutant htt, we wanted to examine whether levels of S16 phosphorylation are increased in striatal tissues. To this end, we synthesized two biotin-tagged peptides of the N-terminal 17 amino acids of htt containing either S16 (N17-S16) or A16 (N17-A16). The synthesized peptides were incubated with mouse striatal, cortical and cerebellar lysates as the kinase source and radiolabeled ATP as the phosphate source. The peptides were then isolated from the in vitro phosphorylation reaction with streptavidin beads and spotted on a nitrocellulose membrane for autoradiography or subjected to liquid scintillation counting to assess the phosphorylation levels (Fig. 5A). This assay revealed that mouse brain lysates do contain kinases or other cellular factors that can selectively increase the phosphorylation of S16, but not A16 peptides (Fig. 5B). Although liquid scintillation counting results (c.p.m.) varied because of the variable radioactivity of 32P used, this assay also verified that incubation with striatal lysates resulted in much more N17-S16 phosphorylation than incubation with the cortical or cerebellar lysates under the same in vitro assay conditions (lower panel in Fig. 5C). To test whether N-terminal htt also shows an increased ability to be phosphorylated in the striatum, we expressed N208-23Q containing either S16 (N208-S16) or A16 (N208-A16) (upper panel in Fig. 5D), as these proteins can be more efficiently immunoprecipitated by EM48 than aggregated htt. We used N208-S16 or N208-A16 as the substrates for the in vitro phosphorylation assay. As we saw with the peptides, the immunoprecipitated N208-S16 htt also showed more phosphorylation by the striatal tissue lysates than the cortical or cerebellar lysates (lower panel in Fig. 5D).
Figure 5.
Mouse striatal tissue lysates phosphorylate S16 of N-terminal htt at a higher level than lysates from other brain regions. (A) Lysates of the striatum, cortex and cerebellum from wild-type mice were incubated with N-terminal htt peptides or transfected N-terminal htt. The in vitro phosphorylation with 32P-ATP was analyzed by dot blotting and quantified by liquid scintillation counting. (B) The N-terminal 17 amino acid peptide of htt containing S16, but not A16, was phosphorylated in vitro using the method described in (A). (C) Western blotting for γ-tubulin was used to verify equal amounts of lysates (upper panel). Dot blot analysis (middle panel) and liquid scintillation counting (lower panel) of the in vitro phosphorylation reactions showed that striatal lysates significantly phosphorylated S16 of htt to a greater extent than the lysates from the cortex or cerebellum. (D) N208-23Q with S16 or A16 was expressed in transfected HEK293 cells and immunoprecipitated with EM48. Western blotting with mEM48 verified the precipitated htt (upper panel). Dot blot analysis (middle panel) and liquid scintillation counting (lower panel) from three independent experiments show that the precipitated N208-S16 htt was significantly more phosphorylated by the striatal lysates than the lysates from the cortex or cerebellum. (E) Total, nuclear and cytosolic fractions from the mouse striatum and cortex were used as the source of kinase to phosphorylate N17-16 and N17-A16. Dot blot analysis shows that the striatal nuclear fraction produced a higher level of S16 phosphorylation than the cytosolic fraction. (F) Liquid scintillation counting also revealed a significantly higher level of phosphorylated S16 by the striatal nuclear lysate, though the fractionation decreased the activity compared with total cell lysates. Control is the c.p.m. in the absence of peptides. *P < 0.05; **P < 0.01; ***P < 0.001.
We also used nuclear and cytosolic fractions from mouse brain tissues to assess their activity on S16 phosphorylation. Although the fractionation seemed to reduce nuclear S16 phosphorylation activity compared with S16 phosphorylation activity by the total lysates, the nuclear fraction from the striatal tissue showed higher S16 phosphorylation activity than either the cytosolic fraction or those fractions from the cortex (Fig. 5E and F). The normal function of S16 phosphorylation in the cytosolic fraction remains to be investigated. However, it is likely that the nuclear environment in striatal neurons favors S16 phosphorylation. The higher S16 phosphorylation activity in mouse striatal tissues is consistent with S16 phosphorylation promoting the nuclear localization of mutant htt and the preferential localization of N-terminal mutant htt in the mouse striatum.
Mutant htt with phosphorylated S16 is enriched in the striatal nuclei and shows decreased binding to Tpr
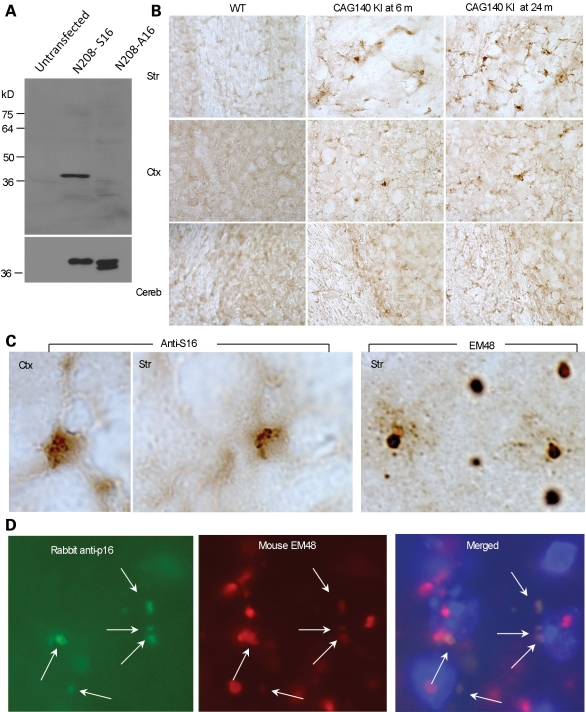
To further examine S16-phosphorylated htt in the mouse brain, we generated a phospho-specific antibody against phosphorylated S16 of htt (anti-S16). Western blot analysis of transfected htt showed that anti-S16 is specific to N208-S16, but not to N208-A16 (Fig. 6A). We next asked whether the striatum is enriched for htt phosphorylated at S16 in the mouse brain that expresses full-length mutant htt by performing immunohistochemistry on brain sections from CAG140 KI mice. Compared with the cortex and cerebellum, there was indeed more intense staining of the striatum by anti-S16. Also, there was an increase in anti-S16 staining in a 24-month-old mouse compared with a 6-month-old mouse (Fig. 6B). Under the same staining conditions, the wild-type mouse striatum showed no obvious anti-S16 staining. It is possible that phosphorylation of S16 in the normal mouse brain is transient or unstable, so that this phosphorylation can only be detected when the phosphorylated htt accumulates in aged HD mouse brains. High-magnification micrographs show that anti-S16 staining appeared to be diffuse in cells and in small aggregates, whereas EM48 intensely labeled large nuclear inclusions (Fig. 6C). We then performed double immunofluorescent staining to define the colocalization of proteins in distinct aggregates. Some small perinuclear EM48-positive aggregates, but not large nuclear aggregates, were labeled by anti-S16 (Fig. 6D), suggesting that S16 phosphorylation may be dynamic, becoming lost in the N-terminal htt in large inclusions. The staining of the small perinuclear htt aggregates by anti-S16 is consistent with previously published data showing that N-terminal htt accumulates in clusters at the perinuclear region prior to translocation into the nucleus (39).
Figure 6.
htt with phosphorylated S16 is more abundant in the striatal neurons and is partly colocalized with EM48-positive small aggregates at the perinuclear region. (A) Generation of a phospho-S16 (anti-S16)-specific antibody. The specificity of anti-S16 for phosphorylated S16 was verified by western blotting of lysates from HEK293 cells expressing either N208-23Q-S16 or N208-23Q-A16 (upper panel). The same blot was also probed with the mEM48 antibody (lower panel). (B) Immunohistochemistry with anti-S16 was used to compare the levels of S16-phosphorylated htt in the striatum (Str), cortex (Ctx) and cerebellum (Cereb) of full-length htt (CAG140) KI mice at 6 or 24 months of age. S16-phosphorylated htt in the striatum increased with age and was more abundant than in the cortex. (C) Anti-S16 labeled diffuse htt and small aggregates, whereas EM48 intensively labeled large nuclear htt inclusions in the striatum of a CAG140 KI mouse. (D) Double immunostaining of the striatum of a CAG140 KI mouse with anti-S16 (green) and mEM48 (red) revealed that S16-phosphorylated htt was localized to mEM48-positive small perinuclear aggregates (arrows) in the perinuclear region. Hoechst dye was used to stain the nucleus.
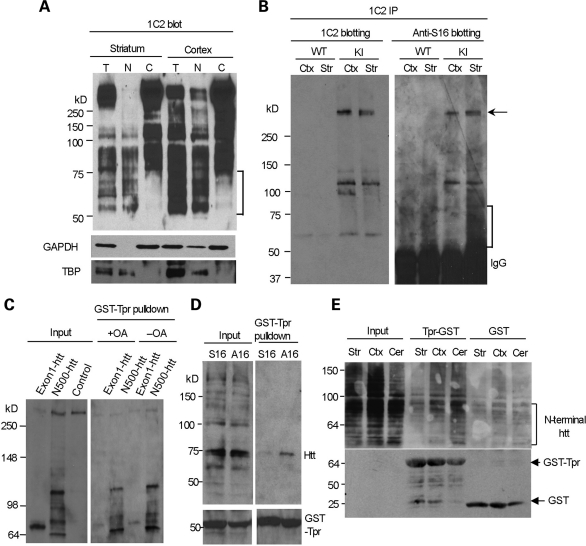
The 1C2 antibody, which reacts with the expanded polyQ domain and can specifically recognize mutant htt, was used to perform immunoblotting of subcellular fractions from CAG140 KI mice. The results revealed that htt fragments <75 kDa were enriched in the nuclear fraction (Fig. 7A). These fragments are comparable with degraded products from N208-143Q in transfected cells. To examine whether the striatum contains more S16-phosphorylated htt fragments in CAG140 KI mice, we performed immunoprecipitation with 1C2 to enrich for the mutant htt fragments, as transient or unstable phosphorylation may lead to low phosphorylated htt levels. The immunoprecipitates were then probed with anti-S16 to reveal phosphorylated htt, which showed that the striatum of the CAG140 KI mice contained more phosphorylated N-terminal htt fragments than the cortex (the bracket in Fig. 7B). Phosphorylation of full-length mutant htt (the arrow in Fig. 7B) also appeared to be more abundant in the striatum than the cortex.
Figure 7.
N-terminal mutant htt with phosphorylated S16 is enriched in the nuclear fraction of striatal neurons and shows decreased binding to Tpr. (A) 1C2 western blotting revealing that N-terminal mutant htt fragments <75 kDa (marked by the bracket) are present only in the total and nuclear fractions, but not in the cytosolic fractions, of the mouse striatum and cortex. The same blot was also probed with antibodies to the cytoplasmic protein GAPDH and the nuclear protein TBP. (B) Mutant htt in CAG140 KI mouse striatum and cortex was immunoprecipitated by 1C2. Western blots with anti-S16 and 1C2 showed more abundant N-terminal fragments (marked by the bracket) in the striatum that were labeled by anti-S16. Densitometry analysis (lower panel) of the relative percentage of phosphorylated full-length htt by measuring the intensity of htt labeled by anti-S16 relative to htt labeled by 1C2. (C) HEK293 cells were cotransfected with GST-Tpr and exon1-150Q or N500-htt-140Q htt. More N500-htt-140Q and its degraded products than exon1-150Q were precipitated by GST-Tpr. Note that very little full-length normal htt (arrow) was precipitated and that increased phosphorylation by OA treatment reduces the interaction of N-terminal htt with GST-Tpr. (D) HEK293 cells were cotransfected with GST-Tpr (bracket) and N208-143Q containing S16 or A16. A GST pulldown followed by western blotting with mEM48 showed that phosphorylation elimination by A16 substitution increased the association of htt with Tpr. (E) Striatal, cortical and cerebellar lysates were prepared from CAG140 KI mice and incubated with GST-Tpr or GST alone. Western blotting of the GST pulldown samples with 1C2 showed that the binding of N-terminal htt fragments (marked by the bracket) to Tpr was reduced in the striatum. The blot was also probed with an antibody to GST to reveal GST and GST-Tpr used for the precipitation of mouse brain mutant htt.
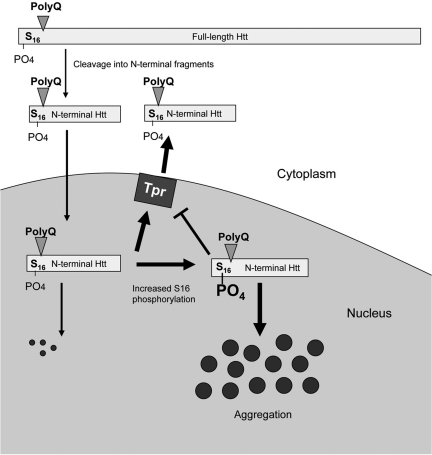
S16 phosphorylation may regulate htt's association with nuclear proteins to promote its nuclear accumulation. Since the nuclear export of htt involves the nuclear pore complex protein called Tpr (25), we explored whether S16 phosphorylation could alter the association of Tpr with N208-143Q. polyQ-expanded exon1 htt or N500-htt was cotransfected with GST-Tpr into HEK293 cells. A GST pulldown was used to show that more N500-htt and its degraded products than exon1 htt were pulled down. Importantly, endogenous full-length human htt did not bind GST-Tpr (Fig. 7C), suggesting that protein context influences the interaction of N-terminal mutant htt with Tpr. Increased cellular phosphorylation by OA treatment reduced the association of N-terminal mutant htt with GST-Tpr (Fig. 7C), further suggesting that phophorylation of S16 is important for the binding of N-terminal htt to Tpr. We then used transfected proteins in HEK293 cells to assess the binding of GST-Tpr to N208-143Q containing either S16 or A16. A GST pulldown experiment also revealed a decreased interaction between GST-Tpr and transfected N208-143Q htt containing S16 compared with N208-143Q containing the A16 substitution (Fig. 7D). This result suggests that when N-terminal mutant htt is phosphorylated at S16, its association with Tpr is decreased, which may lead to increased localization in the nucleus. Because mouse striatal tissues show increased S16 phosphorylation, we wanted to test whether N-terminal htt fragments in the striatum have a decreased association with Tpr; so we performed GST-Tpr pulldown to compare the association of N-terminal htt fragments with Tpr in different brain regions. As revealed by 1C2 staining of the pulldown samples, GST-Tpr associated with fewer N-terminal htt fragments in the striatum than in the cortex or cerebellum (Fig. 7E). These findings support the idea that striatal tissues are enriched for mutant htt phosphorylated at S16, which reduces the association of N-terminal htt with the nuclear export protein Tpr and increases the nuclear accumulation and aggregation of mutant htt, leading to the preferential nuclear accumulation of mutant htt in the striatum. A schematic diagram for this idea is also depicted in Figure 8.
Figure 8.
Schematic diagram of increased S16 phosphorylation and nuclear accumulation of mutant htt. In the cytoplasm, full-length mutant htt is proteolytically degraded into small N-terminal htt fragments, which can enter the nucleus. Increased phosphorylation of S16 in N-terminal mutant htt enhances its aggregation and reduces its association with the nuclear pore complex protein Tpr, resulting in the accumulation of mutant htt in the nucleus. The size of N-terminal htt does not represent its actual proportion of full-length htt.
DISCUSSION
Nuclear translocation of N-terminal htt fragments is an important process in HD pathogenesis, yet the mechanisms underlying this nuclear localization, especially the preferential nuclear accumulation of mutant htt in striatal neurons, remain unclear. Here we found that this nuclear localization is associated with increased phosphorylation of S16 of N-terminal htt and decreased interaction with the nuclear pore protein Tpr, which is involved in nuclear export of proteins (23–26). These results provide new insights into the selective neuropathology of HD and add to recent findings about the important roles of post-translational modification on HD neuropathology.
Although full-length htt is known to localize in the nucleus (20–22), various HD KI mouse models clearly show that N-terminal mutant htt can preferentially accumulate in the nuclei of striatal neurons (8–10). Understanding this phenomenon is important to explain the selective neurodegeneration seen in HD. Here we show that phosphorylation of S16 can promote the nuclear localization of N-terminal mutant htt. This is consistent with a recent report that phosphorylation of both S13 and S16 can increase nuclear htt accumulation (18), but our new finding reveals that S16 is more important than S13 for regulating the nuclear localization of mutant htt and that striatal tissues show increased S16 phosphorylation, providing a more selective target for future therapeutics.
Our results also support the idea that the nuclear import of htt is a passive diffusion process that depends on the size of imported proteins, since smaller N-terminal htt is more readily distributed in the nucleus. The fact that mutant htt shows preferential localization in the nuclei of striatal neurons in HD KI mice (8–10) suggests that tissue-specific factors are responsible for this preferential accumulation; however, this preferential accumulation is absent in transgenic R6/2 mice that express exon1 mutant htt under the control of the human htt promoter (40) and N171-82Q mice that express the first 171 amino acids of htt under the control of the mouse prion promoter (28). Analysis of various HD mouse models expressing full-length mutant htt demonstrates the presence of multiple N-terminal fragments at higher molecular weights than those of exon1 htt and N171-82Q (29,41–43). PolyQ-containing peptides become more aggregated when they contain fewer amino acids (other than the polyQ tract) and when they are expressed at higher levels (44). Also, we found that exon1 mutant htt binds to Tpr weakly compared with larger N-terminal mutant htt fragments. Thus, overexpression of small N-terminal htt, such as exon1 htt, can facilitate its aggregation in neuronal nuclei, thereby preventing its nuclear export and making its accumulation in the striatum no longer cell type dependent. When mutant htt is expressed at an endogenous level, N-terminal htt fragments that are larger than exon1 htt and can passively enter the nucleus may also be exported via passive diffusion or interaction with other nuclear proteins, such as Tpr. Aging is likely to increase the accumulation of N-terminal htt fragments, because of a reduced cellular capacity to refold or clear these fragments in the neuronal nucleus. Our findings suggest that the export of N-terminal htt fragments from the nucleus is more dependent on the interaction of these fragments with Tpr, which can be inhibited by S16 phosphorylation. Therefore neurons, including striatal neurons, with increased phosphorylation of S16 in htt would show a greater accumulation of mutant htt and formation of htt aggregates in their nuclei during aging.
The role of nuclear aggregates of polyQ proteins in neuronal degeneration remains to be defined. The HD mouse model we investigated in our study does not show obvious neurodegeneration and apoptosis. Species differences may account for the lack of overt neurodegeneration or apoptosis in HD mouse models (33). Despite this, the nuclear localization of htt has been established as a critical step toward HD pathology (34–38). However, recent studies suggest that phosphorylation of N-terminal mutant htt is protective against htt toxicity (18,19), although mimicking S13 and S16 phosphorylation also promotes nuclear htt accumulation (18). Because these studies used mutations of both S13 and S16, whether phosphorylation of S13 and S16 individually mediated different events was unclear. Since there is very little nuclear htt in the BAC transgenic HD mice (19), the protective effects seen in these mice could be due to the effect of phosphorylation on cytoplasmic htt, or it could result from a lower level of transgenic htt due to the replacement of both S13 and S16 by aspartic acid residues. A key insight from the above studies is that the phosphorylation of htt can increase its nuclear localization (18). Our study identified S16 phosphorylation as an important modulator in the generation of small htt fragments. These small N-terminal htt fragments can accumulate in the nucleus to affect gene expression or become inclusions that can also be protective.
Post-translational modifications are known to influence polyQ protein toxicity (45), and the effects of these modifications on the function of full-length htt appear to be diverse and complex (21,46–49). This is perhaps because full-length htt plays vital roles in a variety of cellular functions. Our finding that increased S16 phosphorylation occurs in the striatum uncovers a more specific target for reducing the selective nuclear accumulation of htt in neuronal cells. S16 phosphorylation in htt was not found to be mediated by IKK, which could phosphorylate S13 (18). The molecule(s) responsible for S16 phosphorylation remain to be identified. The increased S16 phosphorylation in striatal neurons could be due to increased activity of a kinase or reduced activity of a phosphatase in striatal neurons. There are many potential pathophysiological triggers that could cause aberrant phosphorylation. For example, age-dependent cellular stress can be the initiating event to alter kinase or phosphatase activity. Transcription dysregulation mediated by mutant htt could result in altered expression of the putative kinase or phosphatase. Identifying the cellular factors or signaling pathways responsible for this increased phosphorylation in striatal neurons would help in the development of effective therapeutics. Moreover, understanding the mechanism behind the selective accumulation of mutant htt in affected neurons also has broader implications for the selective neuropathology seen in other polyQ diseases that feature the nuclear accumulation of polyQ proteins.
MATERIALS AND METHODS
Animals
All animal procedures were approved by the Institutional Animal Care and Use Committee of Emory University. Full-length mutant htt KI (CAG140) mice were provided by Dr Michael Levine at UCLA (27) and were maintained in the animal facility at Emory University in accordance with institutional guidelines.
DNA constructs
PRK-HA vectors expressing htt fragments with N-terminal lengths of 208, 300 or 500 amino acids containing 140-143Q with the HA epitope at the C-terminus were generated. Site-directed mutagenesis was performed to replace S16 with alanine (A16) and aspartic acid (D16) in the htt constructs. PCR primers [reverse: 5′-ATGAAAGCCTTCGAGTCCCTCAAGGCCTTCCAG-3′ (A16) or 5′ATGAAAGCCTTCGAGTCCCTCAAGGACTTCCAG-3′ (D16); forward: 5′-ATTTCCTTATAGAGCTCGAGCTGTAACCTTGGAAGATTAGAATCC-3′] were designed for the first round to introduce the mutation. PCR primers (forward: 5′-ATTTCCTTATAGAGCTCGAGCTGTAACCTTGGAAGATTAGAATCC-3′; reverse: 5′-TAGGATCCGCCATGGCGACCCTGGAAAAGCTGATGAAGGCCTTCGAGTCCCTC-3′) were designed for the second round of PCR to introduce the BamHI cloning site into the construct for insertion into the PRK-HA vector. Constructs were sequenced through Macrogen to confirm the correct mutations.
Antibodies and reagents
The anti-htt antibodies (rabbit EM48 and mouse mEM48) were previously produced in our laboratory (50). A polyclonal antibody against phospho-S16 of htt was generated by AnaSpec, Inc., using a peptide containing the first 5–22 amino acids (CLEKLMKAFESLK-pS-FQQ) of htt with phosphorylated S16 as the antigen. The peptides containing S16 or A16 are labeled with biotin at the N-terminus and used for phosphorylation analysis. The mouse antibody 1C2 was purchased from Millipore (Temecula, CA, USA). The mouse anti-γ-tubulin antibody was purchased from Sigma-Aldrich (St Louis, MO, USA) and used at a 1:50 000 dilution. Additional antibodies used were against GAPDH (Ambion), TBP N-12 (Santa Cruz) and HA (Cell Signaling). Secondary antibodies were peroxidase-conjugated donkey anti-mouse or donkey anti-rabbit IgG (H+L) from Jackson ImmunoResearch (West Grove, PA, USA). OA was obtained from Sigma-Aldrich.
Cell culture, transient transfection and drug treatments
HEK293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS, 100 μg/ml penicillin, 100 units/ml streptomycin and 250 μg/μl fungizone amphotericin B. Cells were incubated at 37°C in 5% CO2. At a confluency of 70%, the cells were transfected with 1–2 μg/well (6-well plate) or 0.5–1 μg/well (12-well plate) of DNA and lipofectamine (Invitrogen) for 48 h. For phosphorylation studies, transfected cells were treated with 1.5 μm OA for 15 min in serum-free media.
Immunocytochemistry
Male and female mice were anesthetized and perfused intracardially with phosphate-buffered saline (PBS, pH 7.2) for 30s followed by 4% paraformaldehyde in 0.1 m phosphate buffer (PB) at pH 7.2. Brains were removed, cryoprotected in 30% sucrose at 4°C and sectioned at 40 µm using a freezing microtome. Free-floating sections were in 4% paraformaldehyde in 0.1 m PB for 10 min and preblocked in 4% normal goat serum in PBS, 0.1% Triton X, and then incubated with the phospho-S16 or mEM48 antibody at 4°C for 48 h. The immunoreactive product was visualized with the Avidin–Biotin Complex Kit (Vector ABC Elite, Burlingame, CA, USA).
Transfected HEK293 cells were fixed with 4% paraformaldehyde for 15 min and then blocked for 1 h with 3% BSA and 0.2% Triton X-100 in 1× PBS. Cells were incubated overnight with mEM48 (1:100 dilution) in 3% BSA in 1× PBS. The nucleus was visualized using Hoechst staining (Molecular Probes) at a dilution of 1:5000. Fluorescent images were obtained using a Zeiss microscope (Axiaovert 200 MOT) with a digital camera (Hamamatsu Orca-100) and Openlab software (Improvision, Inc.).
Primary neuron culture and transfection
Neurons from the striatum and cortex of E18 rats were isolated and washed in Hank's balanced salt solution (HBSS; Invitrogen). Neurons were treated with 2.5% trypsin in HBSS at 37°C for 10 min and then cultured in sterile-filtered neurobasal medium (2 mm l-glutamine, 2% B-27 supplement, 100 U/ml streptomycin and 100 U/ml penicillin) on poly-d-lysine-coated plates. At 50–70% confluence in a 12-well plate, the cells were transfected with 2 μg of DNA/well and lipofectamine 2000 (Invitrogen). After 5 h, the transfection reagent was removed and replaced with half fresh neurobasal medium and half pre-transfection neurobasal medium.
Cell viability assays
An MTS assay kit (Promega G3850) was used to measure cell viability. Transfected HEK293 cells were collected in cold serum-free medium and washed twice. The cells were resuspended in serum-free medium, and 75 μl of the cells and the MTS reagent were added to a well of a 96-well plate in triplicate. After 30 min at 37°C, a SpectraMax Plus plate reader (Molecular Devices) was used to quantify color changes corresponding to cell viability.
Immunoprecipitation
Transfected cells or mouse brain tissue lysates were collected and lysed in Nonidet P-40 lysis buffer [50 mm Tris–HCl (pH 7.4), 50 mm NaCl, 0.2% Triton X-100, 1% Nonidet P-40, 1 mm PMSF, 1:100 protease inhibitor cocktail (P8340, Sigma), 10 mm NaF and 0.2 mm NaVO3]. Lysates were cleared with 50 μl of protein A beads (Sigma) for 1 h at 4°C. Cleared lysates were incubated overnight with mEM48 at a dilution of 1:10 at 4°C while rocking. Twenty-five microliters of fresh protein A beads were added for 2 h and then washed three times with Nonidet P-40 buffer. 1% SDS was added to the beads which were then boiled and analyzed by western blotting.
GST pulldown
Transfected cells were lysed in Nonidet P-40 lysis buffer. GSH beads (50 μl) were added to each reaction. The reactions were incubated at 4°C for 3 h of rocking. The beads were isolated by centrifuging at 1000r.p.m. for 30 s. The beads were washed three times with NP-40 buffer and resusupended in 1× SDS-protein-loading dye.
Nuclear fractionation and western blotting
To isolate the nuclear fraction from transfected HEK293 cells, the cells were collected and incubated on ice for 10 min in buffer A [20 mm Tris–HCl, pH 7.4, 10 mm NaCl, 3 mm MgCl2, 1 mm PMSF, 10 mm NaF, 0.2 mm NaVO3 and 1:100 protease inhibitor cocktail (P8340, Sigma)]. Cells were passed through a 22-g syringe to release the cytoplasmic protein. Cells were centrifuged at 12 000g for 30s to collect the intact nuclei. The supernatant was clarified at 16 000r.p.m. for 5 min and saved as the cytoplasmic fraction. The nuclei were washed with buffer A three times and lysed in PBS lysis buffer [1× PBS, 1% Triton X-100 and 1:100 protease inhibitor cocktail (P8340, Sigma)].
Brain tissues from mice at 4–8 months of age were homogenized for 20 strokes with a glass-on-glass homogenizer in a homogenizing buffer at 0.1 g/ml [0.25 m sucrose, 15 mm Tris–HCl, pH 7.9, 60 mm KCl, 5 mm EDTA, 1 mm EGTA, 100 μg/ml PMSF, 10 mm NaF, 0.2 mm NaVO3 and 1:100 protease inhibitor cocktail (P8340, Sigma)]. After incubating on ice for 10 min, the intact nuclei were isolated by centrifuging at 2000g for 5 min. The supernatant was clarified at 16 000 r.p.m. for 5 min and saved as the cytoplasmic fraction. The nuclear pellet was washed twice with homogenizing buffer and then lysed in 1× PBS lysis buffer.
In vitro phosphorylation assay
The striatum, cortex and cerebellum from a wild-type male and female C3/BL6 mouse at 4–8 months of age were homogenized in Nonidet P-40 lysis buffer [50 mm Tris–HCl (pH 7.4), 50 mm NaCl, 0.2% Triton X-100, 1% Nonidet P-40, 1 mm PMSF, 1:100 protease inhibitor cocktail (P8340, Sigma), 10 mm NaF and 0.2 mm NaVO3] at 2 mg/ml. The S16 and A16 peptides or transfected N208-S16 and N208-A16 immunoprecipitated from HEK293 cells were incubated in 1× kinase buffer [20 mm HEPES (pH 7.6), 5 mm MgCl2, 55 mm KCl and 10 μm unlabeled ATP], lysate from the striatum, cortex or cerebellum and 2 μCi of [γ-32P] ATP (Perkin Elmer) in a total reaction volume of 20 μl for 1 h at 30°C. The peptides were separated from the rest of the reaction by adding 100 μl of cold 1× PBS and 20 μl of high-capacity streptavidin beads (Pierce) to the reaction and by incubating at 4°C while rocking for 2 h. The beads were washed with 1× PBS and the peptide was eluted with 8 m guanidine at pH 1.5. For the immunoprecipitated htt, the kinase reaction was performed with htt attached to the beads. The beads were isolated from the reaction and washed with 1× PBS. Htt was eluted from the beads with 0.1 m glycine (pH 2.8). Two microliters of the eluate was spotted onto a nitrocellulose membrane and exposed to autoradiography film. Phosphorylation levels were quantified by adding 2 μl of each reaction to 5 ml of scintillation fluid (Ecolume) and by counting the radioactive units with a liquid scintillation counter (Beckman LS 6500). All counts were measured in triplicate.
Statistical analysis
Results generated from three or more independent experiments are expressed as the mean ± SD and were analyzed for statistical significance using a two-tailed Student's t-test.
FUNDING
This work was supported by grants from the National Institutes of Health (AG019206 and NS041669 to X.-J.L.; NS045016 and AG031153 to S.H.L.).
ACKNOWLEDGEMENTS
We thank Dr Scott Zeitlin for providing a neo-stop vector, Ashley O'Neill for assisting with the mouse genotyping analysis and Cheryl Strauss for critical reading of the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. doi:10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 2.Gutekunst C.A., Li S.H., Yi H., Mulroy J.S., Kuemmerle S., Jones R., Rye D., Ferrante R.J., Hersch S.M., Li X.J. Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J. Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. doi:10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 4.Van Raamsdonk J.M., Murphy Z., Slow E.J., Leavitt B.R., Hayden M.R. Selective degeneration and nuclear localization of mutant huntingtin in the YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 2005;14:3823–3835. doi: 10.1093/hmg/ddi407. doi:10.1093/hmg/ddi407. [DOI] [PubMed] [Google Scholar]
- 5.Landles C., Bates G.P. Huntingtin and the molecular pathogenesis of Huntington's disease. Fourth in molecular medicine review series. EMBO Rep. 2004;5:958–963. doi: 10.1038/sj.embor.7400250. doi:10.1038/sj.embor.7400250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadri-Vakili G., Cha J.H. Mechanisms of disease: histone modifications in Huntington's disease. Nat. Clin. Pract. Neurol. 2006;2:330–338. doi: 10.1038/ncpneuro0199. doi:10.1038/ncpneuro0199. [DOI] [PubMed] [Google Scholar]
- 7.Havel L.S., Li S., Li X.J. Nuclear accumulation of polyglutamine disease proteins and neuropathology. Mol. Brain. 2009;2:21. doi: 10.1186/1756-6606-2-21. doi:10.1186/1756-6606-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler V.C., White J.K., Gutekunst C.A., Vrbanac V., Weaver M., Li X.J., Li S.H., Yi H., Vonsattel J.P., Gusella J.F., et al. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum. Mol. Genet. 2000;9:503–513. doi: 10.1093/hmg/9.4.503. doi:10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- 9.Li H., Li S.H., Johnston H., Shelbourne P.F., Li X.J. Amino-terminal fragments of mutant huntingtin show selective accumulation in striatal neurons and synaptic toxicity. Nat. Genet. 2000;25:385–389. doi: 10.1038/78054. doi:10.1038/78054. [DOI] [PubMed] [Google Scholar]
- 10.Lin C.H., Tallaksen-Greene S., Chien W.M., Cearley J.A., Jackson W.S., Crouse A.B., Ren S., Li X.J., Albin R.L., Detloff P.J. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum. Mol. Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. doi:10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 11.Gusella J.F., MacDonald M.E., Ambrose C.M., Duyao M.P. Molecular genetics of Huntington's disease. Arch. Neurol. 1993;50:1157–1163. doi: 10.1001/archneur.1993.00540110037003. [DOI] [PubMed] [Google Scholar]
- 12.Vonsattel J.P., DiFiglia M. Huntington disease. J. Neuropathol. Exp. Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. doi:10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Menalled L.B., Chesselet M.F. Mouse models of Huntington's disease. Trends Pharmacol. Sci. 2002;23:32–39. doi: 10.1016/s0165-6147(00)01884-8. doi:10.1016/S0165-6147(00)01884-8. [DOI] [PubMed] [Google Scholar]
- 14.Heng M.Y., Detloff P.J., Albin R.L. Rodent genetic models of Huntington disease. Neurobiol. Dis. 2008;32:1–9. doi: 10.1016/j.nbd.2008.06.005. doi:10.1016/j.nbd.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Wellington C.L., Leavitt B.R., Hayden M.R. Huntington disease: new insights on the role of huntingtin cleavage. J. Neural. Transm. Suppl. 2000;58:1–17. doi: 10.1007/978-3-7091-6284-2_1. [DOI] [PubMed] [Google Scholar]
- 16.Imarisio S., Carmichael J., Korolchuk V., Chen C.W., Saiki S., Rose C., Krishna G., Davies J.E., Ttofi E., Underwood B.R., et al. Huntington's disease: from pathology and genetics to potential therapies. Biochem. J. 2008;412:191–209. doi: 10.1042/BJ20071619. doi:10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 17.Miller J.P., Holcomb J., Al-Ramahi I., de Haro M., Gafni J., Zhang N., Kim E., Sanhueza M., Torcassi C., Kwak S., et al. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington's disease. Neuron. 2010;67:199–212. doi: 10.1016/j.neuron.2010.06.021. doi:10.1016/j.neuron.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson L.M., Aiken C.T., Kaltenbach L.S., Agrawal N., Illes K., Khoshnan A., Martinez-Vincente M., Arrasate M., O'Rourke J.G., Khashwji H., et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J. Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. doi:10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu X., Greiner E.R., Mishra R., Kodali R., Osmand A., Finkbeiner S., Steffan J.S., Thompson L.M., Wetzel R., Yang X.W. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. doi:10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler V.C., White J.K., Gutekunst C.A., Vrbanac V., Weaver M., Li X.J., Li S.H., Yi H., Vonsattel J.P., Gusella J.F., et al. Early phenotypes that presage late-onset neurodegenerative disease allow testing of modifiers in Hdh CAG knock-in mice. Hum. Mol. Genet. 2000;11:633–640. doi: 10.1093/hmg/11.6.633. doi:10.1093/hmg/11.6.633. [DOI] [PubMed] [Google Scholar]
- 21.Jeong H., Then F., Melia T.J., Jr, Mazzulli J.R., Cui L., Savas J.N., Voisine C., Paganetti P., Tanese N., Hart A.C., et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. doi:10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godin J.D., Colombo K., Molina-Calavita M., Keryer G., Zala D., Charrin B.C., Dietrich P., Volvert M.L., Guillemot F., Dragatsis I., et al. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron. 2010;67:392–406. doi: 10.1016/j.neuron.2010.06.027. doi:10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 23.Bangs P., Burke B., Powers C., Craig R., Purohit A., Doxsey S. Functional analysis of Tpr: identification of nuclear pore complex association and nuclear localization domains and a role in mRNA export. J. Cell Biol. 1998;143:1801–1812. doi: 10.1083/jcb.143.7.1801. doi:10.1083/jcb.143.7.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frosst P., Guan T., Subauste C., Hahn K., Gerace L. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J. Cell Biol. 2002;156:617–630. doi: 10.1083/jcb.200106046. doi:10.1083/jcb.200106046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cornett J., Cao F., Wang C.E., Ross C.A., Bates G.P., Li S.H., Li X.J. Polyglutamine expansion of huntingtin impairs its nuclear export. Nat. Genet. 2005;37:198–204. doi: 10.1038/ng1503. doi:10.1038/ng1503. [DOI] [PubMed] [Google Scholar]
- 26.Ben-Efraim I., Frosst P.D., Gerace L. Karyopherin binding interactions and nuclear import mechanism of nuclear pore complex protein Tpr. BMC Cell Biol. 2009;10:74. doi: 10.1186/1471-2121-10-74. doi:10.1186/1471-2121-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickey M.A., Kosmalska A., Enayati J., Cohen R., Zeitlin S., Levine M.S., Chesselet M.F. Extensive early motor and non-motor behavioral deficits are followed by striatal neuronal loss in knock-in Huntington's disease mice. Neuroscience. 2008;157:280–295. doi: 10.1016/j.neuroscience.2008.08.041. doi:10.1016/j.neuroscience.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schilling G., Becher M.W., Sharp A.H., Jinnah H.A., Duan K., Kotzuk J.A., Slunt H.H., Ratovitski T., Cooper J.K., Jenkins N.A., et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum. Mol. Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. doi:10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 29.Woodman B., Butler R., Landles C., Lupton M.K., Tse J., Hockly E., Moffitt H., Sathasivam K., Bates G.P. The Hdh(Q150/Q150) knock-in mouse model of HD and the R6/2 exon 1 model develop comparable and widespread molecular phenotypes. Brain Res. Bull. 2007;72:83–97. doi: 10.1016/j.brainresbull.2006.11.004. doi:10.1016/j.brainresbull.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Heng M.Y., Tallaksen-Greene S.J., Detloff P.J., Albin R.L. Longitudinal evaluation of the Hdh(CAG)150 knock-in murine model of Huntington's disease. J. Neurosci. 2007;27:8989–8998. doi: 10.1523/JNEUROSCI.1830-07.2007. doi:10.1523/JNEUROSCI.1830-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Z.X., Li S.H., Evans J., Pillarisetti A., Li H., Li X.J. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington's disease. J. Neurosci. 2003;23:2193–2202. doi: 10.1523/JNEUROSCI.23-06-02193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradford J., Shin J.Y., Roberts M., Wang C.E., Li X.J., Li S. Expression of mutant huntingtin in mouse brain astrocytes causes age-dependent neurological symptoms. Proc. Natl Acad. Sci. USA. 2009;106:22480–22485. doi: 10.1073/pnas.0911503106. doi:10.1073/pnas.0911503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang D., Wang C.E., Zhao B., Li W., Ouyang Z., Liu Z., Yang H., Fan P., O'Neill A., Gu W., et al. Expression of Huntington's disease protein results in apoptotic neurons in the brains of cloned transgenic pigs. Hum. Mol. Genet. 2010;19:3983–3994. doi: 10.1093/hmg/ddq313. doi:10.1093/hmg/ddq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hackam A.S., Singaraja R., Wellington C.L., Metzler M., McCutcheon K., Zhang T., Kalchman M., Hayden M.R. The influence of huntingtin protein size on nuclear localization and cellular toxicity. J. Cell Biol. 1998;141:1097–1105. doi: 10.1083/jcb.141.5.1097. doi:10.1083/jcb.141.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters M.F., Nucifora F.C., Jr, Kushi J., Seaman H.C., Cooper J.K., Herring W.J., Dawson V.L., Dawson T.M., Ross C.A. Nuclear targeting of mutant Huntingtin increases toxicity. Mol. Cell. Neurosci. 1999;14:121–128. doi: 10.1006/mcne.1999.0773. doi:10.1006/mcne.1999.0773. [DOI] [PubMed] [Google Scholar]
- 36.Chan E.Y., Luthi-Carter R., Strand A., Solano S.M., Hanson S.A., DeJohn M.M., Kooperberg C., Chase K.O., DiFiglia M., Young A.B., et al. Increased huntingtin protein length reduces the number of polyglutamine-induced gene expression changes in mouse models of Huntington's disease. Hum. Mol. Genet. 2002;11:1939–1951. doi: 10.1093/hmg/11.17.1939. doi:10.1093/hmg/11.17.1939. [DOI] [PubMed] [Google Scholar]
- 37.Schilling G., Becher M.W., Sharp A.H., Jinnah H.A., Duan K., Kotzuk J.A., Slunt H.H., Ratovitski T., Cooper J.K., Jenkins N.A., et al. Nuclear-targeting of mutant huntingtin fragments produces Huntington's disease-like phenotypes in transgenic mice. Hum. Mol. Genet. 1999;13:1599–1610. doi: 10.1093/hmg/ddh175. doi:10.1093/hmg/ddh175. [DOI] [PubMed] [Google Scholar]
- 38.Bangs P., Burke B., Powers C., Craig R., Purohit A., Doxsey S. Contribution of nuclear and extranuclear polyQ to neurological phenotypes in mouse models of Huntington's disease. Hum. Mol. Genet. 1998;14:3065–3078. doi: 10.1093/hmg/ddi340. [DOI] [PubMed] [Google Scholar]
- 39.Sawa A., Nagata E., Sutcliffe S., Dulloor P., Cascio M.B., Ozeki Y., Roy S., Ross C.A., Snyder S.H. Huntingtin is cleaved by caspases in the cytoplasm and translocated to the nucleus via perinuclear sites in Huntington's disease patient lymphoblasts. Neurobiol. Dis. 2005;20:267–274. doi: 10.1016/j.nbd.2005.02.013. doi:10.1016/j.nbd.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 40.Davies S.W., Turmaine M., Cozens B.A., DiFiglia M., Sharp A.H., Ross C.A., Scherzinger E., Wanker E.E., Mangiarini L., Bates G.P. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. doi:10.1016/S0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 41.Zhou H., Cao F., Wang Z., Yu Z.X., Nguyen H.P., Evans J., Li S.H., Li X.J. Huntingtin forms toxic NH2-terminal fragment complexes that are promoted by the age-dependent decrease in proteasome activity. J. Cell Biol. 2003;163:109–118. doi: 10.1083/jcb.200306038. doi:10.1083/jcb.200306038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham R.K., Deng Y., Slow E.J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. doi:10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 43.Gray M., Shirasaki D.I., Cepeda C., Andre V.M., Wilburn B., Lu X.H., Tao J., Yamazaki I., Li S.H., Sun Y.E., et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. doi:10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams A.J., Paulson H.L. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. doi:10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duvick L., Barnes J., Ebner B., Agrawal S., Andresen M., Lim J., Giesler G.J., Zoghbi H.Y., Orr H.T. SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010;67:929–935. doi: 10.1016/j.neuron.2010.08.022. doi:10.1016/j.neuron.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anne S.L., Saudou F., Humbert S. Phosphorylation of huntingtin by cyclin-dependent kinase 5 is induced by DNA damage and regulates wild-type and mutant huntingtin toxicity in neurons. J. Neurosci. 2007;27:7318–7328. doi: 10.1523/JNEUROSCI.1831-07.2007. doi:10.1523/JNEUROSCI.1831-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colin E., Zala D., Liot G., Rangone H., Borrell-Pages M., Li X.J., Saudou F., Humbert S. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27:2124–2134. doi: 10.1038/emboj.2008.133. doi:10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aiken C.T., Steffan J.S., Guerrero C.M., Khashwji H., Lukacsovich T., Simmons D., Purcell J.M., Menhaji K., Zhu Y.Z., Green K., et al. Phosphorylation of threonine 3: implications for Huntingtin aggregation and neurotoxicity. J. Biol. Chem. 2009;284:29427–29436. doi: 10.1074/jbc.M109.013193. doi:10.1074/jbc.M109.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y., Lin F., Qin Z.H. The role of post-translational modifications of huntingtin in the pathogenesis of Huntington's disease. Neurosci. Bull. 2010;26:153–162. doi: 10.1007/s12264-010-1118-6. doi:10.1007/s12264-010-1118-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang C.E., Zhou H., McGuire J.R., Cerullo V., Lee B., Li S.H., Li X.J. Suppression of neuropil aggregates and neurological symptoms by an intracellular antibody implicates the cytoplasmic toxicity of mutant huntingtin. J. Cell Biol. 2008;181:803–816. doi: 10.1083/jcb.200710158. doi:10.1083/jcb.200710158. [DOI] [PMC free article] [PubMed] [Google Scholar]