Abstract
Insulin resistance in skeletal muscle is a key phenotype associated with type 2 diabetes (T2D) for which the molecular mediators remain unclear. We therefore conducted an expression analysis of human muscle biopsies from patients with T2D; normoglycemic but insulin-resistant subjects with a parental family history (FH+) of T2D; and family history-negative control individuals (FH–). Actin cytoskeleton genes regulated by serum response factor (SRF) and its coactivator megakaryoblastic leukemia 1 (MKL1) had increased expression in T2D and FH+ groups. Furthermore, striated muscle activator of Rho signaling (STARS), an activator of SRF, was upregulated in T2D and FH+ and was inversely correlated with insulin sensitivity. Skeletal muscle from insulin-resistant mice recapitulated this gene expression pattern and showed reduced G-actin and increased nuclear localization of MKL1, each of which regulates SRF activity. Overexpression of MKL1 or reduction in G-actin decreased insulin-stimulated Akt phosphorylation, whereas reduction of STARS expression increased insulin signaling and glucose uptake. Pharmacological SRF inhibition by CCG-1423 reduced nuclear MKL1 and improved glucose uptake and tolerance in insulin-resistant mice in vivo. Thus, SRF pathway alterations are linked to insulin resistance, may contribute to T2D pathogenesis, and could represent therapeutic targets.
Introduction
Type 2 diabetes (T2D) is increasing at an alarming rate worldwide, affecting a projected 366 million individuals by the year 2030 (1). Despite genome-wide association studies confirming genetic links to T2D susceptibility (2), we do not yet fully understand the molecular basis of either insulin resistance or insulin secretory dysfunction, the key components associated with diabetes pathophysiology.
Insulin resistance is an early detectable metabolic defect in humans with T2D, is present in offspring of diabetic parents, and predicts incident diabetes (3–5). Alterations in insulin signaling (6), mitochondrial dysfunction (7, 8), endoplasmic reticulum stress (9), oxidative stress (10), and inflammation (11) have all been implicated in the induction of cellular insulin resistance. However, while insulin resistance is manifested by reduced glucose uptake into skeletal muscle (12), the underlying molecular defects mediating insulin resistance and diabetes risk in humans remain unknown (2, 13). Genome-wide association studies have identified over 30 polymorphic loci enriched in individuals with T2D, but thus far identification of genetic markers of the insulin-resistant state has proved elusive (2).
Analysis of gene expression can be used to identify signatures or early defects related to disease phenotypes. Previous studies have shown that individuals with T2D have decreased expression of nuclear-encoded genes involved in mitochondrial oxidative phosphorylation in skeletal muscle (14–16). This change in gene expression can be recapitulated in healthy humans by high-fat diet (HFD) or lipid infusion (17, 18) and is also observed in mice fed a HFD (19) or in mice with streptozotocin-induced diabetes (20), indicating that these changes may be related to obesity and/or effects of hyperglycemia. While some studies have shown similar changes in genes of mitochondrial oxidative metabolism in normoglycemic individuals with diabetes family history (14), this pattern has not been consistently observed in either humans (21, 22) or mouse models of insulin resistance (23), indicating that other pathways may also contribute to skeletal muscle insulin resistance and diabetes risk.
In the present study, we aimed to identify transcriptional phenotypes associated with insulin resistance through detailed metabolic and skeletal muscle gene expression analysis in 3 cohorts of individuals: 1 with established T2D, 1 with parental family history of diabetes (FH+) but normoglycemia, and 1 with normoglycemia and no family history of diabetes (FH–). We demonstrate that in skeletal muscle of both humans with T2D and insulin-resistant but normoglycemic FH+ individuals, there is increased expression of genes linked to the actin cytoskeleton and regulated by serum response factor (SRF) and its coactivator megakaryoblastic leukemia 1 (MKL1). We also show that these changes in expression contribute to alterations in skeletal muscle glucose uptake in association with insulin resistance.
Results
Enrichment of SRF target gene expression in humans with insulin resistance and T2D.
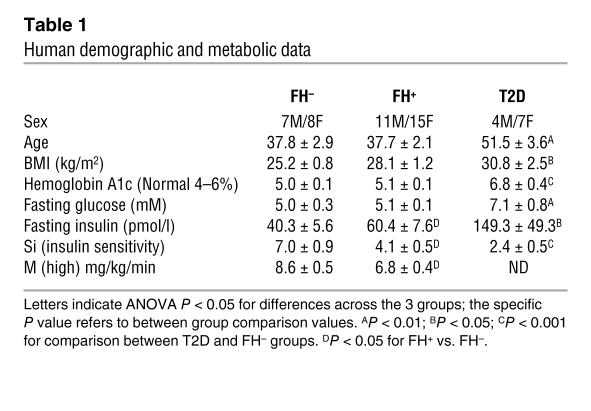
To identify differentially expressed genes associated with insulin resistance, we analyzed gene expression in quadriceps muscle biopsies obtained from 3 groups of metabolically characterized human subjects (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI41940DS1): (a) normoglycemic individuals without diabetes in any first-degree relative (family history negative (FH–, n = 15); (b) normoglycemic individuals with 1 or both parents with T2D (FH+, n = 26); and (c) drug-naive individuals with established T2D (n = 11). FH+ subjects were nondiabetic as defined by fasting glucose and oral glucose tolerance test, had normal hemoglobin A1c, had slightly, but not statistically, higher body mass index (BMI), and were insulin resistant, as demonstrated by a 50% increase in fasting insulin levels and a 41% reduction in insulin sensitivity (SI, assessed by the Bergman minimal model from intravenous glucose tolerance testing) when compared with FH– subjects (P < 0.05). Insulin resistance in FH+ individuals was further demonstrated by a 21% decrease in insulin-stimulated glucose uptake during a euglycemic hyperinsulinemic clamp (P < 0.05) (Table 1). Subjects with established T2D were even more insulin resistant, as indicated by both a 3.7-fold increase in fasting insulin and a 65% reduction in SI (P < 0.05 and P < 0.002, respectively). The T2D subjects were also older, heavier (BMI 30.8 vs. 25.2 kg/m2, P < 0.05), and by definition hyperglycemic, with significantly elevated fasting plasma glucose and hemoglobin A1c (Table 1).
Table 1 .
Human demographic and metabolic data
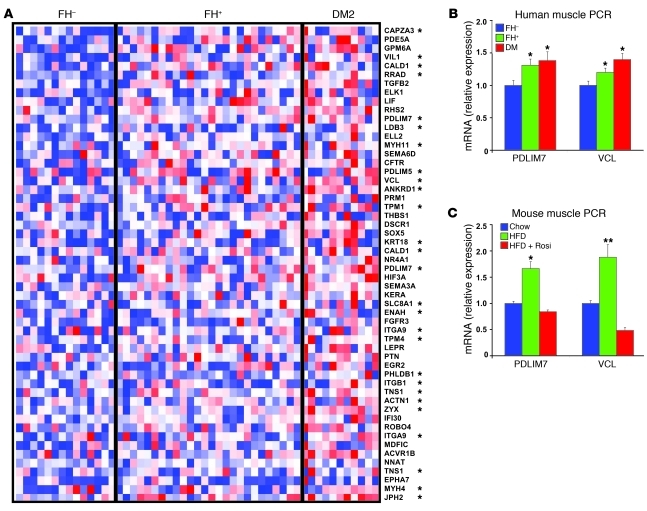
RNA was isolated from quadriceps muscle biopsies obtained in the fasting state. Gene expression was analyzed using high-density oligonucleotide arrays (Affymetrix U133A 2.0). Expression data were analyzed both on an individual and a pathway level. Consistent with previous studies (14, 15), gene set enrichment analysis revealed significant enrichment of pathways involved in oxidative phosphorylation and fatty acid metabolism in downregulated genes in subjects with established T2D. However, this pattern was not observed in normoglycemic FH+ insulin-resistant subjects, suggesting it is not related to the insulin resistance that precedes T2D in first-degree relatives. The top-ranking gene set enriched in both insulin resistant FH+ and T2D human skeletal muscle was a group of genes regulated by the transcription factor SRF and its cofactor MKL1 (originally identified by ref. 24) (normalized enrichment score 1.63, FDR = 0.02 for T2D vs. FH–, enrichment score 0.73 for FH+ vs. FH–). A heat map representation of SRF target genes (24, 25) with greater than 1.3-fold expression changes in T2D vs. controls is provided in Figure 1A and demonstrates increased expression in both individuals with a family history of diabetes and those with established diabetes.
Figure 1. SRF pathway is enriched in skeletal muscle from humans with diabetes or diabetes family history.
(A) Heat map demonstrates expression pattern for SRF target genes, with red color indicating upregulation and blue downregulation, relative to median value. *MKL1-dependent SRF target gene. (B) Expression of a subset of SRF target genes, including PDLIM7 and VCL, was increased in quadriceps muscle from humans with insulin resistance and T2D, as assessed by PCR in a subset of subjects (n = 15 FH–, 25 FH+, 11 T2D). SRF target genes are also upregulated in (C) HFD-fed mice and normalized by rosiglitazone (Rosi) treatment (3 mg/kg/d in chow, n = 6). *P < 0.05; **P < 0.01.
SRF is a transcription factor integrating growth and cytoskeletal pathway responses to both intra- and extracellular signals. SRF regulates expression of immediate early genes, muscle differentiation genes, and actin cytoskeletal genes (26, 27). In turn, the actin cytoskeleton may link to muscle insulin sensitivity, as actin remodeling is critical for both insulin- and contraction-dependent glucose uptake (28, 29). Interestingly, ontology classification of SRF target genes differentially expressed in T2D demonstrated that indeed these were genes linked to the actin cytoskeleton (Figure 1A) (ontology classification results with FDR < 0.05; Supplemental Table 3). Quantitative PCR confirmed the microarray results, with 30%–40% increases in expression of cytoskeleton-related SRF targets, including PDZ and LIM domain 7 (PDLIM7/enigma) and vinculin (VCL) in both diabetes and in FH+ insulin resistant subjects without diabetes (P < 0.05; Figure 1B), and also increased expression of zyxin (ZYX) in T2D (40% increase, P < 0.05; Supplemental Figure 1B). Although the dominant pattern was increased expression of cytoskeleton-related SRF target genes, we also observed increased expression of additional SRF target genes in subjects with T2D; these included the immediate-early genes EGR2 and NR4A1 (Supplemental Figure 1B). We observed no changes in expression of randomly selected non-SRF targets (ACADSB and MYSM1). Changes in expression of SRF target genes were independent of BMI. Thus, upregulation of actin cytoskeleton SRF target genes is an early signature of the prediabetic and insulin-resistant state. Increased expression of many SRF-regulated actin cytoskeleton-related genes was also observed in several insulin-resistant mouse models. Expression of PDLIM7 and VCL was significantly increased by 1.5- to 2-fold in muscle of mice with diet-induced obesity (P < 0.05; Figure 1C), leptin-deficient obesity (lepob/ob, not shown), and genetic inactivation of insulin receptor in muscle (MIRKO) (ref. 30 and Supplemental Figure 2). In both HFD-fed and lepob/ob mice, increases in SRF-regulated genes were normalized by treatment with the insulin sensitizer rosiglitazone (P < 0.05 vs. HFD; Figure 1C). Expression of the SRF target PDLIM7 was also increased at a protein level in HFD-fed mice (30% increase, P < 0.02; Supplemental Figure 1C). Interestingly, despite the recent identification of SRF as an important transcriptional regulator of adipogenesis (31), we did not observe increased SRF-dependent gene expression in either adipose or liver during high-fat feeding in mice (data not shown).
Increased expression of STARS in human diabetes may contribute to SRF target gene expression.
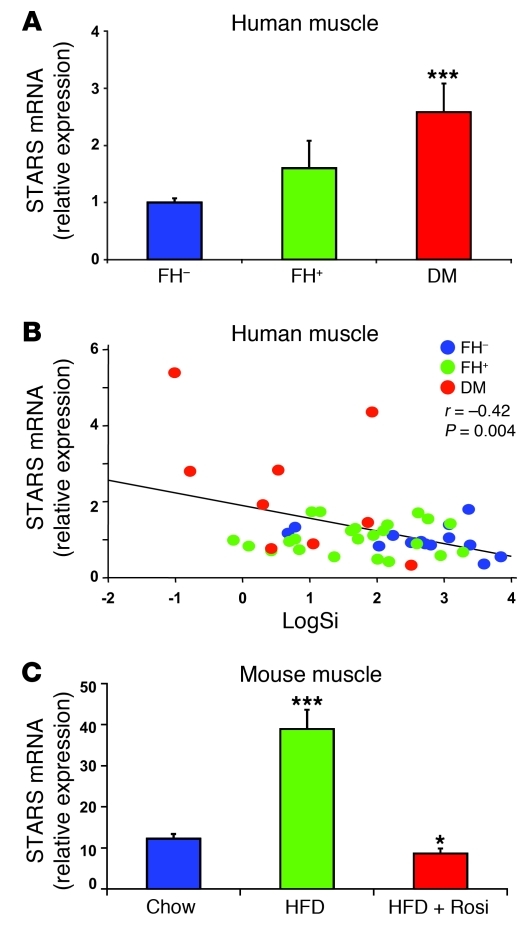
To better understand regulation of SRF-related genes, we examined expression of individual genes as a function of parental history or presence of T2D. Interestingly, a top-ranking differentially expressed gene was striated muscle activator of Rho signaling (STARS; official gene name ABRA, or actin-binding Rho activating protein), a known activator of SRF transcriptional activity (32). Expression of STARS was increased by 2.5-fold in humans with T2D compared with FH– individuals (Figure 2A), with intermediate expression in FH+ individuals (ANOVA P < 0.0001, P < 0.001 T2D vs. both FH– and FH+). While there was substantial interindividual variability, STARS expression robustly and inversely correlated with insulin sensitivity (SI) across the entire cohort (r = –0.42, P = 0.004; Figure 2B); STARS expression did not correlate with BMI. Interestingly, STARS expression also increased in mice made insulin resistant by high-fat feeding (3-fold, P < 0.001) and was normalized by rosiglitazone (Figure 2C). Since STARS is known to activate SRF via actin-dependent induction of nuclear accumulation of MKL1, increased STARS expression may contribute to increased SRF target gene expression in insulin resistance.
Figure 2. Expression of STARS gene is increased in insulin-resistant humans and rodents.
(A) STARS expression is significantly increased in quadriceps muscle of humans with T2D and is (B) strongly and inversely correlated with insulin sensitivity (Si) (r = –0.4, P < 0.004) (n = 15 FH–, 25 FH+, 11 T2D). (C) STARS expression is significantly elevated in skeletal muscle of mice fed a HFD for 5 months and is normalized by concurrent rosiglitazone treatment (3 mg/kg/d in chow; n = 6 mice per group). *P < 0.05; ***P < 0.001.
MKL1 nuclear localization and reduced G-actin contribute to SRF activation and insulin resistance.
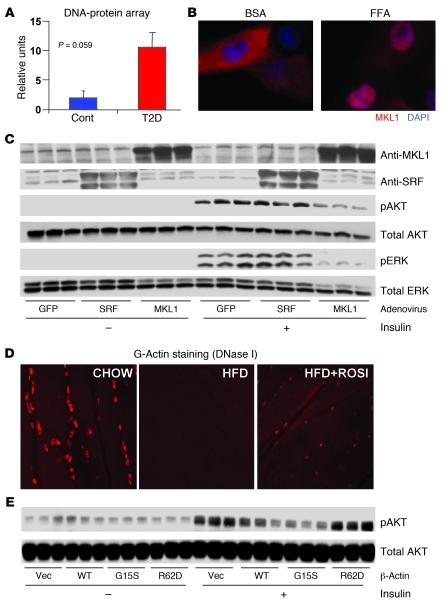
We next determined whether additional mechanisms downstream of STARS could also contribute to increased expression of SRF-related actin genes. We observed no alterations in mRNA expression of SRF itself, its ETS domain cofactor ELK1 (33), or its coactivator MKL1 (34) in muscle from either FH+ or T2D human subjects (Supplemental Figure 3A), or in muscle of insulin-resistant mice at either the mRNA or protein level (data not shown). Similarly, we detected no differences in activity of RhoA, an upstream modulator of SRF activity (Supplemental Figure 3B). DNA binding to serum response elements, assessed via low-density protein-DNA binding array, was 5.2-fold greater in nuclear extracts of primary myotubes from humans with diabetes as compared with controls (P = 0.059; Figure 3A and Supplemental Figure 4) and in myoblasts treated with palmitate (Supplemental Figure 5B), suggesting that enhanced SRF transcriptional activity may contribute to increased SRF target gene expression in insulin-resistant states.
Figure 3. Enhanced MKL1-dependent SRF activity in insulin resistance.
(A) Binding of nuclear extracts from primary myotubes isolated from humans with T2D is increased for SRF-SAP probes, as assessed by Panomics Combo Array (intensity values presented, n = 3/group). Cont, control. (B) MKL1, an SRF cofactor, is predominantly localized in cytosol with vehicle (BSA) treatment, but localizes to the nucleus in FFA-treated myocytes. (C) Adenoviral overexpression of MKL1, but not SRF, decreases Akt and ERK phosphorylation. (D) G-actin staining (DNase I, Texas red) is reduced in mice fed a HFD for 5 months compared with controls (CHOW); these patterns are reversed by rosiglitazone (3 mg/kg/d) for 3 months (HFD+ROSI) (n = 6 animals/group). (E) Overexpression of G15S actin reduces, but R62D actin increases, insulin-stimulated Akt phosphorylation (n = 3 per condition). Original magnification, ×400.
A key modulator of actin cytoskeleton-related SRF transcriptional activity and thus target gene expression is the subcellular localization of the SRF coactivator (MKL1). In turn, localization of MKL1 is regulated by interaction with nuclear G-actin, which results in nuclear export of MKL1 (35). We thus examined whether MKL1 localization was altered in a model of cellular insulin resistance induced by the saturated fatty acid palmitate. In L6 myoblasts, MKL1 was predominantly localized to cytosol in the basal state, but localized to the nucleus after palmitate exposure (Figure 3B and Supplemental Figure 5A).
A key question is whether increased MKL1-dependent SRF transcriptional activity and target gene expression result from or could contribute to insulin resistance. To address this question, we overexpressed either SRF or MKL1 in cultured L6 myotubes using adenoviral-mediated gene transfer (Figure 3C). This resulted in a 5.2-fold increase in SRF and a 4.8-fold increase in MKL1 expression (P < 0.05 and 0.01, respectively). While overexpression of SRF had minimal effect on insulin signaling, overexpression of MKL1 reduced insulin-stimulated Akt phosphorylation by 70% (P = 0.02; Figure 3C) and reduced insulin-stimulated Erk phosphorylation by more than 70% (P < 0.05; Figure 3C).
MKL1 localization and activation of SRF-dependent gene expression, in turn, is regulated by monomeric actin (G-actin) (35). Reduced G-actin, as occurs during F-actin polymerization, permits MKL1 translocation to the nucleus and activation of SRF transcriptional complexes (26, 36). Thus, alterations in G-actin content and/or localization might contribute to upregulation of SRF target gene expression in human insulin resistance. To address these possibilities, we examined G-actin in muscle from mice with diet-induced insulin resistance, by staining sections with rhodamine-conjugated DNase I, a marker of G-actin (35). In muscle from lean controls, G-actin colocalized with nuclear DAPI. In muscle from mice with diet-induced obesity, nuclear G-actin staining was dramatically reduced (Figure 3D). These changes were partially normalized in muscle of obese mice treated with rosiglitazone (Figure 3D). Similar decreases in G-actin staining were observed in rectus abdominis muscle from a cohort of obese, insulin-resistant human subjects as compared with lean controls (Supplemental Table 1 and Supplemental Figure 6).
To further evaluate the potential contribution of reduced nuclear G-actin to observed patterns of SRF transcriptional activity, we determined whether β-actin mutants with altered cellular localization would modulate insulin action. Interestingly, overexpression of the G15S mutant actin, which reduces G-actin via stabilization of F-actin and thus increases MKL1 nuclear localization, stimulated SRF transcriptional activity in myoblasts (refs. 37 and 38; Supplemental Figure 7, A and B). This also resulted in reduced insulin-stimulated Akt phosphorylation compared with cells expressing wild-type actin (Figure 3E). Conversely, the R62D mutant actin, which cannot efficiently form F-actin and thus creates more G-actin, blocked SRF transcriptional activity (Supplemental Figure 7, A and B) and increased Akt phosphorylation by 87% (P = 0.02; Figure 3E). Since insulin-stimulated glucose transport is a key phenotype associated with insulin sensitivity, we next asked whether expression of a constitutively nuclear MKL1 would modulate cellular glucose uptake. In support of this notion, overexpression of nuclear MKL1 (SA-MKL1) reduced the effect of insulin to stimulate glucose uptake (Supplemental Figure 8A). Conversely, knockdown of MKL1 expression by specific siRNA significantly increased insulin-stimulated glucose uptake (Supplemental Figure 8B). Together, these data indicate that experimental reductions in G-actin and associated increases in MKL1 nuclear localization and SRF transcriptional activity can indeed induce insulin resistance in cultured cells.
The STARS-SRF pathway modulates insulin action.
One potential upstream regulator of MKL1- and actin-dependent SRF activation is STARS, an actin-binding protein that can reduce G-actin, increase nuclear MKL1, and activate SRF transcriptional activity (32, 39, 40). Since STARS expression is increased in parallel with insulin resistance in both humans and mice, we wished to further examine relationships between activation of this pathway and metabolic phenotypes. We next asked whether experimental reduction in expression of STARS might have beneficial effects on insulin action and/or glucose uptake in cultured myotubes.
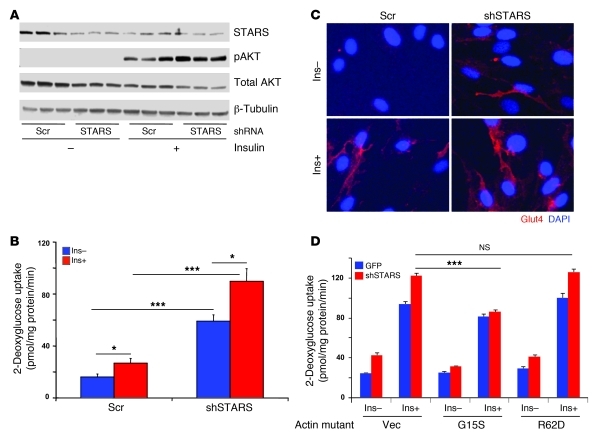
Infection of L6 myotubes with an adenovirus containing a short hairpin RNA targeting STARS resulted in a 54% reduction in STARS protein expression (Supplemental Figure 9B) and a 1.7-fold increase in insulin-stimulated Akt phosphorylation (P < 0.05; Figure 4A). Moreover, reduced STARS expression significantly increased both basal and insulin-stimulated glucose uptake in myotubes, by 3.7-fold and 3.6-fold, respectively (P < 0.001; Figure 4B). This effect occurred without changes in GLUT1 or GLUT4 expression, as assessed by Western blotting (not shown). On the other hand, shRNA-mediated reduction in STARS expression increased plasma membrane GLUT4 localization in both the basal and insulin-stimulated states, indicating effects to promote GLUT4 trafficking to the plasma membrane (Figure 4C). These effects of STARS were modulated by actin; expression of the G15S mutant abolished the impact of STARS shRNA to promote both basal and insulin-stimulated glucose uptake (Figure 4D). Thus, our data indicate that experimental reductions in STARS expression can increase insulin signaling, glucose uptake, and GLUT4 translocation and that this is mediated in part via actin-dependent processes.
Figure 4. shRNA-mediated experimental reduction in STARS expression increases glucose uptake and plasma membrane GLUT4 in L6 myotubes.
(A) shRNA-mediated reduction in STARS expression increases insulin-stimulated Akt phosphorylation. shRNA-mediated reduction in STARS (B) significantly increases glucose uptake in both the basal and insulin-stimulated state (n = 6) and (C) increases GLUT4 localization in the plasma membrane. (D) These effects are mediated, at least in part, via actin rearrangement, as seen by the ability of the G15S actin mutant to reduce effect of shRNA targeting STARS. For all panels, insulin stimulation was 100 nM for 10 minutes. *P < 0.05; ***P < 0.001. Original magnification, ×400.
SRF inhibitor CCG-1423 improves glucose uptake.
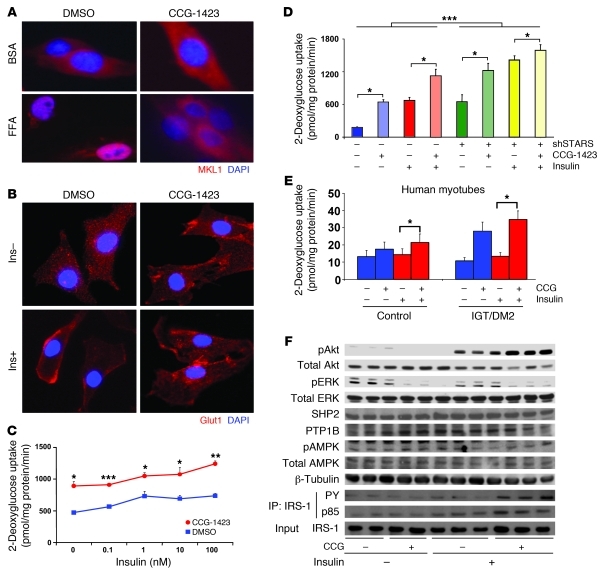
Since reduction in SRF pathway activity by genetic knockdown of STARS enhanced insulin action and glucose transport, we asked whether pharmacological inhibition of SRF transcriptional activity would similarly modulate glucose transport and metabolic phenotypes. CCG-1423 is an SRF transcriptional inhibitor identified by compound library screening (41). We confirmed that CCG-1423 reduced basal SRE promoter activity in L6 myotubes and efficiently blocked serum-stimulated activation (Supplemental Figure 10). Since we hypothesized that these effects of CCG-1423 to reduce SRF activity were mediated via alterations in MKL1 localization, we assessed MKL1 localization in L6 myoblasts in the presence or absence of CCG-1423. As noted, MKL1 was predominantly localized to the nucleus following treatment with the saturated fatty acid palmitate (Figure 5A and Supplemental Figure 11). In contrast, CCG-1423 treatment resulted in predominant localization of MKL1 to the cytosol and reversed palmitate-induced MKL1 nuclear localization, resulting in partial cytoplasmic localization (Figure 5A and Supplemental Figure 11). Thus, CCG-1423 represses SRF activity, at least in part by regulation of MKL1 cytosol/nuclear localization.
Figure 5. SRF inhibitor CCG-1423 improves glucose uptake and insulin action.
(A) MKL1 is predominantly nuclear in FFA-treated cells, but is largely cytosolic following CCG-1423 treatment. (B) GLUT4 translocation is induced by CCG-1423 in both basal and insulin-stimulated states. (C) Both basal and insulin-stimulated 2-deoxyglucose uptake are increased by more than 2-fold in L6 myotubes following CCG-1423 treatment (1 μM, 16 hours), with enhanced insulin sensitivity. *P < 0.05; **P < 0.01; ***P < 0.001 for effects of CCG-1423 versus DMSO. (D) Knockdown of STARS and CCG yield additive effects to promote basal and insulin-stimulated glucose uptake in L6 myotubes (differentiation day 7, CCG 1 μM for 16 hours; insulin 100 nM for 10 minutes). ***P < 0.001 for shSTARS versus shSCR, for all conditions. Effects of CCG are significant in both basal and insulin-stimulated states (*P < 0.001), in both shSCR and shSTARS conditions. (E) CCG-1423 increases basal glucose uptake in primary human myotubes isolated from control subjects with normoglycemia (control, left bars) or from subjects with IGT or T2D (right bars). Note the greater magnitude of CCG-1423 in IGT/T2D than controls. *P < 0.05 for CCG-1423 versus vehicle. (F) CCG-1423 increases insulin-stimulated Akt phosphorylation (100 nM, 10 minutes), blocks ERK phosphorylation, and increases IRS-1 tyrosine phosphorylation and its association with the p85 regulatory subunit of PI3K. Original magnification, ×400.
Since actin rearrangement contributes to both localization of MKL1 and GLUT4 translocation/glucose uptake (42), we next asked whether CCG-1423 had any impact on GLUT4 translocation to the plasma membrane. Indeed, CCG-1423 induced GLUT4 translocation to the plasma membrane in both the basal and insulin-stimulated states (Figure 5B). However, these effects were independent of changes in total GLUT4 or GLUT1 glucose transporter expression, which did not differ following CCG-1423 treatment (not shown). Consistently, in L6 cells incubated with CCG-1423, glucose uptake was increased by 2-fold in the basal state (Figure 5C); CCG-1423 similarly increased the magnitude of glucose uptake at all insulin concentrations (Figure 5C). shRNA-mediated knockdown of STARS expression further increased both basal and insulin-stimulated glucose uptake and enhanced effects of CCG-1423 (Figure 5D).
We observed similar effects of CCG-1423 in primary human myotubes, inducing a 1.3-fold increase in basal glucose uptake (P < 0.05) in myotubes isolated from healthy control subjects (Figure 5E). Interestingly, these effects were substantially greater in myotubes isolated from subjects with T2D or impaired glucose tolerance (IGT) (mean 2.6-fold, P < 0.05 vs. controls; Figure 5E). While these data need to be extended to larger cohorts, the differential responsiveness in subjects with T2D supports the concept that SRF pathways are more robustly activated in humans with insulin resistance and T2D, and therefore potentially more sensitive to SRF inhibition.
CCG-1423 also enhanced insulin action in L6 myotubes, increasing insulin-stimulated Akt phosphorylation by 41% (P = 0.007) (Figure 5F). This occurred in parallel with a 90% increase in IRS-1 tyrosine phosphorylation and its association with the p85 regulatory subunit of PI3K (Figure 5F). In contrast, CCG-1423 inhibited both basal and insulin-stimulated p42/44 ERK phosphorylation (Figure 5F); CCG-1423 did not affect expression of the phosphatases SHP2, PTP1B, or protein content or phosphorylation of AMP kinase (Figure 5F). Thus, CCG-1423 both increased insulin-stimulated signaling through the PI3K/Akt pathway and enhanced GLUT4 translocation and glucose uptake.
Effects of CCG-1423 in vivo.
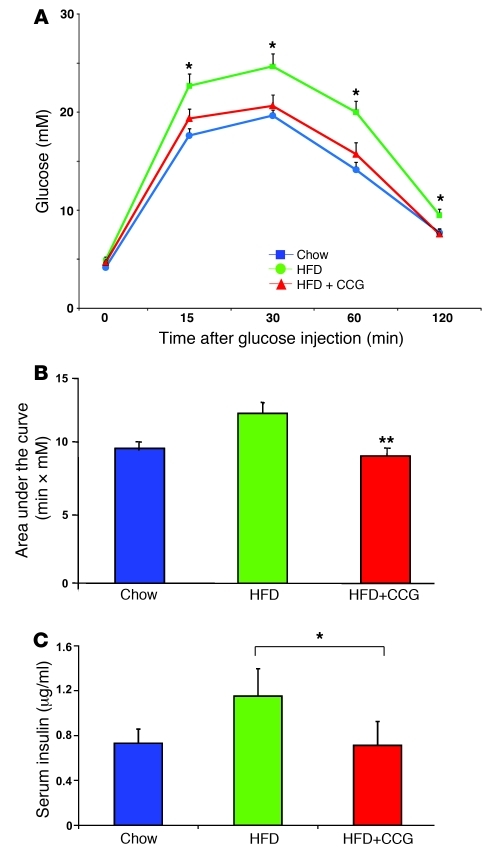
To examine the therapeutic potential of this SRF inhibitor in vivo, we assessed responses to CCG-1423 treatment in mice with HFD-induced obesity. As expected, mice fed a HFD for 10 weeks had significantly elevated glucose as compared with chow-fed control, under both fasting (5.3 ± 0.2 vs. 3.7 ± 0.1 mM, HFD vs. chow) and fed (10.4 ± 0.8 vs. 7.3 ± 0.3 mM) conditions. Treatment of HFD mice with CCG-1423 (0.15 mg/kg body weight intraperitoneally) for 2 weeks produced no significant change in food intake or body weight (not shown) in comparison with vehicle-treated controls. Consistent with its effect to reduce SRF transcription, CCG-1423 treatment for 2 weeks significantly decreased expression of SRF targets in skeletal muscle, e.g., ZYX, PDLIM7, as compared with vehicle-injected controls (Supplemental Figure 12). More importantly, in vivo glucose tolerance in HFD-fed mice treated with CCG-1423 was significantly improved as compared with HFD-fed vehicle-treated controls and was almost superimposable with the glucose tolerance of chow-fed animals (Figure 6A, P < 0.006 for HFD vs. HFD-CCG area under the curve, as quantified in Figure 6B). In parallel, the high insulin levels (reflecting HFD-induced insulin resistance) were normalized by CCG-1423 (Figure 6C). There were no differences in insulin-tolerance testing or in circulating fatty acid or adiponectin levels (not shown). Together, these results support the concept that strategies to reduce SRF transcriptional activity may have beneficial effects to enhance glucose disposal in insulin resistance and T2D.
Figure 6. SRF inhibitor-CCG-1423 improves glucose tolerance in vivo.
(A) Glucose tolerance improved following in vivo CCG-1423 treatment in HFD-fed mice (HFD+CCG) compared with vehicle-treated HFD or chow-fed controls. (B) Area under curve for glucose tolerance test. **P < 0.01 for HFD vs. HFD-CCG AUC. (C) Insulin levels at 30 minutes after glucose injection are significantly reduced in CCG-1423–treated mice. *P < 0.05.
Discussion
Identification of the primary molecular defects contributing to the pathophysiology of human T2D is an important scientific and clinical goal. We demonstrate increased expression of MKL1-dependent SRF target genes in skeletal muscle to be a novel pattern in diabetes, and importantly, in normoglycemic insulin-resistant individuals with a parental history of diabetes. Although the changes in SRF target gene expression are modest, they are similar in magnitude to the 20%–30% reduction in expression of OXPHOS genes recognized in patients with established diabetes (14, 15) and could be physiologically important due to the critical requirement for actin cytoskeletal proteins for GLUT4 translocation and glucose uptake in peripheral tissues and consistent patterns of regulation within this pathway (28, 43). Moreover, at least 2 cytoskeleton-related SRF target genes overexpressed in humans with insulin resistance (PDLIM7/enigma and RAD) have been experimentally shown to inhibit insulin-stimulated GLUT4 translocation and glucose uptake in other studies (44, 45). Thus, increased MKL1-dependent SRF activation and increased expression of cytoskeleton-related SRF target genes may contribute to reduced glucose uptake in insulin resistance and T2D.
While the exact mechanism leading to increases in SRF target genes may be multifold, our data suggest that these patterns are linked to increased expression of the SRF upstream regulatory gene STARS, sequestration of monomeric G-actin, and nuclear localization of the SRF coactivator MKL1 (Figure 7). In support of this model, we find that expression of STARS is increased 2.5-fold in patients with established T2D with intermediate expression (1.6-fold higher) in family history–positive subjects; across the entire cohort, expression of STARS also correlates closely with the degree of insulin resistance. Moreover, G-actin, which is an inhibitor of MKL1 activity, is reduced in both humans and mice with obesity-associated insulin resistance, even in the absence of established diabetes.
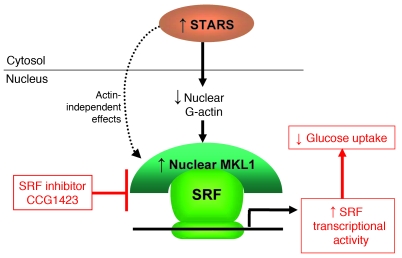
Figure 7. Schematic model.
Skeletal muscle insulin resistance is associated with increased expression of STARS and decreased G-actin, both of which induce MKL1 nuclear accumulation, resulting in activation of MKL1-dependent SRF transcriptional activity and reduced glucose uptake. CCG-1423 blocks SRF transcriptional activity by inhibiting MKL1 nuclear localization.
SRF is a ubiquitous transcription factor, binding to a specific 10-bp DNA sequence (CC[A/T]6GG, the so-called CArG box) within target promoters to regulate expression of genes involved in growth, proliferation, differentiation, and the actin cytoskeleton (31, 46). SRF transcriptional activity can be modulated by phosphorylation (47) and requires at least 2 major classes of cofactors (48): (a) a ternary complex factor (TCF) family of Ets domain proteins (e.g., ELK1, Sap1, and Net), predominantly regulating expression of immediate early genes, but also repressing MKL1-dependent expression, and (b) myocardin-related transcription factors (including MKL1), which are activated by Rho-actin signaling and nuclear translocation and involved predominantly in regulating cytoskeleton-related genes (25, 27, 49). The mechanisms by which nuclear MKL1 activates actin-related subsets of SRF-dependent genes remains unclear, but recent studies implicate ATP-dependent chromatin remodeling by Brg1 (50). While we observed no differences in mRNA expression of SRF, ELK1, or MKL1 in human muscle, we were unable to fully assess posttranscriptional modifications, e.g., phosphorylation and sumoylation, which could contribute to SRF activation. For example, MAPK- and ELK1-dependent phosphorylation events could also alter coactivator/corepressor recruitment and thus contribute to SRF activation in the setting of insulin resistance and/or oxidative stress (6, 51); however, we did not observe alterations in ELK1 DNA binding in human primary myotubes in either the basal or insulin-stimulated state. We did observe increased expression of some immediate early genes in fasting muscle biopsy samples from subjects with diabetes (e.g., EGR2, NR4A1). While these data would traditionally suggest activation of MAPK- and TCF-dependent pathways, it has recently been demonstrated that MKL1/2 can regulate serum-induced expression of subsets of immediate early genes, including EGR2, NR4A1, and VCL, among others (48). Thus, while we cannot exclude the possibility of ternary complex-dependent effects, our data more strongly support a more dominant role for MKL1-dependent mechanisms (24) in mediating activation of the subset of SRF target genes altered with insulin resistance in skeletal muscle.
G-actin has recently been recognized as an important negative regulator of MKL1-dependent SRF transcriptional activity (35). Reductions in G-actin lead to retention of MKL1 in the nucleus and enhancement of its SRF coactivator activity. This is consistent with our findings. Thus, there is reduced G-actin in muscle from humans and rodents with insulin resistance, and expression of mutant actin molecules that alter actin dynamics modulates SRF transcriptional activity in myotubes.
Our data suggest that increased expression of STARS, as observed in both human and rodent insulin resistance, may be the key mediator of reduced nuclear G-actin, increased nuclear MKL1, and SRF transcriptional activation (39). STARS is a muscle-specific protein that sequesters actin monomers, promoting dissociation of MKL1 from actin, and thereby inducing MKL1 nuclear accumulation (34). Our data indicate that effects of STARS are likely to be primarily mediated by actin-dependent mechanisms, as actin-dependent activation of SRF (via expression of the G15S actin mutant) abolished beneficial effects of STARS protein reduction. However, we cannot exclude a role for additional upstream regulators of actin dynamics potentially altering MKL1-related SRF transcription, such as members of the Rho family GTPases (26, 52); however, we did not detect differences in RhoA (Supplemental Figure 3A) or CDC42 activity (not shown) in skeletal muscle homogenates from obese, insulin-resistant humans as compared with lean controls. Additional studies will be required to examine mechanisms mediating increased STARS expression in insulin resistance and diabetes and to examine the role of other factors, such as posttranscriptional modification of MKL1 (49) and other SRF-interacting proteins in contributing to the altered expression of SRF target gene expression.
A key and as yet unanswered question is whether increased SRF transcriptional activity in muscle is a cause or effect of insulin resistance. SRF-null mice die during early embryogenesis (53) and muscle-specific SRF deletion results in hypotrophic myofibers (54), indicating a possible role of SRF in muscle development or differentiation. However, we observed no significant differences in fiber type–linked expression in our human cohort, and the induction of similar SRF-related patterns by high-fat feeding or deletion of insulin receptor in muscle indicates that these changes can occur secondary to insulin resistance. Increased STARS expression, alterations in F/G-actin balance (28, 55), nuclear localization of MKL1, and increased SRF activation observed in this study likely reflect an integrated transcriptional response to the insulin-resistant metabolic environment. Which factors mediate this effect remain unknown, but potential contributors linked to insulin sensitivity include obesity, inactivity, intramyocellular lipids, and age. We recognize that subjects with established T2D in our cohort were older and more obese than family history–negative individuals. Although we are unable to fully exclude the possible contribution of aging and adiposity to STARS and SRF target gene expression, we did not observe a correlation between BMI or age and gene expression patterns in our cohort. Moreover, the observation that patterns of increased SRF-related gene expression were reversed by rosiglitazone in rodents again suggests a link with systemic insulin sensitivity.
It is difficult to determine from human tissues whether alterations in STARS and MKL1-dependent SRF activation could be a cause or effect of insulin resistance. However, both genetic and pharmacologic manipulation of the STARS/MKL1/SRF pathway produce significant alterations in insulin action, indicating that this pathway may contribute to the pathophysiology of T2D. Reduced STARS expression, mediated by shRNA, enhances insulin-stimulated Akt phosphorylation, glucose transport, and GLUT4 translocation. Likewise, chemical inhibition of MKL1-dependent SRF pathway activity by the SRF inhibitor CCG-1423 improves many facets of disordered metabolism associated with insulin resistance. These include increased IRS1 phosphorylation, increased activation of PI3K and Akt, increased GLUT4 translocation, increased glucose transport in cultured myotubes, and improved glucose tolerance in insulin-resistant mice. Our data suggest that CCG-1423 inhibition of SRF activity is mediated downstream of STARS, as efficacy remains with knockdown of STARS expression. In agreement, Evelyn et al. have demonstrated that CCG-1423 and related compounds alter SRF/MKL1 complex formation and/or nuclear localization (41, 56). Indeed, we show that CCG-1423 reversed MKL1 nuclear localization in lipid-mediated cellular insulin resistance. The effects of CCG-1423 in enhancing glucose uptake in human myotubes occurred both in normal myotubes and in myotubes derived from subjects with IGT or T2D, and appeared to be greater in the latter. While these findings need to be confirmed, these data suggest that the MLK1-dependent SRF pathway may be a target for future therapy.
In conclusion, we demonstrate that upregulation of SRF transcriptional activity is a signature of insulin resistance and T2D in skeletal muscle. These changes are mediated by increased expression of STARS, altered levels of G-actin, and enhanced MKL1 coactivation of SRF. More importantly, we demonstrate that reductions in SRF transcriptional activity and target gene expression can have beneficial effects on muscle and whole-body metabolism, suggesting that this pathway is an important target for the development of novel therapeutics for insulin resistance and T2D.
Methods
Human subjects and human primary myotubes.
Human studies were approved by the Institutional Review Board of the Joslin Diabetes Center. Written informed consent was obtained from all subjects. 52 human volunteers were recruited on the basis of established diabetes or parental history of diabetes. A 100-g oral glucose tolerance test (OGTT) and intravenous glucose tolerance tests were performed in all subjects, and values for insulin sensitivity (Si) were calculated using the Bergman minimal model, Minmod (57). Subjects with diabetes had diagnosed disease of greater than 1-year duration and were treated with educational interventions to improve diet and exercise, but did not receive pharmacologic therapy.
Muscle biopsies of the vastus lateralis muscle were performed after local anesthesia with 1% lidocaine, using a modified Bergstrom needle with suction in the fasting state. For actin staining, open biopsies of rectus abdominis muscle were obtained during elective abdominal surgery.
Primary human myotubes were isolated from skeletal muscle biopsy samples from human volunteers and differentiated as described (58). Data for these subjects are presented in Supplemental Table 2.
Microarray and PCR expression analysis.
RNA was isolated using Trizol, followed by RNeasy columns with DNase I treatment (QIAGEN). For array analysis, cRNA was generated using standard protocols (14). Per-gene expression analysis and gene-set enrichment analysis were performed in the Gene Pattern software package ( http://www.broad.mit.edu/cancer/software/genepattern/). For both individual gene analysis and gene set enrichment analysis, a false discovery rate of less than 0.25 was considered significant. Primary data from microarrays have been deposited into GEO (GSE25462).
cDNA was synthesized using random hexamers (Advantage) for subsequent real-time quantitative PCR analysis (ABI Prism 7900; Applied Biosystems Inc.). PCR products were detected using Sybr Green and normalized to cyclophilin expression. Primers were designed using Primer Quest (Integrated DNA Technologies Inc). Sequences are provided in Supplemental Table 4.
Animal care and metabolic assessment.
Mice were housed 4 per cage in an Office of Laboratory Animal Welfare–certified animal facility, with a 12-hour light/12-hour dark cycle. The Joslin Institutional Animal Care and Use Committee approved all experimental plans. Age-matched C57BL/6 males fed a chow (10% calories from fat) or HFD (60% calories from fat; Research Diets) from age 6 weeks were obtained from The Jackson Laboratory.
For gene expression and G-actin staining, mice were fed an HFD (Research Diets) for 5 months. Some animals were treated with rosiglitazone for 5 months (HFD+ROSI, 3 mg/kg/d). Glucose and insulin tolerance were assessed following intraperitoneal glucose (2 g/kg glucose) or insulin injection (0.75 units/kg Humulin R). Serum insulin levels were measured during the glucose tolerance test at baseline and at 30 minutes after glucose injection (ELISA; Crystal Chem).
For SRF inhibitor experiments, 16-week-old HFD-fed mice were treated with CCG-1423 (0.15 mg/kg/d, intraperitoneally) or vehicle alone (DMSO) for 2 weeks.
Cell culture.
Mouse C2C12 and rat L6 myoblasts (ATCC) were maintained in DMEM supplemented with 20% and 10% FBS (Invitrogen), respectively, at a confluency of 60%–70%. To initiate differentiation, cells were allowed to reach 100% confluency, and medium was changed to DMEM containing 2% horse serum (Invitrogen) and changed every 2 days. Full differentiation, with myotube fusion and spontaneous twitching, was observed at 5 days.
For human myotube culture, materials were purchased from Irvine Scientific except for skeletal muscle basal medium (SkGM). The methods for muscle cell isolation from freshly biopsied tissue and growth have been described in detail previously (58). At 80%–90% confluency, cells were fused for 4 days in α-MEM containing 2% FBS, 1% FUNGI-BACT, 100 U/ml penicillin, and 100 g/ml streptomycin. Fusion medium was changed every other day. Within 4 days, approximately 90% of the cells take on the multinucleated morphology characteristic of mature, differentiated myotubes (58).
Immunoprecipitation and Western blotting.
Protein was harvested from serum-starved cells treated as described in lysis buffer containing protease inhibitor mixture (Sigma-Aldrich), sodium orthovanadate, and Triton X-100. For immunoprecipitations, whole-cell lysates (200 μg for IR, 500 μg for IRS1) were incubated with antibodies (anti-insulin receptor or anti-IRS1; Santa Cruz Biotechnology Inc.) and protein A–agarose beads at 4°C overnight, washed, and solubilized. Protein concentrations were determined using a BCA assay kit (Pierce). Proteins were separated by SDS-PAGE for subsequent Western blotting. Membranes were blocked for 1 hour in 5% skim milk in PBS/Tween-20, incubated with primary antibodies (1:1000) overnight, incubated with horseradish peroxidase–conjugated anti-rabbit IgG (1:2000) at room temperature for 1 hour, and visualized using chemiluminescence (Western Lightning Kit; PerkinElmer Life Sciences).
Transfection and reporter analysis.
L6 cells were cotransfected with plasmid DNA encoding the 4X CArG promoter (32) using Mirus transfection reagent (Mirus Bio). Cells were treated from 2 hours to overnight with experimental treatments, as specified, in serum-free DMEM 24 hours after DNA transfection and lysed (Promega); luciferase activity was determined using the dual luciferase assay kit (Promega) and normalized to that of pRL-TK.
DNA binding arrays.
DNA binding arrays (ComboArray, MA1215) were purchased from Panomics; manufacturers’ protocols were used together with nuclear extracts from human primary myotubes.
Immunohistochemistry.
Muscle samples were fixed in 10% formalin, embedded in paraffin, and sectioned at 5-μm intervals. Texas Red–conjugated DNase I was used as a marker of G-actin following manufacturers’ protocols (Molecular Probes).
For MKL1 staining, myoblasts were fixed with 2% PFA after incubation with palmitate (500 μM, complexed to BSA) or BSA alone. Cells were permeabilized in 0.1% Triton X-100 with PBS for 10 minutes and blocked with 5% skim milk for 1 hour at room temperature. Cells were incubated with anti-MKL1 primary antibody (H-140; Santa Cruz Biotechnology Inc.) and subsequently with FITC-conjugated goat anti-rabbit secondary antibody (SC-2012; Santa Cruz Biotechnology Inc.) and counterstained with DAPI.
For plasma membrane GLUT4 staining, Myc-GLUT4–overexpressing cells were provided by Amira Klip, University of Toronto (Toronto, Ontario, Canada). Myc-GLUT4–overexpressing L6 myoblasts were grown on sterile 8-well chamber slides (Lab-Tek II) overnight at 37°C. Cells were incubated with serum-free medium overnight with anti-Myc antibody (1 μg/ml, 9E-10; Santa Cruz Biotechnology Inc.), then stimulated with 100 nM insulin for 10 minutes and fixed with 2% PFA for 10 minutes. After washing with PBS, cells were incubated with FITC-conjugated anti-mouse IgG for 45 minutes in the dark and mounted with DAPI-containing mounting medium (VECTASHIELD) for visualization.
Adenoviral shRNA-mediated reductions in gene expression.
shSTARS target sequences were cloned into pAdtrack adenoviral vector. L6 myotubes were infected with amplified adenovirus (8 × 107 pfu/well/6-well plate, or 2 × 107 pfu/well for chamber slides) at 3 days after initiation of differentiation. Efficiency of knockdown was assessed by quantitative RT-PCR (qRT-PCR) (Supplemental Figure 9) 48 hours after infection. For Western blot, qRT-PCR, or GLUT4 translocation and glucose transport experiments, cells were harvested at 48 hours after infection. For glucose transport experiments, cells were fasted overnight in serum-free medium containing 0.1% BSA; the following morning, experiments were performed as indicated below. shSTARS target sequence is as follows: forward, 5′-GATCCCCCTGTAAGGCCCATCGGAAATTCAAGAGATTTCCGATGGGCCTTACAGTTTTTGGAAA-3′; reverse, 5′-AGCTTTTCCAAAAACTGTAAGGCCCATCGGAAATCTCTTGAATTTCCGATGGGCCTTACAGGGG-3′.
Glucose uptake.
Following overnight serum starvation, L6 myotubes were treated with insulin as specified for 20 minutes and then incubated with transport solution (140 mM NaCl, 20 mM Hepes, 5 mM KCl, 2.5 mM MgSO4, 1.0 mM CaCl2, 10 μM 2-deoxyglucose, 0.5 μCi/ml 3H 2-deoxyglucose) for 10 minutes. Cells were lysed in 0.05N NaOH and radioactivity determined by scintillation counting.
RhoA activity.
RhoA activity was measured according to manufacturer’s protocol (BK124; Cytoskeleton Inc.). In brief, tissue samples were washed with cold PBS 3 times and homogenized gently in ice-cold cell lysis buffer. Tissue lysates were harvested after centrifugation at 8,200 g at 4°C for 15 minutes; 20 μl was removed for protein quantification in order to adjust sample concentration to 0.5 mg/ml. After adding an equal volume of binding buffer, triplicate assays were performed using 1.5 μg protein/well. Samples were incubated for 30 minutes and then washed 3 times with washing buffer. Antigen-presenting buffer was added for exactly 2 minutes before removal; samples were then incubated with 1:250 dilution of anti-RhoA antibody at room temperature for 45 minutes, washed 3 times, and incubated with secondary antibodies for another 45 minutes. HRP detection reagent was added and signal was read by measuring absorbance at 490 nm using a microplate spectrometer.
Reagents.
AICAR was from Biomol International (Plymouth Meeting). Rosiglitazone was provided by GlaxoSmithKline. CCG-1423 was purchased from Cayman Chemical. All other chemicals were purchased from Sigma-Aldrich. Antibodies were obtained from Cell Signaling Technology and Santa Cruz Biotechnology Inc. Human SRF adenoviral construct was provided by Techung Lee (University of Buffalo, Buffalo, New York, USA); HA-tagged MKL1 adenoviral construct was obtained from Paul Herring (Indiana University, Indianapolis, Indiana, USA).
Statistics.
Microarray data were analyzed as described above. For all other comparisons, ANOVA or Student’s t tests were performed, and P less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We gratefully acknowledge research support from NIH DK062948 (to M.E. Patti), DK02795 (to A.B. Goldfine), DK070648 (to A.B. Goldfine), DK060837 (Diabetes Genome Anatomy Project to M.E. Patti), LM008748 (i2b2), 5P30 DK 36836 (DERC Animal Core and Specialized Assay Core), ADA-06-CD-07 (to A.B. Goldfine), and the Graetz Fund. We thank Amira Klip (University of Toronto) for the gift of L6 myoblasts expressing GLUT4 and thank Amira Klip, Shunsuke Ishii (Riken Institute, Japan), and C. Ronald Kahn (Joslin Diabetes Center) for valuable suggestions. We thank Guido Posern (Max-Planck-Institute, Germany), Paul Herring (Indiana University, USA), Techung Lee (University of Buffalo, USA), and Ron Prywes (Columbia University, USA) for DNA plasmids and adenovirus materials.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(3):918–929. doi:10.1172/JCI41940.
References
- 1. Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27(5):1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 2. Doria A, Patti ME, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008;8(3):186–200. doi: 10.1016/j.cmet.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eriksson J, et al. Early metabolic defects in persons at increased risk for non-insulin-dependent diabetes mellitus. N Engl J Med. 1989;321(6):337–343. doi: 10.1056/NEJM198908103210601. [DOI] [PubMed] [Google Scholar]
- 4. Martin BC, Warram JH, Rosner B, Rich SS, Soeldner JS, Krolewski AS. Familial clustering of insulin sensitivity. Diabetes. 1992;41(7):850–854. doi: 10.2337/diabetes.41.7.850. [DOI] [PubMed] [Google Scholar]
- 5. Tabak AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimaki M, Witte DR. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet. 2009;373(9682):2215–2221. doi: 10.1016/S0140-6736(09)60619-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cusi K, et al. Insulin resistance differentially affects the PI 3-kinase-and MAP kinase - mediated signaling in human muscle. J Clin Invest. 2000;105(3):311–320. doi: 10.1172/JCI7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51(10):2944–2950. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- 8. Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004;350(7):664–71. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eizirik DL, Cardozo AK, Cnop M. The role for endoplasmic reticulum stress in diabetes mellitus. Endocr Rev. 2008;29(1):42–61. doi: 10.1210/er.2007-0015. [DOI] [PubMed] [Google Scholar]
- 10. Houstis N, Rosen ED, Lander ES. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440(7086):944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- 11. Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116(7):1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rothman DL, et al. Decreased muscle glucose transport/phosphorylation is an early defect in the pathogenesis of non-insulin-dependent diabetes mellitus. Proc Natl Acad Sci U S A. 1995;92(4):983–987. doi: 10.1073/pnas.92.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jin W, Patti ME. Genetic determinants and molecular pathways in the pathogenesis of Type 2 diabetes. Clin Sci (Lond). 2009;116(2):99–111. doi: 10.1042/CS20080090. [DOI] [PubMed] [Google Scholar]
- 14. Patti ME, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100(14):8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 16. Sreekumar R, Halvatsiotis P, Schimke JC, Nair KS. Gene expression profile in skeletal muscle of type 2 diabetes and the effect of insulin treatment. Diabetes. 2002;51(6):1913–1920. doi: 10.2337/diabetes.51.6.1913. [DOI] [PubMed] [Google Scholar]
- 17. Sparks LM, et al. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes. 2005;54(7):1926–1933. doi: 10.2337/diabetes.54.7.1926. [DOI] [PubMed] [Google Scholar]
- 18. Richardson DK, et al. Lipid infusion decreases the expression of nuclear encoded mitochondrial genes and increases expression of extracellular matrix genes in human skeletal muscle. J Biol Chem. 2004;280(11):10290–10297. doi: 10.1074/jbc.M408985200. [DOI] [PubMed] [Google Scholar]
- 19. Crunkhorn S, et al. Peroxisome proliferator activator receptor gamma coactivator-1 expression is reduced in obesity: potential pathogenic role of saturated fatty acids and p38 mitogen-activated protein kinase activation. J Biol Chem. 2007;282(21):15439–15450. doi: 10.1074/jbc.M611214200. [DOI] [PubMed] [Google Scholar]
- 20. Yechoor VK, Patti ME, Saccone R, Kahn CR. Coordinated patterns of gene expression for substrate and energy metabolism in skeletal muscle of diabetic mice. Proc Natl Acad Sci U S A. 2002;99(16):10587–10592. doi: 10.1073/pnas.142301999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morino K, et al. Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. J Clin Invest. 2005;115(12):3587–3593. doi: 10.1172/JCI25151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nair KS, et al. Asian Indians have enhanced skeletal muscle mitochondrial capacity to produce ATP in association with severe insulin resistance. Diabetes. 2008;57(5):1166–1175. doi: 10.2337/db07-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hancock CR, et al. High-fat diets cause insulin resistance despite an increase in muscle mitochondria. Proc Natl Acad Sci U S A. 2008;105(22):7815–7820. doi: 10.1073/pnas.0802057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Selvaraj A, Prywes R. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol Biol. 2004;5:13. doi: 10.1186/1471-2199-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun Q, et al. Defining the mammalian CArGome. Genome Res. 2006;16(2):197–207. doi: 10.1101/gr.4108706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sotiropoulos A, Gineitis D, Copeland J, Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98(2):159–169. doi: 10.1016/S0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 27. Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16(11):588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 28. Zaid H, Antonescu CN, Randhawa VK, Klip A. Insulin action on glucose transporters through molecular switches, tracks and tethers. Biochem J. 2008;413(2):201–215. doi: 10.1042/BJ20080723. [DOI] [PubMed] [Google Scholar]
- 29. Sakamoto K, Holman GD. Emerging role for AS160/TBC1D4 and TBC1D1 in the regulation of GLUT4 traffic. Am J Physiol Endocrinol Metab. 2008;295(1):E29–E37. doi: 10.1152/ajpendo.90331.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bruning JC, et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell. 1998;2(5):559–569. doi: 10.1016/S1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 31. Mikkelsen TS, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143(1):156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuwahara K, Barrientos T, Pipes GC, Li S, Olson EN. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol Cell Biol. 2005;25(8):3173–3181. doi: 10.1128/MCB.25.8.3173-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harris RA, et al. Regulation of the branched-chain alpha-ketoacid dehydrogenase and elucidation of a molecular basis for maple syrup urine disease. Adv Enzyme Regul. 1990;30:245–263. doi: 10.1016/0065-2571(90)90021-S. [DOI] [PubMed] [Google Scholar]
- 34. Du KL, Chen M, Li J, Lepore JJ, Mericko P, Parmacek MS. Megakaryoblastic leukemia factor-1 transduces cytoskeletal signals and induces smooth muscle cell differentiation from undifferentiated embryonic stem cells. J Biol Chem. 2004;279(17):17578–17586. doi: 10.1074/jbc.M400961200. [DOI] [PubMed] [Google Scholar]
- 35. Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316(5832):1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- 36. Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113(3):329–342. doi: 10.1016/S0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 37. Posern G, Sotiropoulos A, Treisman R. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol Biol Cell. 2002;13(12):4167–4178. doi: 10.1091/mbc.02-05-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Posern G, Miralles F, Guettler S, Treisman R. Mutant actins that stabilise F-actin use distinct mechanisms to activate the SRF coactivator MAL. EMBO J. 2004;23(20):3973–3983. doi: 10.1038/sj.emboj.7600404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuwahara K, et al. Modulation of adverse cardiac remodeling by STARS, a mediator of MEF2 signaling and SRF activity. J Clin Invest. 2007;117(5):1324–1334. doi: 10.1172/JCI31240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arai A, Spencer JA, Olson EN. STARS, a striated muscle activator of Rho signaling and serum response factor-dependent transcription. J Biol Chem. 2002;277(27):24453–24459. doi: 10.1074/jbc.M202216200. [DOI] [PubMed] [Google Scholar]
- 41. Evelyn CR, et al. CCG-1423: a small-molecule inhibitor of RhoA transcriptional signaling. Mol Cancer Ther. 2007;6(8):2249–2260. doi: 10.1158/1535-7163.MCT-06-0782. [DOI] [PubMed] [Google Scholar]
- 42. Kanzaki M, Pessin JE. Insulin-stimulated GLUT4 translocation in adipocytes is dependent upon cortical actin remodeling. J Biol Chem. 2001;276(45):42436–42444. doi: 10.1074/jbc.M108297200. [DOI] [PubMed] [Google Scholar]
- 43. Brozinick JT, Jr, Berkemeier BA, Elmendorf JS. “Actin”g on GLUT4: membrane & cytoskeletal components of insulin action. Curr Diabetes Rev. 2007;3(2):111–122. doi: 10.2174/157339907780598199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barres R, et al. Enigma interacts with adaptor protein with PH and SH2 domains to control insulin-induced actin cytoskeleton remodeling and glucose transporter 4 translocation. Mol Endocrinol. 2006;20(11):2864–2875. doi: 10.1210/me.2005-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moyers JS, Bilan PJ, Reynet C, Kahn CR. Overexpression of Rad inhibits glucose uptake in cultured muscle and fat cells. J Biol Chem. 1996;271(38):23111–23116. doi: 10.1074/jbc.271.38.23111. [DOI] [PubMed] [Google Scholar]
- 46. Miano JM. Role of serum response factor in the pathogenesis of disease. Lab Invest. 2010;90(9):1274–1284. doi: 10.1038/labinvest.2010.104. [DOI] [PubMed] [Google Scholar]
- 47. Iyer D, et al. Serum response factor MADS box serine-162 phosphorylation switches proliferation and myogenic gene programs. Proc Natl Acad Sci U S A. 2006;103(12):4516–4521. doi: 10.1073/pnas.0505338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee SM, Vasishtha M, Prywes R. Activation and repression of cellular immediate early genes by serum response factor cofactors. J Biol Chem. 2010;285(29):22036–22049. doi: 10.1074/jbc.M110.108878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Muehlich S, Wang R, Lee SM, Lewis TC, Dai C, Prywes R. Serum-induced phosphorylation of the serum response factor coactivator MKL1 by the extracellular signal-regulated kinase 1/2 pathway inhibits its nuclear localization. Mol Cell Biol. 2008;28(20):6302–6313. doi: 10.1128/MCB.00427-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang M, Fang H, Zhou J, Herring BP. A novel role of Brg1 in the regulation of SRF/MRTFA-dependent smooth muscle-specific gene expression. J Biol Chem. 2007;282(35):25708–25716. doi: 10.1074/jbc.M701925200. [DOI] [PubMed] [Google Scholar]
- 51. Krook A, et al. Characterization of signal transduction and glucose transport in skeletal muscle from type 2 diabetic patients. Diabetes. 2000;49(2):284–292. doi: 10.2337/diabetes.49.2.284. [DOI] [PubMed] [Google Scholar]
- 52. Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac 1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81(7):1159–1170. doi: 10.1016/S0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 53. Arsenian S, Weinhold B, Oelgeschlager M, Ruther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17(21):6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Charvet C, et al. New role for serum response factor in postnatal skeletal muscle growth and regeneration via the interleukin 4 and insulin-like growth factor 1 pathways. Mol Cell Biol. 2006;26(17):6664–6674. doi: 10.1128/MCB.00138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McCarthy AM, Spisak KO, Brozinick JT, Elmendorf JS. Loss of cortical actin filaments in insulin-resistant skeletal muscle cells impairs GLUT4 vesicle trafficking and glucose transport. Am J Physiol Cell Physiol. 2006;291(5):C860–C868. doi: 10.1152/ajpcell.00107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Evelyn CR, et al. Design, synthesis and prostate cancer cell-based studies of analogs of the Rho/MKL1 transcriptional pathway inhibitor, CCG-1423. Bioorg Med Chem Lett. 2010;20(2):665–672. doi: 10.1016/j.bmcl.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bergman RN, Prager R, Volund A, Olefsky JM. Equivalence of the insulin sensitivity index in man derived by the minimal model method and the euglycemic glucose clamp. J Clin Invest. 1987;79(3):790–800. doi: 10.1172/JCI112886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Henry RR, Abrams L, Nikoulina S, Ciaraldi TP. Insulin action and glucose metabolism in nondiabetic control and NIDDM subjects. Comparison using human skeletal muscle cell cultures. Diabetes. 1995;44(8):936–946. doi: 10.2337/diab.44.8.936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.