Abstract
Kaposi sarcoma herpesvirus (KSHV) is specifically associated with Kaposi sarcoma (KS) and 2 B cell lymphoproliferative diseases, namely primary effusion lymphoma (PEL) and multicentric Castleman disease (MCD). KS, PEL, and MCD are largely incurable and poorly understood diseases most common in HIV-infected individuals. Here, we have revealed the role of viral FLICE-inhibitory protein (vFLIP) in the initiation of PEL and MCD by specifically expressing vFLIP at different stages of B cell differentiation in vivo. Mice showed MCD-like abnormalities and immunological defects including lack of germinal centers (GCs), impaired Ig class switching, and affinity maturation. In addition, they showed increased numbers of cells expressing cytoplasmic IgM-λ, a thus far enigmatic feature of the KSHV-infected cells in MCD. B cell–derived tumors arose at high incidence and displayed Ig gene rearrangement with downregulated expression of B cell–associated antigens, which are features of PEL. Interestingly, these tumors exhibited characteristics of transdifferentiation and acquired expression of histiocytic/dendritic cell markers. These results define immunological functions for vFLIP in vivo and reveal what we believe to be a novel viral-mediated tumorigenic mechanism involving B cell reprogramming. Additionally, the robust recapitulation of KSHV-associated diseases in mice provides a model to test inhibitors of vFLIP as potential anticancer agents.
Introduction
Understanding how viruses subvert host molecular pathways and cause cellular transformation has been a fascinating and challenging task since Peyton Rous’ pioneering work in 1911 that gave birth to the concept of viral oncogenesis (1). Kaposi sarcoma herpesvirus (KSHV) is one of the most recently discovered human cancer viruses (2), and, besides being recognized as the etiological agent of Kaposi sarcoma (KS), it is also associated with the pathogenesis of 2 lymphoproliferative diseases, namely primary effusion lymphoma (PEL) and multicentric Castleman disease (MCD) (2–4). KSHV-associated lymphoproliferative diseases are rare even in regions with high KSHV seroprevalence (5), suggesting that KSHV alone is not sufficient for their development. Nonetheless, the specific and consistent association of KSHV with these distinct clinical entities (3, 6) as well as the presence of multiple putative oncogenes within its genome strongly indicate that KSHV is necessary for the underlying oncogenic process.
PEL is an aggressive subtype of non-Hodgkin B cell lymphoma characterized by body cavity lymphomatous effusions with features of both immunoblasts (CD30+, CD39+, IRF4+) and plasma cells (BLIMP1+, CD138+), even if it is clearly distinct from both (7, 8). Although PEL commonly lacks expression of B cell–specific markers including CD19, Oct2, Pax5, and surface Ig, the presence of Ig gene rearrangements (6, 9) confirms its derivation from the B cell lineage, and the presence of somatic hypermutation (SHM) in the rearranged Ig variable (V) genes indicates transition through the germinal center (GC) (9). Therefore, PEL might originate from antigen-selected GC B cells prior to their becoming terminally differentiated plasma cells. MCD is a polyclonal lymphoproliferative disorder (10) characterized by the presence of KSHV-infected monotypic cytoplasmic IgM-λ–expressing plasmablasts residing primarily in the mantle zone (11), dissolution of the follicles, and prominent interfollicular vascular proliferation (12). Although negative for certain B cell–associated markers such as Pax5, CD20, CD30, and CD138, MCD cells resemble mature B cells, as they express the preplasma cell markers IRF4 and BLIMP1, the memory B cell marker CD27, Oct2, and Ki67 (13). However, MCD plasmablasts lack SHM in their rearranged IgV genes (11), suggesting that KSHV may preferentially target IgM-λ–expressing naive B cells and sustain their differentiation into plasmablasts bypassing the GC reaction.
KSHV conforms to the replication paradigm shared by all herpesviruses, displaying both lytic and latent modes of infection. Although lytic KSHV infection has been documented to contribute to Kaposi sarcomagenesis (14–16), KSHV genes important in viral genomic persistence and cell transformation are found among those transcribed during latency (i.e., LANA, v-cyclin, viral FLICE-inhibitory protein [vFLIP]), as KSHV infection is, indeed, predominantly latent in KSHV-induced lymphoid tumors (17). Many data indicate a role for vFLIP in KSHV pathogenesis, as it is a latent gene capable of activating NF-κB (18, 19), a hallmark cellular pathway constitutively active in PEL and indispensable for the maintenance of the tumor phenotype (20–22). FLIP proteins are a group of cellular and viral proteins identified as inhibitors of death receptor–induced (DR-induced) apoptosis (23, 24). They contain 2 death effector domains (DED) capable of inhibiting DED-DED interactions between FAS-associated protein with death domain (FADD) and procaspases 8 and 10 within the death-inducing signaling complex (DISC), otherwise responsible for DR-induced apoptosis (25). KSHV vFLIP becomes part of the DISC and prevents the recruitment and processing of procaspase 8 and, thereby, FAS-induced apoptosis (24). vFLIP can control cell death also by inhibiting autophagy (26). Another established function of vFLIP is its binding to IκB kinase γ (IKKγ), which induces IKKα/β phosphorylation, IκBα degradation, and p100 cleavage, thus resulting in both classical and alternative NF-κB activation (18, 19, 27). The specific elimination of vFLIP by RNA interference results in significantly decreased NF-κB activity and apoptosis, confirming that vFLIP is essential for the survival of PEL cells (22).
However, the role of vFLIP in the initiation of KSHV-related diseases is largely unknown. Moreover, even if there is evidence that the PEL cell of origin is GC experienced (i.e., mutated Ig genes), while MCD develops from earlier precursors that eventually mature into plasmablasts in a GC-independent manner, still unclear is the precise stage-specific histiogenesis of these diseases. One previously reported vFLIP Tg mouse model displayed a broad range of lymphoma subtypes at low incidence and did not recapitulate PEL (28). Thus, in the attempt to more faithfully recapitulate KSHV-associated diseases, we used a recombinant activation approach to express vFLIP in a stage-specific manner within the B cell compartment, either in all CD19+ B cells or, more restrictedly, in GC B cells. Both Tg lines showed defects in B cell differentiation as well as MCD-like abnormalities and developed tumors of B cell origin at high incidence. These mice also revealed a previously unknown function for vFLIP in inducing expansion of the macrophage/DC compartment via both cellular transdifferentiation and paracrine mechanisms. Our findings have important implications for the pathogenesis of all KSHV-associated malignancies that invariably display a rich infiltrate of histiocytes, whose origin and function have long been enigmatic. Moreover, we believe our study is the first comprehensive characterization of the immunological outcomes of in vivo expression of vFLIP, which potentially contributes to immune dysfunction during tumor development and maintenance in vivo.
Results
Generation of mice conditionally expressing vFLIP in B cells.
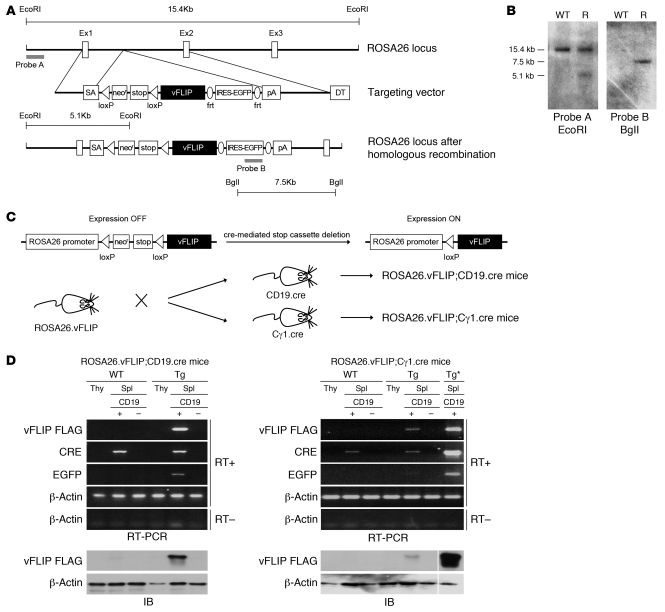
To study the role of vFLIP in the pathogenesis of KSHV-associated lymphoproliferative disorders, we generated Tg mice expressing vFLIP in a stage-specific manner during B cell development. We introduced a cDNA-encoding FLAG-tagged vFLIP preceded by a loxP-flanked neoR-STOP cassette and followed by frt-flanked IRES-EGFP sequence into the ubiquitously expressed ROSA26 locus (Figure 1, A and B). The elimination of the STOP cassette and subsequent activation of Tg expression were achieved by crossing ROSA26.vFLIP knockin mice with mice expressing cre recombinase under the control of either CD19 or Cγ1 promoter, resulting in vFLIP expression in all CD19+ B cells or selectively in IgG1+ GC B cells, respectively (Figure 1C). EGFP coexpressed with vFLIP from a common transcript due to the insertion of an IRES between the 2 gene sequences was used to identify cells expressing the vFLIP Tg.
Figure 1. Generation of ROSA26.vFLIP;CD19.cre and ROSA26.vFLIP;Cγ1.cre mice.
(A) Schematic representation of the ROSA26 locus before (top) and after (bottom) homologous recombination with the targeting vector (middle) carrying a triple-flagged vFLIP encoding sequence. (B) A representative Southern blot analysis of EcoRI- or BglI-digested WT and recombinant (R) ES cell DNA. Probes used and expected genomic fragment sizes are also indicated. (C) The strategy for recombinant activation of vFLIP expression in vivo. ROSA26.vFLIP knockin mice were bred with either CD19.cre or Cγ1.cre mice to obtain Tg expression from early precursor B cell stage or from GC stage, respectively. (D) Tg expression was specifically detected, both by RT-PCR (upper panel) and anti-FLAG immunoblotting (lower panel) in splenic CD19+ B cells derived from both ROSA26.vFLIP;CD19.cre and ROSA26.vFLIP;Cγ1.cre mice. Sample lanes separated by thin white lines were run on the same gel but were noncontiguous. Tg*, ROSA26.vFLIP;CD19.cre mice used for positive control; Spl, spleen; Thy, thymus.
Tg mice (carrying both cre and vFLIP) were born at the expected Mendelian frequency and were indistinguishable from their WT littermate controls (carrying only cre) in terms of fertility and developmental features. Expression of vFLIP was evaluated in 2- to 3-month-old mice after immunization with sheep red blood cells (SRBC). vFLIP mRNA and protein were specifically found in splenic CD19+ B cells isolated from both ROSA26.vFLIP;CD19.cre and ROSA26.vFLIP;Cγ1.cre Tg mice, but not in the thymus or in the splenic CD19– cell fraction nor in any tissue from control mice (Figure 1, D and E). ROSA26.vFLIP;CD19.cre Tg mice expressed EGFP in the vast majority of splenic CD19+ B cells (80% ± 1.9%), but not in T cells. Moreover, the Tg, as monitored by EGFP detection, was expressed throughout the various stages of B cell differentiation comprising immature, mature, follicular, marginal zone, GC B cells, and plasma cells (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI44417DS1). Transitional B cells did not show significant EGFP expression, suggesting an unfavorable effect of the Tg at this particular stage of B cell differentiation. Consistent with the activation pattern of the Cγ1 promoter, in ROSA26.vFLIP;Cγ1.cre Tg mice, the majority of splenic GC IgG1+ B cells (67.6% ± 3.9%) and only a minor percentage of CD19+ B cells (5.6% ± 0.2%) expressed EGFP, showing that the Tg was specifically expressed upon activation of the Cγ1 promoter (Supplemental Figure 1B). Considering that in the ROSA26.vFLIP;Cγ1.cre Tg mice, only a small proportion of B cells express the Tg, vFLIP expression level was comparable at the single-cell level in the targeted B cells (i.e., CD19+ B cells and IgG1+ GC B cells) of the 2 Tg mouse lines. Taken together, these data show that vFLIP had an expected B cell–restricted pattern of expression in the 2 vFLIP Tg mouse lines generated.
vFLIP expression in B cells impairs GC formation, Ig class switch recombination, and affinity maturation.
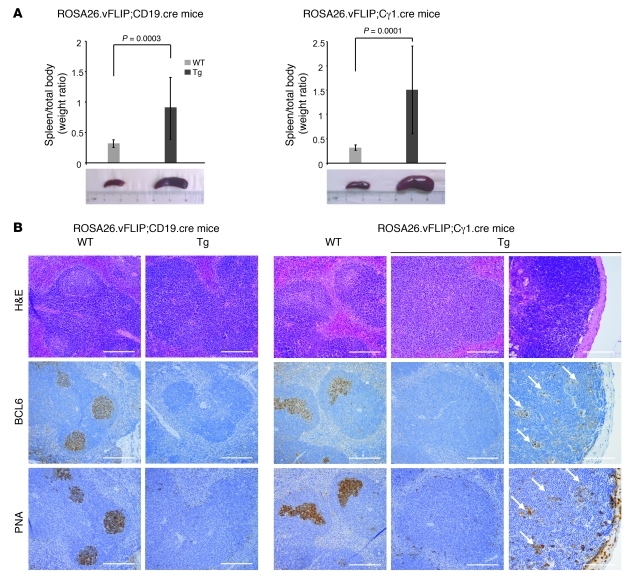
Splenomegaly was consistently observed in both immunized and unimmunized ROSA26.vFLIP;CD19.cre and ROSA26.vFLIP;Cγ1.cre Tg mice starting from 2–3 months of age (Figure 2A). Although ROSA26.vFLIP;Cγ1.cre Tg mice expressed vFLIP in a lower percentage of cells, splenomegaly was consistently more pronounced in this line. Staining of spleen sections showed that the overall architecture was intact, with clear demarcation between normally formed T- and B-zones (Supplemental Figure 2). In WT mice upon immunization with SRBC, H&E staining showed numerous well-developed GCs positive for 2 well-characterized GC markers, namely the receptor for peanut agglutinin (PNA) and BCL6. In contrast, both Tg mouse lines displayed complete absence of GCs within the follicular areas, even upon immunization, and the receptors for PNA and BCL6 were not expressed (Figure 2B). Small clusters of BCL6+PNA+Ki67+ GC-like cells were occasionally observed in ROSA26.vFLIP;Cγ1.cre Tg mice (Figure 2B), indicating abortive GC reactions. This suggests that GC reactions were only possible when the expression of vFLIP was turned on after the GC reactions had already begun.
Figure 2. Splenomegaly and lack of GC formation.
(A) Splenomegaly was observed in both ROSA26.vFLIP;CD19.cre (left) and ROSA26.vFLIP;Cγ1.cre mice (right); at least 20 Tg and control animals were analyzed (error bars indicate SEM). (B) H&E, PNA, and BCL6 staining revealed the lack of GCs in both Tg mouse lines. Abortive GCs in Peyer patches of ROSA26.vFLIP;Cγ1.cre mice are shown (arrows). Scale bars: 200 μm.
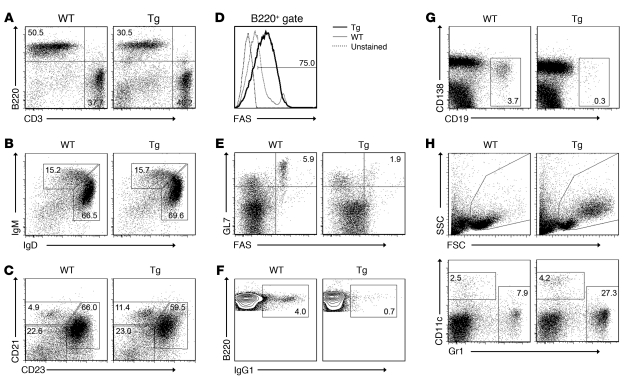
To gain insights into the mechanism underlying the splenomegaly and the effect of the Tg expression in vivo, the major splenic B cell populations were analyzed by flow cytometry in immunized mice (Figure 3 and Supplemental Figure 1). Consistent with the splenomegaly, the total splenic B cell number was increased (data not shown) in spite of a slightly reduced splenic B to T cell ratio (Figure 3A). No significant changes were observed in immature B cell, mature B cell, and follicular B cell numbers, while marginal zone and FAS+ B cells were increased (Figure 3, B–D). The increase of FAS+ B cells might represent a consequence of vFLIP-mediated NF-κB activation, leading to a paradoxical proapoptotic effect (29), which, however, remains abortive since vFLIP inhibits FAS-mediated apoptosis. Indeed, while WT and Tg EGFP–CD19+ B cells as well as WT and Tg T cells that do not express the Tg underwent apoptosis in response to anti-FAS treatment, Tg EGFP+CD19+ B cells were resistant to FAS-induced apoptosis despite having upregulated FAS on their surfaces (Supplemental Figure 3). The reduction in GC B cells (Figure 3E), IgG1+ B cells (Figure 3F), and plasma cells (Figure 3G) was in agreement with immunohistochemical data. Similar results were observed in ROSA26.vFLIP;Cγ1.cre mice (Supplemental Figure 1B). Comparison with unimmunized mice showed that immunization failed to promote GC formation and production of class-switched IgG1 in both ROSA26.vFLIP;CD19.cre (Supplemental Figure 4) and ROSA26.vFLIP;Cγ1.cre Tg mice (data not shown). Of interest, vFLIP Tg mice displayed the expansion of side/forward scatter-high (SSC/FSChi) cells that expressed the macrophage antigen Gr1 and the DC antigen CD11c (Figure 3H), but not EGFP (Supplemental Figure 1). Collectively, these results demonstrate the multiple abnormalities in the B cell lineage, including reduction in GC and IgG1+ B cells, increase in FAS+ B cells, and the expansion of a macrophage/DC-like population in the 2 B cell–specific vFLIP Tg mouse lines. The expansion of the macrophage/DC-like cells may result from positive survival signals derived from vFLIP-expressing B cells, as it has been reported that vFLIP is capable of activating the expression of monocyte/macrophage-activating chemokines (30, 31).
Figure 3. Abnormal splenic cell populations in ROSA26.vFLIP;CD19.cre mice.
Flow cytometry analysis displayed (A) decrease of B versus T cell ratio, as shown by B220 and CD3 staining; (B) no changes in immature (B220+IgD–IgM+) and mature (B220+IgD+IgM–) B cells; (C) no changes in follicular B cells (B220+CD21–CD23+) and increase of marginal zone B cells (B220+CD21+CD23–); (D) increase of FAS+ B cells; (E) reduction of GC B cells (B220+GL7+FAS+); (F) reduction of IgG1-expressing B cells; (G) reduction of plasma cells; (H) increase of side/forward scatter–high macrophage/DCs (SSC/FSChiB220– and Gr1+ or CD11c+). Data are representative of at least 3 experiments with similar results (error bars indicate SEM); at least 3 Tg and control animals were analyzed in each experiment.
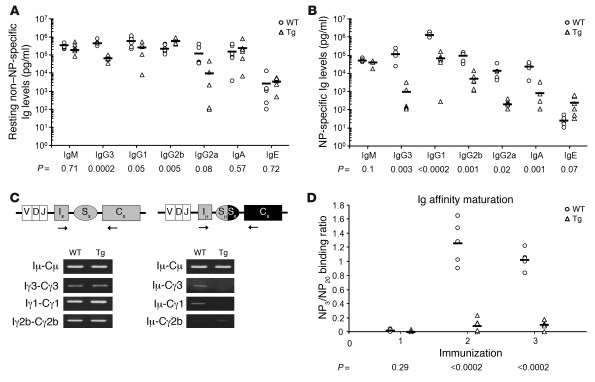
We hypothesized that class switch recombination (CSR) and SHM may be differentially affected in the 2 vFLIP Tg lines due to diverse Tg activation patterns, allowing us to predict different pathological outcomes seen in PEL and MCD (i.e., PEL would be favored if CSR and SHM are permitted, while MCD would develop in their absence). To test this hypothesis and to investigate the effects of the lack of GCs on CSR and SHM, we analyzed resting and the T cell–dependent antigen nitrophenol-specific (NP-specific) Ig production as well as Ig affinity maturation in both mouse Tg lines. In ROSA26.vFLIP;CD19.cre mice, while both resting and NP-specific IgM levels were unaffected, resting IgG3, IgG1, and IgG2a levels as well as NP-specific Igs of all isotypes except IgE were decreased (Figure 4, A and B), consistent with a marked reduction of IgG3 and IgG1 post-switch transcripts (Figure 4C) as well as IgG1+ B cells (Figure 3F) in immunized mice. Affinity maturation was evaluated by measuring the binding of NP-specific IgG1 antibodies to NP3- and NP20-coated plates. Higher NP3/NP20 binding ratio indicates the presence of higher-avidity (i.e., affinity-matured) antibodies. Interestingly, both vFLIP Tg mice failed to develop any antibody affinity maturation upon sequential immunizations with NP (Figure 4D). Similar results were observed also in ROSA26.vFLIP;Cγ1.cre mice (Supplemental Figure 5), although we cannot exclude a local ongoing CSR and SHM from which a mutated clone can arise, given the sporadic formation of small clusters of GC-like cells. Taken together, these data ascribe an important role to vFLIP as a negative regulator of both CSR and Ig affinity maturation in vivo, in addition to a suppressor of GC formation.
Figure 4. Serum antibody concentration and Ig affinity maturation in ROSA26.vFLIP;CD19.cre mice.
Antibody levels were analyzed by ELISA. P values derived from Student’s t test on the means (bars) of WT versus Tg mice are given below each antibody subclass in the graphs. (A) Resting serum antibody levels evaluated on preimmune serum samples. (B) NP-specific antibody response referred to the second boost. (C) Post-switch transcripts were significantly reduced (right panel), except for Iμ-Cγ2b, although germline transcripts were unaffected (left panel). (D) Tg mice showed lack of affinity maturation upon sequential immunizations performed with NP24-KLH on days 0, 21, and 42, respectively. Higher NP3/NP20 binding ratios indicate the presence of higher-avidity (i.e., affinity-matured) IgG1 antibodies.
vFLIP Tg mice develop MCD-like pathological abnormalities.
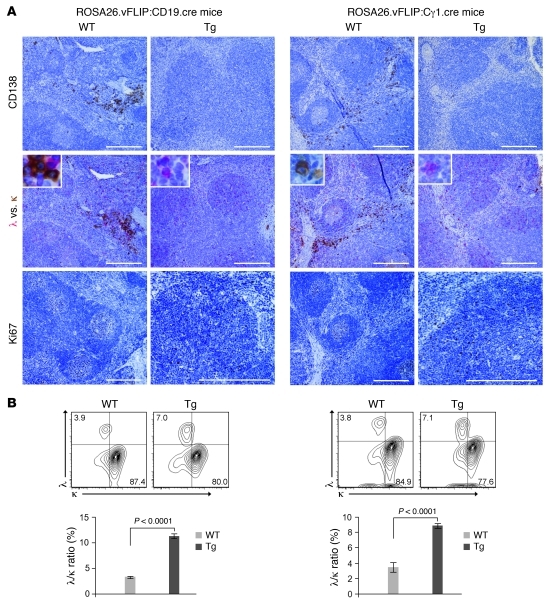
One of the unique features of MCD is the presence of monotypic cytoplasmic IgM-λ–expressing plasmablasts residing in the mantle zone. Although the mechanism underlying this bias for Ig-λ locus rearrangement remains obscure, the recent understanding that the generation of λ+ B cells depends on NF-κB signals (32) suggests that vFLIP, as an NF-κB activator, may have a role in this process. As expected, WT mice predominantly expressed the κ light chain (κ:λ ratio, 90:3) (Figure 5B). B cells of vFLIP Tg mice showed significantly increased λ light chain expression (κ:λ ratio, 78:7) (Figure 5B). Double immunohistochemistry for cytoplasmic κ and λ showed that these numerous λ-expressing cells were CD138– and morphologically resembled the plasmablasts observed in MCD, being smaller, less bright, and with a much less abundant cytoplasm as compared with normal plasma cells (Figure 5A). Only 2- to 2.5-fold enrichment of Ig-λ chain–expressing plasmablasts was seen, which differs quantitatively from the almost exclusive expression of λ chain in KSHV-infected cell seen in MCD. Nevertheless, vFLIP is likely to contribute to this phenotype, as it was consistently observed in a large number of mice analyzed. Therefore, vFLIP expression during the natural history of KSHV infection might be responsible, although perhaps only partially, for this hallmark of MCD. In addition, more BLIMP1-expressing cells were found in the spleen (data not shown), consistent with BLIMP1 expression in MCD plasmablasts. Furthermore, while the proliferation marker Ki67 only stained the GCs in WT mice, numerous Ki67+ cells reminiscent of the Ki67+ plasmablasts observed in MCD were found scattered throughout the entire follicle in vFLIP Tg mice (Figure 5A). Collectively, vFLIP Tg mice developed several pathological abnormalities typical of MCD, supporting a role for vFLIP in the pathogenesis of this disease.
Figure 5. Expansion of Ki67+λ+ B cells.
(A) Spleen sections immunostained with CD138, λ (red), κ (brown), and Ki67. Insert shows higher magnification of cells expressing cytoplasmic λ and κ. Scale bars: 200 μm; 20 μm (insert). (B) Flow cytometry showing percentage of B cells expressing either intracellular λ or κ light chain (upper panel); λ versus κ ratio is also reported (lower panel). Data represent 1 of 3 experiments with similar results (error bars indicate SEM); at least 3 Tg and control animals were analyzed in each experiment.
vFLIP expression leads to B cell reprogramming and tumor development.
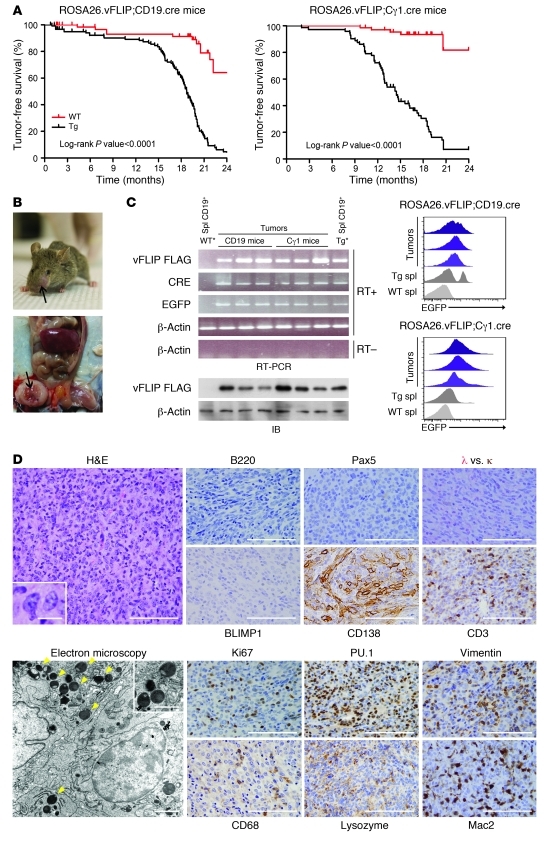
Both vFLIP Tg lines developed tumors at high incidence, and nearly all Tg mice succumbed to disease between 7 and 19 months of age (Figure 6A). Tumors arose in various locations rich in lymphoid tissue including nasopharynx, armpit, and inguinal region (Figure 6B). In a considerable number of cases, mice developed multiple tumors and in some cases tumor metastases to lymph nodes were found, which supported the malignant nature of the growth. Macroscopically, tumors had white to tan color and soft consistency; abundant necrosis was occasionally present. The tumors expressed the Tg at both RNA and protein levels and, therefore, were derived from cells expressing either CD19- or Cγ1-driven cre recombinase (Figure 6C). EGFP expression was detected in the tumor cells by flow cytometry (Figure 6C) and immunohistochemistry (Supplemental Figure 6). In agreement with the ability of vFLIP to activate NF-κB pathway, phospho-p65 and phospho-IκBα were detected in the tumor samples expressing the Tg, similarly to what was observed in the Tg B cells (Supplemental Figure 7). Histologically, tumors were characterized by sheets of large cells with abundant cytoplasm, oval to irregular nuclei, and prominent nucleoli, and spindling of cells was focally noted (Figure 6D). Surrounding normal lymphoid tissue was present in some cases, suggesting that the tumors arose in the context of lymphoid organs (data not shown). Unexpectedly, immunohistochemistry failed to detect B cell–specific markers B220, Pax5, BLIMP1, and λ and κ, but revealed positivity for CD138 (Figure 6D). Endothelial and stromal/mesenchymal origins of the tumors were excluded by negative CD34 and SMA staining, respectively (data not shown), and tumor-infiltrating lymphocytes were mainly CD3+ T cells (Figure 6D). The proliferation index (Ki67+ cells) was approximately 30%–40% for the neoplastic cells (Figure 6D). Surprisingly, tumor cells were positive for PU.1, a marker expressed by both B cells and macrophages, Vimentin, and other macrophage/DC markers (CD68, Lysozyme, and Mac2) (Figure 6D). Ultrastructural examination further confirmed the histiocytic features of the tumor cells, including irregularly folded nuclei and abundant cytoplasm containing numerous electron-dense vesicles indicative of lysosomes (Figure 6D). Collectively, these findings led to a pathological diagnosis of histiocytic/DC sarcoma.
Figure 6. Tumor characterization.
(A) Statistical analysis of event-free survival by Kaplan-Meier cumulative survival curve and the log-rank test to evaluate statistical significance. More than 100 mice in each group were followed up for up to 2 years. (B) A representative tumor in the nose and one in the abdominal cavity are shown. (C) Tg expression was detected in the tumors by RT-PCR (upper left panel), immunoblot (lower left panel), and flow cytometry (right panel). WT* and Tg*, ROSA26.vFLIP;CD19.cre mice were used for negative and positive controls, respectively. (D) Tumor sections were stained with H&E, B cell–specific markers (B220, Pax5, λ and κ), BLIMP1, and CD138 (upper panel). Tumors express markers indicative of histiocytic/DC sarcoma (Pu.1, Vimentin, CD68, Lysozyme, Mac2) (lower panels). Electron microscopy revealed electron-dense vesicles suggestive of lysozymes (arrows) (lower panels). Scale bars: 100 μm, 14 μm (inset), immunohistochemistry images; 2 μm, 1.44 μm (inset), electron microscopy images.
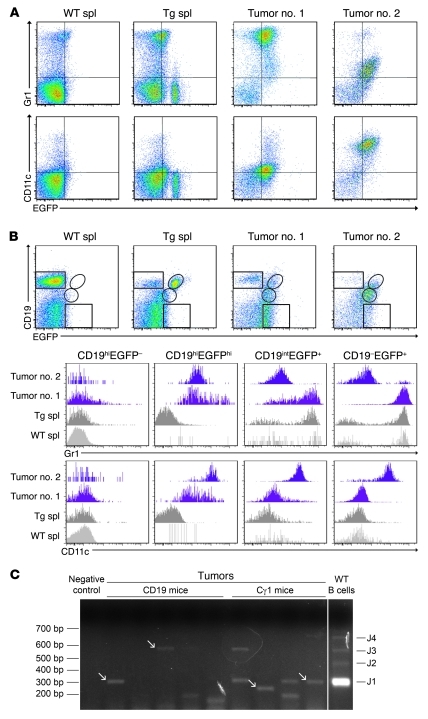
Given the unexpected tumor phenotype, additional flow cytometry analyses were performed to further characterize the tumor population in relation to the B cell lineage where the Tg was constitutively expressed. All tumor cases expressed either Gr1 or CD11c (Figure 7A). Although we could not detect B cell antigens by immunohistochemistry, most tumor cells, defined as Tg-expressing EGFP+ cells, expressed intermediate levels of CD19 (Figure 7B). Of interest, while WT or Tg B cells expressed high levels of CD19 and did not express Gr1 or CD11c, the tumor cells were characterized by downregulation of CD19 and acquisition of Gr1 or CD11c expression (Figure 7B), a biphenotypic feature that further suggested plasticity between B cell and macrophage/DC lineages during tumor formation.
Figure 7. Biphenotypic features of the B cell–derived transdifferentiated tumors.
(A) Flow cytometry showing Gr1 and/or CD11c expression in the EGFP+ tumor cells. Analysis of 2 representative tumors and control WT and Tg spleens is shown. (B) Many tumor cells express intermediate levels of CD19 and are, therefore, biphenotypic (i.e., CD19intGr1+ or CD19intCD11c+). (C) Tumor clonality analysis by RT-PCR with a set of forward primers covering the most commonly used IgVH gene families and reverse primers located in the JH1-4 gene segments. Polyclonal B cells show 4 bands, indicating JH1, JH2, JH3, and JH4 segments were rearranged, while many tumors gave 1 or 2 bands of different size, indicating monoclonal rearrangements. Arrows indicate monoclonal bands. Sample lanes separated by thin white lines were run on the same gel but were noncontiguous.
The expression of the Tg within the tumor cells, which was dependent on CD19- or Cγ1-driven expression of cre recombinase, strongly supported the B cell origin of the tumors. To confirm the B cell origin of the tumor cells, analysis of Ig gene rearrangement was performed by RT-PCR with a set of forward primers covering the most commonly used IgV heavy (H) gene families and reverse primers located in the JH1–4 gene segments (33). A fraction of the analyzed tumor cases (about 35%) displayed monoclonal IgH rearrangements, as evidenced by the emergence of a single band yielding an in-frame sequence (Figure 7C and Supplemental Figure 6). In most of the remaining cases, loss of heterogeneity in the PCR signal was also found, demonstrating the presence of oligoclonality.
In summary, our data showed that both ROSA26.vFLIP;Cγ1.cre and ROSA26.vFLIP;CD19.cre mice developed tumors at high incidence with morphological and immunophenotypical features typical of histiocytic/DC sarcoma (i.e., Ki67+PU.1+Vimentin+CD68intLysozyme+Mac2+CD11c+Gr1+CD19int) and favored transdifferentiative ontogenesis from vFLIP-expressing B cells. The paracrine effect these B cells exert on surrounding macrophage/DCs may also contribute to tumor development as a variable proportion of CD11c+Gr1+, but EGFP– cells were found in some tumors (Figure 7A). These findings are reminiscent of the cell heterogeneity within the tumor lesions as observed in human KS (34, 35) and other KSHV-associated diseases.
Discussion
In this study, we have investigated the effects of stage-specific vFLIP expression during B cell differentiation in the attempt to recapitulate KSHV-associated diseases (PEL and MCD). Mice developed pathological abnormalities mimicking MCD and showed immunological defects (i.e., lack of GC as well as impairment in Ig class switching and affinity maturation). These findings unveiled for what we believe is the first time the in vivo immunological effects of vFLIP expression. Most surprisingly, in vivo vFLIP expression in B cells led to the development of B cell–derived histiocytic/DC sarcoma, which highlights plasticity between B cell and macrophage/DC lineages and explains the reprogramming ability of KSHV during tumor formation.
Given the evidence that PEL has experienced the GC reaction while MCD develops from earlier precursors that eventually mature into plasmablasts in a GC-independent manner (36), we predicted that ROSA26.vFLIP;Cγ1.cre mice would have represented, a priori, a mouse model more prone to develop PEL-like abnormalities, since Tg activation was designed to occur when expression of the Cγ1 promoter is activated in preparation for class switching, which occurs as the cells initiate the GC reaction. Unexpectedly, both Tg mouse lines lacked GCs and developed MCD-like pathological abnormalities. Thus, our results indicate that even the selective B cell stage–specific pattern of expression achieved by activating vFLIP under the control of Cγ1 promoter might have occurred too early in the GC reaction, preventing its progress and an accurate recapitulation of PEL. Instead, it is possible that the activation of vFLIP at a more mature stage of B cell differentiation, such as in post-GC B cells, might permit completion of the GC reaction and perhaps more accurate PEL development. The conditional knockin mouse model developed here will now allow us to test this hypothesis. Nevertheless, tumors occurring in both vFLIP Tg lines did show some features of PEL (i.e., B cell–derived tumors lacking several B cell–associated markers but positive for CD138), indicating that these characteristics of PEL can be induced by vFLIP alone. It is also likely that the expression of vFLIP alone, even if turned on at the most permissive stage of B cell differentiation, is not sufficient to induce a faithful equivalent of KSHV-associated diseases as observed in humans and that cooperation with other latent proteins (e.g., vCyclin, LANA, LANA-2/vIRF-3, Kaposin B, vIL-6) and/or noncoding transcripts (e.g., miR-K12-11) (37) found to be coexpressed in PEL and MCD may be required. In this regard, previously reported Tg mice for LANA and vFLIP also failed to fully recapitulate KSHV-associated diseases, although tumorigenic properties of the viral products were otherwise demonstrated. Our mouse model contrasts with previous Tg models of KSHV-encoded genes in the high frequency of tumors and the presence of several, albeit not all, unique properties of MCD and PEL not seen in other Tg mouse models.
Here, we identified the in vivo immunological functions of vFLIP when selectively expressed in B cells by demonstrating its role in abrogating GC formation and negatively affecting Ig class switching and affinity maturation. These functions may have a role in tumor maintenance by abrogating the host immune response against virus-infected tumor cells, thus expanding the knowledge on the immune evasion mechanisms exploited by KSHV. Similarly to other viruses capable of efficiently establishing a persistent infection, KSHV has evolved several different strategies to achieve evasion of host immune responses, including inhibition of antigen presentation, molecular mimicry of host signaling, and expression of decoys that inhibit cell death (38). In addition to vFLIP’s ability to inhibit FAS-mediated apoptosis (39), the profound GC suppression here ascribed to vFLIP might represent a novel mechanism KSHV developed to achieve immune evasion by hampering humoral immune responses and abrogating the immunological microenvironment where KSHV-infected cells undergo T cell recognition. The aberrant finding of GC suppression seems to be a consequence of vFLIP-mediated activation of the NF-κB pathway; once activated (e.g., as during CD40 activation or LMP1 expression), it leads to IRF4-mediated transcriptional repression of BCL6 (40). In turn, lack of BCL6, a molecule essential for GC formation (41), can account for the lack of GC observed in vFLIP Tg mice.
Based on these observations, one may speculate that the humoral immune response is affected by the presence of KSHV and that Ig affinity maturation is impaired in KSHV-infected individuals, or at least those with MCD or KSHV-associated lymphadenopathies. While cellular responses to KSHV infection have been studied, our understanding of antibody response to KSHV or in the context of KSHV infection is very limited. The general consensus is that KSHV-specific responses are frequent, diverse, and strongly differentiated toward an effector phenotype (CTLs) in patients who control the infection (42–46). Conversely, KSHV-specific CTLs are very rare in patients who progress to KS, supporting the crucial role of cellular immune responses in controlling KSHV replication, preventing malignancies and conferring resistance to persistent infection (42–44, 46). Although in few cases of MCD polyfunctional KSHV-specific CD8 responses have been observed, suggesting that cellular immunity is less protective in MCD than in KS, a level of immune escape is present also in MCD but restricted to fully differentiated effector memory CD8+ T cells (47). In terms of humoral immunity, one study showed that KSHV-specific antibodies do not correlate with protection and actually positively correlate with viral load (48). To the best of our knowledge, the status of affinity maturation of KSHV-specific antibodies has not been tested, but it is likely to be impaired given the inefficiency in controlling viral load and disease progression. The hypothesis that KSHV infection qualitatively affects antibody responses could be tested experimentally through prospective epidemiological studies.
Also, the immunological functions of vFLIP described here highlight the fact that different strategies are exploited by even closely related viruses to manipulate the immune system. Indeed, EBV LMP1, a TNF receptor analog that shares with vFLIP its ability to activate NF-κB, rescues an otherwise severely impaired T cell–dependent Ig class switching when expressed in B cells of Cd40–/– mice (49), even if both GC formation and affinity maturation remained abrogated similarly to vFLIP Tg mice. Lack of GC upon enforced expression of either LMP1 or vFLIP is not surprising, since CD40-activated NF-κB signaling is known to be selectively downregulated in GC cells (50) and, therefore, its constitutive activation is most likely to be unfavorable for GC formation. In addition, the different effects exerted by LMP1 and vFLIP on Ig class switching may be due to the differences in signaling. For example, it is possible that this is partially due to different intrinsic modulation on noncoding RNAs, particularly miR155, an oncogenic noncoding transcript (51) critical for GC formation (52), normal immune function (53), and the generation of class-switched antibodies (54). miR155-deficient mice highly expressed PU.1, a direct target of miR155 that favors macrophage over B-lymphocyte differentiation (55), whose overexpression in WT B cells impairs the emergence of IgG1-switched cells (54). Of interest, LMP1 activates miR155, and reduction in PU.1 might explain the ability of LMP1 to induce Ig class switching (56) and, in contrast to vFLIP Tg mice, lack of propensity to develop histiocytic sarcomas. Therefore, since miR155 is essential for GC formation as well as Ig CSR and SHM, we speculated that vFLIP might downregulate miR155 and cause the abrogation of these immunological functions. However, we found that vFLIP does not affect miR155 expression, despite our observation of the upregulation of the miR155 target gene PU.1 within vFLIP-expressing tumors (Supplemental Figure 8), which implies that these immunological functions can be abrogated even in the presence of normal levels of miR155. The fact that vFLIP does not upregulate miR155 expression may explain why KSHV encodes a miR155 ortholog in its genome (37, 57) that may enable KSHV to complement vFLIP expression to allow Ig class switching in PEL in vivo. Lack of miR155 upregulation upon vFLIP expression may also partially explain the phenotypic differences seen in the LMP1 and vFLIP Tg mice.
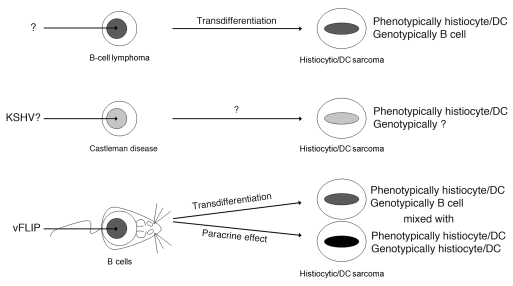
In spite of a histiocytic/DC phenotype, the B cell derivation of the tumor cells is supported by (a) the expression of the Tg, which is driven by 2 independent and different B cell–specific promoters, CD19 and Cγ1, making the possibility of leaky expression in non–B cell lineage unlikely, (b) the intermediate expression of CD19 by the tumor cells, and formally proven by (c) the presence of monoclonal Ig gene rearrangement. Growing evidence supports the existence of an extraordinary plasticity between committed B cells and other cell lineages, particularly macrophage/DC, in both normal and pathological conditions (58). Myeloid conversion of B cells, with retention of Ig gene rearrangement as evidence of B cell ontogenesis, has been obtained by enforced expression of myeloid/macrophage determinants (i.e., CCAAT/enhancer binding protein [C/EBP] α/β and PU.1) and subsequent functional inactivation of Pax5, a major controller of B cell identity (59). Moreover, transdifferentiation can also occur during clonal evolution of B cell malignancies, a concept first demonstrated by the development of histiocytic/DC sarcoma in a patient with a previously diagnosed follicular lymphoma (FL) found to be clonally related, as both neoplasms retained the same IgH rearrangement and identical breakpoint of the FL-specific translocation (60). Of interest, development of follicular/DC sarcoma has also been reported in a patient previously diagnosed with Castleman disease at the same site (61, 62), suggesting KSHV could have a role in this phenomenon. In our study, by expressing only one KSHV gene in B cells, we observed initially pathological alterations mimicking MCD and, eventually, development of histiocytic/DC sarcoma possibly via transdifferentiation mechanism from B cells. Therefore, this surprising phenotype is important since it may recapitulate some peculiar examples of human pathology. This is also the first experimental model in which this transdifferentiation phenomenon is reported as entirely initiated by ectopic expression of a single (KSHV) viral gene product (Figure 8). This potent transdifferentiation is likely due to an epigenetic process resulting in the expansion of macrophage/DC population upon constitutive expression of vFLIP in B cells and is interesting for several reasons. First, it underlines the existence of a general potential plasticity between B cells and other cell lineages (33, 58), particularly macrophage/DCs (59, 60). Second, since KSHV is associated with diseases in which tumor derivation has been made extremely puzzling by the presence of a rich macrophage component, especially KS, our finding suggests this phenomenon is a result of vFLIP-driven reprogramming from B cells to macrophage/DCs and/or a paracrine effect due to the secretion of macrophage/DC-stimulating factors from vFLIP-expressing B cells (Figure 8). Although most macrophages in KS lesions do not contain KSHV, largely favoring a paracrine effect, rare cells coexpress KSHV LANA and histiocytic antigens (63), suggesting that a pathogenetic mechanism also involving cell reprogramming and transdifferentiation might coexist. Furthermore, the highly abnormal immunophenotype of PEL may be partly explained by vFLIP-induced transcriptional reprogramming.
Figure 8. Model of vFLIP-mediated tumorigenesis, cell reprogramming, and paracrine stimulation of histiocytic/DCs.
The expression of KSHV vFLIP alone in B cells initially results in pathological alterations mimicking MCD and eventually leads to the development of B cell–derived tumors via transdifferentiation, as evidenced by the biphenotypic features of the tumor cells (i.e., phenotypically macrophage/DCs, genotypically B cells). This surprising phenotype has a correspondence in human pathology (i.e., development of follicular/DC sarcoma in a patient with previously diagnosed Castleman disease) and underlines an existing plasticity between B cell and macrophage/DC lineages, as recently reported also for other B cell malignancies (e.g., FL). Moreover, the expansion of phenotypically and genotypically genuine macrophage/DCs, as observed in the Tg mouse spleen, suggests that vFLIP-expressing B cells can sustain the proliferation of this compartment also via a paracrine mechanism. This process may contribute to the cell heterogeneity seen in KS, which is typically characterized by abundant histiocytic infiltrate of unknown origin and function.
Although coordinated silencing of B cell transcription factors (e.g., Pax5) and activation of transcriptional program related to myeloid-macrophage lineage (e.g., PU.1 and C/EBPα/β) seem to orchestrate the B-to-macrophage cell lineage transition, the molecular mechanisms underlying this fascinating phenomenon are far from being fully understood. The cell reprogramming observed in vFLIP Tg mice makes this mouse model an ideal system to obtain insights into the complexity of these mechanisms, culminating not only in lineage diversion but also tumor formation.
In conclusion, we have revealed a function for vFLIP in inducing expansion of the macrophage/DC compartment via both transdifferentiation and paracrine mechanism. This has important implications for the pathogenesis of all KSHV-associated malignancies that invariably display a rich infiltrate of histiocytes of unknown origin. For what we believe is the first time, immunological functions have been ascribed to the in vivo expression of vFLIP, which may contribute to host immune dysfunction during tumor development and maintenance. Moreover, we believe these are the first Tg mice expressing a KSHV gene that recapitulates the cytoplasmic IgM-λ expression of the KSHV-infected cells in MCD and the abnormal B cell genotypic/phenotypic properties of PEL. Finally, these mice develop tumors with high frequency and reproducibility, providing a robust model to test inhibitors of vFLIP as potential anticancer agents.
Methods
Generation of ROSA26.vFLIP;CD19.cre and ROSA26.vFLIP;Cγ1.cre Tg mouse lines.
We cloned a C-terminal flagged vFLIP sequence into the ROSA26 locus by using a modified version of pROSA1 targeting vector (64), in which a unique AscI restriction site was embedded between a loxP-flanked neoR-STOP cassette and a frt-flanked IRES-EGFP sequence followed by a polyadenylation site (65). vFLIP was inserted into the AscI site. The final targeting vector was electroporated into 129/Sv-derived ES cells, following standard protocols in the Transgenic Mouse Facility at Columbia University. The targeted ES cells were screened for homologous recombination by Southern blot analysis using diagnostic digestions and probes as shown in Figure 1. Correctly targeted ES cells were injected into C57BL/6-derived blastocysts to obtain chimeric mice. The ROSA26.vFLIP Tg mouse line was developed by crossing the chimera males with C57BL/6 WT females for a few generations before being made homozygous.
To generate mice expressing the Tg in a B cell–specific manner, heterozygous ROSA26.vFLIP Tg mice were crossed with homozygous CD19.cre knockin mice (66) or Cγ1.cre knockin mice (67) of either C57BL/6 or BALB/c genetic background; therefore, all experimental mice were on 129/Sv-C57BL/6 or 129/Sv-C57BL/6-BALB/c mixed genetic background and age-matched littermates were used as controls. Genotyping was performed by PCR analysis on mouse tail DNA. All animal studies were approved by the Institutional Animal Care and Use Committee at Weill Cornell Medical College. Mice were monitored for tumor development biweekly and sacrificed when visibly ill, according to approved protocols. Statistical analysis of event-free survival was performed by GraphPad Prism v.5 using Kaplan-Meier cumulative survival curve and the log-rank test to evaluate statistical significance.
B cell purification, RT-PCR, and immunoblotting.
CD19+ and CD19– splenic cells were isolated by magnetic cell separation using MACS columns (Miltenyi Biotech), following the manufacturer’s instructions. RNA extraction and RT-PCR were performed using standard protocols as detailed in Supplemental Methods. Total protein extracts were prepared from thymus as well as CD19+ and CD19– splenocytes using RIPA buffer, gel electrophoresed on 12% SDS-PAGE gel, transferred to a polyvinylidene difluoride membrane (Millipore), and immunostained according to standard methods using anti-FLAG (M2; Sigma-Aldrich), anti–phospho-p65 (93H1; Cell Signaling), anti–phospho-IκBα (5A5; Cell Signaling), and anti–β-actin (Sigma-Aldrich) antibodies.
Flow cytometry.
Single-cell suspensions prepared from spleen and tumor biopsies were stained using standard procedures with a panel of fluorescent-labeled antibodies (see Supplemental Methods). 7AAD was used for the exclusion of dead cells. Data were acquired on FACSCalibur or LSRII flow cytometer (BD) and analyzed using FlowJo software (Tree Star).
Immunohistochemistry.
Formalin-fixed, paraffin-embedded sections of 4-μm thickness were stained for H&E or immunostained with the following antibodies: anti-B220 (RA3-6B2; BD Biosciences — Pharmingen), anti-CD3 (Dako), anti-BCL6 (N3; Santa Cruz Biotechnology Inc.), anti-CD138 (281-2; BD Biosciences — Pharmingen), anti-CD34 (MEC14.7; Abcam), anti-CD68 (FA-11; Abcam), anti-Ki67 (Novocastra), anti-κ (Southern Biotech), anti-λ (Southern Biotech), anti-Lysozyme (BGN/06/961; Abcam), anti-Pax5 (24; BD Transduction Laboratories), PNA (Vector), anti-PU.1 (G148-74; BD Biosciences — Pharmingen), anti-SMA (B4; Santa Cruz Biotechnology Inc.), anti-Vimentin (C20; Santa Cruz Biotechnology Inc.), anti-Mac2 (M3/38; Cedarlane), anti-EGFP (Abcam), and anti-BLIMP1 (63D; Santa Cruz Biotechnology Inc.).
Immunization.
Mice 8–12 weeks of age were immunized intraperitoneally with 0.5 ml of a 2% SRBC suspension in PBS (Cocalico Biologicals) and analyzed after 12 days. For analysis of T cell–dependent antigen-specific immune response, 12-week-old mice were immunized on day 0 with 100 μg of NP24-KLH in complete Freund adjuvant and bled both before immunization and on day 5. On days 21 and 42, mice were boosted with 100 μg NP24-KLH in incomplete Freund adjuvant and bled 5 days after each immunization.
ELISA.
To determine the concentration of resting or NP-specific antibodies of specific Ig isotypes, microtiter plates (NUNC) were coated with either unlabeled anti-mouse Ig antibodies (SouthernBiotech) or NP20-BSA, respectively. AP-labeled goat anti-mouse Ig isotype-specific antibodies (SouthernBiotech) and p-nitrophenyl phosphate (SouthernBiotech) were used as revealing antibodies and AP substrate, respectively. The OD at 405 nm was measured and readings within the assay linear range were used to calculate the serum antibody concentrations against control mouse Ig isotype standards (SouthernBiotech). For measurement of the affinity maturation of NP-specific IgG1 antibodies, each serum sample was titrated on both NP3-BSA– and NP20-BSA–coated plates. Low-affinity antibodies will only bind to the highly haptenated proteins (NP20-BSA), while high-affinity Ig binds equally to both the high and low haptenated proteins. Thus, the ratio of the binding to NP3-BSA and NP20-BSA is a measure of the relative Ig affinity maturation.
IgVH rearrangement analysis.
IgVH rearrangement analysis was performed on cDNA with a set of forward primers that anneal to the framework region of the most abundantly used IgVH gene families and reverse primers located in the JH1-4 gene segments (33). PCR products were gel-purified using the QIAquick purification method (QIAGEN) and directly sequenced. Sequences were compared with those in the IMGT/V-QUEST database ( http://www.imgt.org).
Statistics.
Statistical significance was assessed by a 2-tailed unpaired Student’s t test. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank K. Rajewsky and Y. Sasaki for the modified version of pROSA1 targeting vector and Y.-F. Liu for help with the immunohistochemistry. This work was funded by the National Cancer Institute (R01CA068939 and R01CA103646 grants to E. Cesarman); additional support was provided by the Pathology and Laboratory Medicine Translational Research Program.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(3):1141–1153. doi:10.1172/JCI44417.
Kang Chen’s present address is: Department of Medicine, Division of Clinical Immunology, Mount Sinai Medical Center, New York, New York, USA.
See the related Commentary beginning on page 838.
References
- 1. Rous P. A sarcoma of the fowl transmissible by an agent separable from the tumor cells. J Exp Med. 1911;13(4):397–411. doi: 10.1084/jem.13.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chang Y, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266(5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 3. Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332(18):1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 4. Soulier J, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86(4):1276–1280. [PubMed] [Google Scholar]
- 5. Carbone A, et al. Kaposi’s sarcoma-associated herpesvirus DNA sequences in AIDS-related and AIDS-unrelated lymphomatous effusions. Br J Haematol. 1996;94(3):533–543. doi: 10.1046/j.1365-2141.1996.d01-1826.x. [DOI] [PubMed] [Google Scholar]
- 6. Nador RG, et al. Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi’s sarcoma-associated herpes virus. Blood. 1996;88(2):645–656. [PubMed] [Google Scholar]
- 7. Jenner RG, et al. Kaposi’s sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc Natl Acad Sci U S A. 2003;100(18):10399–10404. doi: 10.1073/pnas.1630810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klein U, et al. Gene expression profile analysis of AIDS-related primary effusion lymphoma (PEL) suggests a plasmablastic derivation and identifies PEL-specific transcripts. Blood. 2003;101(10):4115–4121. doi: 10.1182/blood-2002-10-3090. [DOI] [PubMed] [Google Scholar]
- 9. Matolcsy A, Nador RG, Cesarman E, Knowles DM. Immunoglobulin VH gene mutational analysis suggests that primary effusion lymphomas derive from different stages of B cell maturation. Am J Pathol. 1998;153(5):1609–1614. doi: 10.1016/S0002-9440(10)65749-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Judde JG, et al. Monoclonality or oligoclonality of human herpesvirus 8 terminal repeat sequences in Kaposi’s sarcoma and other diseases. J Natl Cancer Inst. 2000;92(9):729–736. doi: 10.1093/jnci/92.9.729. [DOI] [PubMed] [Google Scholar]
- 11. Du MQ, et al. Kaposi sarcoma-associated herpesvirus infects monotypic (IgM lambda) but polyclonal naive B cells in Castleman disease and associated lymphoproliferative disorders. Blood. 2001;97(7):2130–2136. doi: 10.1182/blood.V97.7.2130. [DOI] [PubMed] [Google Scholar]
- 12. Ballon G, Cesarman E. Castleman’s disease. In: Paul JP, Volberding A, eds.Viral and immunological malignancies . Hamilton, Ontario, Canada: BC Decker, Inc; 2006:108–121. [Google Scholar]
- 13. Chadburn A, et al. Immunophenotypic analysis of the Kaposi sarcoma herpesvirus (KSHV; HHV-8)-infected B cells in HIV+ multicentric Castleman disease (MCD). Histopathology. 2008;53(5):513–524. doi: 10.1111/j.1365-2559.2008.03144.x. [DOI] [PubMed] [Google Scholar]
- 14. Sodhi A, et al. The TSC2/mTOR pathway drives endothelial cell transformation induced by the Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor. Cancer Cell. 2006;10(2):133–143. doi: 10.1016/j.ccr.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 15. Bais C, et al. Kaposi’s sarcoma associated herpesvirus G protein-coupled receptor immortalizes human endothelial cells by activation of the VEGF receptor-2/ KDR. Cancer Cell. 2003;3(2):131–143. doi: 10.1016/S1535-6108(03)00024-2. [DOI] [PubMed] [Google Scholar]
- 16. Grundhoff A, Ganem D. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J Clin Invest. 2004;113(1):124–136. doi: 10.1172/JCI200417803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J Virol. 1998;72(2):1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chaudhary PM, Jasmin A, Eby MT, Hood L. Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene. 1999;18(42):5738–5746. doi: 10.1038/sj.onc.1202976. [DOI] [PubMed] [Google Scholar]
- 19. Field N, et al. KSHV vFLIP binds to IKK-gamma to activate IKK. J Cell Sci. 2003;116(pt 18):3721–3728. doi: 10.1242/jcs.00691. [DOI] [PubMed] [Google Scholar]
- 20. Keller SA, et al. NF-kappaB is essential for the progression of KSHV- and EBV-infected lymphomas in vivo. Blood. 2006;107(8):3295–3302. doi: 10.1182/blood-2005-07-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Keller SA, Schattner EJ, Cesarman E. Inhibition of NF-kappaB induces apoptosis of KSHV-infected primary effusion lymphoma cells. Blood. 2000;96(7):2537–2542. [PubMed] [Google Scholar]
- 22. Guasparri I, Keller SA, Cesarman E. KSHV vFLIP is essential for the survival of infected lymphoma cells. J Exp Med. 2004;199(7):993–1003. doi: 10.1084/jem.20031467. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Irmler M, et al. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388(6638):190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 24. Thome M, et al. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386(6624):517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 25. Krueger A, Baumann S, Krammer PH, Kirchhoff S. FLICE-inhibitory proteins: regulators of death receptor-mediated apoptosis. Mol Cell Biol. 2001;21(24):8247–8254. doi: 10.1128/MCB.21.24.8247-8254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee JS, et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol. 2009;11(11):1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matta H, Chaudhary PM. Activation of alternative NF-kappa B pathway by human herpes virus 8-encoded Fas-associated death domain-like IL-1 beta-converting enzyme inhibitory protein (vFLIP). Proc Natl Acad Sci U S A. 2004;101(25):9399–9404. doi: 10.1073/pnas.0308016101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chugh P, et al. Constitutive NF-kappaB activation, normal Fas-induced apoptosis, and increased incidence of lymphoma in human herpes virus 8 K13 transgenic mice. Proc Natl Acad Sci U S A. 2005;102(36):12885–12890. doi: 10.1073/pnas.0408577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuhnel F, et al. NFkappaB mediates apoptosis through transcriptional activation of Fas (CD95) in adenoviral hepatitis. J Biol Chem. 2000;275(9):6421–6427. doi: 10.1074/jbc.275.9.6421. [DOI] [PubMed] [Google Scholar]
- 30. Punj V, Matta H, Schamus S, Chaudhary PM. Integrated microarray and multiplex cytokine analyses of Kaposi’s sarcoma associated herpesvirus viral FLICE inhibitory protein K13 affected genes and cytokines in human blood vascular endothelial cells. BMC Med Genomics. 2009;2:50. doi: 10.1186/1755-8794-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sakakibara S, Pise-Masison CA, Brady JN, Tosato G. Gene regulation and functional alterations induced by Kaposi’s sarcoma-associated herpesvirus-encoded ORFK13/vFLIP in endothelial cells. . J Virol. 2009;83(5):2140–2153. doi: 10.1128/JVI.01871-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Derudder E, et al. Development of immunoglobulin lambda-chain-positive B cells, but not editing of immunoglobulin kappa-chain, depends on NF-kappaB signals. Nat Immunol. 2009;10(6):647–654. doi: 10.1038/ni.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133(2):250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Browning PJ, et al. Identification and culture of Kaposi’s sarcoma-like spindle cells from the peripheral blood of human immunodeficiency virus-1-infected individuals and normal controls. Blood. 1994;84(8):2711–2720. [PubMed] [Google Scholar]
- 35. Uccini S, et al. Kaposi’s sarcoma cells express the macrophage-associated antigen mannose receptor and develop in peripheral blood cultures of Kaposi’s sarcoma patients. Am J Pathol. 1997;150(3):929–938. [PMC free article] [PubMed] [Google Scholar]
- 36. Du MQ, Bacon CM, Isaacson PG. Kaposi sarcoma-associated herpesvirus/human herpesvirus 8 and lymphoproliferative disorders. J Clin Pathol. 2007;60(12):1350–1357. doi: 10.1136/jcp.2007.047969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Skalsky RL, et al. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81(23):12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coscoy L. Immune evasion by Kaposi’s sarcoma-associated herpesvirus. Nat Rev Immunol. 2007;7(5):391–401. doi: 10.1038/nri2076. [DOI] [PubMed] [Google Scholar]
- 39. French LE, Tschopp J. Inhibition of death receptor signaling by FLICE-inhibitory protein as a mechanism for immune escape of tumors. J Exp Med. 1999;190(7):891–894. doi: 10.1084/jem.190.7.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saito M, et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12(3):280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 41. Ye BH, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16(2):161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 42. Lambert M, et al. Differences in the frequency and function of HHV8-specific CD8 T cells between asymptomatic HHV8 infection and Kaposi sarcoma. Blood. 2006;108(12):3871–3880. doi: 10.1182/blood-2006-03-014225. [DOI] [PubMed] [Google Scholar]
- 43. Osman M, et al. Identification of human herpesvirus 8-specific cytotoxic T-cell responses. J Virol. 1999;73(7):6136–6140. doi: 10.1128/jvi.73.7.6136-6140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Strickler HD, et al. Human herpesvirus 8 cellular immune responses in homosexual men. J Infect Dis. 1999;180(5):1682–1685. doi: 10.1086/315056. [DOI] [PubMed] [Google Scholar]
- 45. Wang QJ, et al. CD8+ cytotoxic T lymphocyte responses to lytic proteins of human herpes virus 8 in human immunodeficiency virus type 1-infected and -uninfected individuals. J Infect Dis. 2000;182(3):928–932. doi: 10.1086/315777. [DOI] [PubMed] [Google Scholar]
- 46. Wilkinson J, et al. Identification of Kaposi’s sarcoma-associated herpesvirus (KSHV)-specific cytotoxic T-lymphocyte epitopes and evaluation of reconstitution of KSHV-specific responses in human immunodeficiency virus type 1-Infected patients receiving highly active antiretroviral therapy. J Virol. 2002;76(6):2634–2640. doi: 10.1128/JVI.76.6.2634-2640.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guihot A, et al. Multicentric Castleman disease is associated with polyfunctional effector memory HHV-8-specific CD8+ T cells. Blood. 2008;111(3):1387–1395. doi: 10.1182/blood-2007-03-080648. [DOI] [PubMed] [Google Scholar]
- 48. Sitas F, Newton R, Boshoff C. Increasing probability of mother-to-child transmission of HHV-8 with increasing maternal antibody titer for HHV-8. N Engl J Med. 1999;340(24):1923. doi: 10.1056/NEJM199906173402414. [DOI] [PubMed] [Google Scholar]
- 49. Uchida J, et al. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science. 1999;286(5438):300–303. doi: 10.1126/science.286.5438.300. [DOI] [PubMed] [Google Scholar]
- 50. Basso K, et al. Tracking CD40 signaling during germinal center development. Blood. 2004;104(13):4088–4096. doi: 10.1182/blood-2003-12-4291. [DOI] [PubMed] [Google Scholar]
- 51. Tam W, Ben-Yehuda D, Hayward WS. bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol Cell Biol. 1997;17(3):1490–1502. doi: 10.1128/mcb.17.3.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316(5824):604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 53. Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vigorito E, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27(6):847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288(5470):1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 56. Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36(20):6608–6619. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gottwein E, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450(7172):1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cobaleda C, Busslinger M. Developmental plasticity of lymphocytes. Curr Opin Immunol. 2008;20(2):139–148. doi: 10.1016/j.coi.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 59. Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117(5):663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 60. Feldman AL, et al. Clonally related follicular lymphomas and histiocytic/dendritic cell sarcomas: evidence for transdifferentiation of the follicular lymphoma clone. Blood. 2008;111(12):5433–5439. doi: 10.1182/blood-2007-11-124792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chan AC, Chan KW, Chan JK, Au WY, Ho WK, Ng WM. Development of follicular dendritic cell sarcoma in hyaline-vascular Castleman’s disease of the nasopharynx: tracing its evolution by sequential biopsies. Histopathology. 2001;38(6):510–518. doi: 10.1046/j.1365-2559.2001.01134.x. [DOI] [PubMed] [Google Scholar]
- 62. Cronin DM, Warnke RA. Castleman disease: an update on classification and the spectrum of associated lesions. Adv Anat Pathol. 2009;16(4):236–246. doi: 10.1097/PAP.0b013e3181a9d4d3. [DOI] [PubMed] [Google Scholar]
- 63. Parravicini C, et al. Differential viral protein expression in Kaposi’s sarcoma-associated herpesvirus-infected diseases: Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. Am J Pathol. 2000;156(3):743–749. doi: 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci U S A. 1997;94(8):3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sasaki Y, et al. Canonical NF-kappaB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity. 2006;24(6):729–739. doi: 10.1016/j.immuni.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 66. Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25(6):1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Casola S, et al. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc Natl Acad Sci U S A. 2006;103(19):7396–7401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.