Abstract
Hypertrophic cardiomyopathy (HCM) is a leading cause of sudden death in children and young adults. Abnormalities in several signaling pathways are implicated in the pathogenesis of HCM, but the role of the RAS-RAF-MEK-ERK MAPK pathway has been controversial. Noonan syndrome (NS) is one of several autosomal-dominant conditions known as RASopathies, which are caused by mutations in different components of this pathway. Germline mutations in RAF1 (which encodes the serine-threonine kinase RAF1) account for approximately 3%–5% of cases of NS. Unlike other NS alleles, RAF1 mutations that confer increased kinase activity are highly associated with HCM. To explore the pathogenesis of such mutations, we generated knockin mice expressing the NS-associated Raf1L613V mutation. Like NS patients, mice heterozygous for this mutation (referred to herein as L613V/+ mice) had short stature, craniofacial dysmorphia, and hematologic abnormalities. Valvuloseptal development was normal, but L613V/+ mice exhibited eccentric cardiac hypertrophy and aberrant cardiac fetal gene expression, and decompensated following pressure overload. Agonist-evoked MEK-ERK activation was enhanced in multiple cell types, and postnatal MEK inhibition normalized the growth, facial, and cardiac defects in L613V/+ mice. These data show that different NS genes have intrinsically distinct pathological effects, demonstrate that enhanced MEK-ERK activity is critical for causing HCM and other RAF1-mutant NS phenotypes, and suggest a mutation-specific approach to the treatment of RASopathies.
Introduction
Cardiac hypertrophy is a major way by which cardiomyocytes respond to various stresses, including abnormal neurohormonal stimuli, hemodynamic overload, and injury. There are 2 general types of cardiac hypertrophy (1, 2): physiological, which is associated with exercise or pregnancy, and pathological, whose primary cause is genetic defects (termed primary hypertrophy); and excessive afterload, resulting from conditions such as hypertension or valvular stenosis (termed secondary hypertrophy). With increased cardiac stress, cardiac hypertrophy may initially represent a compensatory response of the myocardium. However, chronic pathological hypertrophy predisposes to ventricular dilatation, heart failure, arrhythmia, and/or sudden death (3, 4). Physiological hypertrophy is typically concentric, with preservation of chamber shape, absence of inflammation or fibrosis, and normal cardiac gene expression. In contrast, pathological hypertrophy eventually progresses to chamber dilatation (eccentric hypertrophy), is often associated with fibrosis, and typically leads to the reactivation of a fetal gene expression program characterized by increased levels of (among others) atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), and β–myosin heavy chain (β-MHC) (5). Delineating the molecular pathways that distinguish physiological and pathological hypertrophy, and identifying ways to reverse the latter, are of obvious medical importance.
Primary hypertrophic cardiomyopathy (HCM), the prototypic genetic form of pathological hypertrophy, is a leading cause of sudden death in the young (6). The hallmark of HCM is cardiac hypertrophy in the absence of an obvious inciting hypertrophic stimulus (7). Mutations in genes encoding sarcomeric proteins (e.g., β-MHC, cardiac troponin T, and myosin-binding protein C) account for approximately 75% of primary HCM cases. Such mutations usually alter sarcomere structure and function and result in mechanical, biochemical, and/or bioenergetic stresses that activate cardiomyocyte signaling pathways to mediate the hypertrophic phenotype (8–11). Aberrant activation of hypertrophic signaling pathways can themselves result in hypertrophy. For example, germline mutations in AMPK are a rare cause of HCM (12–14). Moreover, genetic and cellular models have identified multiple signaling systems that can cause or contribute to pathological hypertrophy, including the calcineurin-NFAT, PI3K-Akt-mTOR, glycogen synthase kinase–3β (GSK-3β), and JNK pathways (1, 2, 15). The detailed mechanism by which aberrant activation of these pathways evokes pathological hypertrophy remains incompletely understood.
The RAS-RAF-MEK-ERK MAPK pathway (referred to herein as the RAS-ERK pathway) is a central signaling cascade evoked by multiple agonists, including growth factors (e.g., Heregulin, IGF-I, EGF, and PDGF), cytokines (e.g., IL-6, cardiotrophin, and leukemia inhibitory factor [LIF]), G protein–coupled receptor (GPCR) agonists (e.g., angiotensin II [Ang II] and β-adrenergic agonists), and physical stimuli (e.g., mechanical stretch), in cardiomyocytes as well as other cell types (1, 2, 16). The pathway is initiated by activation of RAS, which requires RAS–guanine nucleotide exchange factors (RAS-GEFs) such as SOS1 and, in most cell types, the protein-tyrosine phosphatase SHP2 (encoded by PTPN11). RAS recruits RAF proteins (e.g., RAF1, BRAF, and ARAF) to the cell membrane, where they are activated and subsequently form complexes with MEK1/2 and ERK1/2, aided by scaffolds such as KSR. Activated RAF proteins phosphorylate MEK1/2, which in turn phosphorylates ERK1/2. ERKs phosphorylate cytosolic substrates and also translocate to the nucleus to stimulate diverse gene expression programs by phosphorylating several transcription factors (16, 17).
The role of the RAS-ERK pathway in cardiac hypertrophy has been controversial. Some data argue that excessive activity of this pathway causes HCM, whereas other evidence suggests involvement in physiological, but not pathological, hypertrophy (18, 19). Transgenic mice with cardiac-specific expression of oncogenic HRas (G12V) display significant cardiac hypertrophy, decreased contractility, diastolic dysfunction associated with interstitial fibrosis, induction of cardiac fetal genes, and sudden death (20–22), all of which are consistent with HCM. In cultured cardiomyocytes, depletion of Erk1/2 with antisense oligonucleotides or pharmacological inhibition of Mek1/2 attenuates the hypertrophic response to agonist stimulation (23, 24). Mice with cardiac-specific overexpression of dominant-negative Raf1 have no overt phenotype, but they are resistant to the development of cardiac hypertrophy in response to pressure overload (25), which suggests that signals from Raf1 are necessary for the hypertrophic response. On the other hand, transgenic mice expressing an activated Mek1 allele under the control of the α-MHC promoter have concentric hypertrophy with enhanced contractile performance, show no signs of decompensation over time, and reportedly do not progress to pathological hypertrophy (26). A recent study even argued against any role for ERK1/2 in cardiac hypertrophy, as Erk1–/–Erk2+/– mice, as well as transgenic mice with cardiac-specific expression of dual specificity phosphatase 6 (Dusp6), an ERK1/2-specific phosphatase, showed a normal hypertrophic response to pressure overload and exercise (27).
Over the past 10 years, germline mutations in genes encoding several members of the RAS-ERK pathway have been identified in a set of related, yet distinct, human developmental syndromes (28–32), now collectively termed the RASopathies (31, 32). These disorders, some (but not all) of which include HCM as a syndromic phenotype, present an opportunity to clarify the role of the RAS-ERK pathway in cardiac hypertrophy. Noonan syndrome (NS), a relatively common autosomal-dominant disorder with an occurrence of 1 in about 1,000–2,500 live births, typically presents with proportional short stature, facial dysmorphia, and cardiovascular abnormalities. About 25%–50% of NS patients exhibit some form of myeloproliferative disorder (MPD), which is usually transient and resolves spontaneously; rarely, NS patients develop the severe childhood MPD juvenile myelomonocytic leukemia (JMML) or other forms of leukemia (33). Mutations in PTPN11 that increase SHP2 phosphatase activity account for approximately 50% of NS cases (34); other known NS genes include SOS1 (~10%; refs. 35, 36), RAF1 (3%–5%; refs. 37, 38), KRAS (1%–2%; refs. 39, 40), NRAS (<1%; ref. 41), and SHOC2 (<1%; ref. 42).
Although NS patients typically have valvuloseptal defects, approximately 20% have HCM (43). Moreover, different NS genes are differentially associated with HCM. Only approximately 10% of NS patients with PTPN11 mutations (44) and approximately 20% of those with mutations in SOS1 (35) develop HCM. By contrast, HCM is found in approximately 95% of patients bearing RAF1 mutations that cause increased kinase activity (37, 38). The frequency of HCM also varies in other RASopathies. HCM is the most frequent (~80%) cardiovascular manifestation of LEOPARD syndrome (LS), caused by phosphatase-inactivating mutations of PTPN11 (45–48), but also is common (~50% in each) in Costello syndrome (CS), caused by gain-of-function mutations in HRAS (49, 50), and cardio-facio-cutaneous (CFC) syndrome, caused by BRAF, MEK1, or MEK2 mutations (51–53). Whether these differences represent differential effects of specific RAS-ERK pathway mutations, the effects of modifiers in the outbred human population, or both remains unclear.
Mouse models have begun to address such issues and to provide insight into the detailed pathogenesis and potential therapeutic approaches to these disorders. For example, we previously generated a knockin mouse model of the NS-associated Ptpn11D61G mutation that recapitulates the major features of NS, including short stature, facial dysmorphia, mild MPD, and valvuloseptal defects. These mice, like most PTPN11 mutant NS patients, do not have HCM (54). Transgenic mice expressing a different NS-associated Ptpn11 mutant, Q79R, also show valvuloseptal defects and facial abnormalities seen in NS patients, which are prevented by genetic ablation of Erk1/2 and prenatal pharmacological inhibition of Mek, respectively (55–57). Genetic ablation of Erk1 also prevents the development of valvuloseptal defects in mice expressing a highly activated Ptpn11 mutant in endocardial cells (58). A knockin mouse model of CS caused by the HRasG12V mutation shows HCM, but these mice also have aortic stenosis, making it unclear whether hypertrophy is primary or secondary (59).
Here, we have generated knockin mice expressing the kinase-activating NS mutant Raf1L613V. Similar to Ptpn11 mutant mice, mice expressing this Raf1 allele had short stature, facial dysmorphia, and hematological abnormalities; however, they did not have valvuloseptal defects, but instead developed HCM. Remarkably, nearly all phenotypic abnormalities in Raf1-mutant mice were reversed by postnatal MEK inhibitor treatment. Our results show that different NS genes have intrinsically distinct pathological effects and demonstrate that enhanced MEK-ERK activity is critical for causing HCM and other RAF1-mutant NS phenotypes. Along with the companion study on LS-associated HCM by Marin et al. (60), these findings suggest a mutation-specific approach to the treatment of RASopathies.
Results
Generation of L613V/+ mice.
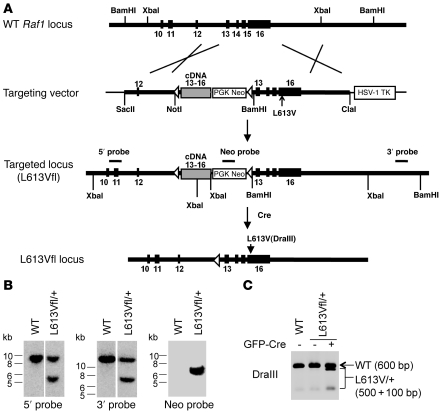
Expression of an activated Raf1 mutant during development might cause embryonic lethality. Therefore, in order to investigate the effects of the NS-associated, kinase-activating RAF1L613V mutant, we designed an inducible knockin Raf1L613V allele (L613Vfl; Figure 1A). The targeting vector included a cassette containing a splice acceptor sequence, a Raf1 cDNA fragment encoding WT exons 13–16, and a pGK-Neo (Neo) gene. The fusion cDNA/Neo cassette was flanked by loxP sites and was positioned upstream of exons 13–16 of the Raf1 gene itself, with an L613V mutation introduced into exon 16 and an HSV-TK cassette for negative selection. In the absence of Cre recombinase (Cre), Raf1 exon 12 should be spliced to the cDNA (exon 13–16), leading to the production of WT Raf1. When Cre is present, the floxed cassette should be excised, evoking transcription of the mutant Raf1 allele.
Figure 1. Generation of inducible Raf1L613V knockin mice.
(A) Targeting strategy. Structures of the Raf1 locus, targeting vector, mutant allele, and location of probes for Southern blotting are shown. (B) Correct targeting of ES cells. Genomic DNA from WT ES cells and PCR-positive L613Vfl/+ ES clones was digested with XbaI (5′ and Neo probe) or BamHI (3′ probe) and subjected to Southern blotting with 5′, 3′, or Neo probes. Blots with 5′ and 3′ probes were run on the same gel but were noncontiguous (white lines). (C) Expression of Raf1L613V allele is inducible. RNA was isolated from WT and L613Vfl/+ ES cells with or without prior transfection of MSCV-Cre-GFP plasmid and reverse transcribed into cDNA. A PCR product, obtained by using primers within exon 11 and at the end of exon 16 of the Raf1 cDNA, was digested with DraIII. Note that the mutant allele was silent until Cre was introduced, and then was expressed efficiently.
The targeting construct was electroporated into G4 ES cells, and correctly targeted clones (L613Vfl/+) were identified by PCR and confirmed by Southern blotting (Figure 1B). We also validated the desired properties of the targeted locus in L613Vfl/+ ES cells (Figure 1C). As expected, expression of the mutant allele was undetectable by RT-PCR in the absence of Cre, but it was induced effectively upon introduction of a Cre expression vector (MSCV-GFP-Cre). Mutant Raf1 protein was also expressed at levels comparable to those of WT Raf1 (data not shown, but see Supplemental Figure 4; supplemental material available online with this article; doi: 10.1172/JCI44929DS1). Chimeras were then generated by outbred morula aggregation, and germline transmission was obtained. L613Vfl/+ progeny were crossed to CMV-Cre mice, which express Cre ubiquitously, and then to WT mice, thereby generating mice with global Raf1L613V expression (referred to herein as L613V/+ mice) on a 129S6 × C57BL/6 mixed background.
L613V/+ mice were obtained at the expected Mendelian ratio at weaning, which indicated that on this mixed background, Raf1L613V expression during development is compatible with embryonic viability. However, similar to mice expressing NS-associated Ptpn11 mutant alleles (54), L613V/+ mice could not be obtained after backcrossing to C57BL/6 mice for more than 3 generations. Consequently, all experiments herein were performed on the 129S6 × C57BL/6 mixed background.
L613V/+ mice show multiple NS phenotypes.
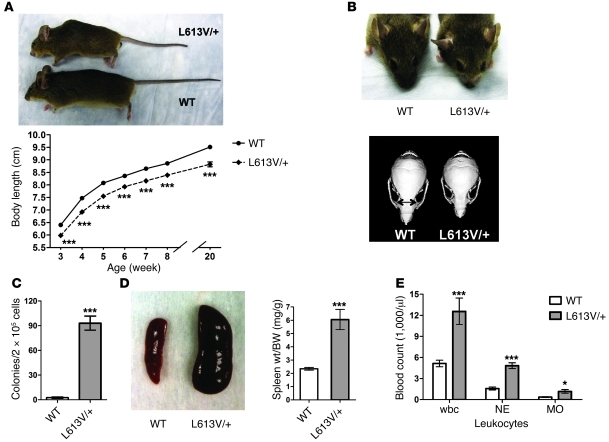
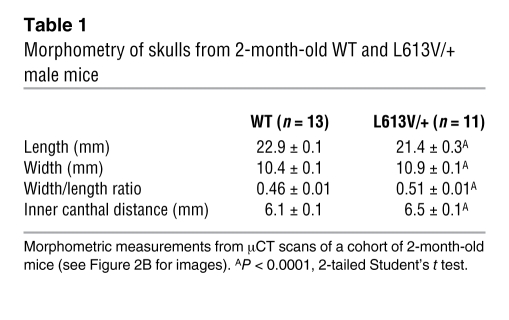
L613V/+ newborns showed normal size at birth (data not shown). At weaning, however, male (Figure 2A) and female (data not shown) L613V/+ mice were significantly smaller than their WT littermates, and they remained shorter throughout their lives. Although their overall body proportions were normal, L613V/+ mice exhibited facial dysmorphia (Figure 2B and Table 1). Consistent with their decreased body size, the skulls of L613V/+ mice were significantly shorter than those of WT mice. Their skull width was increased, however, resulting in a significantly greater width/length ratio. As a result, L613V/+ mice had a “triangular” facial appearance, with a blunter snout and widely set eyes (increased inner canthal distance). These features are reminiscent of the facial phenotype of mice expressing NS-associated Ptpn11 mutations (54, 57, 58) and represent the mouse equivalent of the facial abnormalities seen in NS patients (61).
Figure 2. L613V/+ mice show multiple NS phenotypes.
(A) Short stature in L613V/+ mice. Gross appearance of 2-month-old WT and L613V/+ male mice and growth curves of WT (n = 45) and L613V/+ (n = 45) male mice are shown. Differences were significant at all time points (P < 0.0001, 2-way repeated-measures ANOVA; ***P < 0.0001, Bonferroni post-test). (B) L613V/+ mice have facial dysmorphia. Gross facial appearance and representative μCT scans of skulls from WT and L613V/+ mice. Double-headed arrows indicate inner canthal distance. See Table 1 for morphometric measurements. (C) Cytokine-independent myeloid colonies from BM of 2-month-old mice (n = 6 per genotype). ***P < 0.0001, 2-tailed Student’s t test. (D) Splenomegaly in L613V/+ mice. Representative gross appearance and spleen weight/BW ratio in WT (n = 25) and L613V/+ (n = 25) mice at 4 months. ***P < 0.0001, 2-tailed Student’s t test. (E) Increased total wbcs, neutrophils (NE), and monocytes (MO) in 1-year-old L613V/+ mice (n = 8 per genotype). *P < 0.05, ***P < 0.0001, 2-tailed Student’s t test.
Table 1 .
Morphometry of skulls from 2-month-old WT and L613V/+ male mice
Like mouse models of Ptpn11 mutation–associated NS (54, 58) and many NS patients (62), L613V/+ mice had hematological defects. There was abnormal expansion of myeloid progenitors, and BM from L613V/+ mice yielded factor-independent myeloid colonies (Figure 2C). L613V/+ mice also developed splenomegaly, which became more severe as they aged (Figure 2D). Peripheral blood counts were normal at 4 months of age, but by 1 year, L613V/+ mice had developed subtle but statistically significant leukocytosis, neutrophilia, and monocytosis (Figure 2E) with normal hematocrit and platelet counts (data not shown).
L613V/+ mice show cardiac hypertrophy with chamber dilatation.
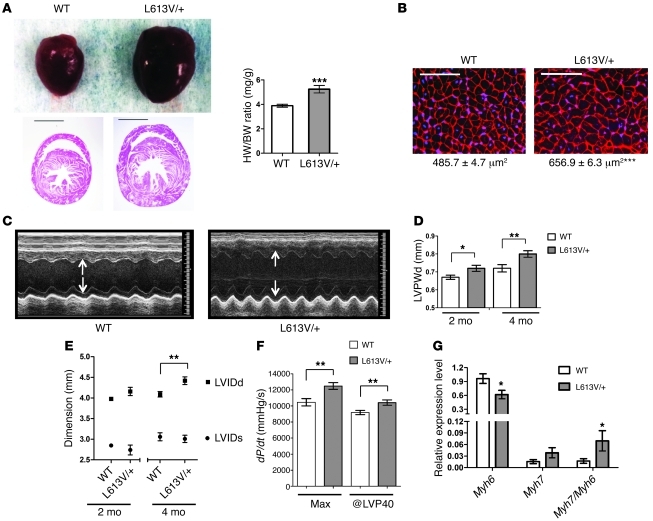
Unlike PTPN11 alleles, which are negatively associated with HCM in NS patients (44) and in mouse models (54, 56, 58), RAF1 mutations that encode proteins with increased kinase activity are strongly associated with HCM (37, 38). Remarkably, L613V/+ mice showed evidence of cardiac hypertrophy as early as 2 weeks after birth, as indicated by an increased heart weight/BW ratio (Supplemental Figure 1A). Cardiac enlargement became even more obvious in adult L613V/+ mice, with histological analysis revealing substantial thickening of the ventricular wall and septum (Figure 3A). Increased heart size can reflect a larger number of cardiomyocytes (e.g., as a consequence of excess proliferation during development) and/or cardiomyocyte hypertrophy. Cardiomyocyte proliferation, as measured by BrdU incorporation assays, was comparable in E16.5 L613V/+ and WT embryos (Supplemental Figure 1B). By contrast, cross-sectional area markedly increased — by about 35% — in cardiomyocytes from 8-week-old L613V/+ compared with WT mice (Figure 3B), indicative of cardiac hypertrophy.
Figure 3. L613V/+ mice show cardiac hypertrophy with chamber dilatation.
(A) Representative gross appearance and H&E-stained cross-sections (original magnification, ×4; scale bars: 2 mm) of WT (n = 25) and L613V/+ (n = 25) hearts at 8 weeks, as well as heart weight/BW ratio (HW/BW) of 4-month-old WT and L613V/+ mice. (B) Cross-sections of cardiomyocytes (original magnification, ×400; scale bars: 100 μm). Cross-sectional area (numbers below) was measured in WGA-strained sections from 8 week-old mice (n = 5 samples per genotype, with 200 cells counted per sample using ImageJ). (C) Representative echocardiograms of hearts from 4-month-old mice. Arrows indicate LV diastolic dimension. (D) LVPWd at 2 and 4 months, measured by echocardiography. n = 13 (WT); 11 (L613V/+). (E) LVIDd and LVIDs of 2- and 4-month-old WT (n = 13) and L613V/+ (n = 11) hearts. (F) Cardiac contractility of 4-month-old WT (n = 13) and L613V/+ (n = 11) hearts, as measured by invasive hemodynamic analysis. (G) Myh6 and Myh7 gene expression in 4-month-old WT (n = 6) and L613V/+ (n = 9) hearts, assessed by quantitative real-time PCR. (B, D, and E–G) *P < 0.05, **P < 0.005, ***P < 0.0001, 2-tailed Student’s t test.
Cardiac hypertrophy can be secondary to pressure overload caused by stenotic valves or hypertension. Notably, mice expressing the NS-associated Ptpn11D61G mutation have severe valvuloseptal abnormalities, including atrial, atrioventricular, or ventricular septal defects and double-outlet RV (54). In contrast, valvuloseptal development, as assessed by histology, appeared normal in 1-week-old L613V/+ mice (Supplemental Figure 1C and data not shown). Invasive hemodynamic studies established that ventricular pressure was actually lower in L613V/+ mice than in WT controls (see below).
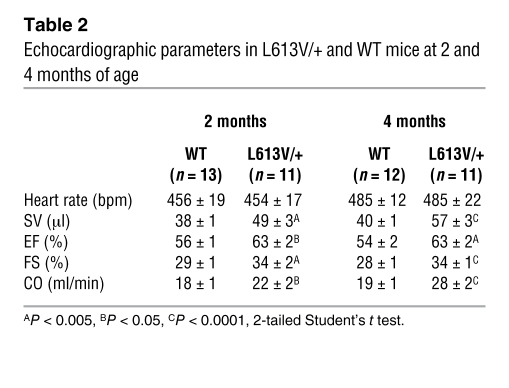
To assess cardiac morphology and function, we performed echocardiography on L613V/+ mice and littermate controls at 2 and 4 months of age. As expected, LV diastolic posterior wall thickness (LVPWd) was increased in L613V/+ mice (Figure 3, C and D). Although chamber size was normal in 2-month-old mice, by 4 months, L613V/+ hearts showed an increase in LV internal end-diastolic dimension (LVIDd). LV internal end-systolic dimension (LVIDs) remained within normal limits (Figure 3, C and E), indicating preserved or enhanced function. Consistent with the latter interpretation, stroke volume (SV), ejection fraction (EF), fractional shortening (FS), and cardiac output (CO) were increased in L613V/+ mice (Table 2).
Table 2 .
Echocardiographic parameters in L613V/+ and WT mice at 2 and 4 months of age
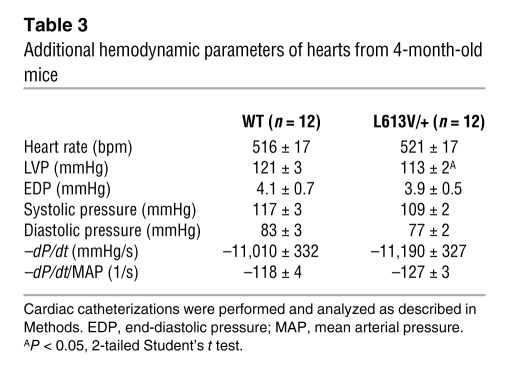
Invasive hemodynamic studies confirmed and extended these conclusions (Figure 3F and Table 3). L613V/+ mice showed increased dP/dtmax, consistent with enhanced contractility, but no change in cardiac relaxation (i.e., –dP/dt). Afterload (systolic pressure) was slightly lower in L613V/+ mice. Although this finding ruled out hypertension as a cause of hypertrophy in L613V/+ mice, it complicated comparison of dP/dtmax values. For this reason, we also compared dP/dt estimated at LV pressure (LVP) of 40 mm Hg (dP/dt@LVP40), therefore reducing or eliminating the influence of afterload (63). Importantly, dP/dt@LVP40 was increased in L613V/+ animals (Figure 3F), providing conclusive evidence of increased contractility. Moreover, there was no pressure gradient across the aortic valves of L613V/+ mice (Table 3), ruling out aortic valve stenosis as a cause of their cardiac hypertrophy. Our finding of eccentric cardiac hypertrophy in the absence of pressure overload was consistent with the conclusion that L613V/+ mice have pathological hypertrophy.
Table 3 .
Additional hemodynamic parameters of hearts from 4-month-old mice
Mice and humans with pathological hypertrophy often reactivate specific fetal genes (5, 6). There are 2 isoforms of cardiac myosin: α-MHC (faster kinetics; encoded by MYH6) and β-MHC (slower kinetics; encoded by MYH7). In rodents, Myh7 is expressed mainly in late fetal life, whereas Myh6 is expressed predominantly in the adult. Reexpression of Myh7 and a shift from α-Mhc to β-Mhc is a marker for phenotypic reprogramming and HCM (5). Indeed, Myh6 mRNA levels decreased significantly in L613V/+ hearts, and there was a trend toward increased Myh7 expression (P = 0.09, 1-tailed Student’s t test; Figure 3G). Consequently, the Myh7/Myh6 ratio increased significantly. Expression of Nppa (encoding Anp) and Nppb (encoding Bnp), 2 other fetal genes often associated with cardiac hypertrophy (5, 64), was unaffected in L613V/+ hearts (data not shown).
Enhanced hypertrophic response and functional decompensation in L613V/+ hearts following pressure overload.
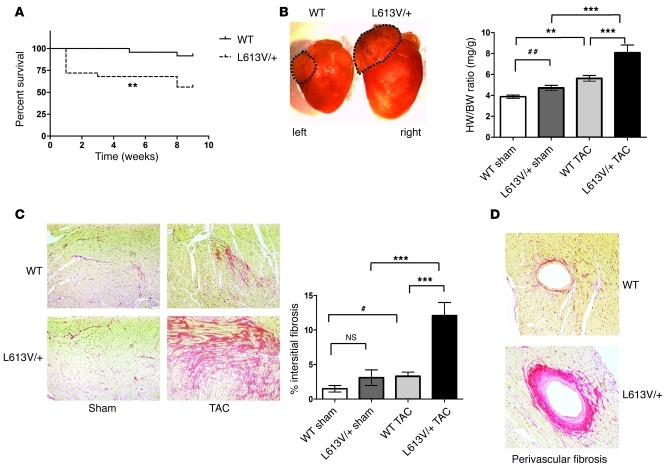
Although L613V/+ mice showed cardiac hypertrophy, they displayed enhanced cardiac function without signs of heart failure for at least a year of life. To gain further insight into the nature of the hypertrophy in L613V/+ mice, we assessed their response to biomechanical stress by transverse aortic constriction (TAC). L613V/+ mice had an unusually high acute death rate after this procedure compared with controls (Figure 4A). Furthermore, the hearts of surviving L613V/+ mice showed dramatic ventricular, as well as left atrial, enlargement compared with WT mice (Figure 4B). Although WT mice had an approximately 45% increase in heart weight/BW ratio following TAC, L613V/+ mice had an approximately 72% increase. L613V/+ mice also developed more severe interstitial fibrosis (Figure 4C) and perivascular fibrosis (Figure 4D and Supplemental Figure 2A) after TAC. We excluded 2 L613V/+ mice from analysis: by 8 weeks of TAC, these mice had sustained a large spontaneous transmural infarct involving approximately 30% of the ventricular free wall, and extensive fibrosis with impaired systolic and diastolic function was evident (Supplemental Figure 2B).
Figure 4. Abnormal response of L613V/+ mice to pressure overload.
(A) Survival curves of WT (n = 25) and L613V/+ (n = 24) mice after TAC. **P < 0.005, log-rank test. (B) Gross appearance of hearts and heart weight/BW ratio at 8 weeks after TAC or sham surgery. Dashed outlines demonstrate markedly enlarged left atrium in L613V/+ compared with WT mice. **P < 0.005, ***P < 0.0001, Bonferroni post-test when ANOVA was significant; ##P < 0.005, 1-tailed Student’s t test. (C) Severe interstitial fibrosis in L613V/+ hearts (PSR staining; original magnification, ×100) at 8 weeks after TAC. Percent pixels staining positive with PSR for interstitial fibrosis was quantified using ImageJ. n = 14 (WT); 13 (L613V/+). ***P < 0.0001, Bonferroni post-test when ANOVA was significant; #P < 0.05, 2-tailed Student’s t test. (D) Perivascular fibrosis in hearts (PSR staining; original magnification, ×200) at 8 weeks after TAC. Similar results were obtained when Masson Trichrome stain was used to assess fibrosis.
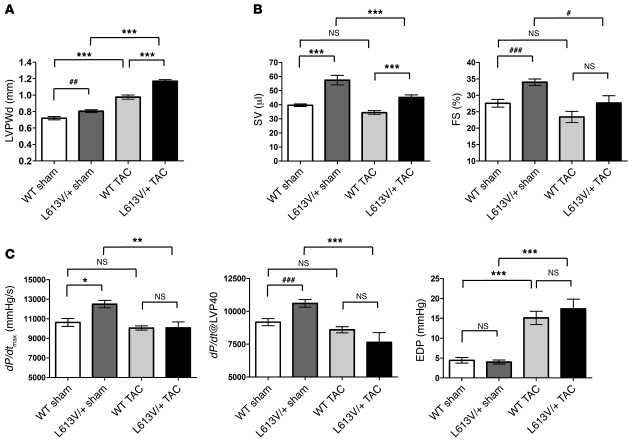
These morphologic and histological findings established that L613V/+ mice have an altered response to pressure overload. Consistent with this, TAC provoked increases in LVPWd in WT and L613V/+ mice; the increase was more pronounced in the L613V/+ mice (Figure 5A). LVIDd did not change after TAC in WT or L613V/+ mice, but remained elevated in the latter (Supplemental Figure 3A). Most importantly, several parameters of cardiac function, including SV and FS, deteriorated in L613V/+ mice, whereas these were unaffected in WT mice (Figure 5B). There also was a trend toward decreased CO in L613V/+ mice subjected to TAC, although this did not reach statistical significance because these mice increased their heart rate sufficiently to compensate for decreased ventricular function (Supplemental Figure 3B). In addition, cardiac contractility (measured as either dP/dtmax or dP/dt@LVP40) decreased in L613V/+ mice, but not in WT mice (Figure 5C). Cardiac relaxation — as assessed by –dP/dt, normalized to mean arterial pressure (i.e., afterload) — was reduced comparably, whereas end-diastolic pressure was increased to a similar extent in WT and L613V/+ mice (Figure 5C and Supplemental Figure 3C). Thus, while WT mice could adapt appropriately to pressure overload, L613V/+ mice exhibited substantial, occasionally fatal, functional decompensation with reductions in SV, FS, and dP/dtmax and dP/dt@LVP40, consistent with early stages of heart failure by 8 weeks of TAC.
Figure 5. Echocardiographic and hemodynamic parameters in WT and L613V/+ mice following pressure overload.
(A) LVPWd at 8 weeks after TAC or sham surgery. (B) Decreased SV and FS in L613V/+ mice after TAC. (C) Decreased cardiac contractility in L613V/+ mice after TAC. Because LVPs were not identical in WT and L613V/+ mice (see Table 3), both dP/dtmax and dP/dt@LVP40 are shown. *P < 0.05, **P < 0.005, ***P < 0.0001, Bonferroni post-test when ANOVA was significant; ##P < 0.005, ###P < 0.0001, 1-tailed Student’s t test. n = 12 (WT sham); 11 (L613V/+ sham); 22 (WT TAC); 13 (L613V/+ TAC).
The Raf1L613V mutant increases Mek and Erk activation in response to multiple stimuli.
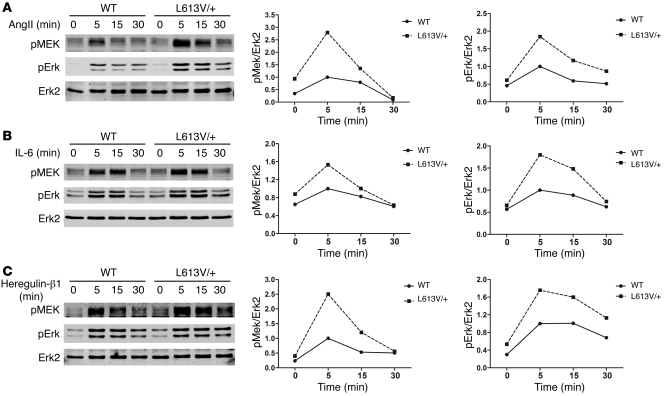
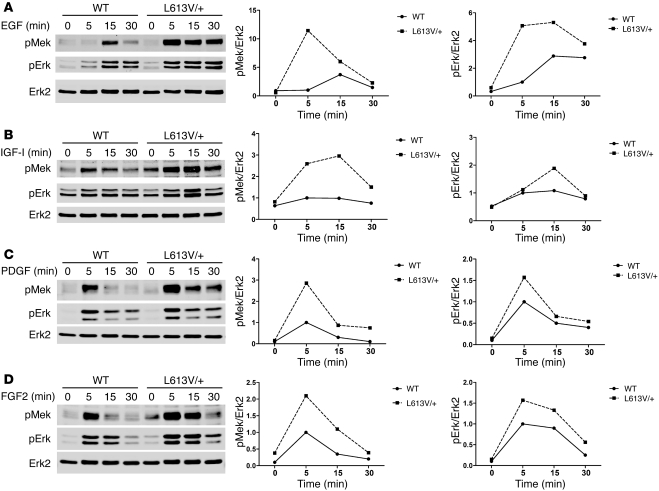
Compared with WT RAF1, Raf1L613V has increased kinase activity in vitro and an enhanced ability to activate MEK-ERK in transfection studies (37, 38). We assessed the effect of Raf1L613V expressed at endogenous levels on the RAS-ERK pathway. Consistent with the earlier overexpression experiments, Mek and Erk activation (as inferred from immunoblots with activation-specific antibodies) was enhanced in multiple cell types expressing Raf1L613V in response to a variety of stimuli, including LIF-stimulated ES cells (Supplemental Figure 4A) and EGF- or PDGF-stimulated mouse embryonic fibroblasts (MEFs; Supplemental Figure 4, B and C). Of direct relevance to the L613V/+ cardiac phenotype, Mek and Erk activation also were higher in L613V/+ than in WT neonatal cardiomyocytes stimulated with receptor tyrosine kinase (heregulin-β1), cytokine receptor (IL-6), or GPCR (Ang II) agonists (Figure 6). Recently, cardiac fibroblasts were implicated in the genesis of cardiac hypertrophy (65, 66); notably, L613V/+ cardiac fibroblasts also showed enhanced agonist-stimulated Mek-Erk activation (Figure 7). Both the quantitative and the qualitative effects of the mutant Raf1 allele on Mek and Erk activation differed in cardiomyocytes versus cardiac fibroblasts (or MEFs) and in response to different stimuli. In some cases, mutant Raf1 affected only the magnitude of activation, in others, solely the duration of activation, and for still others, both magnitude and duration. Such differences might reflect distinct feedback responses to the agonists in various cell types.
Figure 6. Raf1L613V mutant increases Mek and Erk activation in cardiomyocytes.
Cardiomyocytes prepared from neonatal WT and L613V/+ mice were starved for 24 hours and then stimulated for the indicated number of minutes with 1 μg/ml Ang II (A), 10 ng/ml IL-6 (B), and 100 ng/ml heregulin-β1 (C). Cell lysates (15 μg protein) were immunoblotted with the indicated antibodies. Quantification of blots is also shown. 1 of 2 experiments with comparable results is shown.
Figure 7. Raf1L613V mutant increases Mek and Erk activation in cardiac fibroblasts.
Cardiac fibroblasts prepared from neonatal WT and L613V/+ mice were starved for 16 hours and then stimulated for the indicated number of minutes with 50 ng/ml EGF (A), 100 ng/ml IGF-I (B), 100 ng/ml PDGF (C), and 50 ng/ml FGF2 (D). Cell lysates (20 μg protein) were immunoblotted with the indicated antibodies. Quantification of blots is also shown. 1 of 2 experiments with comparable results is shown.
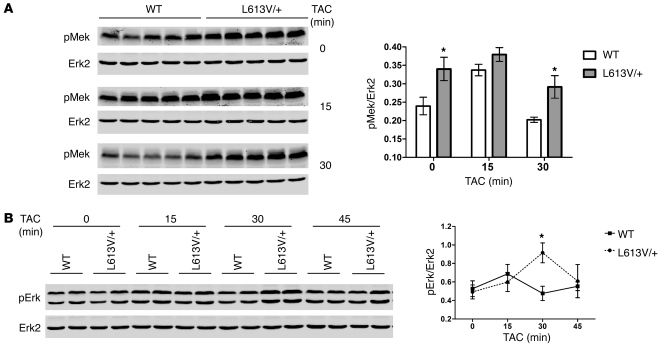
Although it was difficult to detect Erk activation in the adult heart under basal conditions (data not shown), basal Mek activity was significantly higher in adult L613V/+ compared with WT hearts (Figure 8A). To compare Mek and Erk activation in vivo, we monitored the response of WT and L613V/+ mice to pressure overload evoked by TAC for up to 45 minutes. Mek activation remained significantly higher in L613V/+ hearts throughout the period of acute TAC (Figure 8A). Erk activation was significantly higher in L613V/+ hearts after 30 minutes of TAC compared with WT hearts, but was similar to WT at other time points (Figure 8B).
Figure 8. Enhanced Mek and Erk activation in L613V/+ hearts after pressure overload.
Hearts from WT and L613V/+ mice were subjected to TAC for the indicated number of minutes (n = 5 per group per time point), then lysed and analyzed by immunoblotting with the indicated antibodies. Erk2 levels are shown as a loading control. Each lane represents an individual animal; quantification of all samples is also shown. (A) Mek activation, with all samples from a single time point analyzed on the same gel. (B) Representative samples of Erk activation from each time point analyzed on the same gel. (A and B) *P < 0.05, 2-tailed Student’s t test.
We also assayed several signaling pathways implicated in other models of cardiac hypertrophy/HCM by immunoblotting with appropriate phosphospecific antibodies. Activation of the MAPK family members JNK and p38 was comparable in Ang II–stimulated neonatal cardiomyocytes and EGF-stimulated cardiac fibroblasts (Supplemental Figure 5, A and B). Likewise, Akt, GSK-3β, and p70S6K activation in response to the agonists tested were unaffected by Raf1L613V expression in either cell type (Supplemental Figure 5, A and B). Importantly, in the same experiments, Mek and Erk activation were enhanced in L613V/+ cardiomyocytes and cardiac fibroblasts.
MEK inhibitor treatment normalizes NS phenotypes in L613V/+ mice.
The genetics of NS and other RASopathies and the ability of Raf1L613V to selectively enhance Mek and Erk activation by multiple agonists in cardiomyocytes and cardiac fibroblasts strongly implicate enhanced Mek-Erk activation in the pathogenesis of NS phenotypes, including HCM. We asked whether any of these phenotypes could be reversed if Mek-Erk activation was normalized by treatment of L613V/+ mice with a MEK inhibitor. In initial experiments, the ATP-uncompetitive inhibitor PD0325901 (67) or empty vehicle was injected i.p. daily to WT and L613V/+ mice (5 mg/kg BW), beginning at 4 weeks and continuing for the succeeding 6 weeks. Importantly, at the start of the treatment period, L613V/+ mice already showed substantial growth defects, facial dysmorphia, and cardiac hypertrophy.
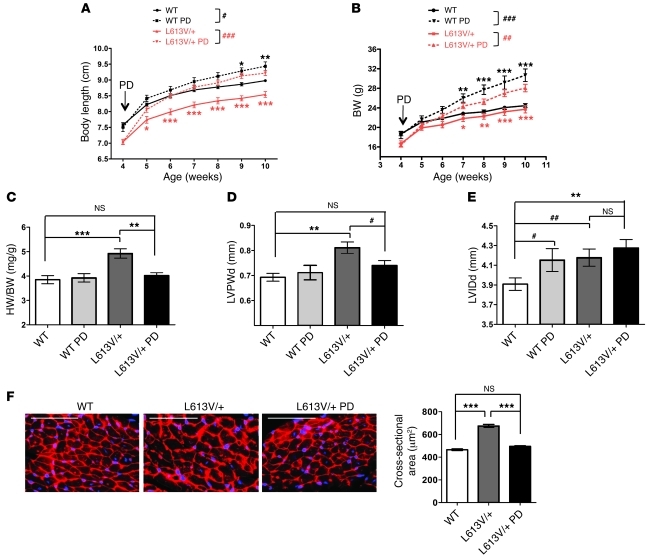
Remarkably, the body length of L613V/+ mice began to catch up with WT mice after 1 week of treatment, and by 2 weeks, PD0325901-treated L613V/+ mice were the same length as untreated WT mice (Figure 9A). PD0325901-treated WT mice also increased their body length such that by the last 2 weeks of treatment, they were significantly longer than control, untreated WT mice. Notably, however, PD0325901-treated L613V/+ mice achieved the same final body length as did treated WT mice, which argues that increased Mek-Erk activity is the primary cause of the growth defect in L613V/+ mice (see Discussion). Inhibitor treatment also increased the BW of L613V/+ mice, but surprisingly, they — as well as treated WT mice — gained substantially more BW than did untreated WT mice (Figure 9B). Increased BW in PD0325901-treated mice was accompanied (and, presumably, in large part caused) by an obvious increase in body fat (data not shown); thus, increased adiposity/body mass was an unanticipated, and to our knowledge previously unreported, side effect of PD0325901 treatment (and possibly MEK inhibitor treatment in general; see Discussion).
Figure 9. MEK inhibitor treatment rescues growth defect and cardiac hypertrophy in L613V/+ mice.
Mice were injected i.p. daily with PD0325901 (PD; 5 mg/kg BW) or vehicle, starting at 4 weeks of age and for the succeeding 6 weeks. Body length (A) and BW (B) were measured weekly. Note the rapid normalization of body length, as well as the increase in BW caused by inhibitor treatment. #P < 0.05, ##P < 0.005, ###P < 0.0001, 2-way repeated-measures ANOVA; *P < 0.05, **P < 0.005, ***P < 0.0001, Bonferroni post-test when ANOVA was significant (black symbols, WT PD vs. WT control; red symbols, L613V/+ PD vs. L613V/+ control). (C) Heart weight/BW ratio and (D) LVPWd were restored to within normal limits in inhibitor-treated mice. **P < 0.005, ***P < 0.0001, Bonferroni post-test when ANOVA was significant; #P < 0.05, 1-tailed Student’s t test. (E) LVIDd. **P < 0.005, Bonferroni post-test when ANOVA was significant; #P < 0.05, ##P < 0.005, 1-tailed Student’s t test. n = 14 (WT); 10 (L613V/+); 6 (WT PD); 14 (L613V/+ PD). (F) Cross-sectional area of cardiomyocytes (original magnification, ×400; scale bar, 100 μm), measured in WGA-strained heart sections (n = 2 samples per group, with 200 cells counted per sample using ImageJ). ***P < 0.0001, Bonferroni post-test when ANOVA was significant. See also Supplemental Figure 6.
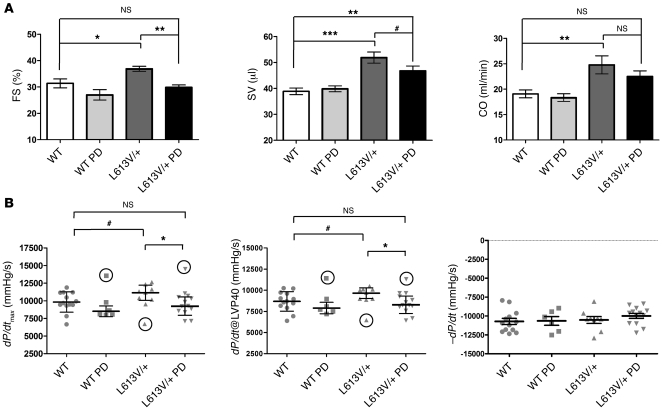
PD0325901 treatment also affected the L613V/+ cardiac phenotype. The heart weight/BW ratio in L613V/+ mice was restored to normal range (i.e., WT control) after treatment, whereas there was no significant change in treated WT mice (Figure 9C). Echocardiography (Figure 9, D and E) and invasive hemodynamic (Figure 10) studies showed significant improvement in multiple parameters of cardiac morphology and function. Furthermore, histological assessment of cross-sectional area of cardiomyocytes confirmed the normalization of cardiomyocyte size in L613V/+ mice after treatment (Figure 9F). The significant increase in body size and body mass caused by PD0325901 treatment potentially complicates echocardiographic and invasive hemodynamic comparisons of WT and L613V/+ mice before and after treatment. Therefore, we compared all parameters using both nominal values and values normalized by cube root of BW (BW1/3) (Supplemental Figure 6); overall, the 2 analyses led to similar conclusions. First, there was a significant reduction (toward normal) in LVPWd in L613V/+ mice after treatment (Figure 9D); this difference was even more significant when normalized by BW1/3 (Supplemental Figure 6A). Nominal LVIDd was unchanged in PD0325901-treated L613V/+ mice (Figure 9E), although when this value was normalized, chamber dilatation improved significantly, becoming comparable to that of WT controls (Supplemental Figure 6B). Inhibitor treatment clearly reduced the abnormal SV and FS in L613V/+ mice toward normal levels (i.e., untreated or treated WT; Figure 10A and Supplemental Figure 6C). There also was a strong trend toward decreased CO in L613V/+ mice after PD0325901 treatment (Figure 10A and Supplemental Figure 6D). Finally, the excessive cardiac contractility (dP/dt and dP/dt@LVP40) in L613V/+ mice was ameliorated by PD0325901 treatment, whereas cardiac relaxation remained unchanged (Figure 10B).
Figure 10. MEK inhibitor treatment normalizes cardiac function in L613V/+ mice.
(A) Echocardiographic parameters of hearts after treatment with PD0325901 as described in Figure 9. Note normalization of SV and FS, with a trend toward CO normalization. *P < 0.05, **P < 0.005, ***P < 0.0001, Bonferroni post-test when ANOVA was significant; #P < 0.05, 1-tailed Student’s t test. (B) Hemodynamic parameters, assessed by cardiac catheterization, after PD0325901 treatment. For calculating statistical significance, significant outliers (circled data points), as assessed by Grubbs test, were removed. *P < 0.05, Bonferroni post-test when ANOVA was significant (P = 0.09 including outliers); #P < 0.05, 1-tailed Student’s t test (P = 0.12 including outliers). n = 14 (WT); 10 (L613V/+); 6 (WT PD); 14 (L613V/+ PD).
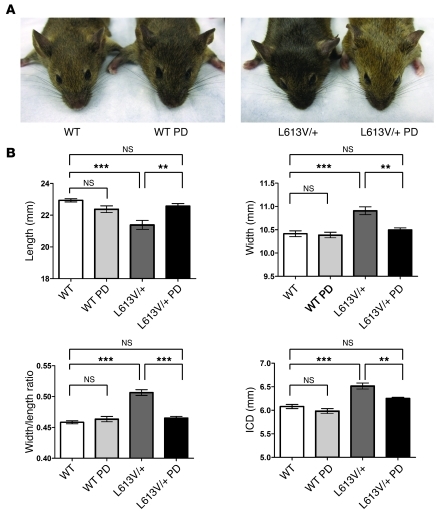
PD0325901 treatment did not improve the facial dysmorphia in L613V/+ mice in the above study, most likely because skull development had already been completed by the onset of drug administration. We tested whether earlier, but still postnatal, MEK inhibitor treatment could prevent or ameliorate L613V/+ facial defects. Lactating female mice were injected i.p. with PD0325901 (5 mg/kg BW) daily, beginning at P0 until weaning at P21. Weaned mice were then injected individually with the same dose of PD0325901 for another 5 weeks. As expected from our initial treatment regimen (Figure 9A), the growth defect in L613V/+ mice was again prevented (data not shown). Remarkably, however, earlier PD0325901 treatment had a dramatic effect on the appearance of L613V/+ mice: they no longer had triangular faces, instead appearing indistinguishable from treated or untreated WT controls (Figure 11A). μCT morphometry confirmed that inner canthal distance was reduced significantly, skull length increased, and skull width and width/length ratio decreased in PD0325901-treated L613V/+ mice (Figure 11B); all values were indistinguishable from those of treated or untreated WT mice by the end of the treatment period.
Figure 11. Early postnatal MEK inhibitor treatment rescues facial dysmorphia in L613V/+ mice.
Lactating female mice were injected i.p. daily with PD0325901 (5 mg/kg BW) or vehicle, starting at P0 until weaning at P21. Weaned mice were then injected i.p. individually with PD0325901 (5 mg/kg BW) or vehicle daily for another 5 weeks. (A) Gross facial appearance. (B) Morphometric measurements of skulls from μCT scans. ICD, inner canthal distance. **P < 0.005, ***P < 0.0001, Bonferroni post-test when ANOVA was significant. n = 11 (WT); 10 (L613V/+); 6 (WT PD); 7 (L613V/+ PD).
Discussion
We describe here a knockin mouse model for NS caused by a Raf1 gain-of-function mutation. Similar to mouse models of Ptpn11 mutation–associated NS, Raf1L613V heterozygosity caused proportional short stature, facial dysmorphia, and hematological defects. Unlike phosphatase-activating Ptpn11 alleles, which cause valvuloseptal abnormalities (54, 55, 58), L613V/+ mice had normal valvuloseptal development and instead exhibited eccentric cardiac hypertrophy that decompensated upon pressure overload. Agonist-evoked Mek-Erk activation was enhanced in multiple cell types without changes in several other signaling pathways implicated in cardiac hypertrophy/HCM. Remarkably, postnatal MEK inhibition normalized the growth, facial, and cardiac defects in L613V/+ mice, demonstrating that continued MEK-ERK activity is critical for causing HCM and other NS phenotypes and identified MEK inhibitors as potential therapeutic agents for the treatment of NS.
RASopathies are a class of human genetic syndromes caused by germline mutations in genes that encode components of the RAS-ERK pathway (31, 32). Not surprisingly, these disorders share several features, albeit with varying degrees of penetrance, yet each also exhibits unique and characteristic phenotypes. Conceivably, the specific mutant gene, possibly as a consequence of its position in the pathway and susceptibility to feedback regulation, could direct the phenotype. Alternatively, genetic modifiers in the highly outbred human population could be determinative.
Previous mouse models suggest that both the gene and the genetic background are important to the ultimate RASopathy phenotype. Clearly, different mutations in the same RASopathy gene can result in distinguishable phenotypes: gain-of-function Ptpn11 mutations, depending on the degree of their phosphatase activity, cause a variable spectrum of NS phenotypes (54, 55, 58). The current study, along with a parallel analysis of knockin mice expressing a NS-associated Sos1E846K mutant (68), shows that mutations in different genes that cause the same RASopathy syndrome yield different phenotypes: mice with phosphatase-activating Ptpn11 mutations have valvuloseptal defects, but not HCM (54, 55); Sos1E846K/+ mice develop LV hypertrophy with incompletely penetrant aortic stenosis; and L613V/+ mice exhibited HCM with normal valvuloseptal development.
On the other hand, mutations associated with different RASopathies also have distinct effects in mice. In contrast to the NS mice discussed above, an HRasG12V knockin mouse model of CS shows abnormal cranial dimensions, papillomas, and angiosarcomas. These mice have cardiac hypertrophy, but also aortic stenosis, making it unclear whether the hypertrophy is primary or secondary (32, 59). As described in the companion paper by Marin et al. (60), a mouse model of LS caused by Ptpn11Y279C indicates that, unlike Ptpn11 alleles with increased catalytic activity, catalytically impaired mutants develop HCM and skeletal abnormalities as well as short stature and facial dysmorphia.
While the specific mutation plays a major role in determining RASopathy phenotype, modifier loci also clearly contribute: just as there is considerable phenotypic variation between family members carrying the same NS or LS allele (69), there are differences in disease spectrum and severity of mice with Ptpn11 (54, 58) and Raf1 (data not shown) mutations on different strain backgrounds. Ptpn11D61G/+ mice show incomplete penetrance of valvuloseptal defects on mixed background and various penetrance of embryonic lethality on different strain backgrounds (54, 58). L613V/+ mice were obtained at the expected Mendelian ratio on mixed background, whereas on the C57BL/6 background, this mutant allele almost always was lethal (data not shown). All of these data suggest that incomplete penetrance reflects strain-specific modifiers. Genomic scans using SNP panels should help to determine whether cloneable modifiers exist or whether heterosis accounts for the variable penetrance.
The role of the RAS-ERK pathway in cardiac hypertrophy has been controversial. Overexpression of MAPK phosphatase 1 (MKP-1) blocks both agonist-induced hypertrophy in vitro and pressure overload–associated hypertrophy in vivo (70). However, MKP-1 inactivates all 3 major MAPKs, so the study could not address the specific effects of Ras-Erk pathway activation. Depletion of ERK1/2 with antisense oligonucleotides or pharmacological inhibition of MEK1/2 attenuates the hypertrophic response to agonist stimulation of cultured cardiomyocytes (23, 24), consistent with a requirement for MEK-ERK activation in the hypertrophic response. Transgenic mice with cardiac-specific expression of HRasG12V display HCM associated with interstitial fibrosis and sudden death (20–22). Cardiac-specific Nf1-deleted mice develop marked cardiac hypertrophy, progressive cardiomyopathy, and fibrosis as adults (71).
Conversely, other studies suggest that MEK-ERK activity is dispensable for cardiomyocyte hypertrophy. Transgenic mice with cardiac-restricted expression of activated Mek1 exhibit concentric hypertrophy without signs of cardiomyopathy (26). Although hypertrophy in this model was interpreted as physiological, these mice also have impaired diastolic function and reactivated cardiac fetal gene expression, which is more consistent with pathological hypertrophy. A recent study showed that Erk1–/–Erk2+/– mice or transgenic mice with cardiac-specific expression of Dusp6, an Erk1/2-specific phosphatase, showed a normal hypertrophic response to pressure overload and exercise (27). In both of these lines of mice, however, residual Erk activity cannot be excluded. Also, it is possible that Dusp6 has other targets besides Erk1/2, which could complicate interpretation of these results. Moreover, most of these earlier studies involved cardiomyocyte-specific expression or deletion of potential hypertrophy-related genes, which excludes the potential contribution of other cell types in the heart to the hypertrophic response. Recent studies show that cardiac fibroblasts play key roles in myocardial development and function (72, 73). Embryonic cardiac fibroblasts induce myocyte proliferation, whereas adult cardiac fibroblasts promote myocyte hypertrophy (72) and evoke pathological hypertrophy and fibrosis in response to disease stimuli (65, 66). Of particular note, enhanced Ras-Erk activation in cardiac fibroblasts is implicated in pathological hypertrophy and fibrosis caused by overexpression of the β-adrenergic receptor in cardiomyocytes (66).
Our mouse model, in which a NS-associated Raf1 mutant was expressed globally under normal promoter control, supports the conclusion that excessive Ras-Erk pathway activity causes HCM. Several lines of evidence indicated that L613V/+ mice have pathological cardiac hypertrophy. Hypertrophy was eccentric in these mice, and they showed the characteristic shift from Myh6 to Myh7 expression seen in pathological hypertrophy. In response to pressure overload by TAC, they had an unusually high death rate, presumably due to inability to adapt to this stress or arrhythmia, while surviving mice showed clear evidence of functional decompensation. Importantly, in our model, unlike many previous studies (see above), the mutant allele was expressed in both cardiomyocytes and cardiac fibroblasts, as well as multiple other cell types. Moreover, Mek-Erk activation was enhanced in response to multiple agonists in these cells. It will be important to determine whether mutant expression in cardiomyocytes, cardiac fibroblasts, or both is important for HCM in L613V/+ mice; our inducible Raf1 allele should facilitate such analyses. Most importantly, postnatal MEK inhibitor treatment substantially normalized the cardiac defects in L613V/+ mice, providing strong evidence for the critical role of the RAS-ERK pathway in initiating and maintaining the cardiac hypertrophic response.
Postnatal MEK inhibitor treatment also normalized the growth defects and, if administered early enough, the facial dysmorphia in L613V/+ mice. Notably, MEK inhibitor treatment also increased the body length of WT mice, but there was no difference between the mutant and WT treated groups in terms of final body length after treatment. Likewise, MEK inhibitor treatment at doses that effectively normalized L613V/+ cardiac anatomy and function had little effect on cardiac function in WT mice. These results strongly suggest that all of these NS phenotypes are caused by excessive MEK-ERK activity, as opposed to the MEK inhibitor acting on a parallel pathway to mitigate syndromic features.
Unexpectedly, we found that PD0325901 treatment caused a significant increase in BW with an obvious increase in body fat. Although we cannot exclude the possibility that this is an idiosyncratic (i.e., off-target) effect of this specific MEK inhibitor, other evidence points to a potential obesity-promoting effect of MEK-ERK inhibition. For example, leptin activates Erk via an Shp2-dependent pathway (74, 75), and deletion of Shp2 in postmitotic forebrain neurons causes early-onset obesity with decreased ERK activation and evidence of leptin resistance (76). We suspect that MEK inhibition may act in analogous ways to promote obesity in our mice.
In summary, our data demonstrate a critical role of the RAS-ERK pathway in the genesis of HCM in NS and show that NS phenotypes can be rescued by pharmacological inhibition of MEK1/2. Previous studies showed that genetic ablation of Erk1/2 (55, 58) or prenatal treatment with a MEK inhibitor (57, 68) can prevent some NS phenotypes. Although these studies provided evidence for the key role of Mek-Erk hyperactivity in NS pathogenesis, they did not resolve whether MEK inhibition can reverse these phenotypes. Conversely, our results suggest that MEK inhibition may be useful for the specific treatment of Raf1 mutant NS, and possibly for other RASopathies associated with increased MEK-ERK pathway activity. Interestingly, the companion paper by Marin et al. shows that LS-associated HCM is associated with hyperactivation of the PI3K-Akt pathway and can be rescued by rapamycin treatment (60). Taken together, these studies argue for a mutation-specific, personalized approach to RASopathy therapy.
Methods
Generation of L613V/+ mice.
To construct the targeting vector for our inducible Raf1L613V knockin mice, a short arm containing Raf1 exon 12 (SacII-NotI genomic fragment) and a long arm including exons 13–16 (BamHI-ClaI genomic fragment) were ligated into the vector pGK Neo-HSV-1 TK (77). The L613V (exon 16) mutation, marked by a unique DraIII site, was introduced by site-directed mutagenesis. A splice acceptor sequence, a Raf1 cDNA fragment encoding wild-type exons 13–16, and a pGK-Neo (Neo) gene were positioned after the first loxP site as a SalI-XbaI fragment. The targeting vector was linearized with SacII and electroporated into G4 ES cells (129S6 × C57BL/6 F1 background). Genomic DNA, isolated from doubly G418/1-resistant and (2-deoxy-2-fluoro-β-D-arabinofuranosyl)-5 iodouracil–resistant (FIAU-resistant) ES clones (positive and negative selection, respectively), was screened by PCR using primers outside and inside the targeting vector (Supplemental Table 1), followed by NotI digestion, which marks the targeting vector. Homologous recombinants were confirmed by Southern blotting using Neo and external (5′ and 3′) probes (Supplemental Table 1). For these experiments, genomic DNA was digested with XbaI (5′ and Neo probes) or BamHI (3′ probe).
To validate the desired properties of the targeted locus, correctly targeted ES cells were transfected with a Cre-expressing plasmid (MSCV-GFP-Cre) to excise the cDNA-Neo cassette (see below). Expression of Raf1L613V mRNA was confirmed by RT-PCR (Supplemental Table 1), followed by digestion with DraIII, which marks the L613V allele. Chimeras were generated by outbred morula aggregation (Toronto Centre of Phenogenomics), and germline transmission was obtained (L613Vfl/+ mice). L613Vfl/+ mice (129Sv × C57BL/B6) were crossed to CMV-Cre (C57BL/B6) mice, which express Cre ubiquitously, and then to WT (129S6) mice to generate mice with global Raf1L613V expression (L613V/+ mice; 129Sv × C57BL/B6). Mice on a 129Sv × C57BL/B6 mixed background were used for all experiments. For genotyping, genomic DNA was prepared from tails, then subjected to PCR (Supplemental Table 1) and digestion with DraIII.
All animal studies were approved by the University Health Network Animal Care Committee (Toronto, Ontario, Canada) and performed in accordance with the standards of the Canadian Council on Animal Care.
Cell culture.
ES cells were cultured on γ-irradiated MEF feeders in knockout DMEM (Invitrogen), containing 15% ES-tested (HyClone, Thermo Scientific) FBS, 2 mM l-glutamine (Invitrogen), 0.1 mM nonessential amino acids (Invitrogen), 0.1 mM β-mercaptoethanol (Sigma-Aldrich), 100 U/ml penicillin/streptomycin (Invitrogen), and 500 U/ml LIF (ESGRO, Chemicon). ES cells were transfected with MSCV-GFP-Cre plasmid using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol. GFP-positive cells were purified by fluorescence-activated cell sorting (FACS) at 48 hours after transfection and used for RNA isolation. For biochemical studies, ES cells were removed from feeders, starved in knockout DMEM containing 1% FBS for 6 hours, and then stimulated with 103 U/ml LIF before harvesting.
Primary MEFs were prepared from E13.5 embryos and cultured in DMEM (Invitrogen) containing 10% FBS and 100 U/ml penicillin/streptomycin (Invitrogen), as described previously (78). MEFs were starved in serum-free DMEM for 16 hours before stimulation, and then stimulated with 10 ng/ml EGF or 50 ng/ml PDGF (both from PeproTech) before harvesting.
Neonatal mouse ventricular myocytes (neonatal cardiomyocytes) were isolated using methods adapted from a previous study (79). In brief, 1-day-old mouse hearts were harvested and predigested with 0.15 mg/ml trypsin (Invitrogen) at 4°C for 12–16 hours, followed by 50 U/ml type II collagenase (Worthington Biochemical) and 0.2 mg/ml trypsin in calcium- and bicarbonate-free Hanks buffer with HEPES for 1–2 hours at 37°C. Noncardiomyocytes were depleted by differential plating for 1 hour. Cardiomyocytes were counted, seeded at 2.5 × 105 cells/ml on Falcon Primaria tissue-culture plates (BD Biosciences), and cultured at 37°C in DMEM/Ham’s F12 (1:1 [v/v]; Invitrogen), 10% FBS, and 100 U/ml penicillin/streptomycin (Invitrogen) supplemented with 0.1 mM bromodeoxyuridine (Sigma-Aldrich) and 20 μM arabinosylcytosine (Sigma-Aldrich) to inhibit rapidly proliferating cells. After 24 hours, this medium was replaced with serum-free DMEM/Ham’s F12 (1:1) medium supplemented with 1% insulin-transferrin-selenium supplements-X (Invitrogen). After an additional 24 hours, cardiomyocytes were stimulated with 10 ng/ml IL-6 (PeproTech), 100 ng/ml heregulin-β1 (PeproTech), or 1 μg/ml Ang II (Sigma-Aldrich) before harvesting.
Noncardiomyocytes from the above preparation, mainly cardiac fibroblasts, were cultured in DMEM containing 10% FBS and 100 U/ml penicillin/streptomycin. Cardiac fibroblasts were starved in serum-free DMEM for 16 hours, and then stimulated with EGF (50 ng/ml), IGF-I (100 ng/ml), PDGF (100 ng/ml), or FGF2 (100 ng/ml), all from PeproTech, before harvesting.
Body size analysis and morphometry.
For growth curves, body length (anal-nasal length) and BW were measured weekly. For skeletal morphometry, mice were anesthetized with 2% isoflurane and scanned using a Locus Ultra μCT scanner (GE Healthcare). 3D images of the skeleton were generated and analyzed with GEHC MicroView software (GE Healthcare). Skull measurements were made according to Jackson Laboratory standard protocols and procedures (http://craniofacial.jax.org/standard_protocols.html).
Histology, immunohistochemistry, and BrdU incorporation assays.
Hearts for morphometry and histochemistry were fixed in the relaxed state by infusion of 1% KCl in PBS, followed by 10% buffered formalin. Hearts were then incubated in 10% buffered formalin overnight and embedded in paraffin. Sections (5 μm) were prepared and stained with H&E, picrosirius red (PSR), or Masson Trichrome. Cell membranes were stained with FITC-conjugated wheat germ agglutinin (WGA; Sigma-Aldrich) according to the manufacturer’s protocol. Nuclei were stained with DAPI. Cross-sectional area of cardiomyocytes with centrally located nuclei (to ensure the same plane of sectioning) was measured by using ImageJ. 5 individual samples were analyzed per genotype, with 200 cells measured in each.
For proliferation assays, pregnant mice (E16.5) were injected i.p. with BrdU (100 μg/g BW) 1 hour before sacrifice. Embryos were fixed in 10% buffered formalin overnight and embedded in paraffin. BrdU incorporation was detected using rat anti-BrdU primary antibody (1:50; Abcam). Immune complexes were visualized using F(ab)2 biotin-conjugated donkey anti-rat IgG (1:500; Research Diagnostics Inc.) and the Vectastain Elite ABC Kit (Vector Laboratories). Sections were counterstained with hematoxylin. For each sample, BrdU+ cells were counted in 10 randomly selected fields.
Hematopoietic analysis.
Myeloid colony assays (in the absence of added cytokines) were performed as described previously (80). In brief, BM cells were suspended in MethoCult M3234 without cytokines (Stem Cell Technologies) at 105 cells/ml, and colonies were scored after 7–9 days. Complete blood counts were determined by using a Hemavet 850FS (Drew Scientific).
Echocardiographic and hemodynamic measurements.
For echocardiography and cardiac catheterization, mice were anesthetized with isoflurane/oxygen (2%:100%), and body temperature was maintained at approximately 37.5°C. Transthoracic 2D and M-mode echocardiography was performed from the long axis view of the heart at the level of the papillary muscle with a Visualsonics Vevo 770 imaging system (Visualsonics) equipped with a 30-MHz linear transducer (RMV707B). Measurements of LVIDs, LVIDd, and LVPWd were made under Time Motion-mode (TM-mode). The papillary muscles were excluded from all measurements. Measurements were averaged from at least 3 separate cardiac cycles. From TM-mode measurements, end-diastolic volume (EDV) was calculated as (4.5 × normalized LVIDd2); end-systolic volume (ESV) was calculated as (3.72 × normalized LDIVs2); SV was calculated as EDV — ESV; CO was calculated as SV × heart rate; FS percentage was calculated as (LVIDd – LVIDs)/LVIDd × 100; and EF percentage was calculated as (EDV – ESV)/EDV × 100.
For invasive hemodynamic assessments, a 1.2F catheter (model no. FTS-1211B-0018; Scisense Inc.) was inserted via the right carotid artery into the LV. Hemodynamic signals were digitized at a sampling rate of 1 kHz and recorded by computer using the MP100 imaging system and Acqknowledge software (BIOPAC Systems Inc). Following recording of LV pressure, the catheter was relocated to the ascending aortic for measurement of aortic blood pressure. Mean arterial pressure was calculated as (systolic pressure + diastolic pressure × 2)/3.
TAC.
8- to 9-week-old male mice (25–30 g BW) were anesthetized with 2% isoflurane, intubated, connected to a ventilator (Harvard Apparatus), and ventilated at a tidal volume of approximately 230 μl and 135 breaths/min. A parasternal thoracotomy was performed to expose the transverse aorta, which was then constricted with a 7/0 silk suture tied around a 27-gauge needle. Pressure overload was maintained for various times as indicated in Results and the figure legends.
MEK inhibitor treatment.
N-([R]-2,3-dihydroxy-propoxy)-3,4-difluoro-2-(2-fluoro-4-iodo-phenylamino)-benzamide (PD0325901) was synthesized according to the disclosure in document WO2007042885(A2) (67). All chemicals necessary for the synthesis were purchased from Sigma-Aldrich.
PD0325901 was dissolved in DMSO at a concentration of 50 mg/ml, then resuspended in vehicle (0.5% hydroxypropyl methylcellulose with 0.2% Tween 80) at a concentration of 0.5 mg/ml, and injected i.p. (5 mg/kg BW) daily for the indicated times. Control mice were injected with vehicle. The same protocol was used to inject lactating females for early postnatal treatment.
Quantitative real-time RT-PCR.
Total RNA from the LV was prepared using the RNeasy minikit (Qiagen). RNA (2 μg) was reverse transcribed using SuperScriptIII (Invitrogen). TaqMan probe-based gene expression analyses (Applied Biosystems) for Myh7, Myh6, Nppa, and Nppb (Supplemental Table 1) were conducted according to the manufacturer’s instructions. Each sample was measured in triplicate, and the relative expression was normalized to GAPDH.
Biochemical analysis.
Total protein extracts from cells or tissues were prepared by homogenization in RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.5% Na deoxycholate, and 0.1% SDS) containing a protease and phosphatase inhibitor cocktail (40 μg/ml PMSF, 20 mM NaF, 1 mM Na3VO4, 10 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 2 μg/ml antipain, 2 μg/ml pepstatin A, 20 μg/ml leupeptin, and 20 μg/ml aprotinin). Homogenates were centrifuged at 16,100 g for 15 minutes at 4°C, and the supernatants were collected. Lysates (10–25 μg protein) were resolved by SDS-PAGE and analyzed by immunoblotting. Antibodies for immunoblots included Raf1 (BD Biosciences); SH-PTP2 and ERK2 (C-18 and D2, respectively; Santa Cruz Biotechnology Inc.); and phospho-MEK1/2, MEK1/2, phospho-p44/42 MAPK, phospho-S6 (Ser235/236), p38, phospho-p38, phospho-JNK1/2, Akt1, phospho-Akt (Ser473), phospho–GSK-3β (Ser9), and phospho-P70S6K (all from Cell Signaling Technology). Primary antibody binding was visualized by IRDye infrared secondary antibodies using the Odyssey Infrared Imaging System (LI-COR Biosciences). Quantification of immunoblots was performed using Odyssey version 3.0 software.
Statistics.
All data are presented as mean ± SEM. Statistical significance was determined using 1- or 2-tailed Student’s t test, 1-way ANOVA, or 2-way repeated measure ANOVA, as appropriate. If ANOVA was significant, individual differences were evaluated using Bonferroni post-test. Deviation of progeny from Mendelian frequency was assessed by χ2 test. Kaplan-Meier survival curves were analyzed using the log-rank test. For experiments in Figure 10B, significant outliers were identified using Grubbs test. All statistical analyses were performed with GraphPad Prism 5. For all studies, a P value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Angel Sing (Ontario Cancer Institute) for technical assistance. This work was supported by NIH grants HL083273 and R37CA49152 (to B.G. Neel) and Canadian Institutes of Health Research grants 153103 (to P.H. Backx) and 106526 (to T. Araki). This research also was funded in part by the Ontario Ministry of Health and Long Term Care (OMOHLTC). The views expressed do not necessarily reflect those of the OMOHLTC. P.H. Backx is supported by Career Investigator Awards (I-6891) of the Heart and Stroke Foundation of Ontario. X. Wu was supported by Frederick Banting and Charles Best Canada Graduate Scholarship. K.-H. Kim was supported in part by graduate studentships from the Ontario Graduate Scholarship in Science and Technology program.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(3):1009–1025. doi:10.1172/JCI44929.
See the related Commentary beginning on page 844.
References
- 1. Dorn GW 2nd, Force T. Protein kinase cascades in the regulation of cardiac hypertrophy. J Clin Invest. 2005;115(3):527–537. doi: 10.1172/JCI24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7(8):589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 3. Haider AW, Larson MG, Benjamin EJ, Levy D. Increased left ventricular mass and hypertrophy are associated with increased risk for sudden death. J Am Coll Cardiol. 1998;32(5):1454–1459. doi: 10.1016/S0735-1097(98)00407-0. [DOI] [PubMed] [Google Scholar]
- 4. Berenji K, Drazner MH, Rothermel BA, Hill JA. Does load-induced ventricular hypertrophy progress to systolic heart failure? Am J Physiol Heart Circ Physiol. 2005;289(1):H8–H16. doi: 10.1152/ajpheart.01303.2004. [DOI] [PubMed] [Google Scholar]
- 5. Schoenfeld JR, et al. Distinct molecular phenotypes in murine cardiac muscle development, growth, and hypertrophy. J Mol Cell Cardiol. 1998;30(11):2269–2280. doi: 10.1006/jmcc.1998.0787. [DOI] [PubMed] [Google Scholar]
- 6. Soor GS, et al. Hypertrophic cardiomyopathy: current understanding and treatment objectives. J Clin Pathol. 2009;62(3):226–235. doi: 10.1136/jcp.2008.061655. [DOI] [PubMed] [Google Scholar]
- 7. Maron BJ. Hypertrophic cardiomyopathy: a systematic review. Jama. 2002;287(10):1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 8. Anan R, et al. Prognostic implications of novel beta cardiac myosin heavy chain gene mutations that cause familial hypertrophic cardiomyopathy. J Clin Invest. 1994;93(1):280–285. doi: 10.1172/JCI116957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marian AJ, Roberts R. Recent advances in the molecular genetics of hypertrophic cardiomyopathy. Circulation. 1995;92(5):1336–1347. doi: 10.1161/01.cir.92.5.1336. [DOI] [PubMed] [Google Scholar]
- 10. Schwartz K, Carrier L, Guicheney P, Komajda M. Molecular basis of familial cardiomyopathies. Circulation. 1995;91(2):532–540. doi: 10.1161/01.cir.91.2.532. [DOI] [PubMed] [Google Scholar]
- 11. Niimura H, et al. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N Engl J Med. 1998;338(18):1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 12. Blair E, et al. Mutations in the gamma(2) subunit of AMP-activated protein kinase cause familial hypertrophic cardiomyopathy: evidence for the central role of energy compromise in disease pathogenesis. Hum Mol Genet. 2001;10(11):1215–1220. doi: 10.1093/hmg/10.11.1215. [DOI] [PubMed] [Google Scholar]
- 13. Oliveira SM, Ehtisham J, Redwood CS, Ostman-Smith I, Blair EM, Watkins H. Mutation analysis of AMP-activated protein kinase subunits in inherited cardiomyopathies: implications for kinase function and disease pathogenesis. J Mol Cell Cardiol. 2003;35(10):1251–1255. doi: 10.1016/S0022-2828(03)00237-2. [DOI] [PubMed] [Google Scholar]
- 14. Murphy RT, et al. Adenosine monophosphate-activated protein kinase disease mimicks hypertrophic cardiomyopathy and Wolff-Parkinson-White syndrome: natural history. J Am Coll Cardiol. 2005;45(6):922–930. doi: 10.1016/j.jacc.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 15. Rohini A, Agrawal N, Koyani CN, Singh R. Molecular targets and regulators of cardiac hypertrophy. Pharmacol Res. 2010;61(4):269–280. doi: 10.1016/j.phrs.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 16. Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3(6):459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 17. McCubrey JA, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773(8):1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bueno OF, Molkentin JD. Involvement of extracellular signal-regulated kinases 1/2 in cardiac hypertrophy and cell death. Circ Res. 2002;91(9):776–781. doi: 10.1161/01.RES.0000038488.38975.1A. [DOI] [PubMed] [Google Scholar]
- 19. Lorenz K, Schmitt JP, Vidal M, Lohse MJ. Cardiac hypertrophy: targeting Raf/MEK/ERK1/2-signaling. Int J Biochem Cell Biol. 2009;41(12):2351–2355. doi: 10.1016/j.biocel.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 20. Hunter JJ, Tanaka N, Rockman HA, Ross J, Jr, Chien KR. Ventricular expression of a MLC-2v-ras fusion gene induces cardiac hypertrophy and selective diastolic dysfunction in transgenic mice. J Biol Chem. 1995;270(39):23173–23178. doi: 10.1074/jbc.270.39.23173. [DOI] [PubMed] [Google Scholar]
- 21. Zheng M, et al. Sarcoplasmic reticulum calcium defect in Ras-induced hypertrophic cardiomyopathy heart. Am J Physiol Heart Circ Physiol. 2004;286(1):H424–H433. doi: 10.1152/ajpheart.00110.2003. [DOI] [PubMed] [Google Scholar]
- 22. Mitchell S, et al. Distinct gene expression profiles in adult mouse heart following targeted MAP kinase activation. Physiol Genomics. 2006;25(1):50–59. doi: 10.1152/physiolgenomics.00224.2005. [DOI] [PubMed] [Google Scholar]
- 23. Glennon PE, Kaddoura S, Sale EM, Sale GJ, Fuller SJ, Sugden PH. Depletion of mitogen-activated protein kinase using an antisense oligodeoxynucleotide approach downregulates the phenylephrine-induced hypertrophic response in rat cardiac myocytes. Circ Res. 1996;78(6):954–961. doi: 10.1161/01.res.78.6.954. [DOI] [PubMed] [Google Scholar]
- 24. Clerk A, Michael A, Sugden PH. Stimulation of the p38 mitogen-activated protein kinase pathway in neonatal rat ventricular myocytes by the G protein-coupled receptor agonists, endothelin-1 and phenylephrine: a role in cardiac myocyte hypertrophy? J Cell Biol. 1998;142(2):523–535. doi: 10.1083/jcb.142.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harris IS, Zhang S, Treskov I, Kovacs A, Weinheimer C, Muslin AJ. Raf-1 kinase is required for cardiac hypertrophy and cardiomyocyte survival in response to pressure overload. Circulation. 2004;110(6):718–723. doi: 10.1161/01.CIR.0000138190.50127.6A. [DOI] [PubMed] [Google Scholar]
- 26. Bueno OF, et al. The MEK1-ERK1/2 signaling pathway promotes compensated cardiac hypertrophy in transgenic mice. EMBO J. 2000;19(23):6341–6350. doi: 10.1093/emboj/19.23.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Purcell NH, et al. Genetic inhibition of cardiac ERK1/2 promotes stress-induced apoptosis and heart failure but has no effect on hypertrophy in vivo. Proc Natl Acad Sci U S A. 2007;104(35):14074–14079. doi: 10.1073/pnas.0610906104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tartaglia M, Gelb BD. Noonan syndrome and related disorders: genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2005;6:45–68. doi: 10.1146/annurev.genom.6.080604.162305. [DOI] [PubMed] [Google Scholar]
- 29. Bentires-Alj M, Kontaridis MI, Neel BG. Stops along the RAS pathway in human genetic disease. Nat Med. 2006;12(3):283–285. doi: 10.1038/nm0306-283. [DOI] [PubMed] [Google Scholar]
- 30. Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29(8):992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- 31. Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19(3):230–236. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rauen KA, et al. Proceedings from the 2009 genetic syndromes of the Ras/MAPK pathway: From bedside to bench and back. Am J Med Genet A. 2010;152A(1):4–24. doi: 10.1002/ajmg.a.33183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chan G, Kalaitzidis D, Neel BG. The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev. 2008;27(2):179–192. doi: 10.1007/s10555-008-9126-y. [DOI] [PubMed] [Google Scholar]
- 34. Tartaglia M, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29(4):465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 35. Roberts AE, et al. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007;39(1):70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- 36. Tartaglia M, et al. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39(1):75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 37. Pandit B, et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007;39(8):1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 38. Razzaque MA, et al. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet. 2007;39(8):1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- 39. Schubbert S, et al. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38(3):331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 40. Zenker M, et al. Expansion of the genotypic and phenotypic spectrum in patients with KRAS germline mutations. J Med Genet. 2007;44(2):131–135. doi: 10.1136/jmg.2006.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cirstea IC, et al. A restricted spectrum of NRAS mutations causes Noonan syndrome. Nat Genet. 2010;42(1):27–29. doi: 10.1038/ng.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cordeddu V, et al. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat Genet. 2009;41(9):1022–1026. doi: 10.1038/ng.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nishikawa T, Ishiyama S, Shimojo T, Takeda K, Kasajima T, Momma K. Hypertrophic cardiomyopathy in Noonan syndrome. Acta Paediatr Jpn. 1996;38(1):91–98. doi: 10.1111/j.1442-200x.1996.tb03445.x. [DOI] [PubMed] [Google Scholar]
- 44. Sznajer Y, et al. The spectrum of cardiac anomalies in Noonan syndrome as a result of mutations in the PTPN11 gene. Pediatrics. 2007;119(6):e1325–1331. doi: 10.1542/peds.2006-0211. [DOI] [PubMed] [Google Scholar]
- 45. Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J Biol Chem. 2006;281(10):6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 46. Hanna N, et al. Reduced phosphatase activity of SHP-2 in LEOPARD syndrome: consequences for PI3K binding on Gab1. FEBS Lett. 2006;580(10):2477–2482. doi: 10.1016/j.febslet.2006.03.088. [DOI] [PubMed] [Google Scholar]
- 47. Tartaglia M, et al. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am J Hum Genet. 2006;78(2):279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Porciello R, Divona L, Strano S, Carbone A, Calvieri C, Giustini S. Leopard syndrome. Dermatol Online J. 2008;14(3):7. [PubMed] [Google Scholar]
- 49. Aoki Y, et al. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37(10):1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 50. Gripp KW, et al. HRAS mutation analysis in Costello syndrome: genotype and phenotype correlation. Am J Med Genet A. 2006;140(1):1–7. doi: 10.1002/ajmg.a.31047. [DOI] [PubMed] [Google Scholar]
- 51. Niihori T, et al. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38(3):294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- 52. Gripp KW, et al. Further delineation of the phenotype resulting from BRAF or MEK1 germline mutations helps differentiate cardio-facio-cutaneous syndrome from Costello syndrome. Am J Med Genet A. 2010;143A(13):1472–1480. doi: 10.1002/ajmg.a.31815. [DOI] [PubMed] [Google Scholar]
- 53. Rodriguez-Viciana P, et al. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311(5765):1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- 54. Araki T, et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10(8):849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- 55. Krenz M, Gulick J, Osinska HE, Colbert MC, Molkentin JD, Robbins J. Role of ERK1/2 signaling in congenital valve malformations in Noonan syndrome. Proc Natl Acad Sci U S A. 2008;105(48):18930–18935. doi: 10.1073/pnas.0806556105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nakamura T, et al. Mediating ERK 1/2 signaling rescues congenital heart defects in a mouse model of Noonan syndrome. J Clin Invest. 2007;117(8):2123–2132. doi: 10.1172/JCI30756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nakamura T, Gulick J, Pratt R, Robbins J. Noonan syndrome is associated with enhanced pERK activity, the repression of which can prevent craniofacial malformations. Proc Natl Acad Sci U S A. 2009;106(36):15436–15441. doi: 10.1073/pnas.0903302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Araki T, Chan G, Newbigging S, Morikawa L, Bronson RT, Neel BG. Noonan syndrome cardiac defects are caused by PTPN11 acting in endocardium to enhance endocardial-mesenchymal transformation. Proc Natl Acad Sci U S A. 2009;106(12):4736–4741. doi: 10.1073/pnas.0810053106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen X, et al. Endogenous expression of Hras(G12V) induces developmental defects and neoplasms with copy number imbalances of the oncogene. Proc Natl Acad Sci U S A. 2009;106(19):7979–7984. doi: 10.1073/pnas.0900343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Marin TM, et al. Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of LEOPARD syndrome–associated PTPN11 mutation. . J Clin Invest. 2011;121(3):1026–1043. doi: 10.1172/JCI44972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Allanson JE, et al. The face of Noonan syndrome: Does phenotype predict genotype. Am J Med Genet A. 2010;152A(8):1960–1966. doi: 10.1002/ajmg.a.33518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hasle H. Malignant diseases in Noonan syndrome and related disorders. Horm Res. 2009;72 suppl 2:8–14. doi: 10.1159/000243773. [DOI] [PubMed] [Google Scholar]
- 63. Ross J, Jr, Sobel BE. Regulation of cardiac contraction. Annu Rev Physiol. 1972;34:47–90. doi: 10.1146/annurev.ph.34.030172.000403. [DOI] [PubMed] [Google Scholar]
- 64. Nakagawa O, et al. Rapid transcriptional activation and early mRNA turnover of brain natriuretic peptide in cardiocyte hypertrophy. Evidence for brain natriuretic peptide as an “emergency” cardiac hormone against ventricular overload. J Clin Invest. 1995;96(3):1280–1287. doi: 10.1172/JCI118162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Takeda N, et al. Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. J Clin Invest. 2010;120(1):254–265. doi: 10.1172/JCI40295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Thum T, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456(7224):980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 67. Barrett SD, et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg Med Chem Lett. 2008;18(24):6501–6504. doi: 10.1016/j.bmcl.2008.10.054. [DOI] [PubMed] [Google Scholar]
- 68. Chen PC, et al. Activation of multiple signaling pathways causes developmental defects in mice with a Noonan syndrome–associated Sos1 mutation. J Clin Invest. 2010;120(12):4353–4365. doi: 10.1172/JCI43910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bertola DR, Pereira AC, de Oliveira PS, Kim CA, Krieger JE. Clinical variability in a Noonan syndrome family with a new PTPN11 gene mutation. Am J Med Genet A. 2004;130A(4):378–383. doi: 10.1002/ajmg.a.30270. [DOI] [PubMed] [Google Scholar]
- 70. Bueno OF, et al. The dual-specificity phosphatase MKP-1 limits the cardiac hypertrophic response in vitro and in vivo. Circ Res. 2001;88(1):88–96. doi: 10.1161/01.res.88.1.88. [DOI] [PubMed] [Google Scholar]
- 71. Xu J, Ismat FA, Wang T, Lu MM, Antonucci N, Epstein JA. Cardiomyocyte-specific loss of neurofibromin promotes cardiac hypertrophy and dysfunction. Circ Res. 2009;105(3):304–311. doi: 10.1161/CIRCRESAHA.109.201509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ieda M, et al. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 2009;16(2):233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Miragoli M, Gaudesius G, Rohr S. Electrotonic modulation of cardiac impulse conduction by myofibroblasts. Circ Res. 2006;98(6):801–810. doi: 10.1161/01.RES.0000214537.44195.a3. [DOI] [PubMed] [Google Scholar]
- 74. Margetic S, Gazzola C, Pegg GG, Hill RA. Leptin: a review of its peripheral actions and interactions. Int J Obes Relat Metab Disord. 2002;26(11):1407–1433. doi: 10.1038/sj.ijo.0802142. [DOI] [PubMed] [Google Scholar]
- 75. Bjorbaek C, et al. Divergent roles of SHP-2 in ERK activation by leptin receptors. J Biol Chem. 2001;276(7):4747–4755. doi: 10.1074/jbc.M007439200. [DOI] [PubMed] [Google Scholar]
- 76. Zhang EE, Chapeau E, Hagihara K, Feng GS. Neuronal Shp2 tyrosine phosphatase controls energy balance and metabolism. Proc Natl Acad Sci U S A. 2004;101(45):16064–16069. doi: 10.1073/pnas.0405041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Su IH, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4(2):124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 78. Zhang SQ, et al. Shp2 regulates SRC family kinase activity and Ras/Erk activation by controlling Csk recruitment. Mol Cell. 2004;13(3):341–355. doi: 10.1016/S1097-2765(04)00050-4. [DOI] [PubMed] [Google Scholar]
- 79. Zobel C, Kassiri Z, Nguyen TT, Meng Y, Backx PH. Prevention of hypertrophy by overexpression of Kv4.2 in cultured neonatal cardiomyocytes. Circulation. 2002;106(18):2385–2391. doi: 10.1161/01.CIR.0000033970.22130.93. [DOI] [PubMed] [Google Scholar]
- 80. Chan G, et al. Leukemogenic Ptpn11 causes fatal myeloproliferative disorder via cell-autonomous effects on multiple stages of hematopoiesis. Blood. 2009;113(18):4414–4424. doi: 10.1182/blood-2008-10-182626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.