Abstract
Malnutrition substantially increases susceptibility to Entamoeba histolytica in children. Leptin is a hormone produced by adipocytes that inhibits food intake, influences the immune system, and is suppressed in malnourished children. Therefore we hypothesized that diminished leptin function may increase susceptibility to E. histolytica infection. We prospectively observed a cohort of children, beginning at preschool age, for infection by the parasite E. histolytica every other day over 9 years and evaluated them for genetic variants in leptin (LEP) and the leptin receptor (LEPR). We found increased susceptibility to intestinal infection by this parasite associated with an amino acid substitution in the cytokine receptor homology domain 1 of LEPR. Children carrying the allele for arginine (223R) were nearly 4 times more likely to have an infection compared with those homozygous for the ancestral glutamine allele (223Q). An association of this allele with amebic liver abscess was also determined in an independent cohort of adult patients. In addition, mice carrying at least 1 copy of the R allele of Lepr were more susceptible to infection and exhibited greater levels of mucosal destruction and intestinal epithelial apoptosis after amebic infection. These findings suggest that leptin signaling is important in mucosal defense against amebiasis and that polymorphisms in the leptin receptor explain differences in susceptibility of children in the Bangladesh cohort to amebiasis.
Introduction
The biological mechanisms by which malnutrition contributes to an estimated one-third of all deaths among children (1) and 60% of deaths due to diarrhea (2) are poorly understood (1). Leptin is a product of the LEP gene on chromosome 7 that functions to signal satiety and is implicated in regulation of the immune system and the gastrointestinal tract (3, 4). Circulating leptin concentrations are low in malnourished infants and are associated with a suppression of proinflammatory cytokine production (5). Leptin promotes differentiation, proliferation, survival, and function of both innate and adaptive immune cells (6–10) and acts as a proinflammatory cytokine that orchestrates the inflammatory response toward a Th1 phenotype while suppressing Th2 and regulatory T cell functions (5, 11, 12). In addition to the proinflammatory effects on the immune system, leptin may also modulate intestinal barrier function by promoting regeneration and inhibiting apoptosis of intestinal epithelium (13, 14) as well as stimulating mucin secretion (15) and maintaining intestinal morphology (16).
Amebiasis, a potentially fatal enteric infection caused by the parasite Entamoeba histolytica, is exacerbated by malnutrition (17–19). We tested common polymorphisms in the leptin and leptin receptor genes in children exposed to E. histolytica, hypothesizing that any polymorphism altering leptin or its action might increase susceptibility to infection. Subjects were a cohort of 185 children living in an urban slum in Dhaka, Bangladesh, prospectively observed by home visits every other day for amebiasis over 9 years, as well as 86 adult males with amebic liver abscess (ALA). We discovered that a common polymorphism in the leptin receptor was associated with susceptibility to amebiasis. The role of this polymorphism in amebiasis was demonstrated in mice segregating for the susceptible allele.
Results
After 9 years of prospective observation by household visits every other day, 220 of the original 289 children remained enrolled in the cohort. Of these children, 185 represented nuclear families and were included in this genetic analysis. Ninety percent of the children had been infected at least once with E. histolytica in 9 years. Forty percent of the children were either malnourished or stunted at baseline (ref. 20, Supplemental Table 1, and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI45294DS1).
We genotyped 15 SNPs in leptin (LEP) and 77 SNPs in the leptin receptor (LEPR); 1 SNP showed significant deviation (P < 0.01) from Hardy-Weinberg equilibrium and was excluded. Five SNPs in LEPR and 5 SNPs in LEP were either monomorphic or had a minor allele frequency (MAF) of less than 5% in this population and were not considered in further analyses. A test of association was performed between the remaining SNPs and E. histolytica infection, and 6 SNPs of LEPR had an empiric P value of less than 0.05 (Supplemental Figure 2 and Supplemental Table 2). All markers showing significant associations were in LEPR, located between introns 5 and 19. A single haplotype block (haplotype block 7) including 12 SNPs spanned intron 2 through intron 6, of which rs4655537 in intron 6 was significant for the association both as a single SNP and as part of this haplotype (Supplemental Figure 2A).
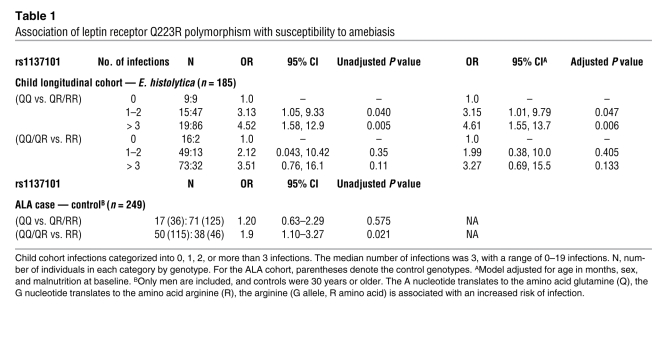
To determine whether this region harbored a potential causal variant, known coding SNPs within this significant haplotype block or near individual significant SNPs were identified. Five nonsynonymous SNPs were genotyped: rs1137100 (K109R, exon 4), rs1137101 (Q223R, exon 6), rs13306526 (I503V, exon 11), rs8179183 (K656N, exon 15), and rs34499590 (T699M, exon 15). Two SNPs, rs13306526 and rs34499590, were both monomorphic and could not be analyzed, which is consistent with HapMap data for individuals of European ancestry (CEU). No significant association was seen for rs1137100, K109R, with risk of amebiasis (odds ratio [OR] = 0.98; P = 0.674). However, a strong association was identified with coding SNP rs1137101, Q223R, which appeared to have a dominant effect. Individuals segregating for the G allele, which translates to arginine at position 223 of LEPR, were nearly 4 times more likely to have an E. histolytica infection than those homozygous for the ancestral A allele (glutamine; Q223) (OR = 3.91, P = 0.007, 95% CI 1.44, 10.6, empirical P = 0.009, population attributable risk = 69%). This association persisted after adjusting for age, sex, and malnutrition at baseline (OR = 3.98, P = 0.009, 95% CI 1.4, 11.3). In addition, we adjusted for HLA class II alleles DQB1*0601 and DQB1*1501, which were previously shown to be associated with infection in this same cohort (21), and the strength of the association remained (OR = 4.19, P = 0.01, 95% CI 1.39, 12.6). When we stratified the outcome by number of E. histolytica infections, the risk attributable to 223R was pronounced among those with greater than 3 infections (Table 1). Since this Q223R LEPR allele had been associated with adiposity in some but not all human studies (reviewed in ref. 22), we compared the baseline malnutrition and stunting of children with different genotypes at this locus and saw no association (Supplemental Table 3). We concluded the effect of the Q223R leptin receptor polymorphism on amebiasis was not confounded by the nutritional status at baseline.
Table 1 .
Association of leptin receptor Q223R polymorphism with susceptibility to amebiasis
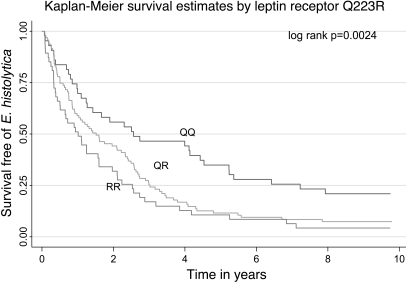
To determine whether this genetic variant altered not only the risk of having an E. histolytica infection, but also the time to an infection, we performed a Kaplan-Meier analysis (Figure 1). The median time from study entry to an infection for individuals homozygous for the glutamine allele (QQ) was 2.56 years, compared with 1.51 years for those with only 1 glutamine (QR) allele and 1.01 years for individuals with no glutamine allele (RR) (log rank P = 0.002). The estimated hazard ratio for individuals segregating for at least one copy of the arginine allele (QR or RR) was 1.82 (P = 0.002, 95% CI 1.1.24–2.67). To insure these effects were not confounded by nutritional status, we compared the time to infection stratified on malnutrition status at baseline. The median time to infection for those malnourished at baseline was 1.23 years compared with 1.61 years for those not malnourished. This difference was not statistically significant (log rank P = 0.17, RH = 1.25, P = 0.17, 95% CI 0.91–1.72). We concluded that the 223Q allele of LEPR was associated with protection from E. histolytica infection and a delay in time to E. histolytica infection.
Figure 1. Kaplan-Meier analysis of time to E. histolytica infection for the RR, QR, and QQ variants.
The importance of LEPR was also seen in an ALA cohort in Bangladesh. ALA results from extension of E. histolytica infection from the intestine. Thus, it was reasonable to expect the Q223R LEPR polymorphism might also affect susceptibility to liver abscess if it predisposed children to E. histolytica infection. Because ALA occurs predominantly in men, we used 86 adult male cases and 161 adult male blood donor controls. ALA was nearly 2 times more likely in individuals carrying RR compared with those who were QR or QQ (OR = 1.9, P = 0.021, 95% CI 1.10–3.27) (Table 1). Interestingly, the association with liver abscess was restricted to those segregating for 2 arginines (RR), whereas susceptibility to E. histolytica infection in the child cohort was conveyed by even a single copy of the arginine allele (QR or RR).
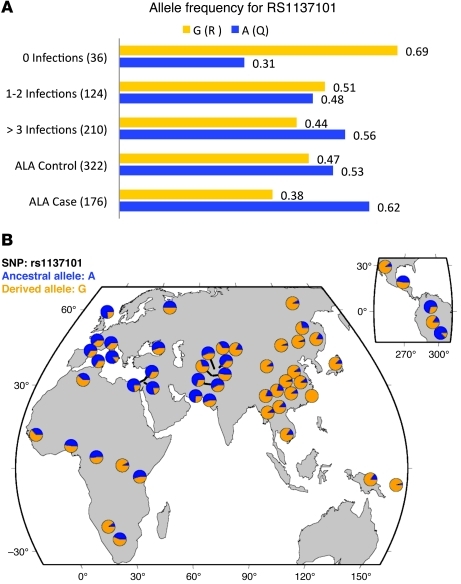
The allele frequencies for rs1137101 were different among children who never had an E. histolytica infection (0.31) and those with ALA (Figure 2). Controls from the ALA study had frequencies of arginine (G nucleotide, R amino acid) that were not significantly different from those observed in the overall child cohort (0.52) or in the HapMap phase II Gujarati Indians from Houston, Texas (GHI) sample (0.52). These controls, as expected, reflect the general population allele frequencies, indicating that this common polymorphism is not unique to this study population. However, frequency of the R allele was substantially increased in liver abscess cases (0.62). These data suggest there is a dose effect with 1 (or more) arginines increasing susceptibility to common infection acquired during childhood and 2 arginines increasing susceptibility to ALA among adults. Figure 2B also outlines the allele frequencies for each of the populations worldwide. Differences in allele frequencies for rs1137101 are observed with the most striking differences between Indo-European countries and East Asian countries where the allele is nearly fixed. Thus, the Q223R LEPR polymorphism appears to be associated with susceptibility to amebiasis in 2 independent studies, and differences in allele frequencies may result in differences in susceptibility to amebiasis and ALA via this pathway.
Figure 2. The allele frequencies for the rs1137101 allele.
The frequencies are presented for the Q and R alleles. The A nucleotide translates to the amino acid glutamine (Q); the G nucleotide translates to the amino acid arginine (R). International HapMap samples frequencies were 0.53 (Q) for the CEU population and 0.52 (Q) for the GHI. (A) The child cohort is represented by 0, 1–2, or more than 3 infections among all genotyped children in the cohort. The overall allele frequency for the child cohort is 0.52 and 0.49 (Q). Adult ALA patients are subdivided into cases and controls. Parentheses denote the number of alleles for each category. The allele frequencies of the HapMap populations reflect the population-based allele frequencies and are similar to children with 1–2 infections (38). Differences in the allele frequencies are most noted for those with 0 infections, or with more than 3 infections, and the ALA cases. (B) Geographic distribution of the rs1137101 alleles as generated by the Human Genome Diversity Project selection browser ( hgdp.uchicago.edu).
We assessed the effect of this SNP on serum concentrations of leptin and soluble leptin receptor. The analysis was stratified by gender and BMI at the time serum was collected. Leptin receptor concentrations were decreased for both boys and girls with the RR genotype (Supplemental Figure 3). This effect was most pronounced in children with BMI less than the median (BMI < 16). The logarithm of serum leptin receptor concentrations was statistically significant for the 3 genotypes (ANOVA, P = 0.0009) and remained significant when stratified by sex (boys, P = 0.02; girls, P = 0.04) or BMI category (low BMI < 16, P = 0.002; high BMI > 16, P = 0.04). No trend in circulating leptin concentrations by genotype was detected (ANOVA, P = 0.065; Supplemental Figure 3), although differences by sex (P < 0.00001) and BMI levels (P < 0.00001) without regard to genotype were present, as expected due to known effects of sex and adiposity on circulating leptin concentrations. Soluble leptin receptor levels have been shown to play a role in levels of total leptin and free leptin, although the relationship is complex (23, 24). These data indicate that differences in leptin receptor expression rather than circulating leptin levels were associated with the SNP.
Recent studies have shown that both adipose tissue and circulating peripheral blood levels of LEPR are highly heritable in expression-QTL studies (25). We evaluated mRNA levels for LEPR in resting PBMCs, using quantitative RT-PCR (qRT-PCR) for both the long and short leptin receptor isoforms. Individuals segregating for 2 copies of the arginine allele (RR) had a significant decrease in expression of the long isoform (LEPR isoform 1 as measured by Lepr_vb_2 primer) compared with those segregating for least 1 glutamine (QR, P = 0.015 or QQ, P = 0.027), respectively (Supplemental Figure 4).
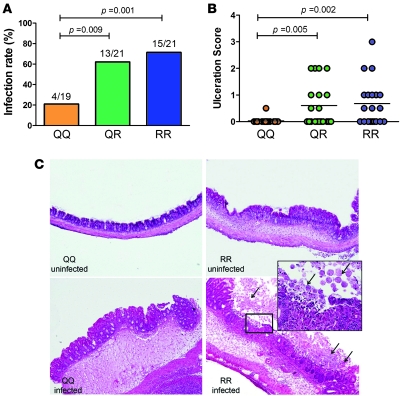
To determine whether the Q223R polymorphism specified the altered phenotype, we challenged 129P3/J mice carrying the QQ, QR, and RR alleles with E. histolytica. Mice with at least 1 copy of arginine (R) were more susceptible to infection, consistent with the human genotype-phenotype relationship (Figure 3A). In addition, the infected RR and QR mice exhibited significantly more severe epithelial ulceration compared with QQ mice that were successfully infected (Figure 3, B and C). These data demonstrate that a single copy of the 223R allele (QR) significantly increases susceptibility to amebiasis and disease severity in animals that are otherwise genetically identical.
Figure 3. The 223R allele of the leptin receptor increases susceptibility to intestinal amebic infection and mucosal destruction in the murine model of intestinal amebiasis.
(A) Mice carrying 1 or 2 copies of the arginine allele at the 223 codon (QR or RR) were more susceptible to intestinal infection with E. histolytica than those homozygous for the glutamine (QQ) allele, as assessed by culture and parasite antigen detection (QQ versus QR P = 0.009, QQ versus RR; P = 0.001 by c2 test). (B) RR and QR mice exhibited more severe epithelial ulceration than the QQ mice by histological examination (QQ versus QR, P = 0.005; QQ versus RR, P = 0.002 by Mann-Whitney test; n = 19, 21, and 21 for QQ, QR and RR mice, respectively). (C) Representative cecal pathology of E. histolytica–challenged but uninfected at day 3 animals showed nearly normal-looking morphology or a mild submucosal edema (top left and right). While infected QQ mice mainly exhibited submucosal edema and epithelial hyperplasia (bottom left), the infected RR cecum displayed marked mucosal destruction/ulceration and inflammation. E. histolytica trophozoites are indicated by arrows (bottom right and insert). Original magnification, ×40; ×200 (inset).
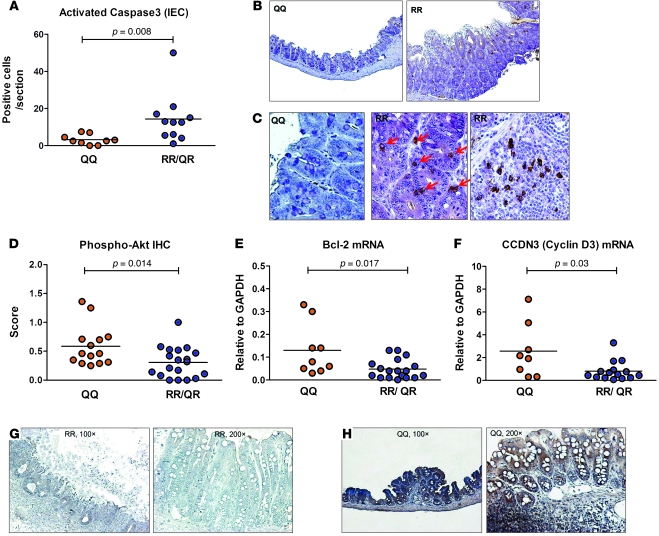
It has been demonstrated that E. histolytica kills host cells by activating caspase-3, which leads to host cell apoptosis and allows amebic invasion (26, 27). Expression of the leptin receptor is known to protect both immune cells and epithelial cells from apoptotic stimuli (6, 13). We hypothesized that the R223 allele of the leptin receptor was less effective than the Q223 allele in protecting the intestinal epithelium from apoptosis induced by E. histolytica. To test this, cecal sections from E. histolytica–challenged QQ, RR, and QR mice were stained for activated caspase-3. The RR and QR mice displayed significantly more active caspase-3 than the QQ mice in intestinal epithelial cells (IECs) (Figure 4, A and B) and mononuclear cells (Figure 4C), suggesting a greater level of apoptosis induced by E. histolytica in mice with the R allele.
Figure 4. Increased caspase-3 activity and decreased expression of phosphorylated Akt and antiapoptotic genes in the ceca of RR and QR mice.
(A) Mice carrying 1 or 2 copies of the arginine allele (QR or RR, n = 9) had increased caspase-3 activity in IECs compared with the QQ mice (n = 11) by day 3 after infection. (B) Representative sections of the cecum showed increased levels of active caspase-3 in infected RR mice. Original magnification, ×100. (C) Higher magnification demonstrated undetectable active caspase-3 staining in QQ IECs (left), but cytoplasmic and perinuclear staining in RR IECs (middle, arrows) and infiltrating cells (right). Original magnification, ×400. (D) Compared with the RR or Q/R animals, QQ mice had significantly greater levels of Akt phosphorylation (n = 14 and 19 for QQ and RR/QR mice respectively) and (E and F) antiapoptotic gene expression including Bcl-2 (E, P = 0.017) and cyclin D3 (F, P = 0.03) in the cecum (n = 9 and 18 for QQ and RR/QR mice respectively). (G) Representative IHC staining for phospho-Akt showed minimal expression in the cecum of the 223R mouse. (H) Intense phospho-Akt expression was present throughout the crypts and villi, and also in the mononuclear cells in the QQ cecum (left and right).
PI3K/Akt signaling is implicated in leptin receptor–mediated antiapoptotic and proproliferative effects by upregulating the expression of survival factors (i.e., Bcl-2, cyclin D3, etc.) (6, 13, 28). We examined phosphorylated Akt expression in the cecum from infected and uninfected QQ, RR, and QR mice and observed a significant decrease in Akt phosphorylation in RR and QR mice relative to QQ littermates regardless of their infectious status. Representative sections showed minimal phospho-Akt staining in the infected RR cecum with mucosal ulcerations and invading trophozoites and uninfected RR cecum with mucosal hyperplasia (Figure 4G). In contrast, pAkt was abundantly present in most of the QQ animals throughout the crypts and the villi, as well as within the lymphoid compartment (Figure 4H). RR and QR mice also had decreased expression of downstream survival factors Bcl-2 and cyclin D3 compared with their QQ littermates (Figure 4, D–F). These data support a role for the PI3K/Akt pathway in controlling amebic-induced IEC apoptosis and suggest that the PI3K/Akt pathway is impaired by the Q223R polymorphism.
Discussion
The most important finding of this study is that the nutritional hormone leptin is important in immunity to the enteric pathogen E. histolytica. The Q223R polymorphism in the leptin receptor was associated in children with decreased expression of soluble and cellular leptin receptors and increased susceptibility to intestinal amebiasis and, in adults, ALA. The Q223R polymorphism is a remarkable example of a common genetic polymorphism having a major effect on disease susceptibility. The importance of this polymorphism in amebiasis was unambiguously validated in mice engineered to differ only by this single amino acid. This observation of a link of adipocytokines with host defense may offer insight into one mechanism by which children are susceptible to diarrhea.
A role for leptin in host resistance to infection had been suggested in children with congenital deficiency of the leptin receptor (29) and in studies using leptin and leptin receptor–deficient mice, which exhibit enhanced susceptibility to infection (3, 30, 31). Most of the prior work in mouse models has implicated the proinflammatory effect of leptin on cell-mediated immunity in control of infection. However, as a pleiotropic hormone, leptin also regulates the neuroendocrine axis centrally and immune/epithelial homeostasis peripherally (3, 31, 32). While the proinflammatory cellular response may contribute to the clearance of certain infections, we hypothesized that leptin contributed to host defense from E. histolytica by preventing epithelial apoptosis and promoting tissue repair. Indeed, the caspase-3–mediated apoptotic pathway is central in E. histolytica killing of host cells (26). Recent in vivo work using caspase-3–knockout and bcl-2–overexpressing mice demonstrated that epithelial cell apoptosis facilitates E. histolytica intestinal infection (30). Interestingly, a loss of resistance to amebiasis was seen in mice with specific deletion of the leptin receptor in the intestinal epithelium. In addition both leptin-deficient (ob/ob) and leptin receptor–deficient (db/db) mice had increased susceptibility to amebiasis (33). These findings, together with the data presented here showing increased epithelial damage in Q223R mice, indicate that antiapoptotic leptin signaling at the intestinal epithelium may be required for resistance to amebiasis.
Two limitations of our study should be considered in evaluating our results. First, the mouse model of amebic colitis does not completely reflect human disease, as the mice are intracecally infected with trophozoites, and not orally with cysts, as occurs in human infection. Second, our study in children required evaluation every other day for 9 years for signs of intestinal amebic infection. This intensive follow-up and cohort design necessary to identify individuals resistant to amebiasis precluded a large sample size or a replicate population. Therefore we used for validation an adult manifestation of amebic disease in the same ethnic population.
Prior to this study there was little understanding of the host factors that control susceptibility to amebiasis. This is despite the fact that E. histolytica infection is estimated by the WHO to result in 34 to 50 million symptomatic cases worldwide each year and as many as 100,000 deaths (34). The impact of amebiasis is underscored by this cohort from Dhaka, Bangladesh, where diarrheal diseases are a leading cause of childhood death: the annual incidence of infection in preschool children was 40% (20). Knowledge of the role of host genetic polymorphisms in the leptin receptor in susceptibility may allow for targeted interventions to prevent or treat amebiasis.
The importance of this work lies in the demonstration of the association between a genetic polymorphism in an adipocytokine receptor with susceptibility to enteric infection in children. This link between genotype and resistance to infection suggests new approaches to prevention or treatment of intestinal infections.
Methods
Study participants.
In 1999, preschool children, 2–5 years old, were enrolled in a natural history prospective cohort to evaluate E. histolytica infection. Initially, 289 children were enrolled from Mirpur, an urban slum in Dhaka, Bangladesh. The participants are of Bihari ethnic origin, and their families settled in Mirpur after the liberation of Bangladesh from Pakistan in 1971. The area is densely populated, with poor sanitary and hygienic conditions. The parents and children were visited and interviewed every other day by health care workers for details about any diarrheal episodes of the child as well as other related health questions. Children with diarrhea were also detected when their parents contacted project personnel at the field clinic. When diarrheal disease was detected, the child was examined and a stool sample was collected for detailed investigation of enteropathogens including E. histolytica. In addition, surveillance stool specimens were obtained every month for detection of E. histolytica infection by antigen detection and culture. Stool samples were also cultured for Entamoeba species in Robinson’s medium within 6 hours of collection. Blood samples were collected from these children every 4 months and tested for serum anti-parasite antibodies. All enrolled children and their family members received free primary health care services, including medications, from the project office in Mirpur, Dhaka, Bangladesh. Episodes of diarrhea were treated with oral rehydration and antibiotics or anti-amebic medications as needed. Participants and their parents were interviewed to determine if there were relatives enrolled in the cohort. First-degree relatives were identified and removed for genetic analyses, resulting in 185 children representing nuclear families.
In addition, 86 ALA patients and 161 age- and sex-matched blood donor controls from 2 public hospitals in Bangladesh were included in this study for replication. Cases and controls were all male due to known differences in ALA frequencies between the sexes. Clinical diagnosis of ALA was based on fever with right upper quadrant abdominal pain and a space-occupying lesion in the liver suggestive of abscess as diagnosed by ultrasonogram. Suspected cases of ALA were confirmed by a positive anti–E. histolytica IgG in serum and in some cases by the presence of E. histolytica DNA in liver abscess pus specimens by PCR. Venous blood was collected from ALA patients and controls and liver abscess pus specimens were collected from ALA patients. Liver abscess pus was aspirated only for clinical purposes as judged by the clinicians caring for the patients and not for the purpose of this study.
Informed consent for all study aspects including genetic testing was obtained (in the case of children from the parents or guardians). The human experimentation guidelines of the US Department of Health and Human Services were evaluated and approved for the conduct of this research by the following institutional review boards: the University of Virginia; the Centre for Health and Population Research; the International Center for Diarrheal Disease Research, Bangladesh; and Johns Hopkins University.
Case definitions.
A “new episode” of E. histolytica infection during the period of observation was defined as a positive E. histolytica stool antigen and/or culture result preceded by more than 2 monthly surveillance stool samples with negative results. This definition was validated by PCR genotyping of a subset of samples. E. histolytica–associated diarrhea/dysentery was defined as 3 or more unformed stools in a 24-hour period accompanied by a positive stool-antigen detection test for E. histolytica. Amebic dysentery was defined as a diarrheal stool sample containing occult or gross blood that was positive for E. histolytica antigen (20). Initial analysis defined cases and controls as children without E. histolytica infection and children with E. histolytica infection, respectively. Additional categories of number of positive E. histolytica infections resulted in 3 categories: never infected, 1–2 infections, and greater than 3 infections over 9 years of follow-up. Malnourished and stunted were defined as a weight-for-age Z-score (WAZ) of less than –2 and height for age Z-score (HAZ) of less than –2, respectively (35). BMI was measured as weight in kilograms divided by height in meters squared at the time of serum collection. The median BMI in the child cohort was 15.7; low and high BMI were determined as those below or above the median, respectively.
Nutritional assessment.
Trained research assistants took anthropometric measurements. Each child was weighed in light clothes with an electronic weighing scale to the nearest value of 100 grams. The standing heights of children were measured to the nearest 0.1 cm using a height stick. Nutritional status was assessed by comparing the weight and height of the study children with those of a National Center for Health Statistics reference population of the same age and sex with the help of Epi6 computer package program.
Sample preparation for genotyping.
Patient DNA samples originated from whole blood and were whole genome amplified using the GenomiPhi DNA Amplification Kit (Amersham Biosciences). Whole genome amplification was performed with a 300 ng input of DNA. We followed the manufacturer’s protocol for the amplification. The amplified product was diluted with 80 μl 1× TE Buffer. Samples were heated to 99°C for 5 minutes and then diluted with 100 μl 1× TE Buffer.
SNP selection.
SNPs within the LEP and LEPR genes and 10 kb upstream and downstream of the genes were identified. These tagSNPs were selected using SNPbrowser software detailing the HapMap (NCBI Build 36) LD map database. Selection criteria for tagSNPs were set at a minimum MAF of 5% for the HapMap Ceph Utah (CEU) population. Redundant SNPs (those with r2 = 1) were removed. In addition to selecting tagSNPs, SNPs were included for every 0.5 LDU unit to insure coverage for any tagSNPs that might fail to genotype. These methods were combined to generate a final SNP list.
Genotyping.
Patient DNA samples originated from whole blood. SNPs were genotyped using the SNPlex platform (Applied Biosystems). This platform allowed for the multiplexed genotyping of up to 48 SNPs per assay. Genotyping for targeted nsSNPs was accomplished using TaqMan SNP Genotyping Assays (Applied Biosystems).
Quantification of leptin and leptin receptor.
Leptin and soluble leptin receptor were assayed in serum from blood samples collected from the study children in the morning before their first meal. Measurements were done in 2008 from 141/185 children ages 10 to 13 years. Leptin was measured with the ELISA kit Human LEPTIN (Biosource), and leptin receptor with the Quantikine Human Leptin sR Immunoassay (R&D Systems).
qRT-PCR methods.
RNA was isolated from peripheral blood mononuclear cells using the PicoPure RNA Isolation Kit (Arcturus). The concentration of RNA was measured using the NanoDrop 1000 (Thermo Scientific) prior to reverse-transcription using Superscript II RT (Invitrogen). Quantitative real-time PCR was performed using the iCycler (BIO-RAD) and 2× QuantiFast SYBR Green PCR Master Mix (QIAGEN) with a total reaction volume of 25 μl. The PCR primers used for the LEPR gene were Hs_LEPR_1_SG QuantiTect Primer Assay (QIAGEN), which detects Homo sapiens leptin receptor (LEPR), transcript variant 1 (NM_002303), transcript variant 2 (NM_001003679), and transcript variant 3 (NM_001003680), and HS_LEPR_vb.2_SG QuantiTect Primer Assay (QIAGEN), which detects transcript variant 1 (NM_002303). Detailed transcript information is accessible via NCBI. Hypoxanthine phosphoribosyltransferase 1 (HPRT1) and peptidylprolyl isomerase A (PPIA) were used as endogenous controls. Resulting data were corrected for relative amplification efficiency via standard curves, and the relative cDNA concentration values were standardized and expressed in proportion to the ARG/GLN frequencies in the sample mean. The LEPR_1_SG and HS_LEPR_vb.2_SG values were then normalized to the mean of PPIA and HPRT1. A nonparametric ANOVA (Kruskal-Wallis test) was used to compare the 3 genotype groups for the 2 LEPR isoforms using GraphPad Prism (version 4.0a).
Statistics.
Allele frequencies for SNPs were estimated from the entire cohort by counting methods. Allelic association was calculated using a c2 test in the statistical program Haploview (v4.0), and only SNPs that had a P value of less than 0.05 after permutation testing (10,000 permutations) were considered significant. Haplotypes and pairwise linkage disequilibrium were also estimated in Haploview using a solid spine of LD (D′ = 0.80). Genotypic association for each SNP was compared between individuals for each clinical category using c2. The magnitude of the association between SNPs and the occurrence of E. histolytica infection was measured by the OR using polytomous logistic regression, which reflects the likelihood of having a particular clinical outcome given the observed genotype. Potential confounders of age, sex, and area within Mirpur were included in the multiple logistic regression models. Permutations in which the case category was shuffled (n = 10,000) were calculated for simple logistic regression analysis only to minimize the type 1 error, and these empirical P values are presented. All regression and Kaplan-Meier analyses were computed using Stata software (version 10.0; StataCorp). Population attributable risk was calculated using the equation 1 – ∑allG (ρG/ORG), where ORG represents the odds ratio and ρG represents cases with the genotype (i.e., AG/GG). The statistical package STATA v9.0 was used for the analysis. P < 0.05 was considered significant.
Animal model.
Mice carrying the humanized LEPR allele at codon 223 were generated as previously described (22). Briefly, the embryonic stem cell clone was injected into C57BL/6J blastocysts, and the founder progeny were crossed to 129P3/J mice. The F1 animals segregating for the targeted Lepr allele were intercrossed to generate the 223Q/Q, 223Q/R, and 223R/R animals used for E. histolytica infection. Animals were maintained under specific-pathogen–free conditions at the University of Virginia. All protocols were approved by the Institutional Animal Care and Use Committee. Mice were monitored closely for general well-being; those who were judged to be in a moribund state (seemingly sick, severely dehydrated, and complete loss of activity) were euthanized before the scheduled termination time, while the rest were sacrificed at day 3 after challenge. Mouse ceca were harvested and placed in Bouin’s fixative (Sigma-Aldrich) for H&E staining. To evaluate infection, 200 μl of cecal contents were assayed for amebic antigen using the E. histolytica II ELISA kit (TechLab) and 300 μl cultured in complete TYI-S-33 medium for 5 days. Statistical comparisons for infection rates were done using Fisher’s exact test; P ≤ 0.05 was considered significant.
Parasites and intracecal inoculation.
Trophozoites for intracecal injection were originally derived from laboratory strain HM1: IMSS (ATCC) that were sequentially passaged through the mouse cecum. Cecal contents were cultured in trypsin-yeast-iron (TYI-S-33) medium supplemented with Diamond Vitamin (JRH Biosciences), bovine serum (Sigma-Aldrich), and 100 U/ml penicillin plus 100 μg/ml streptomycin. For all intracecal inoculations, trophozoites were grown to the log phase and 2 × 106 trophozoites in 150 μl were injected intracecally after laparotomy, as described previously (36).
Immunohistochemical staining and histopathological examination.
Immunohistochemical staining for phosphorylated Akt and activated caspase-3 was performed with an ABC kit (Vector Laboratories), using anti-phospho-specific Akt (Ser473) (Cell Signaling Technology) and anti-human/mouse activated caspase-3 polyclonal antibody (R&D Systems).
To measure the amount of the p-Akt protein, all sections of each sample were scored according to the following criteria: 0, as no positive signal; 1, as a low but detectable signal, with a frequency less than 10% of the mucosal area; 2, as a moderate signal, with frequencies reaching 10%–50% of the mucosal area; and 3, as an intense signal with a frequency of over 50%. Samples were read in a blinded fashion and the score was averaged from all sections (5–9 sections per mouse) examined.
For apoptosis quantification, the total activated caspase-3–positive cells were counted and positive cells from IECs versus infiltrating cells were reported as positive cells per section on average. Histological scoring for mucosal destruction/ulceration was performed in a blinded fashion as previously published (36, 37): no mucosal damage was scored as 0, mild surface mucosal erosion scored as 1, focal ulceration or disrupted architecture scored as 2, and extensive mucosal damage and extension into deeper structures of the bowel wall as 3.
Quantitative RT-PCR.
Total RNA was extracted from the mouse cecum using the RNeasy Mini kit (QIAGEN) following the manufacturer’s instruction. DNase-treated RNA was reversely transcribed into first strand cDNA using SuperScript III reverse transcriptase (Invitrogen Life Technologies). qRT-PCR was performed with the iCycler iQ System (Bio-Rad Laboratories). GAPDH was used as an internal control. The following primers were used in this study: GAPDH, forward: 5′-GCTAAGCAGTTGGTGGTGCA-3′, reverse: 5′-TCACCACCATGGAGAAGGC-3′; BcL-2, forward: 5′-TGCACCTGACGCCCTTCAC-3′, reverse: 5′-AGACAGCCAGGAGAAATCAAACAG-3′; cyclin D3, forward: 5′-CTGGCCATGAACTACCTGGA-3′, reverse: 5′-CCAGGAAATCATGTGCAATC-3′.
Supplementary Material
Acknowledgments
We thank the children and their families in Mirpur, Dhaka, Bangladesh for their participation in this study. We acknowledge the insight and assistance of Joan E. Bailey-Wilson. This work was supported by NIH grants AI043596 (to W.A. Petri), R01 AI071373 (to E. Houpt), R01 DK056731 (to M.G. Myers), R01 DK52431-17 (to R.L. Leibel), and 5P30 DK26687-29 New York Obesity Research Center, Molecular Biology/Molecular Genetics Core (to R.L. Leibel and S.C. Chua Jr.), and in part by the intramural division of the National Human Genome Research Institute, the NIH, and the National Institutes of Allergy and Infectious Disease (to W.A. Petri).
Footnotes
Conflict of interest: William A. Petri Jr. has a licensing agreement for amebiasis diagnostics with TechLab Inc.; however, all royalties from this agreement are donated to the American Society of Tropical Medicine and Hygiene without benefit to William A. Petri Jr.
Citation for this article: J Clin Invest. 2011;121(3):1191–1198. doi:10.1172/JCI45294.
References
- 1. Black RE, et al. Maternal and child undernutrition: global and regional exposures and health consequences. Lancet. 2008;371(9608):243–260. doi: 10.1016/S0140-6736(07)61690-0. [DOI] [PubMed] [Google Scholar]
- 2. Caulfield LE, Richard SA, Black RE. Undernutrition as an underlying cause of malaria morbidity and mortality in children less than five years old. Am J Trop Med Hyg. 2004;71(2 suppl):55–63. [PubMed] [Google Scholar]
- 3. Tilg H, Moschen AR. Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat Rev Immunol. 2006;6(10):772–783. doi: 10.1038/nri1937. [DOI] [PubMed] [Google Scholar]
- 4. Myers MG, Cowley MA, Münzberg H. Mechanisms of leptin action and leptin resistance. Annu Rev Physiol. 2008;70:537–556. doi: 10.1146/annurev.physiol.70.113006.100707. [DOI] [PubMed] [Google Scholar]
- 5. Palacio A, Lopez M, Perez-Bravo F, Monkeberg F, Schlesinger L. Leptin levels are associated with immune response in malnourished infants. J Clin Endocrinol Metab. 2002;87(7):3040–3046. doi: 10.1210/jc.87.7.3040. [DOI] [PubMed] [Google Scholar]
- 6. Bruno A, Conus S, Schmid I, Simon HU. Apoptotic pathways are inhibited by leptin receptor activation in neutrophils. J Immunol. 2005;174(12):8090–8096. doi: 10.4049/jimmunol.174.12.8090. [DOI] [PubMed] [Google Scholar]
- 7. Fernández-Riejos P, Goberna R, Sánchez-Margalet V. Leptin promotes cell survival and activates Jurkat T lymphocytes by stimulation of mitogen-activated protein kinase. Clin Exp Immunol. 2008;151(3):505–518. doi: 10.1111/j.1365-2249.2007.03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lam QL, Liu S, Cao X, Lu L. Involvement of leptin signaling in the survival and maturation of bone marrow-derived dendritic cells. Eur J Immunol. 2006;36(12):3118–3130. doi: 10.1002/eji.200636602. [DOI] [PubMed] [Google Scholar]
- 9. Lo CK, et al. Leptin signaling protects NK cells from apoptosis during development in mouse bone marrow. Cell Mol Immunol. 2009;6(5):353–360. doi: 10.1038/cmi.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Papathanassoglou E, El-Haschimi K, Li XC, Matarese G, Strom T, Mantzoros C. Leptin receptor expression and signaling in lymphocytes: kinetics during lymphocyte activation, role in lymphocyte survival, and response to high fat diet in mice. J Immunol. 2006;176(12):7745–7752. doi: 10.4049/jimmunol.176.12.7745. [DOI] [PubMed] [Google Scholar]
- 11. De Rosa, V, et al. A key role of leptin in the control of regulatory T cell proliferation. Immunity. 2007;26(2):241–255. doi: 10.1016/j.immuni.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 12. Otero M, et al. Towards a pro-inflammatory and immunomodulatory emerging role of leptin. Rheumatology (Oxford). 2006. 45 8 944 950 . 10.1093/rheumatology/kel157 [DOI] [PubMed] [Google Scholar]
- 13. Ogunwobi OO, Beales IL. The anti-apoptotic and growth stimulatory actions of leptin in human colon cancer cells involves activation of JNK mitogen activated protein kinase, JAK2 and PI3 kinase/Akt. Int J Colorectal Dis. 2007;22(4):401–409. doi: 10.1007/s00384-006-0181-y. [DOI] [PubMed] [Google Scholar]
- 14. Sukhotnik I, et al. Leptin affects intestinal epithelial cell turnover in correlation with leptin receptor expression along the villus-crypt axis after massive small bowel resection in a rat. Pediatr Res. 2009;66(6):648–653. doi: 10.1203/PDR.0b013e3181be9f84. [DOI] [PubMed] [Google Scholar]
- 15. El Homsi M, et al. Leptin modulates the expression of secreted and membrane-associated mucins in colonic epithelial cells by targeting PKC, PI3K, and MAPK pathways. Am J Physiol Gastrointest Liver Physiol. 2007;293(1):G365–G373. doi: 10.1152/ajpgi.00091.2007. [DOI] [PubMed] [Google Scholar]
- 16. Brun P, et al. Increased intestinal permeability in obese mice: new evidence in the pathogenesis of nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2007;292(2):G518–G525. doi: 10.1152/ajpgi.00024.2006. [DOI] [PubMed] [Google Scholar]
- 17. Haque R, et al. Correlation of interferon-gamma production by peripheral blood mononuclear cells with childhood malnutrition and susceptibility to amebiasis. Am J Trop Med Hyg. 2007;76(2):340–344. [PubMed] [Google Scholar]
- 18. Petri WA, Mondal D, Peterson KM, Duggal P, Haque R. Association of malnutrition with amebiasis. Nutr Rev. 2009;67 suppl 2:S207–S215. doi: 10.1111/j.1753-4887.2009.00242.x. [DOI] [PubMed] [Google Scholar]
- 19. Mondal D, Haque R, Sack RB, Kirkpatrick BD, Petri WA. Attribution of malnutrition to cause-specific diarrheal illness: evidence from a prospective study of preschool children in Mirpur, Dhaka, Bangladesh. Am J Trop Med Hyg. 2009;80(5):824–826. [PMC free article] [PubMed] [Google Scholar]
- 20. Haque R, et al. Entamoeba histolytica infection in children and protection from subsequent amebiasis. Infect Immun. 2006;74(2):904–909. doi: 10.1128/IAI.74.2.904-909.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duggal P, et al. Influence of human leukocyte antigen class II alleles on susceptibility to Entamoeba histolytica infection in Bangladeshi children. J Infect Dis. 2004;189(3):520–526. doi: 10.1086/381272. [DOI] [PubMed] [Google Scholar]
- 22. Stratigopoulos G, et al. Functional consequences of the human leptin receptor (LEPR) Q223R transversion. Obesity (Silver Spring). 2009;17(1):126–135. doi: 10.1038/oby.2008.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang G, Ge H, Boucher A, Yu X, Li C. Modulation of direct leptin signaling by soluble leptin receptor. Mol Endocrinol. 2004;18(6):1354–1362. doi: 10.1210/me.2004-0027. [DOI] [PubMed] [Google Scholar]
- 24. Huang L, Wang Z, Li C. Modulation of circulating leptin levels by its soluble receptor. J Biol Chem. 2001;276(9):6343–6349. doi: 10.1074/jbc.M009795200. [DOI] [PubMed] [Google Scholar]
- 25. Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452(7186):423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 26. Boettner DR, et al. Entamoeba histolytica phagocytosis of human erythrocytes involves PATMK, a member of the transmembrane kinase family. PLoS Pathog. 2008;4(1):e8. doi: 10.1371/journal.ppat.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huston CD, Houpt ER, Mann BJ, Hahn CS, Petri WA. Caspase 3-dependent killing of host cells by the parasite Entamoeba histolytica. Cell Microbiol. 2000;2(6):617–625. doi: 10.1046/j.1462-5822.2000.00085.x. [DOI] [PubMed] [Google Scholar]
- 28. Bijian K, Takano T, Papillon J, Khadir A, Cybulsky AV. Extracellular matrix regulates glomerular epithelial cell survival and proliferation. Am J Physiol Renal Physiol. 2004;286(2):F255–F266. doi: 10.1152/ajprenal.00259.2003. [DOI] [PubMed] [Google Scholar]
- 29. Farooqi IS, et al. Clinical and molecular genetic spectrum of congenital deficiency of the leptin receptor. N Engl J Med. 2007;356(3):237–247. doi: 10.1056/NEJMoa063988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Becker SM, et al. Epithelial cell apoptosis facilitates Entamoeba histolytica infection in the gut. Am J Pathol. 2010;176(3):1316–1322. doi: 10.2353/ajpath.2010.090740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lord GM, Matarese G, Howard JK, Baker RJ, Bloom SR, Lechler RI. Leptin modulates the T-cell immune response and reverses starvation-induced immunosuppression. Nature. 1998;394(6696):897–901. doi: 10.1038/29795. [DOI] [PubMed] [Google Scholar]
- 32. Matarese G, Moschos S, Mantzoros CS. Leptin in immunology. J Immunol. 2005;174(6):3137–3142. doi: 10.4049/jimmunol.174.6.3137. [DOI] [PubMed] [Google Scholar]
- 33. Guo X, et al. Leptin signaling in intestinal epithelium mediates resistance to enteric infection by Entamoeba histolytica [published online ahead of print December 1, 2010].Mucosal Immunol . doi:10.1038/mi.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. WHO. Amebiasis. Weekly Epidemiologic Record. 1997;72:97–99. [Google Scholar]
- 35. Mondal D, Petri WA, Sack RB, Kirkpatrick BD, Haque R. Entamoeba histolytica-associated diarrheal illness is negatively associated with the growth of preschool children: evidence from a prospective study. Trans R Soc Trop Med Hyg. 2006;100(11):1032–1038. doi: 10.1016/j.trstmh.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 36. Houpt ER, et al. The mouse model of amebic colitis reveals mouse strain susceptibility to infection and exacerbation of disease by CD4+ T cells. J Immunol. 2002;169(8):4496–4503. doi: 10.4049/jimmunol.169.8.4496. [DOI] [PubMed] [Google Scholar]
- 37. Hamano S, Asgharpour A, Stroup SE, Wynn TA, Leiter EH, Houpt E. Resistance of C57BL/6 mice to amoebiasis is mediated by nonhemopoietic cells but requires hemopoietic IL-10 production. . J Immunol. 2006;177(2):1208–1213. doi: 10.4049/jimmunol.177.2.1208. [DOI] [PubMed] [Google Scholar]
- 38. Consortium IH. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.