Abstract
The activity, localization and fate of many cellular proteins are regulated through ubiquitination, a process whereby one or more ubiquitin (Ub) monomers or chains are covalently attached to target proteins. While Ub-conjugated and Ub-associated proteomes have been described, we lack a high-resolution picture of the dynamics of ubiquitination in response to signaling. In this study, we describe the epidermal growth factor (EGF)-regulated Ubiproteome, as obtained by two complementary purification strategies coupled to quantitative proteomics. Our results unveil the complex impact of growth factor signaling on Ub-based intracellular networks to levels that extend well beyond what might have been expected. In addition to endocytic proteins, the EGF-regulated Ubiproteome includes a large number of signaling proteins, ubiquitinating and deubiquitinating enzymes, transporters and proteins involved in translation and transcription. The Ub-based signaling network appears to intersect both housekeeping and regulatory circuitries of cellular physiology. Finally, as proof of principle of the biological relevance of the EGF-Ubiproteome, we demonstrated that EphA2 is a novel, downstream ubiquitinated target of epidermal growth factor receptor (EGFR), critically involved in EGFR biological responses.
Keywords: EGF, network, proteomics, signaling, ubiquitin
Introduction
Post-translational modifications (PTMs) control the function of many proteins and are crucial to the regulation of many cellular processes, as exemplified by the role of phosphorylation in signaling. The reversible addition of a small phosphate group to protein substrates allows the propagation of information through multiple mechanisms, including activation/deactivation of the enzymatic properties of phosphorylated substrates, regulation of their subcellular localization, and their recognition by specific domains present in partner proteins (Seet et al, 2006). In another instance of PTM, a more complex molecule, ubiquitin (Ub), is appended by E3 ligases to a multitude of substrates, thereby modulating their function, localization and protein/protein interaction abilities (Mukhopadhyay and Riezman, 2007; Ravid and Hochstrasser, 2008). Deubiquitinating enzymes (DUBs) revert Ub conjugation, thus ensuring a dynamic equilibrium between pools of ubiquitinated and deubiquitinated proteins (Amerik and Hochstrasser, 2004). Particularly relevant to signaling is the ability of the Ub modification to induce de novo protein/protein interactions, similarly to phosphorylation, through the recognition of ubiquitinated proteins by proteins harboring Ub-binding domains (Hicke et al, 2005; Hurley et al, 2006). This mechanism sits at the heart of several signaling cascades (Mukhopadhyay and Riezman, 2007; Woelk et al, 2007; Chen and Sun, 2009), and is tightly controlled within the cell by endogenous and exogenous signals, such as DNA damage and growth factor stimulation, respectively (Chen and Sun, 2009).
In this latter instance, one of the best-characterized model systems is represented by the epidermal growth factor (EGF)-induced pathway. Upon EGF stimulation, a variety of proteins are subject to Ub modification. These include the EGF receptor (EGFR), which undergoes both multiple monoubiquitination (Haglund et al, 2003) and K63-linked polyubiquitination (Huang et al, 2006), as well as components of the downstream endocytic machinery, which are modified by monoubiquitination (Polo et al, 2002; Mukhopadhyay and Riezman, 2007). The impact of ubiquitination on receptor internalization, intracellular sorting and ultimate metabolic fate has been characterized in detail for various receptors, including the EGFR (Acconcia et al, 2009).
Little is known, however, about the wider impact of EGF-induced ubiquitination on cellular homeostasis and on the pleiotropic biological functions of the EGFR. A decisive step in this direction would be the acquisition of the repertoire of proteins that are ubiquitinated upon EGF stimulation; i.e., the EGF-Ubiproteome. This study was undertaken to address this issue. Recent advances in quantitative mass spectrometry have allowed the study of PTMs on a global scale (Jensen, 2006; Choudhary et al, 2009). In this study, we combined two different purification procedures with high resolution, high accuracy MS, coupled to an efficient quantitation strategy, to obtain the first view of the EGF-induced Ubiproteome.
Results
Purification of Ub-conjugated proteins
Owing to the low abundance and labile nature of ubiquitinated proteins, the most critical step in their identification is the enrichment and purification procedure. This is particularly relevant in our case, as we are interested in the EGF-induced Ubiproteome. Under these conditions, ubiquitination is a rapid, dynamic process, and ubiquitinated substrates are present in the cell at low stoichiometries and in a time-limited manner. To maximize the recovery of ubiquitinated substrates, therefore, we used an integrated approach based on two different purification strategies and two cellular model systems (see schematic representation in Supplementary Figure S1).
In the first purification scheme, a mouse monoclonal antibody (FK2) that recognizes both mono- and polyubiquitinated species, but not free Ub, was used to immunopurify ubiquitinated proteins from HeLa cell lysates. This approach (hereafter, the ‘endogenous’ approach) allows the purification of proteins modified by endogenous Ub, in the absence of any manipulation of the cellular system (see Supplementary information and legend to Supplementary Figure S1 for details).
In an alternative strategy (hereafter, the tandem affinity purification ‘TAP’ approach), we exploited TAP. We developed a tandem affinity tag, consisting of a hexahistidine and a FLAG sequence fused to Ub (FLAG-His-Ub). This construct was transfected into B82L-EGFR cells, a mouse fibroblast cell line expressing human EGFR that has been widely used to study EGF-dependent signaling (Chen et al, 1989). The TAP method should allow the isolation of highly purified ubiquitinated proteins, as fully denaturing conditions (8M urea) are used, which dissolve most weak protein/protein interactions.
To overcome possible non-physiological and/or toxic effects of the overexpression of Ub (Tagwerker et al, 2006), we chose a TET-on inducible system (see Supplementary information for details). The level of expression of tagged Ub at different time points was assessed by immunofluorescence and immunoblot analysis (Figure 1A and B). Detection of high-molecular-weight Ub signals confirmed that the tagged Ub is functional and is conjugated to proteins (Figure 1B). Tagged Ub was expressed at one-tenth of the level of endogenous Ub (Figure 1C), sufficient to maintain the inducibility of the EGF-mediated process, as monitored by monoubiquitination of Eps15 (Figure 1D and Supplementary information). Finally, B82L-EGFR cells overexpressing FLAG-His-Ub displayed the same growth rate as untransfected cells (data not shown), indicating that the expression of the tagged Ub had no major toxic effect. We note that the ‘endogenous’ approach and the ‘TAP’ approach are not directly comparable as they are performed in different cellular systems. This was due to our inability to select a stable HeLa cell line expressing this tagged version of Ub. However, we reasoned that the use of a second cellular system, which differs both in terms of species and of tissue origin, might in some respects be advantageous, as it could lead to the identification of a common repertoire of ubiquitinated substrates.
Figure 1.
Preparation of samples for MS-based proteomics analysis. (A) B82L-EGFR cells stably transfected with FLAG-6His-Ub were fixed at the indicated time points after doxycycline induction (4 μg/ml) and stained with anti-FLAG antibody (red). Nuclei were DAPI stained (blue). (B) Parallel samples, treated as in A, were lysed and immunoblotted (IB) as indicated. (C) Lysates as in B, were IB with anti-Ub antibody to compare the levels of expression of endogenous and tagged Ub. (D) B82L-EGFR cells stably transfected with FLAG-6His-Ub were induced with doxycycline for 48 h and stimulated with EGF or left untreated. Lysates (2 mg) were immunoprecipitated (IP) and IB as indicated. (E, F) Quality control of a representative ‘TAP’ purification (experiment FW1, see Experimental Procedures and Results). B82L-EGFR cells induced for 48 h with doxycycline were stimulated with EGF (100 ng/ml) for 10 min or left untreated. Total cell lysates were analyzed by IB with the indicated antibodies before mixing them (1:1) for the purification (E). Aliquots from each step of the purification procedure were analyzed by IB with anti-FLAG antibody. The amount of loaded samples is expressed as % of total sample volume (F). (G, H) Quality control of a representative ‘endogenous’ purification (experiment FW2, see Experimental Procedures and Results). Lysates from EGF-treated (100 ng/ml, 10 min) or untreated HeLa cells were analyzed as in E (G). The purification procedure was controlled as in F, except that an anti-Ub antibody (P4D1) was used (H). Note that the EGF-induced increase in the total ubiquitinated protein content is not proportional to the increase in protein tyrosine phosphorylation. This is expected as the vast majority of Ub-modified species in the cell is represented by constitutively ubiquitinated proteins, destined to proteasomal degradation.
For both approaches, the purification procedure was carefully set up and the yield was calculated to be 8% for the ‘endogenous’ approach and 20% for the ‘TAP’ approach (see Supplementary information and legend to Supplementary Figure S1 for details). Representative quality control experiments are presented in Figure 1E–H.
Of note, we performed a mock purification for the ‘endogenous’ approach by omitting the FK2 Ab in the purification scheme, and an I-DIRT experiment in the case of the ‘TAP’ approach (see ‘control endogenous’ and ‘I-DIRT TAP’ sheets in Supplementary Table S1 and Supplementary Figure S2). These experiments were used to filter out proteins that were non-specifically recovered during the purification procedures (see Supplementary information for details).
Identification of steady-state Ubiproteomes
Our ultimate goal was the identification of the EGF-induced Ubiproteome. Thus, we employed high resolution, high accuracy MS (Olsen et al, 2005) combined with stable isotope labeling with amino acids in cell culture (SILAC) (Ong and Mann, 2006) to distinguish the EGF-dependent ubiquitination events from the high background of steady-state ubiquitinated proteins. This strategy allowed the identification and quantitation of the steady-state HeLa and B82L-EGFR Ubiproteomes and the specific EGF-Ubiproteomes in a single experiment. We chose a single time point of EGF stimulation (10 min), and performed three biological replicates of both the ‘endogenous’ and the ‘TAP’ purifications. Remarkably, 73.2% (endogenous) and 85.2% (TAP) of proteins were identified in at least two experiments, indicating a high level of reproducibility. Experiment size and features are reported in Supplementary Table S1 and in Supplementary Figure S2.
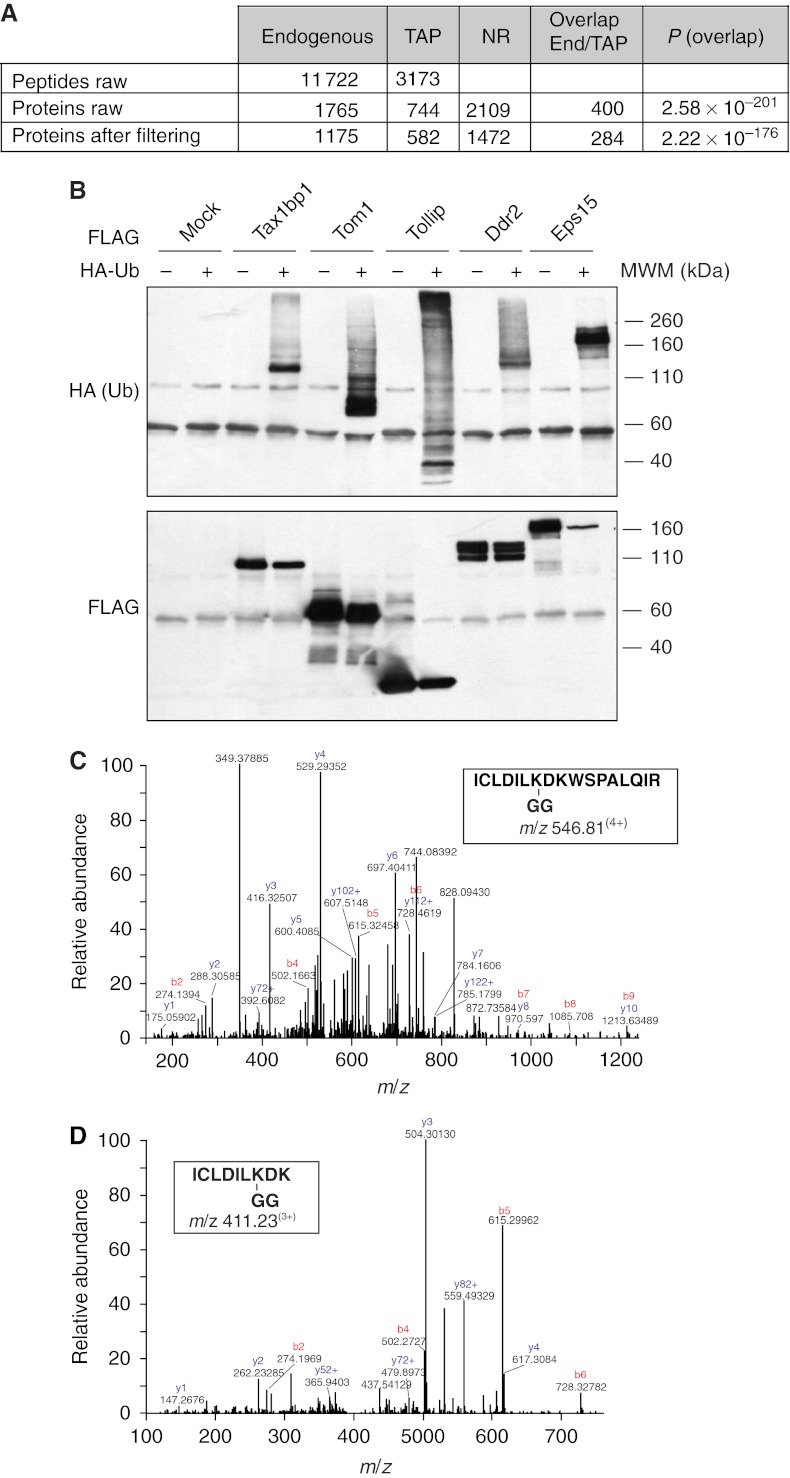
As an initial step in our analysis, we employed a multilayered strategy to define the size and specificity of the identified Ubiproteomes (Figure 2A). To obtain ‘high-confidence data sets’ of quantified proteins, we adopted four filtering criteria described in detail in the Experimental Procedures. By these criteria, from 11 722 non-redundant (NR) peptide sequences (corresponding to 1765 proteins) identified in the endogenous approach, we assembled a SILAC-based proteome comprising 1175 unambiguously identified and quantified proteins (Figure 2A and Supplementary Table S1). In the case of the TAP approach, we unambiguously identified 582 proteins (from 3173 NR peptides, corresponding to 744 proteins; Figure 2A and Supplementary Table S1). In both cases, the identification of short-lived proteins and monoubiquitinated proteins demonstrates that the two strategies are effective. In addition, the retrieval of nuclear proteins together with histones argues for the efficiency of the solubilization procedure. This said, we acknowledge that the identified Ubiproteomes are unlikely to represent the full repertoire of ubiquitinated proteins found in mammalian cell, although they are the most complete set of ubiquitinated proteins identified so far (Supplementary Figure S3).
Figure 2.
The steady-state Ubiproteome. (A) Size and features of steady-state Ubiproteomes identified with the endogenous and TAP approaches. NR, number of non-redundant proteins from the two approaches. Overlap End/TAP, number of proteins identified in common between the two approaches. P, significance P-value of the overlap (Fisher's exact test). (B) Validation of candidates (see also additional examples in Supplementary Figure S3). Cells (293T) were transfected with the indicated constructs and lysed after 24 h. Lysates were IP anti-FLAG (candidates) and IB with anti-HA (Ub) antibody to evaluate the ubiquitination level of the candidates. (C, D) MS/MS spectra of the identified ubiquitination site of ubiquitin-conjugating enzyme Ube2N (Ubc13). (C) Fragmentation pattern of the quadruple-charged tryptic peptide of Ube2N derived from HeLa cellular extracts. (D) Fragmentation pattern of the triply charged tryptic peptide of Ube2N derived from B82L-EGFR cellular extracts. The y-series of ions (C terminus containing fragments) are labeled in blue and b-series (N terminus containing fragments) are labeled in red.
To validate our Ubiproteomes, we tested several candidates by direct immunoprecipitation (IP; Figure 2B and Supplementary Figure S3). On the basis of these results, we estimated a false-positive rate of ∼5% for the ‘endogenous’ approach (2 out of 38 tested candidates were not validated by IP) and ∼3% for the TAP approach (1 out of 30 candidates was not validated, see legend to Supplementary Figure S3 for details). The non-validated proteins possibly represent the fraction of Ub-interacting proteins co-purified during the procedure, as they were absent in the control purifications (Supplementary Table S1).
We also analyzed the overlap between the endogenous and the TAP Ubiproteomes. One limitation of our study is that the two approaches were optimized in two different cell types. Nevertheless, 284 proteins were identified in common between the two approaches (P=2.22 × 10−176, Figure 2A and Supplementary Table S1). This high degree of overlap is remarkable given that the two cellular settings are very different (a human cervical cancer cell line versus a mouse fibroblast cell line).
Of note, we identified 31 ubiquitination sites in 21 target proteins, some of which were previously unknown. The complete list of identified sites is shown in Supplementary Table S1. A representative instance of these findings is the E2-conjugating enzyme Ubc13, for which the same modified K was found in both the human and the mouse cell line (Figure 2C and D, see also legend to Supplementary Table S1 for details).
The EGF-Ubiproteome
We next exploited the quantitative information embedded in the SILAC data to identify the EGF-regulated Ubiproteome. Protein quantitation was performed automatically using MaxQuant, as described previously (Cox and Mann, 2008). Ratios for proteins were determined as the median of all measured peptide ratios for a given protein, to minimize the effect of outliers (Supplementary Figure S4). To identify proteins that were most significantly regulated by EGF, we employed a stringent three-tiered selection process. Starting from the two steady-state Ubiproteomes, we discarded proteins with a P-value >0.1 (Significance B, see Cox and Mann, 2008) and a coefficient of variability >10 (filter A in Figure 3A, see Supplementary information for details). Finally, we applied a manual curation step selecting only those proteins displaying the same trend of regulation in the experimental replicates (filter B, Figure 3A). Additionally, we verified that protein levels did not change upon EGF stimulation in the whole proteomes (Supplementary Figure S5). By these stringent criteria, we concluded that ∼15% of the steady-state Ubiproteome was EGF regulated at 10 min after stimulation; 176 of 1175 proteins in the endogenous approach and 105 of 582 proteins in the TAP approach (Figure 3A and Supplementary Table S2). Interestingly, both hyper- and hypoubiquitinated proteins were detected (134 hyper- and 42 hypoubiquitinated in the endogenous approach, 58 hyper- and 47 hypoubiquitinated in the TAP approach; Figure 3B and C and Supplementary Table S2), indicating that EGFR-mediated signaling can modulate the Ub network in both directions. This finding is supported by the presence of both ubiquitinating and deubiquitinating enzymes in the EGF-Ubiproteome (see below).
Figure 3.

The EGF-regulated Ubiproteome. (A) Determination of EGF-regulated ubiquitinated proteins. We discarded from the steady-state Ubiproteomes, proteins with a significance B>0.1 and a coefficient of variability across replicates >10 (Filter A), and proteins showing a different trend of regulation in replicate experiments (Filter B). NR, number of non-redundant proteins from the two approaches. Overlap End/TAP, number of proteins identified in common between the two approaches. P, significance P-value of the overlap (Fisher's exact test). Hyper- and hypoubiquitinated proteins in the EGF-Ubiproteomes. (B, C) The 176 human (B) and the 105 mouse (C) EGF-regulated proteins were clustered across the three experimental replicates (FW1, FW2, REV). Side color bar represents the log2 of the normalized protein ratio (H, EGF treated versus L, untreated). Red, positive ratio (hyperubiquitinated); blue, negative ratio (hypoubiquitinated); and Black, missing data. (D) Kinetics of Eps15 monoubiquitination in HeLa and B82L-EGFR cells. Serum-starved HeLa (left panel) or B82L-EGFR (right panel) cells were stimulated with EGF (100 ng/ml) for the indicated time points or left untreated. Lysates (1 mg) were IP and IB with the indicated antibodies. Arrows on the lower panels indicate the ubiquitinated form of Eps15. (E, F) PANTHER analysis of the human (E) and the mouse (F) EGF-Ubiproteomes. Enrichment plots of ontology terms (Biological process, x axis) built on the list of EGF-Ubiproteome proteins. The percentage enrichment, based on the ratio between the number of SILAC proteins and the total number of proteins grouped in each term, is plotted on the right y axis (red histograms). The expected enrichment (gray histograms) is obtained from the ratio between the total number of genes in the term and the total number of genes in the genome. The statistical significance between observed and expected enrichments (scattered line) was determined by Fisher's exact test and is plotted on the left y axis as −log10 of P-values; the higher the value, the more significant the enrichment (−log10 of P=0.05 is 1.30). In bold, the 12 pathways found in both EGF-Ubiproteomes (three are closely related: protein/amino-acid biosynthesis; intracellular/other intracellular protein traffic; fatty acid biosynthesis/fatty acid and steroid metabolism).
The major limitation of our study is that our EGF-Ubiproteomes were not time resolved. Indeed, while we identified virtually all proteins known to be ubiquitinated upon EGF stimulation in both Ubiproteomes, only 16 out of 92 proteins were commonly classified as regulated at 10 min of EGF stimulation (when the proteomic analysis was performed). Of these, 14 proteins (10 hyperubiquitinated and 4 hypoubiquitinated) were regulated in the same direction in both approaches (Supplementary Table S1). Twenty-seven additional proteins were found to be regulated only in the endogenous approach (24 hyperubiquitinated and 3 hypoubiquitinated), while 49 additional proteins were identified only in the TAP approach (23 hyperubiquitinated and 26 hypoubiquitinated; see Supplementary Table S1). While cellular specificity may account for some of these variations (Figure 3A), different kinetics of ubiquitination in the two cellular systems might also affect the extent of the overlap. As a case in point, the endocytic adaptor protein Eps15 is ubiquitinated following EGF stimulation (Polo et al, 2002). Eps15 was present in the B82L-EGFR (TAP) but not in the HeLa (endogenous) EGF-Ubiproteome (Supplementary Table S2). Immunoblot analysis revealed that the kinetics of Eps15 ubiquitination upon EGF stimulation differed in the two cellular systems: at 10 min Eps15 was ubiquitinated in B82L-EGFR, but not in HeLa, cells (Figure 3D). Similar results were obtained for Rabex-5 and Hgs (data not shown).
The concept that the two EGF-Ubiproteomes might represent different time-resolved snapshots of the same network in two cellular systems is further supported by results derived from protein ontology analysis. We classified the EGF-regulated Ubiproteomes by PANTHER (Protein ANalysis THrough Evolutionary Relationships; Wiesner et al, 2007), and analyzed the enrichment of ontology terms. We identified 29 biological process (BP) terms as enriched (P-value <0.05) in the endogenous data set and 17 BP terms in the TAP data set (Figure 3E and F). Nine identical and three closely related BP terms were found in both EGF-Ubiproteomes: 41 and 70% of BP terms in the endogenous and TAP Ubiproteomes, respectively (reported in bold in Figure 3E and F). Therefore, regardless of the different experimental strategies employed and, more importantly, of the different experimental models, the two EGF-regulated Ubiproteomes show a high level of conservation in the cellular mechanisms that they represent.
Chain topology of the EGF-regulated Ubiproteome
To evaluate possible changes in the relative abundance of the different chains upon EGF stimulation, we quantified the Ub ‘signature’ peptides by SILAC (see Meierhofer et al, 2008 and Supplementary Table S3). With the TAP approach, MS analysis revealed an increase in the K63-, K11- and K6-chain modifications after EGF stimulation (Supplementary Table S3 and Supplementary Figure S6), whereas with the endogenous approach only K63 linkages accumulated, although to a lesser degree (Supplementary Table S3, see its legend for details). A possible explanation for this discrepancy is that the tagged Ub could affect the activity of specific E3s or DUBs, due to the extra N terminus present in the molecule. This would cause indirect changes in the level of specific chain linkages. To evaluate this possibility, we quantified the Ub ‘signature’ peptides before and after FLAG-His-Ub induction. No significant changes were evident when comparing the two conditions, indicating that, at least in our controlled settings, the expression of tagged Ub per se does not change the level of specific Ub chains (Supplementary Figure S7).
These initial, yet not conclusive, results prompted us to validate the MS analysis using the recently developed K48 and K63 linkage-specific antibodies (Newton et al, 2008). A strong colocalization of EGFR-containing vesicles and the anti-K63 antibody was observed upon EGF stimulation, whereas no colocalization was visible with the K48-specific antibody (Figure 4A). The FK2 antibody, which recognizes all types of poly-Ub chains equally well (Supplementary Figure S8), displayed an intermediate phenotype (Figure 4A).
Figure 4.
EGF induces K63-specific ubiquitination. (A) HeLa cells grown on coverslips were serum-starved for 4 h and treated for 10 min at 37°C with rhodamine-EGF (0.5 μg/ml, red) or left untreated (left panels). Ub (green) was detected with the FK2 antibody that recognizes both mono- and poly-Ub (upper panel) or antibodies that specifically recognize K63- or K48-linked Ub chains (middle and lower panels, respectively). Confocal images are shown. Blue, DAPI staining. Bar, 18 μm. (B) Lysates (1 mg) from HeLa cells stimulated with EGF (100 ng/ml) for the indicated times were subjected to IP with K63-Ub or K48-Ub-chain-specific antibodies or the FK2 antibody. IPs and lysates (50 μg/lane) were IB with the indicated antibodies. P4D1 antibody was used for the anti-Ub IB. Asterisk, unspecific band. Note that both spliced forms of Nedd4L appear to be ubiquitinated.
We then assessed some validated hits from our EGF-Ubiproteome by IP with linkage-specific antibodies and immunoblot analysis. Upon EGF stimulation, EGFR is almost exclusively modified by K63-linked chains (Figure 4B), in agreement with previous findings (Huang et al, 2006). Cbl, which is degraded upon EGF stimulation (Magnifico et al, 2003), is also strongly modified by K63 chains (Figure 4B). Notably, we also validated two novel EGF-induced ubiquitinated targets, namely, Nedd4L and SLC3A2, and showed that they carry almost exclusively K63-linked chains (Figure 4B). These results strengthen the idea that K63 might not only be the preferred signal for receptor internalization as previously suggested (Lauwers et al, 2010), but also be the major Ub-based signal transmitted by the active EGFR.
Network analysis of the EGF-regulated Ubiproteome
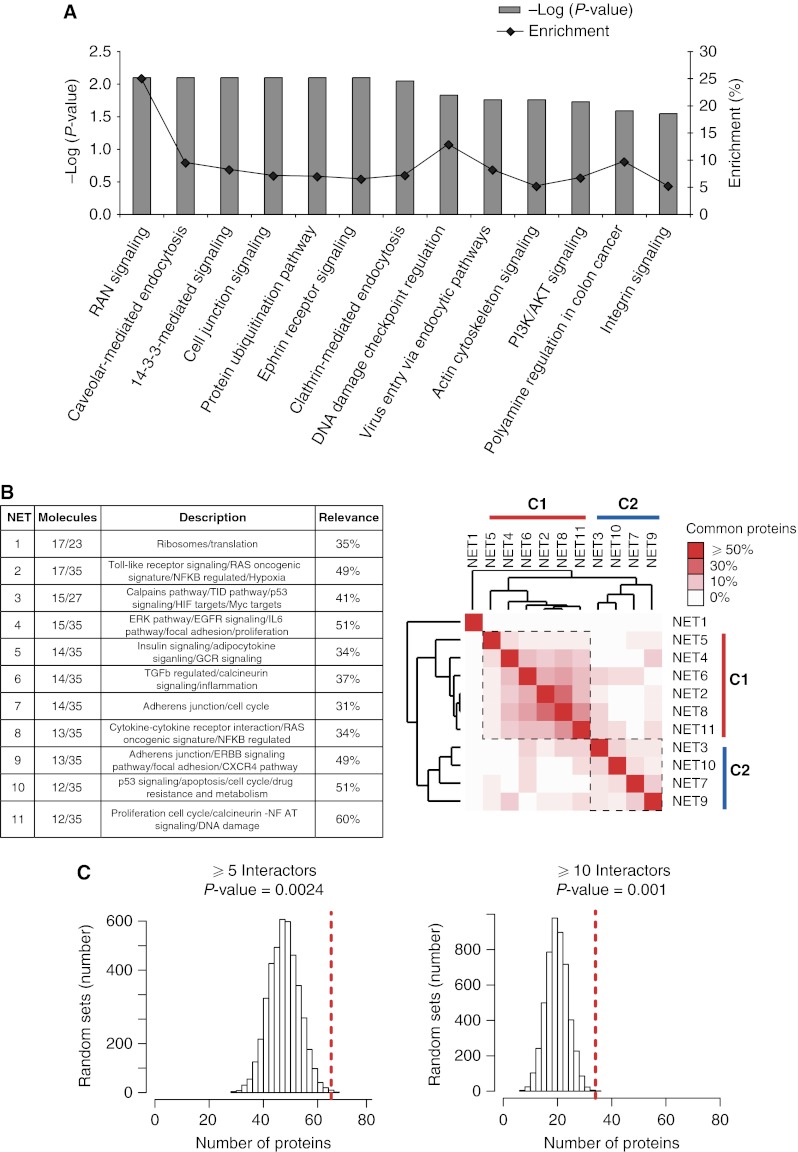
To obtain a higher-resolution molecular picture of the EGF-regulated Ub network, we analyzed the NR-EGF-Ubiproteome (265 proteins, Supplementary Table S2) through the Ingenuity Pathways Knowledge Software (Ingenuity® Systems, http://www.ingenuity.com) and looked for enrichment of canonical pathways. We identified 85 proteins that were significantly enriched in 39 pathways (P-value <0.05, Fisher's exact test), of which 13 pathways (67 proteins) remained significant after multiple test correction (P-value <0.05, Benjamini and Hochberg False Discovery Rate; Figure 5A). In addition to well-established liaisons (such as those with clathrin and caveolar endocytosis, or virus entry pathways), the EGF-Ubiproteome intersects many circuitries of intracellular signaling, suggesting crosstalk between EGFR-activated pathways and other signaling pathways through the Ub network.
Figure 5.
Pathway analysis of the EGF-Ubiproteome. (A) Ingenuity pathway analysis. Enrichment plots of canonical pathways (x axis) built on the list of 265 EGF-regulated proteins Ubiproteome (sum of the endogenous and the TAP approaches). The percentage enrichment, based on the ratio between the number of SILAC regulated proteins and the total number of proteins annotated in each pathway, is plotted on the right y axis (black line). The significance of the enrichment (gray bars) was calculated using the Benjiamini Hochberg multiple testing correction and is plotted on the left y axis as −log10 of P-values; the higher the value, the more significant the enrichment (−log10 of P=0.05 is 1.30). (B) Network analysis of the EGF-Ubiproteome. Left panel, list of the 11 networks that reach high statistical significance (P<0.0001, see Supplementary information). Network, network ID number. Molecules, number of EGF-Ubiproteome proteins/total number of network proteins. Description, functional category based on MSigDB analysis. Relevance, percentage of proteins in functional categories relative to total proteins in the network. Right panel, unsupervised hierarchical clustering of the identified networks. Clusters were generated based on the number of proteins in common between networks divided by the total number of proteins present in each network (Supplementary Table S3). The percentage of shared proteins is indicated by color code (white, 0%; red, ⩾50%). (C) Hub analysis. The number of random sets of proteins containing hubs (proteins with more ⩾5, 10 interactors as annotated in the BioGRID human interaction database (Stark et al, 2006)) is displayed. The red dashed line indicates the position in the distribution of the EGF-Ubiproteome (265 proteins). y axis, number of random sets containing hubs (defined as above), x axis number of proteins defined as hubs. P-values indicate significance of the enrichment of hubs in the EGF-Ubiproteome based on this ‘null’ distribution.
We further organized the EGF-Ubiproteome into distinct interaction networks through the Ingenuity Pathways Knowledge Software to predict how the Ub modification might influence the molecular crosstalk between proteins that interact biochemically and/or genetically. The proteins of the EGF-Ubiproteome are grouped into 30 networks, of which 11 reached statistical significance (P<0.0001, random permutation test, Figure 5B). Interestingly, unsupervised clustering of these 11 networks, based on the number of common proteins, revealed two main clusters (C1 and C2, Figure 5B). Functional annotation of molecules in these networks through the Molecular Signature Database (MSigDB) (Subramanian et al, 2007) revealed that cluster C1 is enriched in proliferation and inflammation signatures, whereas cluster C2 contains networks enriched in apoptosis, adhesion and cell cycle signatures. Interestingly, network 1 does not belong to any cluster (i.e., no proteins are shared with other networks) and is specifically enriched in ribosomal proteins (see Figure 5B and Supplementary information for details).
The ability of EGF-Ubiproteome proteins to nucleate clusters of interactions involved in diverse functions suggests that these proteins might act as organizational ‘hubs’, proteins that can establish multiple protein/protein interactions and thereby regulate the organization of networks. This is indeed the case, as proteins of the EGF-Ubiproteome displayed significantly higher connectivity than randomly generated lists of proteins (5000 lists were tested; Figure 5C and Supplementary Table S3). In total, 65 hubs (defined as proteins with ⩾5 interactors) were identified in the EGF-Ubiproteome (Supplementary Table S3). Among them, Hgs and Cbl, which have already been demonstrated to be critical for many intracellular signaling networks (Schmidt and Dikic, 2005; Zwang and Yarden, 2009), displayed the highest connectivity.
Intersection of the EGF-induced Ubi- and phosphotyrosine proteomes
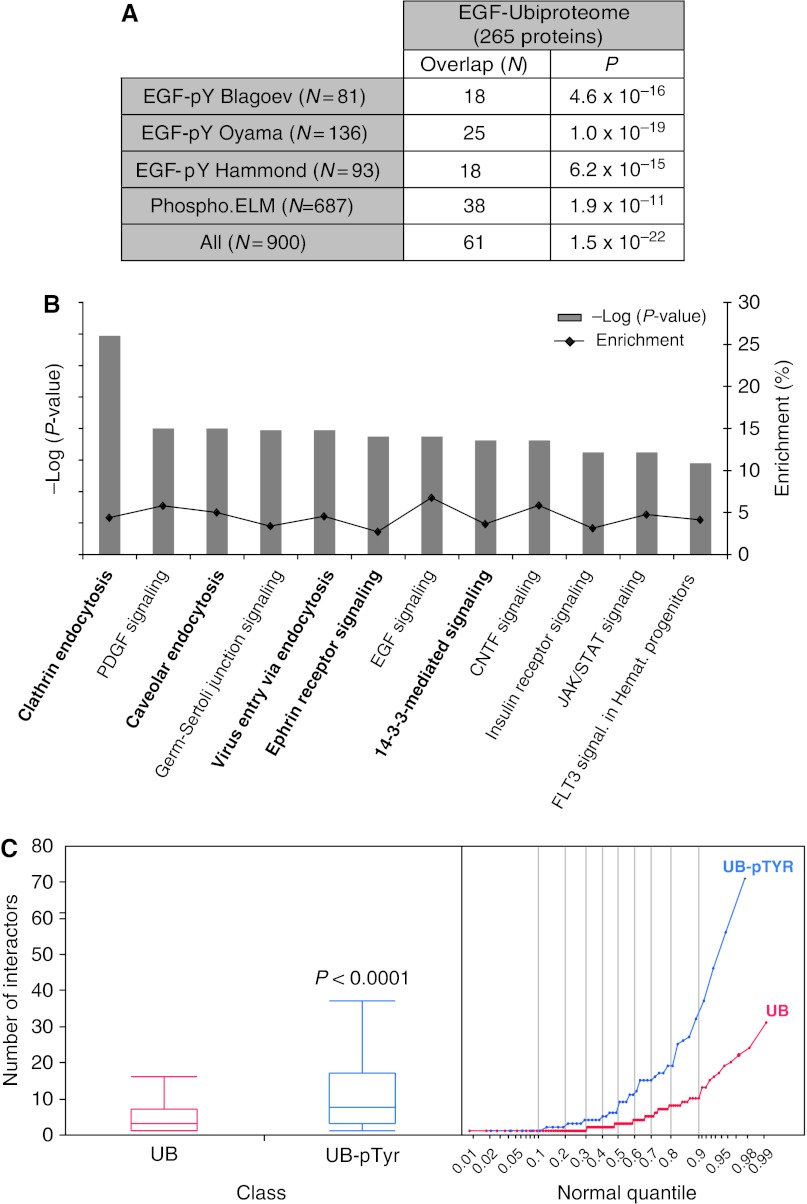
EGF binding to its receptor triggers a series of phosphorylation events that culminates in transcriptional activation and the mitogenic response. Proteins that undergo EGF-triggered phosphorylation have recently been described in three EGF-induced phosphotyrosine (pY) proteomes (Blagoev et al, 2004; Oyama et al, 2009; Hammond et al, 2010). A comparison of our EGF-Ubiproteome with these EGF-pY proteomes, as well as with the phospho.ELM database (Diella et al, 2008) that contains experimentally validated pY-containing proteins, revealed a significant overlap between ubiquitinated and pY proteins (Figure 6A, Supplementary Table S3). In total, 23% (61 of 265) of the EGF-Ubiproteome proteins are also tyrosine phosphorylated (Figure 6A, Supplementary Table S3).
Figure 6.
EGF-Ubiproteome and EGF-pYproteome. Comparison of the EGF-Ubiproteome (265 proteins) with two EGF-induced phosphotyrosine (pY) proteomes (Blagoev et al, 2004; Oyama et al, 2009; Hammond et al, 2010), and a pY-proteome database, phosho.ELM database (Diella et al, 2008). (A) The number of overlapping proteins (N) and the significance of the overlap (P: P-values calculated by Fisher's exact test) are shown. List of overlapping proteins is reported in Supplementary Table S3. (B) Ingenuity pathway analysis. Enrichment plots of canonical pathways (x axis) built on the list of 55 EGF-induced Ub and pY-modified proteins. The percentage of enrichment (black line) is plotted on the right y axis. The significance of the enrichment (gray bars) was calculated using the Benjiamini Hochberg multiple testing correction and is plotted on the left y axis as the −log10 of P-values. In bold, enriched pathways found also in the list of 265 EGF-Ubiproteome (Figure 5). (C) Enrichment of hubs in the list of 55 EGF-induced Ub and pY-modified proteins, or 265 EGF-induced Ub proteins. Only proteins with five interactors or more, as annotated in the BioGRID human interaction database (Stark et al, 2006), were considered for the analysis. Box plots and normal quantile plots are shown. Statistical significance of the difference in enrichment was calculated using Wilcoxon–Kruskal Wallis tests, one-way χ2-approximation (JMP 5.1 software, SAS).
Pathway analysis of these 61 Ub/pY-containing proteins revealed a significant enrichment in endocytic and signal-transduction pathways (Figure 6B). Finally, ‘hub analysis’ revealed that Ub/pY-containing proteins are enriched in highly connected proteins to an even greater extent than Ub-containing proteins alone (Figure 6C). These data point to a complex interplay between the Ub and pY networks and suggest that the flow of information from the receptor to downstream signaling molecules is driven by two complementary and interlinked enzymatic cascades: kinases/phosphatases and E3 ligases/DUBs.
Crosstalk between the EGFR and EphA2 signaling receptors
To provide a proof of principle of the biological relevance of our findings, we focused on EphA2, a receptor tyrosine kinase involved in development and often overexpressed in cancer (Pasquale, 2008). We started from the observation that proteins of the EGF-Ubiproteome are indeed enriched in the Ephrin receptor signaling pathway(s) (Figure 5A) and that EphA2 displayed a ratio of 2.67 (Supplementary Table S2).
As an initial step, we validated the MS data. Upon EGF stimulation, we observed an increase in both the tyrosine phosphorylation and ubiquitination of EphA2, indicating a crosstalk between the two receptors (Figure 7A and Supplementary Figure S9A). Interestingly, the crosstalk was unidirectional, as stimulation with ephrin-A1 or -A5/Fc did not lead to EGFR activation (Figure 7A).
Figure 7.

EphA2 is a novel EGF-induced ubiquitinated target. (A) EphA2 validation. Serum-starved B82L-EGFR cells were stimulated with EGF (100 ng/ml), ephrinA1-Fc (1 μg/ml) or ephrinA5–Fc (1 μg/ml) for 10 min or left untreated. Lysates (1 mg) were IP and IB with the indicated antibodies. (B) Quantification of cell proliferation assessed by BrdU incorporation in control and EphA2 KD cells. Data are expressed as the mean±s.e.m. of three independent experiments. Lower blot shows the efficiency of RNAi in EphA2 KD cells. (C) EphA2 KD reduces cell migration. The lower panels show DAPI-stained representative fields for each sample. Data are expressed as the mean±s.e.m. of three independent experiments.
We then assessed how the signal is transmitted from EGFR to EphA2. The two receptors do not appear to stably physically interact, as determined by co-IP (data not shown). However, the kinase activity, but not the cytoplasmic tail of the EGFR, is required for both modifications of EphA2 (Supplementary Figure S9A). One possibility is that EphA2 phosphorylation might be a prerequisite for Cbl recruitment, as this E3 ligase has previously been shown to be involved in ligand-mediated EphA2 degradation (Walker-Daniels et al, 2002; Wang et al, 2002). While this hypothesis deserves further investigation, we observed that prolonged EGF stimulation did not result in EphA2 degradation, indicating that EGF-induced EphA2 ubiquitination does not signal for degradation (Supplementary Figure S9C and data not shown).
What is then the functional consequence of EphA2 ubiquitination? One obvious possibility is internalization. Thus, we investigated the effect of EGF stimulation on the localization of EphA2 by confocal microscopy. No strong changes were visible on the total level or distribution of EphA2, although a partial co-internalization with EGFR upon EGF activation was clearly detectable (Supplementary Figure S9B).
While these data clearly demonstrate that EphA2 is a novel, downstream ubiquitinated target of EGFR, the role exerted by EphA2 ubiquitination on EGF signaling remains to be established. To start to assess the relevance of EphA2 to EGFR biology, we turned to the normal human breast epithelial cell line MCF10A that expresses both EphA2 and EGFR at significant levels (Supplementary Figure S9C). siRNA knockdown (KD) of EphA2 resulted in reduced EGF-induced proliferation (Figure 7B) and migration (Figure 7C), indicating that this receptor is critically involved in these EGFR biological readout. These results, although preliminary, set the stage for future ‘in depth’ molecular studies and highlight the ‘resource’ feature of our EGF-Ubiproteome.
Discussion
Although the Ub system has been intensively investigated in the past two decades, its impact on cellular homeostasis remains largely unexplored. This is particularly true for the signaling functions of ubiquitination, which have emerged as a major regulatory mechanism of signal transduction (Chen and Sun, 2009). Here, we report the first analysis of the EGF-Ubiproteome, which reveals an unexpected degree of pervasiveness of growth factor-induced ubiquitination across several signaling pathways, and a similarly unanticipated level of integration between two distinct types of PTM-based signaling.
The steady-state Ubiproteome
We initially defined the steady-state Ubiproteomes in HeLa and B82-EGFR cells. By combining the two, we defined a list of 1472 NR proteins, which constitutes the largest collection of ubiquitinated proteins reported so far in mammals (Figure 2A and Supplementary Figure S3). The comparison with previously published Ubiproteomes shows a significant overlap, ranging from ∼60% for data sets obtained in non-stringent conditions to ∼75% for those obtained under stringent denaturing conditions (Supplementary Figure S3). A distinct feature of our Ubiproteome is that it has been extensively validated. The effectiveness of our strategy is underscored by the fact that >95% of the tested candidates were shown to be bona fide ubiquitinated proteins (see Supplementary Figure S3 and its legend). In addition, there is a substantial overlap between the HeLa (endogenous) and B83L-EGFR (TAP) Ubiproteomes, with ∼50% of ‘mouse proteins’ being present in the list of ‘human proteins’.
The EGF-Ubiproteome
We used a stringent statistical analysis to identify 265 proteins, whose ubiquitination was regulated by activated EGFR. Thus, ∼18% of the steady-state Ubiproteome (265 of 1472) is modulated by an exogenous signal. As a kinetic analysis of the EGF-Ubiproteome was not performed, the above percentage likely underestimates the impact of EGFR on the Ub network.
A first obvious question is how EGFR, a receptor tyrosine kinase, transmits signals to the ubiquitination machinery to execute the modification of such a vast number of proteins. There is scarce literature on this topic. One known circuitry involves the E3 ligase Cbl, which binds to pY-sites on the EGFR (Levkowitz et al, 1999) and is then phosphorylated and activated (Kassenbrock and Anderson, 2004). This leads to EGFR ubiquitination and might also facilitate ubiquitination of other receptor-associated molecules, among which Cbl itself (Magnifico et al, 2003). Of note, the presence of non-proteolytic Ub chains on this E3 ligase (Figure 4B) suggests the existence of additional modes of regulation for this key Ub network player. Other effector enzymes (E2s, E3s, DUBs) might also be regulated though EGFR-mediated tyrosine phosphorylation. However, only three E3s (Cbl, Cbl-b and Huwe-1) and no E2s or DUBs have been identified in EGFR pY proteomes (Blagoev et al, 2004; Oyama et al, 2009). Conversely, we identified scores of Ub machinery enzymes in the EGF-Ubiproteome. These include E3s (Cbl, Cbl-b, Nedd4-2, RNF149, Rnf25, Huwe1, ZNF207, Znf319, Hectd1 and HERC4), E2s (Ube2O, UBE2Z, UBA2) and DUBs (JOSD1, USP15, ATXN3, USP11, USP5, USP34). The involvement of such a large number of effectors was unexpected. It appears, therefore, that regardless of the initial triggering mechanism (which must necessarily involve the kinase activity of the EGFR), the Ub signal is rapidly transmitted to, and amplified through, the Ub machinery. Similar to the phosphorylation cascade, in which critical enzymes such as kinases and phosphatases are often activated by phosphorylation, Ub enzymes may be regulated by ubiquitination (see for example Woelk et al, 2006; Ring1B (Ben-Saadon et al, 2006)). The impact of EGFR-mediated ubiquitination on the activity of E3s, E2s and DUBs warrants further investigation.
A second line of EGF-regulated ubiquitination events impinges on endocytic and signaling proteins. While these pathways are known factors in the Ub network, the magnitude of their involvement is somewhat surprising. EGF-regulated ubiquitination is involved in both clathrin-dependent and -independent endocytosis, in dozens of intracellular signaling circuitries, in cell-to-cell and cell-to-substrate adhesion mechanisms, and in actin remodeling. It seems, therefore, that EGFR-dependent ubiquitination intersects all aspects of signaling; the biochemical circuitries (such as PI3K-, 14-3-3-, JAK/STAT- and PTEN-related); the components that confer spatial and temporal dimensions to signaling (endocytosis); the coordinated modifications in cyto-architecture and relationships with the external milieu (actin, integrins, ephrins) that are necessary for the execution of complex signaling programs. As a proof of principle we investigated the functional link between EGFR and EphA2 and we demonstrated that activation of this signaling receptor is indeed required for EGF-mediated proliferation and migration (Figure 7). This result is particularly relevant considering that downregulation of EphA2 has been shown to decrease malignant phenotypes of cancer cells in vitro and to inhibit tumor growth in several mouse cancer models (Pasquale, 2008 and references therein). How this and other crosstalk are achieved, in mechanistic terms, remains to be elucidated. The emerging picture, however, is that the impact of ubiquitination on receptor-activated pathways might be as profound and as vast as the canonical pY-based network.
Finally, a third line of ubiquitinated proteins connects EGFR activity to many other aspects of cellular physiology, including DNA repair, nuclear transport, mRNA processing, various metabolic pathways and ribosome biogenesis. In the latter case, recent literature suggests that ubiquitination and degradation of ribosomal proteins might be a general mechanism adopted by mammalian cells to control ribosome production that can be adjusted according to cellular needs (Caldarola et al, 2009 and references therein). It remains to be established whether the EGFR-dependent ubiquitination of ribosomal proteins, uncovered herein, serves to regulate their degradation or has other, yet to be discovered, non-proteolytic functions.
One intriguing connection concerns EGF regulation of the solute carriers (SLCs)/transporters (validation of SLC3A2 is reported in Figure 4). These molecules are gatekeepers for cells and organelles, and control the uptake and efflux of important metabolites such as sugars, amino acids, nucleotides and inorganic ions. Almost all categories of SLCs are represented in our EGF-induced Ubiproteome, and all are hyperubiquitinated upon EGF stimulation (Supplementary Table S2). A growing body of biochemical and biophysical evidence suggests that these transporters are modulated by trafficking, and that the Ub modification is the signal for their internalization and/or lysosomal degradation (Miranda and Sorkin, 2007). Moreover, while transactivation of EGFR elicited by activation of the Na(+)/K(+)-ATPase has been described (Xie, 2003), our data demonstrate, for the first time, the EGF-induced regulation of SLC proteins. In this context, it is worth mentioning that EGFR is able to interact directly with SLC5A1/SGLT1, stabilizing the sodium/glucose cotransporter and facilitating glucose transport into cells (Weihua et al, 2008). The mechanisms through which EGFR can trigger the ubiquitination of SLCs, and the ultimate functional significance of the modification remain to be elucidated: a line of investigation that may have important implications also for neurological diseases (Tzingounis and Wadiche, 2007) and cancer (Engelman and Cantley, 2008; Nicklin et al, 2009), where alterations of SLCs have an important pathogenetic role.
Connectivity of the EGF-Ubiproteome
The EGF-Ubiproteome displayed remarkable connectivity—a possible indication of a wide pervasiveness of this network in cell regulation—organized into three levels. The first level is represented by intra-network connectivity. Indeed, the proteins of the EGF-Ubiproteome grouped into two major clusters, enriched in proliferation/inflammation and apoptosis/adhesion/cell cycle signatures (Figure 5). This result suggests that the EGF-regulated Ub network is a rather compact infrastructure, which allows coordinated control by EGFR of a multiplicity of signaling mechanisms. This ‘core’ regulatory network then reaches out to intersect (level 2 of connectivity) virtually every aspect of intracellular signaling, as discussed in the previous section.
Finally, a third level of connectivity is represented by the considerable overlap between the EGF-induced Ubiproteome and pY proteome. These two PTM-based networks can be conceptualized as two overlapping, diffusely interconnected, matrices through which the EGFR transduces signals to make them readable to the cell. How this is achieved remains to be established and will require high-resolution studies, probably on a protein-by-protein basis. In principle, ubiquitination might control the stability and/or degradation of pY-containing proteins (Hunter, 2007). In this context, the EGFR might exert dual control on the activation (through pY) and deactivation (through Ub) of signaling pathways. We note, however, that the EGFR mostly induces K63-linked ubiquitination (Figure 4). This modification has been linked to the signaling ability of Ub, rather than to its degradation properties (Woelk et al, 2007). It is thus possible that EGFR-induced ubiquitination adds a layer of signaling complexity to the canonical pY-based circuitry of EGFR signaling.
The ability of the proteins of the EGF-Ubiproteome to nucleate clusters of interactions was mirrored by their enrichment in ‘hubs’, which became even more evident when dually modified (pY and Ub) proteins were considered. Hubs are proteins that form critical interconnections between signaling pathways and are points of fragility of signaling networks (Amit et al, 2007). As such, they represent ideal targets for pharmacological intervention. However, detailed molecular knowledge of the mechanisms of interconnectivity of hubs is indispensable to predict the results of ‘hub interference’. Our results might thus be relevant for the identification of therapeutic targets, and to determine appropriate strategies of intervention in pathological conditions in which subversion of signaling by EGFR (and other receptor tyrosine kinases) is relevant, such as cancer.
Materials and methods
Reagents, constructs and cell culture
EGF was purchased from Intergen (Oxford, UK), rhodamine-EGF was from Molecular Probes. Puromycin, doxycyline, FLAG peptide, trypsin (proteomic grade), DTT, iodacetamide, chloroacetamide and N-ethylmaleimide, ephrinA1/A5 extracellular domain/Fc chimera, L-Arg 12C6, 14N4-HCl, L-Arg 13C6, 15N4-HCl, L-Lys 12C6, 14N2-HCl and L-Lys 13C6, 15N2-HCl were from Sigma. Imidazole and urea were from Carlo Erba. EDTA-free protease inhibitor cocktail tablets were from Roche. Ni-NTA agarose beads were from Qiagen. (K specific only)Ubn chains were from ENZO Life Sciences. Antibodies used were as follows: monoclonal anti-FLAG (M2, Sigma), M2-agarose affinity gel (Sigma), monoclonal anti-HA (clone 16B12, Babco), anti-vinculin (Sigma), monoclonal anti-Ub (P4D1, Santa Cruz and FK2, ENZO Life Sciences), anti-K63 and K48 Ub-chain-specific antibodies (Genentech), polyclonal anti-EGFR (directed against aa 1172–1186 of human EGFR, produced in-house), anti-eps15 (monoclonal, produced in-house), monoclonal anti-EphA2 (clone D7, Upstate) polyclonal anti-EphA2 (clone C-20, Santa Cruz), polyclonal anti-SLC3A2 (H-300, Santa Cruz), monoclonal anti-Cbl (BD), anti-phospho-ERK (Cell Signaling), anti-phospho-AKT (Cell Signaling) and anti-pY (Clone 4G10, Upstate Biotechnology).
The engineering of FLAG-6His-Ub and GST-S5a is reported in Supplementary information.
GST–S5a fusion protein and FK2 antibody were crosslinked using AminoLink® and CarboLink™ Coupling Gel (Pierce Biotechnology), respectively, according to the manufacturer's instructions. Avidin-agarose beads used in the control purification were from Pierce Biotechnology.
B82L fibroblasts expressing the human wild-type EGFR (B82L-wt), kinase-defective EGFR (B82L-Kin−) and the COOH terminally truncated EGFR at Tyr-958 (B82L-958) have been described previously (Welsh et al, 1994). B82L-EGFR cells stably expressing FLAG-6His-Ub were obtained by transfecting pSG213-FLAG-6His-Ub using LipofectAMINE™ (Invitrogen). Cells were selected in medium containing 5 μg/ml puromycine. Expression of FLAG-6His-Ub was induced by adding doxycycline to the culture medium at a final concentration of 4 μg/ml. MCF10A cells were cultured in Dulbecco's modified Eagle's medium and F12 medium (DMEM-F12) supplemented with 5% horse serum, hydrocortisone (0.5 μg/ml), insulin (10 μg/ml), cholera toxin (50 ng/ml), EGF (20 ng/ml) and penicillin-streptomycin (100 μg/ml each). When serum-starved, cells were cultured in the same medium devoid of serum and EGF.
Cell lysis and IP were performed as previously described (Penengo et al, 2006). For the IPs with the anti-Ub antibodies, 500 μg of HeLa lysate was subjected to IP either with 20 μg of FK2 antibody or 5 μg of K63 Ub-chain-specific antibody for 2 h at 4°C, followed by 1 h incubation with Protein G-conjugated sepharose beads (Zymed). Soluble EphrinA1 and A5 fusion proteins (EphrinA1-, EphrinA5–Fc) were obtained by mixing EphrinA1/5 with the Fc portion of human IgG in a molar ratio 1:10. Preclustering was performed on ice for 1 h. Immunofluorescence procedures, ablation of EphA2, BrdU incorporation and migration assays are in the Supplementary information.
The procedures for SILAC labeling and for the TAP and endogenous (FK2) purifications are described in the Supplementary information.
LC-MS/MS analysis
Peptides were separated on a 15 cm C18-reversed phase column (75 μm inner diameter) packed in-house with Reprosil (ReproSil-Pur C18-AQ 3-μm resin, Dr Maisch), using a nanoflow HPLC system (Agilent Technologies, Waldbronn, Germany or Proxeon, Proxeon Biosystems, Odense, Denmark). The HPLC was coupled online via a nanoelectrospray ion source (Proxeon Biosystems) to an LTQ-Orbitrap mass spectrometer (Thermo Scientific). We used a 140 min gradient from 2 to 60% acetonitrile in 0.5% acetic acid at a flow of 250 nl/min. The LTQ-Orbitrap was operated in the positive ion mode, with the following acquisition cycle: a full scan (from m/z 300–2000) recorded in the orbitrap analyzer at resolution R=60 000 followed by sequential isolation and fragmentation of the five most-intense peptide ions in the LTQ analyzer by collisionally induced dissociation. The ‘lock mass’ option was enabled in all full scans to improve mass accuracy of precursor ions (Olsen et al, 2005). Up to 500 sequenced ions were dynamically excluded for 60 s after sequencing. The maximum allowed fill time for an Orbitrap survey scan (ion target 1 000 000) was 1 s, and for an LTQ MS/MS scan (ion target 5000), 150 ms.
Data analysis
The raw data files were analyzed with the in-house developed quantitative proteomics software MaxQuant, version 1.0.11.5 (see Cox and Mann (2008) for details), which was used for peak list generation, identification and quantitation of SILAC pairs, and filtering. The software is supported by Mascot (version 2.2, Matrix Science) as the database search engine for peptide identification (Perkins et al, 1999). MS/MS spectra were searched against the human (endogenous Ub, HeLa cells) or mouse (FLAG-6His-Ub, B82L-EGFR cells) International Protein Index (IPI) databases (versions 3.37). False discovery rates (FDR) were controlled by searching in a concatenated database consisting of the original protein sequences plus their reversed versions, in which all Ks and Rs have been exchanged with each other. Protein sequences of common contaminants, e.g., human keratins and proteases used, were added to the database. Proteins and peptides with an FDR>1% were discarded. Posterior error probability for peptides were calculated as described previously (Cox and Mann, 2008) and set to 1%. The initial mass tolerance in MS mode was set to 7 p.p.m. and MS/MS mass tolerance was 0.5 Da. For the Mascot search, cysteine carbamidomethylation was searched as a fixed modification, whereas N-acetyl (Protein), Oxidation (M) and GlyGly (K) were searched as variable modifications. Labeled arginine (13C6, 15N4) and lysine (13C6, 15N2) were specified as fixed or variable modifications, depending on prior knowledge of the parent ion for the Mascot searches. Full tryptic specificity with up to two missed cleavages was required and only peptides of at least six amino acids were considered. By default, it was required that each protein group was identified by at least one peptide unique to the assigned protein group.
Three independent biological replicates were measured for the endogenous Ub and the overexpressed FLAG-6His-Ub experiments. Two experiments were identical (forward: FW1, FW2) and one was with swapped labels (reverse: REV). Raw files of technical and biological replicates were analyzed together, whereas the ratios of the REV experiments were inverted (1/ratio) in order to have a median ratio over all three experiments. To obtain ‘high-confidence data sets’ of quantified proteins, we adopted four filtering criteria, according to which proteins should (i) be represented by at least two peptides, one of which should be unique for the protein sequence; (ii) be identified in at least two of the three biological replicates; (iii) be quantified on at least three peptide pairs (heavy and light versions of the same peptide), referred to as ratio counts; (iv) not be present to a similar degree in the control purification (see Supplementary Table S1 and its legend for list of contaminants present in the control purification).
The filtering procedure to define potential regulated proteins, the identification of ubiquitination sites and the quantitation of the ‘signature peptides’ are described in the Supplementary information.
Clustering and functional analysis
Bioinformatics analyses were performed with online tools or tools available in-house, as described in the Supplementary information.
Supplementary Material
Acknowledgments
We thank Rosalind Gunby for critically reading the manuscript. Andrea Cocito and Giovanni d’Ario for statistical analysis. Pietro De Camilli for generously providing anti-Nedd4L antibody. This work was supported by grants from AIRC (Italian Association for Cancer Research) and The European Community (VI Framework, Rubicon) to SP, PPDF and MM. TB is a recipient of a European Molecular Biology Organization fellowship.
Author contributions: EA and BO set up and performed the biochemical purifications, TB carried out the mass spectrometry analysis, FB performed the statistical analysis and the meta-analysis, RP and SM carried out the validation experiments. PPDF participated in the initial design of the study and helped to draft the manuscript, MM provided the logistic support for the mass spectrometry analysis. SP designed the experiments, analyzed the data and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acconcia F, Sigismund S, Polo S (2009) Ubiquitin in trafficking: the network at work. Exp Cell Res 315: 1610–1618 [DOI] [PubMed] [Google Scholar]
- Amerik AY, Hochstrasser M (2004) Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta 1695: 189–207 [DOI] [PubMed] [Google Scholar]
- Amit I, Wides R, Yarden Y (2007) Evolvable signaling networks of receptor tyrosine kinases: relevance of robustness to malignancy and to cancer therapy. Mol Syst Biol 3: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A (2006) The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell 24: 701–711 [DOI] [PubMed] [Google Scholar]
- Blagoev B, Ong SE, Kratchmarova I, Mann M (2004) Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotechnol 22: 1139–1145 [DOI] [PubMed] [Google Scholar]
- Caldarola S, De Stefano MC, Amaldi F, Loreni F (2009) Synthesis and function of ribosomal proteins—fading models and new perspectives. FEBS J 276: 3199–3210 [DOI] [PubMed] [Google Scholar]
- Chen WS, Lazar CS, Lund KA, Welsh JB, Chang CP, Walton GM, Der CJ, Wiley HS, Gill GN, Rosenfeld MG (1989) Functional independence of the epidermal growth factor receptor from a domain required for ligand-induced internalization and calcium regulation. Cell 59: 33–43 [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell 33: 275–286 [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Diella F, Gould CM, Chica C, Via A, Gibson TJ (2008) Phospho.ELM: a database of phosphorylation sites—update 2008. Nucleic Acids Res 36: D240–D244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Cantley LC (2008) A sweet new role for EGFR in cancer. Cancer Cell 13: 375–376 [DOI] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I (2003) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol 5: 461–466 [DOI] [PubMed] [Google Scholar]
- Hammond DE, Hyde R, Kratchmarova I, Beynon RJ, Blagoev B, Clague MJ (2010) Quantitative analysis of HGF and EGF-dependent phosphotyrosine signaling networks. J Proteome Res 5: 2734–2742 [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP (2005) Ubiquitin-binding domains. Nat Rev Mol Cell Biol 6: 610–621 [DOI] [PubMed] [Google Scholar]
- Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A (2006) Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell 21: 737–748 [DOI] [PubMed] [Google Scholar]
- Hunter T (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell 28: 730–738 [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G (2006) Ubiquitin-binding domains. Biochem J 399: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen ON (2006) Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol 7: 391–403 [DOI] [PubMed] [Google Scholar]
- Kassenbrock CK, Anderson SM (2004) Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. J Biol Chem 279: 28017–28027 [DOI] [PubMed] [Google Scholar]
- Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, Andre B (2010) The ubiquitin xcode of yeast permease trafficking. Trends Cell Biol 20: 196–204 [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y (1999) Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell 4: 1029–1040 [DOI] [PubMed] [Google Scholar]
- Magnifico A, Ettenberg S, Yang C, Mariano J, Tiwari S, Fang S, Lipkowitz S, Weissman AM (2003) WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J Biol Chem 278: 43169–43177 [DOI] [PubMed] [Google Scholar]
- Meierhofer D, Wang X, Huang L, Kaiser P (2008) Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J Proteome Res 7: 4566–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M, Sorkin A (2007) Regulation of receptors and transporters by ubiquitination: new insights into surprisingly similar mechanisms. Mol Interv 7: 157–167 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205 [DOI] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, Komuves L, French DM, Ferrando RE, Lam C, Compaan D, Yu C, Bosanac I, Hymowitz SG, Kelley RF, Dixit VM (2008) Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134: 668–678 [DOI] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO (2009) Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136: 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics 4: 2010–2021 [DOI] [PubMed] [Google Scholar]
- Ong SE, Mann M (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat Protoc 1: 2650–2660 [DOI] [PubMed] [Google Scholar]
- Oyama M, Kozuka-Hata H, Tasaki S, Semba K, Hattori S, Sugano S, Inoue J, Yamamoto T (2009) Temporal perturbation of tyrosine phosphoproteome dynamics reveals the system-wide regulatory networks. Mol Cell Proteomics 8: 226–231 [DOI] [PubMed] [Google Scholar]
- Pasquale EB (2008) Eph-ephrin bidirectional signaling in physiology and disease. Cell 133: 38–52 [DOI] [PubMed] [Google Scholar]
- Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR (2006) Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell 124: 1183–1195 [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP (2002) A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416: 451–455 [DOI] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M (2008) Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol 9: 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MH, Dikic I (2005) The Cbl interactome and its functions. Nat Rev Mol Cell Biol 6: 907–918 [DOI] [PubMed] [Google Scholar]
- Seet BT, Dikic I, Zhou MM, Pawson T (2006) Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol 7: 473–483 [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34: D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP (2007) GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics 23: 3251–3253 [DOI] [PubMed] [Google Scholar]
- Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P (2006) A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol Cell Proteomics 5: 737–748 [DOI] [PubMed] [Google Scholar]
- Tzingounis AV, Wadiche JI (2007) Glutamate transporters: confining runaway excitation by shaping synaptic transmission. Nat Rev Neurosci 8: 935–947 [DOI] [PubMed] [Google Scholar]
- Walker-Daniels J, Riese DJ II, Kinch MS (2002) c-Cbl-dependent EphA2 protein degradation is induced by ligand binding. Mol Cancer Res 1: 79–87 [PubMed] [Google Scholar]
- Wang Y, Ota S, Kataoka H, Kanamori M, Li Z, Band H, Tanaka M, Sugimura H (2002) Negative regulation of EphA2 receptor by Cbl. Biochem Biophys Res Commun 296: 214–220 [DOI] [PubMed] [Google Scholar]
- Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ, Hung MC (2008) Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell 13: 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh JB, Worthylake R, Wiley HS, Gill GN (1994) Specific factors are required for kinase-dependent endocytosis of insulin receptors. Mol Biol Cell 5: 539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, Forman-Kay JD (2007) Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130: 651–662 [DOI] [PubMed] [Google Scholar]
- Woelk T, Oldrini B, Maspero E, Confalonieri S, Cavallaro E, Di Fiore PP, Polo S (2006) Molecular mechanisms of coupled monoubiquitination. Nat Cell Biol 8: 1246–1254 [DOI] [PubMed] [Google Scholar]
- Woelk T, Sigismund S, Penengo L, Polo S (2007) The ubiquitination code: a signalling problem. Cell Div 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z (2003) Molecular mechanisms of Na/K-ATPase-mediated signal transduction. Ann N Y Acad Sci 986: 497–503 [DOI] [PubMed] [Google Scholar]
- Zwang Y, Yarden Y (2009) Systems biology of growth factor-induced receptor endocytosis. Traffic 10: 349–363 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.