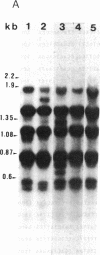
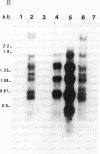
Abstract
The ETn (for "early transposon") family of long repeated sequences in abundantly transcribed in early mouse embryos from retroviral-like long terminal repeats. Nucleotide sequencing of two elements does not reveal any long open reading frame nor significant homology to retroviral proteins. The genetic polymorphism, monitored by Southern blotting within and across mouse species, reflects a concerted mode of evolution for the ETn sequences.
Full text
PDF
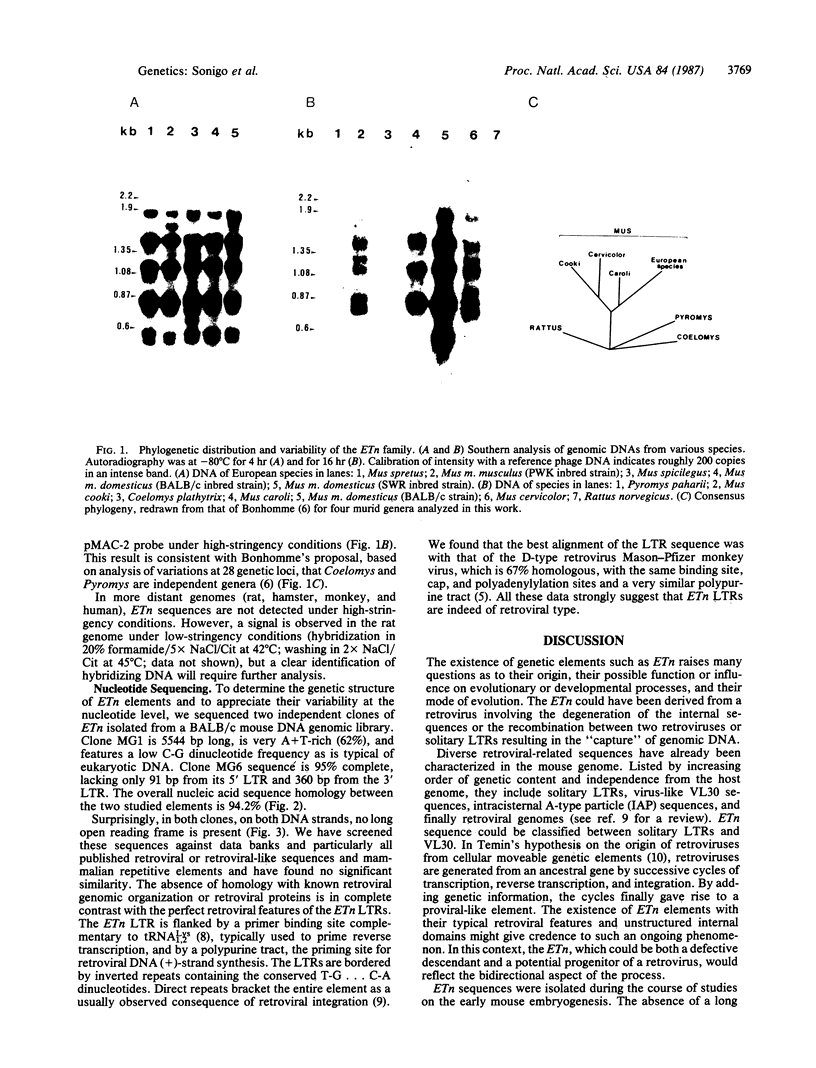


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alizon M., Wain-Hobson S., Montagnier L., Sonigo P. Genetic variability of the AIDS virus: nucleotide sequence analysis of two isolates from African patients. Cell. 1986 Jul 4;46(1):63–74. doi: 10.1016/0092-8674(86)90860-3. [DOI] [PubMed] [Google Scholar]
- Bonhomme F. Evolutionary relationships in the genus Mus. Curr Top Microbiol Immunol. 1986;127:19–34. doi: 10.1007/978-3-642-71304-0_3. [DOI] [PubMed] [Google Scholar]
- Brûlet P., Condamine H., Jacob F. Spatial distribution of transcripts of the long repeated ETn sequence during early mouse embryogenesis. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2054–2058. doi: 10.1073/pnas.82.7.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brûlet P., Kaghad M., Xu Y. S., Croissant O., Jacob F. Early differential tissue expression of transposon-like repetitive DNA sequences of the mouse. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5641–5645. doi: 10.1073/pnas.80.18.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., Posakony J. W. Repetitive sequence transcripts in development. Nature. 1982 Jun 24;297(5868):633–635. doi: 10.1038/297633a0. [DOI] [PubMed] [Google Scholar]
- Doolittle W. F., Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980 Apr 17;284(5757):601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Kaghad M., Maillet L., Brûlet P. Retroviral characteristics of the long terminal repeat of murine E.Tn sequences. EMBO J. 1985 Nov;4(11):2911–2915. doi: 10.1002/j.1460-2075.1985.tb04022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel L. E., Crick F. H. Selfish DNA: the ultimate parasite. Nature. 1980 Apr 17;284(5757):604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Raba M., Limburg K., Burghagen M., Katze J. R., Simsek M., Heckman J. E., Rajbhandary U. L., Gross H. J. Nucleotide sequence of three isoaccepting lysine tRNAs from rabbit liver and SV40-transformed mouse fibroblasts. Eur J Biochem. 1979 Jun;97(1):305–318. doi: 10.1111/j.1432-1033.1979.tb13115.x. [DOI] [PubMed] [Google Scholar]
- Sonigo P., Barker C., Hunter E., Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986 May 9;45(3):375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Origin of retroviruses from cellular moveable genetic elements. Cell. 1980 Oct;21(3):599–600. doi: 10.1016/0092-8674(80)90420-1. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Reverse transcription in the eukaryotic genome: retroviruses, pararetroviruses, retrotransposons, and retrotranscripts. Mol Biol Evol. 1985 Nov;2(6):455–468. doi: 10.1093/oxfordjournals.molbev.a040365. [DOI] [PubMed] [Google Scholar]