Abstract
Ubiquitination plays a role in virtually every cellular signaling pathway ranging from cell cycle control to DNA damage response to endocytosis and gene regulation. The bulk of our knowledge of the ubiquitination system is centered on modification of specific substrate proteins and the enzymatic cascade of ubiquitination. Our understanding of the regulation of the reversal of these modifications (deubiquitination) lags significantly behind. We recently reported a multifaceted study of the fission yeast Schizosaccharomyces pombe DUBs including characterization of their binding partners, in vitro enzymatic activity and subcellular localization.(1) Over half of the 20 fission yeast DUBs have a stable protein partner and some of those partners regulate the localization and/or activity of their cognate DUB. As a next step in understanding how DUBs might otherwise be regulated, we investigated the phosphostatus of the entire fission yeast DUB family using LC−MS/MS, and here we discuss the possible implications of phosphoregulation.
Keywords: ubiquitination, mass spectrometry, LC−MS/MS, phosphorylation
Short abstract
We investigated the phosphostatus of the fission yeast Schizosaccharomyces pombe deubiquitinating enzyme (DUB) complexes using 2D-LC−MS/MS and found 1242 spectra indicative of phosphorylation (neutral loss of 98 Da) corresponding to over 130 sites of modification in 12/20 DUBs and most DUB partners; these results highlight the interconnectivity of ubiquitin and kinase signaling pathways.
Introduction
Ubiquitination of specific cellular proteins serves as a signal for protein degradation, chromatin remodeling, DNA repair, vesicular transport, and changes in protein localization and/or activity depending on the number and structure of the ubiquitin modification.2,3 Protein ubiquitination is highly regulated and requires a cascade of enzymes that culminates in a substrate and site-specific modification. Similarly, deubiquitinating enzymes (DUBs) that remove ubiquitin (or ubiquitin-like modifiers like SUMO or Nedd8) from substrate proteins to allow recycling of ubiquitin and/or modulation of signaling pathways must be tightly controlled.
Ubiquitination and kinase cascades intersect on multiple levels and together they orchestrate key cellular events including endocytosis, cell cycle progression, and growth factor signaling.4−9 Kinases activate E3 ubiquitin ligases (e.g., the anaphase promoting complex/cyclosome) which in turn ubiquitinate kinases (e.g., Polo) or kinase regulatory subunits (e.g., the cyclin subunit of cyclin-dependent kinases (CDK)).10−13 Kinases also regulate protein turnover by marking substrates for phosphorylation-dependent ubiquitin-mediated degradation (e.g., by the SCF ubiquitin ligase).(14) There are many other examples of cross-regulation of ubiquitin and kinase signaling networks, including phosphorylation of deubiquitinating enzymes (e.g., CYLD).(15) Here we set the stage for understanding how DUBs might be regulated by kinases and phosphatases by cataloging phosphorylation sites of all S. pombe DUBs.
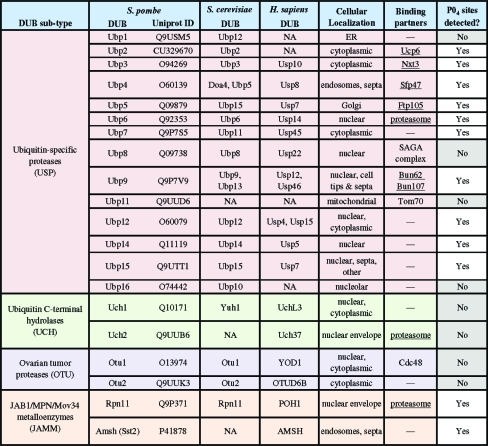
DUBs are a highly conserved family of proteases involved in: (1) processing of ubiquitin precursor proteins, (2) recovery of modified ubiquitin trapped in inactivatable forms, (3) cleavage of ubiquitin from target proteins, and (4) recycling of monoubiquitin from free polyubiquitin chains.16−18 The diversity of DUB functions is reflected in the number of DUBs (95 predicted human DUBs), the variety of catalytic domains—ubiquitin C-terminal hydrolases (UCH), ubiquitin-specific proteases (USP), ovarian tumor proteases (OTU), Machado-Joseph disease proteases (MJD) and JAB1/MPN/Mov34 metalloenzymes (JAMM)(16) and DUB domain architecture.(1)
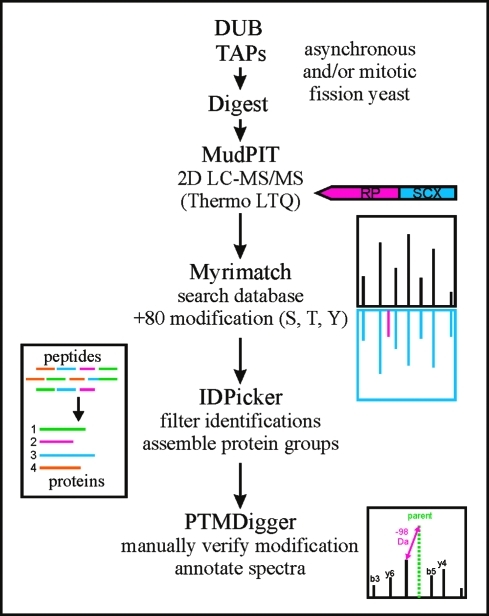
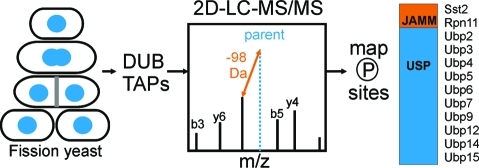
S. pombe is an amenable organism in which to conduct a global study of DUB function and regulation because of the limited number of DUBs containing the required catalytic residues (20), the diversity and conservation of catalytic domains (4 of 5 classes, see Table 1), and the genetic tractability of yeast. We recently reported the cellular localization, enzymatic activity profiles and protein interaction networks of the entire S. pombe DUB family.(1) A few phosphorylation sites for some S. pombe DUBs have been reported in large-scale phosphoproteomics studies,19,20 but a detailed analysis of DUB phosphorylation is lacking. To begin to understand how phosphorylation impacts DUB regulation, we examined the phosphostatus of the entire S. pombe DUB family and their binding partners using tandem affinity purification (TAP) followed by multidimensional LC-MS/MS (MudPIT) from asynchronous and mitotic cell cultures (Figure 1). Here, we present the global phosphorylation status of the S. pombe DUBs and their partners and discuss the implications of these modifications on DUB regulation in eukaryotes.
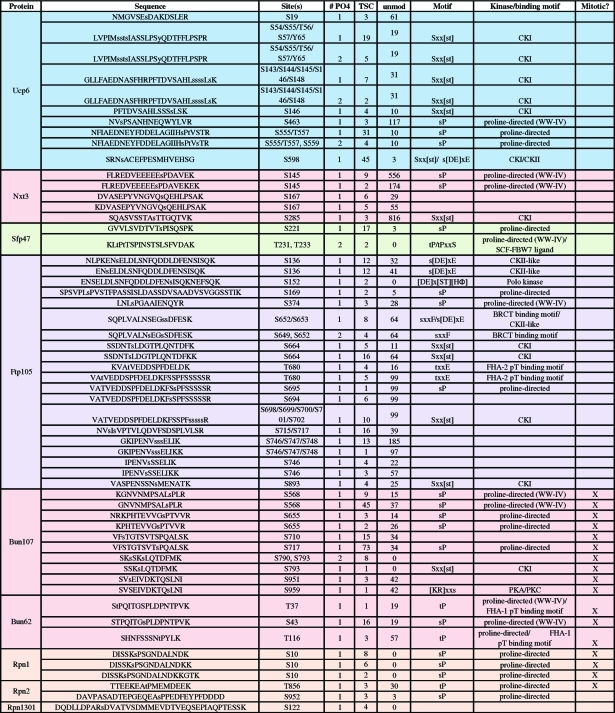
Table 1. S. pombe Deubiquitinating Enzymes (DUBs).
Figure 1.
Experimental scheme for DUB purifications, LC−MS/MS analysis, and phosphopeptide identification and verification.
Experimental Methods
Yeast Strains, Media, Genetic Methods, and Vector Construction
Strain construction and tetrad analysis were accomplished through standard methods. Endogenously tagged strains (Supplemental Table 1, Supporting Information) were grown in yeast extract (YE) media. For expression of N-terminally tagged proteins, strains were transformed with pREP expression vectors, containing a thiamine repressible promoter, using a standard sorbitol transformation procedure.(21) Transformed strains were first grown on minimal media containing thiamine to suppress expression and then, to induce expression, cells were grown in minimal media lacking thiamine for 18 h.(22) Cell cultures used for TAP purifications were grown in 2 L of 4× YE media (C-terminally TAP tagged proteins) or in 8 L of minimal media supplemented with the appropriate nutrients (N-terminally TAP tagged proteins). All 20 DUBs were tagged endogenously at the 3′ end with TAP or linker-TAP as previously described.(23) The linker sequence in the linker-TAP cassettes translates to ILGAPSGGGATAGAGGAGGPAGLI.(24) N-TAP cassettes for Ubp1, Ubp7, and Ubp11 were constructed as previously described.(1)
For mitotic purifications of the nuclear DUBs (Ubp6, Ubp8, Ubp9, Ubp12, Ubp14, Ubp15, Ubp16, Uch1, Uch2, Otu1, and Rpn11), log phase cells containing DUB TAP tags were blocked using a cold sensitive allele of β-tubulin (nda3-KM11, prometaphase) and/or released for 30 min (anaphase). Cells were snap frozen in a dry ice ethanol bath and subjected to TAP/LC-MS/MS as described below.
Protein Methods
Cell pellets were frozen in a dry ice/ethanol bath and lysed by bead disruption in NP-40 lysis buffer under native (Figure 2c) or denaturing conditions (Figure 2a/b) as previously described,(25) except with the addition of 0.1 mM diisopropyl fluorophosphate (Sigma-Aldrich). Proteins were immunoprecipitated by IgG Sepharose beads (GE Healthcare) or anti-GFP (Roche). For phosphatase collapse, immunoprecipitated proteins were incubated with lambda phosphatase (New England Biolabs) in 25 mM HEPES-NaOH pH 7.4, 150 mM NaCl, and 1 mM MnCl2 for 30 min at 30 °C. Immunoblot analysis was performed as previously described(26) except that secondary antibodies were conjugated to Alexa Fluor 680 (Invitrogen) and visualized using an Odyssey Infrared Imaging System (LI-COR Biosciences).
Figure 2.
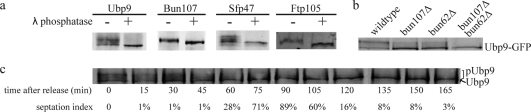
Biochemical analysis of DUB phosphorylation a) Lambda phosphatase collapse for Ubp9, Bun107, Sfp47, and Ftp105 b) phosphorylation status of Ubp9 in the presence or absence of its WD partners and c) block and release experiment illustrating the cell cycle dependency of Ubp9 phosphorylation (see Experimenal Methods for details).
For the block and release experiment, a temperature sensitive strain (cdc25−22 Ubp9-TAP) was grown overnight at 25 °C and then shifted to the nonpermissive temperature (36 °C) for 3 h to block cells in G2. The cells were then released to the permissive temperature and 20 OD pellets were collected every 15 min. Lysates and immunoprecipitations were performed as described above except IgG coated dynabeads (Invitrogen) were used for immunoprecipitation.
DUB purification and LC−MS/MS analysis
Proteins were purified by TAP as described,(27) or using a one step dynabead purification as follows: tosylactivated M-280 Dynabeads were coupled to rabbit IgG (Invitrogen) and used to pull down TAP tagged proteins from native lysates (as in TAP protocol) and then the proteins were eluted using high pH. The purified proteins were then TCA precipitated and digested with trypsin (Promega), chymotrypsin (Princeton Separations), and/or GluC (Thermo) and the resulting peptides were subjected to mass spectrometric analysis on a Thermo LTQ as previously detailed.28,29 Thermo RAW files were converted to MZML files using Scansifter (software developed in-house at the Vanderbilt University Medical Center). Spectra with less than 20 peaks were excluded from our analysis. The S. pombe database (http://www.sanger.ac.uk, October 2009) was searched with the Myrimatch algorithm(30) v1.6.33 on a high performance computing cluster (Advanced Computing Center for Research & Education at Vanderbilt University). We added contaminant proteins (e.g., keratin, IgG) to the complete S. pombe database and reversed and concatenated all sequences to allow estimation of false discovery rates (10 186 entries). Myrimatch parameters were as follows: strict tryptic cleavage; modification of methionine (oxidation, dynamic modification, +16 Da), S/T/Y (phosphorylation, dynamic modification, +80 Da) and cysteine (carboxamidomethylation, static modification, +57 Da) was allowed; precursor ions were required to be within 0.6 m/z of the peptide monoisotopic mass; fragment ions were required to fall within 0.5 m/z of the expected monoisotopic mass. IDPicker31,32 v2.6.126.0 was used to filter peptide matches with the following parameters: max. FDR per result 0.01, max. ambiguous IDs per result 2, min peptide length per result 5, min distinct peptides per protein 3, min additional peptides per protein group 2, minimum number of spectra per protein 3, indistinct modifications M 15.994 Da, C 57.05 Da and distinct modifications S/T/Y 80 Da. IDPicker results were processed in Excel (Microsoft) to generate phosphopeptide lists for the DUBs and their binding partners. Spectra were manually inspected and annotated in SeeMS and a related program called PTMDigger, software developed by in-house (Surendra Dasari, Matthew Chambers, and David Tabb, Vanderbilt University Medical Center). Supplemental Figure 1 (Supporting Information) was generated using software developed in-house (Zeqiang Ma, Surendra Dasari, Matthew Chambers, and David Tabb, Vanderbilt University Medical Center). DUBs and partners were purified with sequence coverage (%) as follows: Otu1 − 51, Otu2 − 35, Ubp1 − 67, Ubp2 − 95, Ubp3 − 64, Ubp4 − 42, Ubp5 − 72, Ubp6 − 56, Ubp7 − 81, Ubp8 − 62, Ubp9 − 67, Ubp11 − 71, Ubp12 − 67, Ubp14 − 80, Ubp15 − 88, Ubp16 − 44, Rpn11 − 63, Sst2 − 65, Uch1 − 84, Uch2 − 82, Ucp6 − 78, Nxt3 − 71, Sfp47 − 46, Ftp105 − 58, Bun62 − 57 and Bun107 − 64. Note that mildly overexpressed N-terminal TAP fusions were used for the low abundance DUBs Ubp1, Ubp7 and Ubp11. For complete protein identification information for each TAP, see a previous publication.(1) Using the stringent filter of FDR < 1%, approximately 1500 mass spectra contained +80 Da shifts, indicative of phosphorylation. These spectra were manually inspected and filtered according to the following criteria: (1) exhibit a prominent (often base) 98 Da (H3PO4) neutral loss peak at the MS2 level and (2) b and y ion intensities >20% of the neutral loss peak (3) contained two or more sequential fragments (b and/or y) bracketing the phosphorylation site(s); 1242 spectra met these criteria. Phosphorylation sites were assigned based on the presence of sequential fragment ions surrounding the modification; if these ions were missing, the phosphorylation site(s) were assigned to multiple sites ambiguously.
Results and Discussion
Deubiquitinating enzymes are present in nearly every cellular compartment(1) (Table 1) and participate in essential cellular processes including regulation of endocytosis, protein degradation, transcription, DNA repair, and protein localization and/or activity.(18) We and others have shown that DUBs are regulated by interaction with protein partners1,18,33 and now we have assessed the phosphostatus of the S. pombe DUB family to set the stage for understanding the interplay of phosphorylation and ubiquitination.
Each of the 20 S. pombe DUBs was purified two or more times from asynchronous cultures using an endogenous C-terminal TAP tag or an inducible N-terminal TAP tag (see Experimental Methods for details).(1) We also performed purifications of the (partially) nuclear DUBs (Ubp6, Ubp8, Ubp9, Ubp12, Ubp14, Ubp15, Ubp16, Uch1, Uch2, Rpn11, and Otu1) from cells arrested in prometaphase using the tubulin mutation, nda3-KM11, and released for 30 min into anaphase to enrich our data set with mitotic phosphorylation events (denoted in Tables 2, 3 and 4). Each purification was precipitated, digested, and analyzed on a Thermo LTQ using a MudPIT protocol (see Experimental Methods for details). The resultant mass spectra were processed using software developed at Vanderbilt Medical Center (Figure 1) to identify phosphorylation sites.
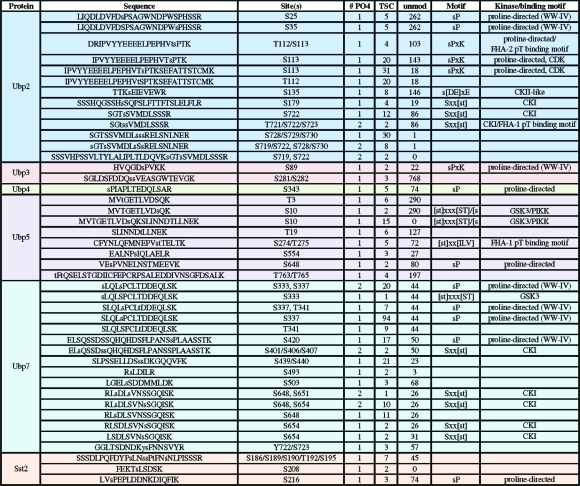
Table 2. Phosphorylation Sites Detected for Cytoplasmic S. pombe DUBs.
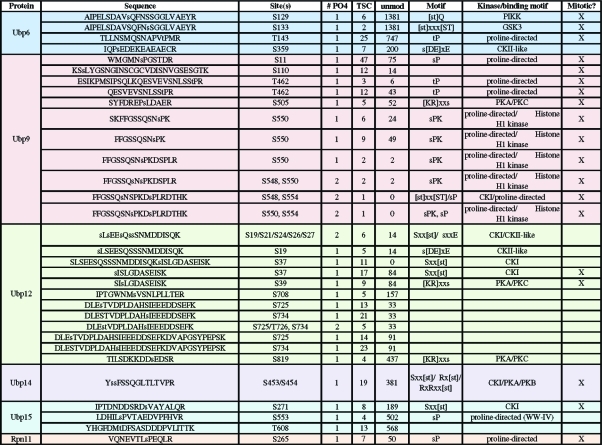
Table 3. Phosphorylation Sites Detected for (Partially) Nuclear S. pombe DUBs.
Table 4. Phosphorylation Sites Detected for Partners of S. pombe DUBs.
Over 1500 mass spectra (FDR > 0.5%) indicative of phosphorylation (+80 Da) were identified from our bioinformatic analysis (Figure 1) and manual validation showed 1242 spectra corresponding to phosphorylation sites (for criteria see Experimental Methods). The overall spectral quality and peptide sequence coverage is illustrated with two examples of parent and daughter spectra (Supplemental Figures 2 and 3, Supporting Information). In total, we identified over 130 phosphorylation sites in over half (12/20) of the S. pombe DUBs and DUB partners (Tables 2, 3, and 4). Only ca. 15% of the phosphosites we identified have been previously reported (see Supplemental Table 2 for details, Supporting Information).19,20 We confirmed biochemically that Upb9, Bun107, Ftp105 and Sfp47 are phosphoproteins by lambda phosphatase collapse and Western blot (Figure 2a). The other DUBs exhibited no discernible gel shift after phosphatase treatment (data not shown), but gel conditions were not optimized for each protein.
Cell Cycle Regulation of DUB Phosphorylation
The mitotic purifications revealed upregulated [st]P proline-directed kinase consensus sites, as one might expect for modification by mitotic CDK. Phosphorylation sites detected in Ubp6, Ubp9 and its partner Bun107 were much more abundant in the mitotic purifications (denoted in Tables 3 and 4), suggesting that these DUBs are cell cycle regulated. Ubp6 is recruited to the proteasome under conditions of ubiquitin stress(34) which was not the case for our experiments, but it is possible that mitotic phosphorylation plays some role in localization or activity of Ubp6.
Phosphorylation of Ubp9 is clearly cell cycle dependent based on block and release experiments (Figure 2c) and enrichment of S11 phosphopeptides identified from mitotic cells; thus, phosphorylation may alter the affinity of Ubp9 for its substrates and/or enhance its catalytic activity rather than affect its cellular localization (which is regulated by its WD partners, see discussion below). All components of the Ubp9 complex are phosphorylated (this study) and conserved throughout eukaryotes.(35) The larger WD partner, Bun107, contains multiple phosphorylation sites consistent with CDK phosphorylation based on amino acid sequence and increased abundance in mitotic purifications (Table 4).
Phosphorylation of DUB Complexes
Cross-regulation between ubiquitination and phosphorylation appears to be a common theme for DUB complexes. Over half of the DUBs interact with protein partners near stoichiometric ratios(1) and most of these DUBs and their partners are phosphorylated (Tables 1−4 and Figure 3), signifying that kinases and phosphosphatases regulate DUBs. Ubp9, a DUB present in the nucleus and at cell tips and septa, is part of a complex containing two WD proteins (Bun62 and Bun107). Both WD partners are required for Ubp9′s DUB activity and regulate its cellular localization.(1) The Ubp9 complex shuttles between the nucleus and cytoplasm, but at steady state, accumulates at active sites of endocytosis (cell tips and septa). When Bun62 is deleted, Ubp9 localizes to cell tips and septa, but not the nucleus, whereas deletion of Bun107 causes retention of Ubp9 in the nucleus.(1) We have discovered that Ubp9 and both of its partners are phosphorylated (Tables 3 and 4 and Figures 2 and 3). To investigate how phosphorylation might impact Ubp9 localization or function, we examined the phosphostatus of Ubp9 in strains where each partner had been deleted individually and in combination (Figure 2b). When either WD partner is lost, Ubp9 is no longer efficiently phosphorylated (Figure 2b), suggesting that Ubp9 is not competent for phosphorylation unless it is in complex with its partners. Both partners of four other DUB complexes are phosphorylated, including two cytoplasmic DUBs Ubp2 and Ubp3 and their partners Ucp6 and Nxt3, respectively, and two endocytic DUBs Ubp4 and Ubp5 and their partners Sfp47 and Ftp105, respectively (Tables 2 and 4, Figures 2 and 3). Sfp47, an SH3 domain protein, and Ftp105, a putative transmembrane protein, recruit their respective DUB partners to specific cellular locations (endosomes for Ubp4 and the Golgi for Ubp5).(1) Phosphorylation and dephosphorylation cycles may modulate complex formation, cellular localization, DUB activity and/or substrate specificity.
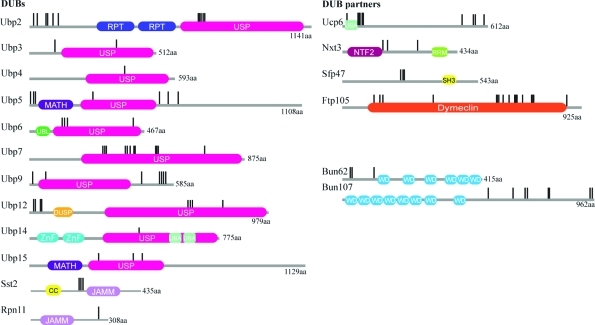
Figure 3.
Domain architecture and mapping of detected phosphorylation sites within the S. pombe DUBs and their partners. Domain architectures were retrieved using the SMART and Pfam databases. The following domains were found: USP (ubiquitin-specific proteases) JAMM (JAB1/MPN/Mov34 metalloenzymes), DUSP (Domain in Ubiquitin-specific proteases), MATH (Meprin and TRAF homology), UBL (Ubiquitin-like), ZnF (Ubiquitin Carboxyl-terminal hydrolase-like zinc finger), UBA (Ubiquitin-associated). RPT are internal repeats. Phosphosites are denoted by vertical black lines.
Location of Phosphosites within DUBs and Their Partners
Surprisingly, most of the DUB phosphorylation sites map to the catalytic DUB domains (Figure 3). In fact, all detected sites for Ubp7 are within its extended USP domain, suggesting that its catalytic activity and/or structure could be regulated by phosphorylation. Ubp2 and Ubp12 each have two clusters of phosphosites—one within their USP domain and one near the N-terminus; perhaps this arrangement allows tuning of DUB cellular localization, substrate binding or catalytic activity by kinases and phosphatases. Finally, two endocytic DUBs Ubp5 and Ubp9 have two clusters of sites at their N- and C- termini, respectively, predominately outside the USP domains. As discussed above, the cellular localization of these two DUBs is regulated by protein partners(1) and so phosphorylation may add another layer of regulation for substrate binding and/or catalytic activity. The phosphosites detected for the DUB partners also cluster within or very near domains (e.g., Ubcp6 and Ftp105) or in regions predicted to be intrinsically disordered (e.g., Sfp47 and Bun107) (Figure 3). These sites may regulate the availability of specific protein domains for interaction with other partners or the catalytic activity of the holo DUB complex.
DUB Phosphorylation Consensus Motifs
Given the diversity of DUB cellular localization and function, it is not surprising that the DUB phosphosites match consensus sequences for multiple protein kinases. The majority of DUB phosphopeptides are products of proline-directed kinases (e.g., MAP kinases or CDK) and many others match consensus sites for casein-type kinases (CKI, CKII, see Tables 1−4 and Supplemental Table 2, Supporting Information). Phosphosites detected in the exclusively cytoplasmic DUBs also include sequences consistent with PIKK and GSK3 consensus sites and the cellular localization of these kinases (Table 2). While the nuclear DUB sites are primarily proline-directed sites, the partially nuclear DUBs, Ubp9 and Ubp12, have phosphopeptides consistent with phosphorylation by PKA/PKC (Table 3, Supplemental Table 2, Supporting Information).
There are also many phosphorylation-dependent WW class IV ligand motifs present in both the cytoplasmic and nuclear DUBs (Tables 2 and 3), suggesting that DUB interactions with WW domain-containing proteins could be controlled by phosphorylation. For instance, the HECT-type E3 Ub-ligases, Pub1, Pub2 and Pub3 and multiple components of the spliceosome contain WW domains and are likely regulated by a combination of kinases and DUBs. The cytoplasmic DUB Ubp2 and endocytic DUBs Ubp5 and Ubp9 are phosphorylated on sites that match the FHA domain consensus binding motif that may function in localization and/or substrate recognition.
Concluding Remarks
Our results show that the majority of DUBs and most DUB partners are phosphorylated, some in a cell cycle-dependent manner. The phosphosites identified for S. pombe DUBs and their partners provide a foundation for understanding the interplay of ubiquitination and phosphorylation in this enzyme class in higher eukaryotes because sites identified in conserved proteins may be conserved or mimicked in higher eukaryotes. Future studies aimed at understanding the intersection of ubiquitination and phosphorylation will be useful for understanding DUB regulation and, more broadly, the cross-regulation of kinase and ubiquitin signaling networks.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (http://www.hhmi.org/), of which K.L.G. is an investigator. Funding for J.R.M. was provided by NIH training grant T32CA009385-25. We thank Rachel Roberts-Galbraith for critical reading of the manuscript and David L. Tabb and members of his lab (especially Zeqiang Ma, Surendra Dasari and Matt Chambers) for use and adaptation of software tools developed in their lab. We also thank Jun-Song Chen for performing MS analysis.
Glossary
Abbreviations
- DUB
deubiquitinating enzyme
- JAMM
JAB1/MPN/Mov34 metalloenzymes
- LC−MS/MS
liquid chromatography−mass spectrometry/mass spectrometry
- MJD
Machado-Joseph disease proteases
- MS
mass spectrometry
- OTU
ovarian tumor proteases
- Ub
ubiquitin
- UCH
ubiquitin C-terminal hydrolases
- USP
ubiquitin-specific proteases
- TSC
total spectral counts
Supporting Information Available
Supplemental tables and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Kouranti I.; McLean J. R.; Feoktistova A.; Liang P.; Johnson A. E.; Roberts-Galbraith R. H.; Gould K. L. A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS Biol. 2010, 8 (9), pii: e1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A.; Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–79. [DOI] [PubMed] [Google Scholar]
- Ikeda F.; Dikic I. Atypical ubiquitin chains: new molecular signals. ’Protein Modifications: Beyond the Usual Suspects’ review series. Embo Rep. 2008, 9 (6), 536–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe E.; Ayscough K. R. The Ark1/Prk1 family of protein kinases. Regulators of endocytosis and the actin skeleton. Embo Rep. 2003, 4 (3), 246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D. O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 1997, 13, 261–91. [DOI] [PubMed] [Google Scholar]
- Citri A.; Yarden Y. EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell. Biol. 2006, 7 (7), 505–16. [DOI] [PubMed] [Google Scholar]
- Peters J. M. SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr. Opin. Cell Biol. 1998, 10 (6), 759–68. [DOI] [PubMed] [Google Scholar]
- Raiborg C.; Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 2009, 458 (7237), 445–52. [DOI] [PubMed] [Google Scholar]
- Sorkin A.; Goh L. K. Endocytosis and intracellular trafficking of ErbBs. Exp. Cell Res. 2009, 315 (4), 683–96. [DOI] [PubMed] [Google Scholar]
- Kraft C.; Herzog F.; Gieffers C.; Mechtler K.; Hagting A.; Pines J.; Peters J. M. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. Embo J. 2003, 22 (24), 6598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahav-Baratz S.; Sudakin V.; Ruderman J. V.; Hershko A. Reversible phosphorylation controls the activity of cyclosome-associated cyclin-ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 1995, 92 (20), 9303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerdt F.; Strebhardt K. Polo-like kinase 1: target and regulator of anaphase-promoting complex/cyclosome-dependent proteolysis. Cancer Res. 2006, 66 (14), 6895–8. [DOI] [PubMed] [Google Scholar]
- Peters J. M. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 2002, 9 (5), 931–43. [DOI] [PubMed] [Google Scholar]
- Petroski M. D.; Deshaies R. J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 2005, 6 (1), 9–20. [DOI] [PubMed] [Google Scholar]
- Reiley W.; Zhang M.; Wu X.; Granger E.; Sun S. C. Regulation of the deubiquitinating enzyme CYLD by IkappaB kinase gamma-dependent phosphorylation. Mol. Cell. Biol. 2005, 25 (10), 3886–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman S. M.; Luna-Vargas M. P.; Velds A.; Brummelkamp T. R.; Dirac A. M.; Sixma T. K.; Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005, 123 (5), 773–86. [DOI] [PubMed] [Google Scholar]
- Komander D.; Clague M. J.; Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell. Biol. 2009, 10 (8), 550–63. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu F. E.; Ventii K. H.; Wilkinson K. D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P.; Trinidad J. C.; Fiedler D.; Roguev A.; Lim W. A.; Shokat K. M.; Burlingame A. L.; Krogan N. J. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009, 7 (6), e1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Grady J. T.; Villen J.; Gygi S. P. Phosphoproteome analysis of fission yeast. J. Proteome Re.s 2008, 7 (3), 1088–97. [DOI] [PubMed] [Google Scholar]
- Prentice H. L. High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 1992, 20 (3), 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S.; Klar A.; Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991, 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Bahler J.; Wu J. Q.; Longtine M. S.; Shah N. G.; McKenzie A. 3rd; Steever A. B.; Wach A.; Philippsen P.; Pringle J. R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 1998, 14 (10), 943–51. [DOI] [PubMed] [Google Scholar]
- Sandblad L.; Busch K. E.; Tittmann P.; Gross H.; Brunner D.; Hoenger A. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell 2006, 127 (7), 1415–24. [DOI] [PubMed] [Google Scholar]
- Gould K. L.; Moreno S.; Owen D. J.; Sazer S.; Nurse P. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. Embo J. 1991, 10 (11), 3297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B. A.; McDonald W. H.; Yates J. R. 3rd; Gould K. L. Phospho-regulation of the Cdc14/Clp1 phosphatase delays late mitotic events in S. pombe. Dev. Cell 2006, 11 (3), 423–30. [DOI] [PubMed] [Google Scholar]
- Tasto J. J.; Carnahan R. H.; McDonald W. H.; Gould K. L. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast 2001, 18 (7), 657–62. [DOI] [PubMed] [Google Scholar]
- McDonald W. H.; Ohi R.; Miyamoto D. T.; Mitchison T. J.; Yates J. R. III. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 2002, 219, 245–51. [Google Scholar]
- Roberts-Galbraith R. H.; Chen J. S.; Wang J.; Gould K. L. The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J. Cell Biol. 2009, 184 (1), 113–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb D. L.; Fernando C. G.; Chambers M. C. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 2007, 6 (2), 654–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.; Chambers M. C.; Tabb D. L. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 2007, 6 (9), 3549–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z. Q.; Dasari S.; Chambers M. C.; Litton M. D.; Sobecki S. M.; Zimmerman L. J.; Halvey P. J.; Schilling B.; Drake P. M.; Gibson B. W.; Tabb D. L. IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res. 2009, 8 (8), 3872–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventii K. H.; Wilkinson K. D. Protein partners of deubiquitinating enzymes. Biochem. J. 2008, 414 (2), 161–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J.; Meides A.; Zhang D. P.; Finley D. A ubiquitin stress response induces altered proteasome composition. Cell 2007, 129 (4), 747–59. [DOI] [PubMed] [Google Scholar]
- Sowa M. E.; Bennett E. J.; Gygi S. P.; Harper J. W. Defining the human deubiquitinating enzyme interaction landscape. Cell 2009, 138 (2), 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.