Abstract
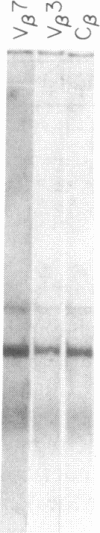
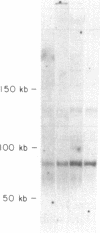
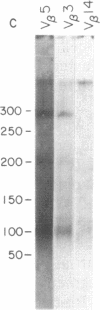
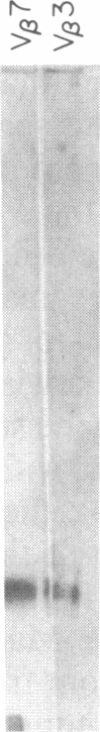
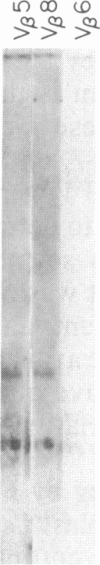
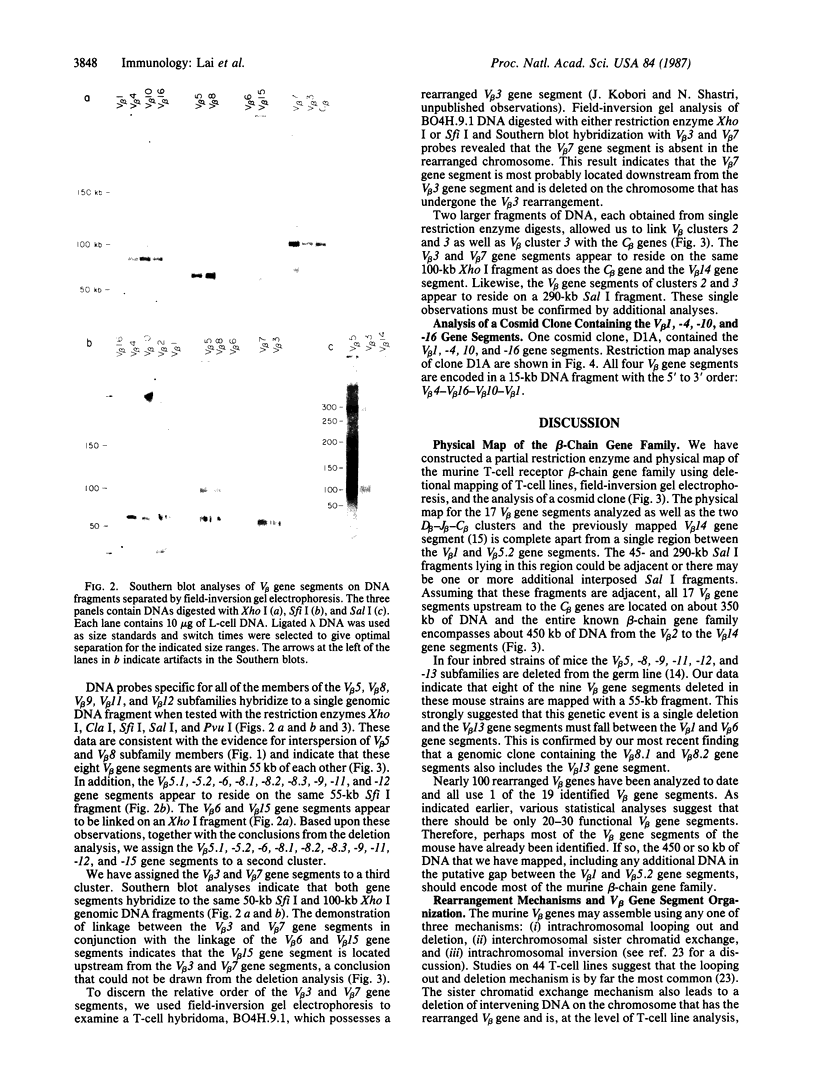
We have combined three different methods, deletion mapping of T-cell lines, field-inversion gel electrophoresis, and the restriction mapping of a cosmid clone, to construct a physical map of the murine T-cell receptor beta-chain gene family. We have mapped 19 variable (V beta) gene segments and the two clusters of diversity (D beta) and joining (J beta) gene segments and constant (C beta) genes. These members of the beta-chain gene family span approximately equal to 450 kilobases of DNA, excluding one potential gap in the DNA fragment alignments.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barth R. K., Kim B. S., Lan N. C., Hunkapiller T., Sobieck N., Winoto A., Gershenfeld H., Okada C., Hansburg D., Weissman I. L. The murine T-cell receptor uses a limited repertoire of expressed V beta gene segments. Nature. 1985 Aug 8;316(6028):517–523. doi: 10.1038/316517a0. [DOI] [PubMed] [Google Scholar]
- Behlke M. A., Chou H. S., Huppi K., Loh D. Y. Murine T-cell receptor mutants with deletions of beta-chain variable region genes. Proc Natl Acad Sci U S A. 1986 Feb;83(3):767–771. doi: 10.1073/pnas.83.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlke M. A., Spinella D. G., Chou H. S., Sha W., Hartl D. L., Loh D. Y. T-cell receptor beta-chain expression: dependence on relatively few variable region genes. Science. 1985 Aug 9;229(4713):566–570. doi: 10.1126/science.3875151. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccia N., Kronenberg M., Saxe D., Haars R., Bruns G. A., Goverman J., Malissen M., Willard H., Yoshikai Y., Simon M. The T cell receptor beta chain genes are located on chromosome 6 in mice and chromosome 7 in humans. Cell. 1984 Jul;37(3):1091–1099. doi: 10.1016/0092-8674(84)90443-4. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Frank M., Olson M. V. Electrophoretic separations of large DNA molecules by periodic inversion of the electric field. Science. 1986 Apr 4;232(4746):65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- Chien Y. H., Gascoigne N. R., Kavaler J., Lee N. E., Davis M. M. Somatic recombination in a murine T-cell receptor gene. Nature. 1984 May 24;309(5966):322–326. doi: 10.1038/309322a0. [DOI] [PubMed] [Google Scholar]
- Chou H. S., Behlke M. A., Godambe S. A., Russell J. H., Brooks C. G., Loh D. Y. T cell receptor genes in an alloreactive CTL clone: implications for rearrangement and germline diversity of variable gene segments. EMBO J. 1986 Sep;5(9):2149–2155. doi: 10.1002/j.1460-2075.1986.tb04478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembić Z., Bannwarth W., Taylor B. A., Steinmetz M. The gene encoding the T-cell receptor alpha-chain maps close to the Np-2 locus on mouse chromosome 14. Nature. 1985 Mar 21;314(6008):271–273. doi: 10.1038/314271a0. [DOI] [PubMed] [Google Scholar]
- Gascoigne N. R., Chien Y., Becker D. M., Kavaler J., Davis M. M. Genomic organization and sequence of T-cell receptor beta-chain constant- and joining-region genes. Nature. 1984 Aug 2;310(5976):387–391. doi: 10.1038/310387a0. [DOI] [PubMed] [Google Scholar]
- Goverman J., Minard K., Shastri N., Hunkapiller T., Hansburg D., Sercarz E., Hood L. Rearranged beta T cell receptor genes in a helper T cell clone specific for lysozyme: no correlation between V beta and MHC restriction. Cell. 1985 Apr;40(4):859–867. doi: 10.1016/0092-8674(85)90345-9. [DOI] [PubMed] [Google Scholar]
- Kavaler J., Davis M. M., Chien Y. Localization of a T-cell receptor diversity-region element. Nature. 1984 Aug 2;310(5976):421–423. doi: 10.1038/310421a0. [DOI] [PubMed] [Google Scholar]
- Kavaler J., Davis M. M., Chien Y. Localization of a T-cell receptor diversity-region element. Nature. 1984 Aug 2;310(5976):421–423. doi: 10.1038/310421a0. [DOI] [PubMed] [Google Scholar]
- Kranz D. M., Saito H., Disteche C. M., Swisshelm K., Pravtcheva D., Ruddle F. H., Eisen H. N., Tonegawa S. Chromosomal locations of the murine T-cell receptor alpha-chain gene and the T-cell gamma gene. Science. 1985 Feb 22;227(4689):941–945. doi: 10.1126/science.3918347. [DOI] [PubMed] [Google Scholar]
- Kronenberg M., Goverman J., Haars R., Malissen M., Kraig E., Phillips L., Delovitch T., Suciu-Foca N., Hood L. Rearrangement and transcription of the beta-chain genes of the T-cell antigen receptor in different types of murine lymphocytes. Nature. 1985 Feb 21;313(6004):647–653. doi: 10.1038/313647a0. [DOI] [PubMed] [Google Scholar]
- Kronenberg M., Siu G., Hood L. E., Shastri N. The molecular genetics of the T-cell antigen receptor and T-cell antigen recognition. Annu Rev Immunol. 1986;4:529–591. doi: 10.1146/annurev.iy.04.040186.002525. [DOI] [PubMed] [Google Scholar]
- Lee N. E., D'Eustachio P., Pravtcheva D., Ruddle F. H., Hedrick S. M., Davis M. M. Murine T cell receptor beta chain is encoded on chromosome 6. J Exp Med. 1984 Sep 1;160(3):905–913. doi: 10.1084/jem.160.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissen M., McCoy C., Blanc D., Trucy J., Devaux C., Schmitt-Verhulst A. M., Fitch F., Hood L., Malissen B. Direct evidence for chromosomal inversion during T-cell receptor beta-gene rearrangements. Nature. 1986 Jan 2;319(6048):28–33. doi: 10.1038/319028a0. [DOI] [PubMed] [Google Scholar]
- Malissen M., Minard K., Mjolsness S., Kronenberg M., Goverman J., Hunkapiller T., Prystowsky M. B., Yoshikai Y., Fitch F., Mak T. W. Mouse T cell antigen receptor: structure and organization of constant and joining gene segments encoding the beta polypeptide. Cell. 1984 Jul;37(3):1101–1110. doi: 10.1016/0092-8674(84)90444-6. [DOI] [PubMed] [Google Scholar]
- Morton C. C., Duby A. D., Eddy R. L., Shows T. B., Seidman J. G. Genes for beta chain of human T-cell antigen receptor map to regions of chromosomal rearrangement in T cells. Science. 1985 May 3;228(4699):582–585. doi: 10.1126/science.3983642. [DOI] [PubMed] [Google Scholar]
- Müller U., Stephan D., Philippsen P., Steinmetz M. Orientation and molecular map position of the complement genes in the mouse MHC. EMBO J. 1987 Feb;6(2):369–373. doi: 10.1002/j.1460-2075.1987.tb04764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popko B., Puckett C., Lai E., Shine H. D., Readhead C., Takahashi N., Hunt S. W., 3rd, Sidman R. L., Hood L. Myelin deficient mice: expression of myelin basic protein and generation of mice with varying levels of myelin. Cell. 1987 Feb 27;48(4):713–721. doi: 10.1016/0092-8674(87)90249-2. [DOI] [PubMed] [Google Scholar]
- Singer P. A., McEvilly R. J., Noonan D. J., Dixon F. J., Theofilopoulos A. N. Clonal diversity and T-cell receptor beta-chain variable gene expression in enlarged lymph nodes of MRL-lpr/lpr lupus mice. Proc Natl Acad Sci U S A. 1986 Sep;83(18):7018–7022. doi: 10.1073/pnas.83.18.7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. H., Goodenow R. S., Hood L. Molecular basis of the dm1 mutation in the major histocompatibility complex of the mouse: a D/L hybrid gene. J Exp Med. 1985 Nov 1;162(5):1588–1602. doi: 10.1084/jem.162.5.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]