Abstract
Background
Several common genetic polymorphisms in the low density lipoprotein receptor (LDL-R) gene have associated with modifications of serum total cholesterol (TC) and low density lipoprotein cholesterol (LDL-C) levels, but the results are not consistent in different populations. Bai Ku Yao is a special subgroup of the Yao minority in China. The present study was undertaken to detect the association of LDL-R gene Ava Ⅱ polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations.
Methods
A total of 1024 subjects of Bai Ku Yao and 792 participants of Han Chinese were randomly selected from our previous stratified randomized cluster samples. Genotyping of the LDL-R gene Ava Ⅱ polymorphism was performed by polymerase chain reaction and restriction fragment length polymorphism combined with gel electrophoresis, and then confirmed by direct sequencing.
Results
The levels of serum TC, high density lipoprotein cholesterol (HDL-C), LDL-C, apolipoprotein (Apo) A1 and the ratio of ApoA1 to ApoB were lower in Bai Ku Yao than in Han (P < 0.01 for all). The frequency of A- and A+ alleles was 65.5% and 34.5% in Bai Ku Yao, and 80.7% and 19.3% in Han (P < 0.001); respectively. The frequency of A-A-, A-A+ and A+A+ genotypes was 42.6%, 45.9% and 11.5% in Bai Ku Yao, and 64.9%, 31.6% and 3.5% in Han (P < 0.001); respectively. There was also significant difference in the genotypic frequencies between males and females in Bai Ku Yao (P <0.05), and in the genotypic and allelic frequencies between normal LDL-C (≤ 3.20 mmol/L) and high LDL-C (>3.20 mmol/L) subgroups in Bai Ku Yao (P < 0.05 for each) and between males and females in Han (P < 0.05 for each). The levels of LDL-C in males and TC and HDL-C in females were different among the three genotypes (P < 0.05 for all) in Bai Ku Yao, whereas the levels of HDL-C in males and HDL-C and ApoA1 in females were different among the three genotypes (P < 0.05-0.001) in Han. The subjects with A+A+ genotype had higher serum LDL-C, TC, HDL-C or ApoA1 levels than the subjects with A-A+ and A-A- genotypes. Spearman rank correlation analysis revealed that the levels of LDL-C in Bai Ku Yao and HDL-C in Han were correlated with genotypes (P < 0.05 and P < 0.01; respectively).
Conclusions
The association of LDL-R gene Ava Ⅱ polymorphism and serum lipid levels is different between the Bai Ku Yao and Han populations. The discrepancy might partly result from different LDL-R gene Ava Ⅱ polymorphism or LDL-R gene-enviromental interactions.
Introduction
Dyslipidemia including high levels of plasma or serum total cholesterol (TC) [1,2], triglycerides (TGs) [3,4], low density lipoprotein cholesterol (LDL-C) [5,6] and apolipoprotein (Apo) B [7,8], and low levels of ApoA1 [7,8] and high density lipoprotein cholesterol (HDL-C) [9,10], is a growing healthcare problem and risk factor for common diseases such as atherosclerosis, coronary heart diseases (CHD) and hypertension [11]. It has been recognized that dyslipidemia is a multifactorial disease involving environmental [12-14] and genetic factors [15-21] and their interactions [22,23]. Family and twin studies have shown that genetic polymorphisms could account for 40-60% of the interindividual variation in plasma lipid concentrations [24-26].
The low density lipoprotein receptor (LDL-R), a transmembrane cellular protein, plays a crucial role in the receptor-mediated pathway of lipoprotein metabolism [27,28]. The LDL-R modulates plasma levels of LDL-C by regulating the uptake of LDL particles by the liver and delivers cholesterol to the adrenal glands and gonads for steroid hormone synthesis and to the liver for bile acid synthesis [28,29]. Mutations in the LDL-R gene that disturb the normal functions of the LDL-R protein can cause familial hypercholesterolemia (FH), which is associated with elevated total and LDL-C and premature CHD [30,31]. FH, however, accounts for only about 5% of patients with CHD, and the contribution of genes to CHD in the remaining 95% of cases is still unknown [32]. Common single nucleotide polymorphisms (SNPs) in genes involved in lipid metabolism are potentially important genetic markers in affecting normal variation in the plasma or serum lipid profiles and thus determining susceptibility or resistance to CHD in the general population. Since the LDL-R gene plays an important role in cholesterol homeostasis [28], it is also possible that genetic variations in this gene may exist that have only a small effect on the function of the receptor. If such variations are common, they may make an important contribution in determining plasma lipid levels in the general population. The human LDL-R gene consists of 18 exons and 17 introns with the length of approximately 45 kb, and is mapped to chromosome 19p13.2. [33,34]. More than 12 restriction fragment length polymorphisms (RFLPs) within and near the LDL-R gene have been reported [35,36]. A common Ava Ⅱ polymorphism has been found to be associate with variation of plasma or serum lipid levels in some studies [37-47] but not in others [48-50].
There are fifty-six ethnic groups in China. Han is the largest group and Yao is the eleventh largest minority among the 55 minority groups according to the population size. Bai Ku Yao (White-trouser Yao), a special and isolated subgroup of the Yao minority, is named so because all the men wear white knee-length knickerbockers. The population size is about 30000. Because of isolation from the other ethnic groups, the special customs and cultures including their clothing, intra-ethnic marriages, dietary habits, and corn wine and rum intakes are still completely preserved to the present day. In several previous epidemiological studies, we found that the serum lipid parameters were lower in Bai Ku Yao than in Han Chinese from the same region [12-14]. This ethnic difference in serum lipid profiles is still not well known. We postulated that there may be significant differences in some lipid metabolism-related genetic polymorphisms between the two ethnic groups. Therefore, the aim of the present study was to detect the association of LDL-R gene Ava Ⅱ polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations.
Materials and methods
Study population
A total of 1024 subjects of Bai Ku Yao who reside in Lihu and Baxu villages in Nandan County, Guangxi Zhuang Autonomous Region, People's Republic of China were randomly selected from our previous stratified randomized cluster samples [12-14]. The ages of the subjects ranged from 15 to 80 years, with an average age of 36.01 ± 18.16 years. There were 468 males (45.70%) and 556 females (54.30%). All subjects were rural agricultural workers. The subjects accounted for 3.41% of total Bai Ku Yao population. During the same period, a total of 792 people of Han Chinese who reside in the same villages were also randomly selected from our previous stratified randomized cluster samples [12-14]. The mean age of the subjects was 35.67 ± 18.63 years (range 15 to 80). There were 330 men (41.67%) and 462 women (58.33%). All of them were also rural agricultural workers. All study subjects were essentially healthy and had no evidence of any chronic illness, including hepatic, renal, or thyroid. The participants with a history of heart attack or myocardial infarction, stroke, congestive heart failure, diabetes or fasting blood glucose ≥ 7.0 mmol/L determined by glucose meter have been excluded. The participants were not taking medications known to affect serum lipid levels (lipid-lowering drugs such as statins or fibrates, beta-blockers, diuretics, or hormones). The present study was approved by the Ethics Committee of the First Affiliated Hospital, Guangxi Medical University. Informed consent was obtained from all subjects after they received a full explanation of the study.
Epidemiological survey
The survey was carried out using internationally standardized methods, following a common protocol [51]. Information on demographics, socioeconomic status, and lifestyle factors was collected with standardized questionnaires. The alcohol information included questions about the number of liangs (about 50 g) of rice wine, corn wine, rum, beer, or liquor consumed during the preceding 12 months. Alcohol consumption was categorized into groups of grams of alcohol per day: ≤ 25 and ≥ 25. Smoking status was categorized into groups of cigarettes per day: ≤ 20 and ≥ 20. At the physical examination, body height, weight, and waist circumference were measured. Sitting blood pressure was measured three times with the use of a mercury sphygmomanometer after the subjects had a 5-minute rest, and the average of the three measurements was used for the level of blood pressure. Systolic blood pressure was determined by the first Korotkoff sound, and diastolic blood pressure by the fifth Korotkoff sound. Body weight, to the nearest 50 grams, was measured using a portable balance scale. Subjects were weighed without shoes and in a minimum of clothing. Height was measured, to the nearest 0.5 cm, using a portable steel measuring device. From these two measurements body mass index (BMI, kg/m2) was calculated. Waist circumference was measured with a nonstretchable measuring tape, at the level of the smallest area of the waist, to the nearest 0.1 cm.
Lipid analysis
A venous blood sample of 8 mL was obtained from all subjects between 8 and 11 AM, after at least 12 hours of fasting, from a forearm vein after venous occlusion for few seconds in a sitting position. A part of the sample (3 mL) was collected into glass tubes and allowed to clot at room temperature, and used to determine serum lipid levels. Another part of the sample (5 mL) was transferred to tubes with anticoagulate solution (4.80 g/L citric acid, 14.70 g/L glucose, and 13.20 g/L tri-sodium citrate) and used to extract DNA. Immediately following clotting serum was separated by centrifugation for 15 minutes at 3000 rpm. The levels of TC, TG, HDL-C, and LDL-C in samples were determined by enzymatic methods with commercially available kits, Tcho-1, TG-LH (RANDOX Laboratories Ltd., Ardmore, Diamond Road, Crumlin Co. Antrim, United Kingdom, BT29 4QY), Cholestest N HDL, and Cholestest LDL (Daiichi Pure Chemicals Co., Ltd., Tokyo, Japan); respectively. Serum ApoA1 and ApoB levels were detected by the immunoturbidimetric immunoassay using a commercial kit (RANDOX Laboratories Ltd.). All determinations were performed with an autoanalyzer (Type 7170A; Hitachi Ltd., Tokyo, Japan) in the Clinical Science Experiment Center of the First Affiliated Hospital, Guangxi Medical University [12-14].
DNA amplification and genotyping
Genomic DNA was extracted from peripheral blood leukocytes using the phenol-chloroform method [15-21]. The extracted DNA was stored at 4°C until analysis. Genotyping of the LDL-R gene Ava Ⅱ polymorphism was performed by polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP) [32,48]. PCR amplification was carried out with forward primer 5'-GTCATCTTCCTTGCTGCCTGTTTAG-3' and reverse primer 5'-GTTTCCACAAGGAGGTTTCAAGGTT-3' (Sangon, Shanghai, People's Republic of China). Each amplification reaction was performed in a total volume of 25 mL, containing 3 μL of genomic DNA, 1.0 μL of each primer (10 μmol/L), 7.5 μL of ddH2O, and 12.5 μL 2 × Taq PCR MasterMix (constituent: 0.1 U Taq polymerase/μL, 500 μM dNTP each and PCR buffer). After initial denaturizing at 94°C for 3 min, the reaction mixture was subjected to 30 cycles of 30 s denaturation at 94°C, 30 s annealing at 55°C and extension 60 s at 72°C, followed by a final 5 min extension at 72°C. After electrophoresis on a 2.5% agarose gel with 0.5 μg/mL ethidium bromide, the amplification products were visualized under ultraviolet light. Then 10 U of Ava Ⅱ enzyme was added directly to the PCR products (8 μL) and digested at 37°C overnight. After restriction enzyme digestion of the amplified DNA, the genotypes were identified by electrophoresis on 2.5% agarose gels and visualized with ethidium-bromide staining ultraviolet illumination. The genotypes were scored by an experienced reader blinded to epidemiological data and serum lipid levels. Six samples (A-A-, A-A+ and A+A+ genotypes in two; respectively) detected by the PCR-RFLP were also confirmed by direct sequencing. The PCR products were purified by low melting point gel electrophoresis and phenol extraction, and then the DNA sequences were analyzed in Shanghai Sangon Biological Engineering Technology & Services Co., Ltd., People's Republic of China.
Diagnostic criteria
The normal values of serum TC, TG, HDL-C, LDL-C, ApoA1, ApoB levels, and the ratio of ApoA1 to ApoB in our Clinical Science Experiment Center were 3.10-5.17, 0.56-1.70, 0.91-1.81, 2.70-3.20 mmol/L, 1.00-1.78, 0.63-1.14 g/L, and 1.00-2.50; respectively. The individuals with TC >5.17 mmol/L and/or TG >1.70 mmol/L were defined as hyperlipidemic [12-14]. Hypertension was diagnosed according to the criteria of 1999 World Health Organization-International Society of Hypertension Guidelines for the management of hypertension [52,53]. The diagnostic criteria of overweight and obesity were according to the Cooperative Meta-analysis Group of China Obesity Task Force. Normal weight, overweight and obesity were defined as a BMI < 24, 24-28, and >28 kg/m2; respectively [54].
Statistical analyses
Epidemiological data were recorded on a pre-designed form and managed with Excel software. All statistical analyses were done with the statistical software package SPSS 13.0 (SPSS Inc., Chicago, Illinois). Quantitative variables were expressed as mean ± standard deviation (SD), and qualitative variables as percentages. Allele frequency was determined via direct counting, and the standard goodness-of-fit test was used to test the Hardy-Weinberg equilibrium. Differences in genotype distribution between the groups were obtained using the chi-square test. The difference in general characteristics between Bai Ku Yao and Han was tested by the Student's unpaired t-test. The association of serum lipid variables with genotypes was tested by analysis of covariance (ANCOVA). Sex, age, BMI, blood pressure, alcohol intake, cigarette smoking were adjusted for the statistical analysis. In order to confirm the association of serum lipid parameters and genotypes (A-A- = 1, A-A+ = 2 and A+A+ = 3), Spearman rank correlation analysis was also performed in the combined population of Bai Ku Yao and Han, Bai Ku Yao, and Han; respectively. A P value of less than 0.05 was considered statistically significant.
Results
General characteristics and serum lipid levels
Table 1 gives the general characteristics and serum lipid levels between the Bai Ku Yao and Han populations. The levels of body height, weight, TC, HDL-C, LDL-C, ApoA1 and the ratio of ApoA1 to ApoB were lower in Bai Ku Yao than in Han (P < 0.01 for all), whereas the percentages of subjects who consumed alcohol or smoked cigarettes were higher in Bai Ku Yao than in Han (P < 0.01 for each). There was no significant difference in the levels of BMI, systolic blood pressure, diastolic blood pressure, pulse pressure, TG, ApoB, age structure, and the ratio of male to female between the two ethnic groups (P >0.05 for all).
Table 1.
The general characteristics and serum lipid levels between the Bai Ku Yao and Han populations
| Parameter | Bai Ku Yao | Han Chinese | t (x2) | P |
|---|---|---|---|---|
| Number | 1024 | 792 | - | - |
| Male/female | 468/556 | 330/462 | 2.954 | 0.085 |
| Age (years) | 36.01 ± 18.16 | 35.67 ± 18.63 | 0.275 | 0.783 |
| Height (cm) | 151.84 ± 7.13 | 156.30 ± 7.82 | -12.670 | 0.000 |
| Weight (kg) | 49.98 ± 7.19 | 53.26 ± 10.30 | -7.982 | 0.000 |
| Body mass index (kg/m2) | 21.64 ± 2.47 | 21.75 ± 3.64 | -0.766 | 0.444 |
| Waist circumference (cm) | 70.80 ± 7.76 | 71.09 ± 10.27 | -0.486 | 0.627 |
| Systolic blood pressure (mmHg) | 116.94 ± 16.68 | 117.36 ± 17.36 | -0.361 | 0.718 |
| Diastolic blood pressure (mmHg) | 74.06 ± 9.41 | 73.77 ± 10.57 | 0.444 | 0.657 |
| Pulse pressure (mmHg) | 42.89 ± 12.09 | 43.59 ± 11.89 | -0.878 | 0.380 |
| Cigarette smoking [n (%)] | ||||
| Nonsmoker | 786 (76.8) | 636 (80.3) | ||
| <20 cigarettes/day | 120 (11.7) | 58 (7.3) | ||
| ≥ 20 cigarettes/day | 118 (11.5) | 98 (12.4) | 9.791 | 0.007 |
| Alcohol consumption [n (%)] | ||||
| Nondrinker | 688 (67.2) | 646 (81.6) | ||
| <25 g/day | 216 (21.1) | 106 (13.4) | ||
| ≥ 25 g/day | 120 (11.7) | 40 (5.1) | 50.079 | 0.000 |
| Total cholesterol (mmol/L) | 4.21 ± 0.96 | 4.51 ± 0.99 | -4.624 | 0.000 |
| Triglyceride (mmol/L) | 1.23 ± 0.80 | 1.25 ± 0.91 | -0.358 | 0.721 |
| HDL-C (mmol/L) | 1.58 ± 0.42 | 1.74 ± 0.49 | -5.350 | 0.000 |
| LDL-C (mmol/L) | 2.48 ± 0.74 | 2.63 ± 0.79 | -2.991 | 0.003 |
| Apolipoprotein (apo) AI (g/L) | 1.24 ± 0.32 | 1.34 ± 0.36 | -4.708 | 0.000 |
| ApoB (g/L) | 0.81 ± 0.24 | 0.79 ± 0.21 | 1.562 | 0.119 |
| ApoAI/ApoB | 1.63 ± 0.65 | 1.78 ± 0.60 | -3.561 | 0.000 |
HDL-C, highdensity lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol
Results of electrophoresis and genotyping
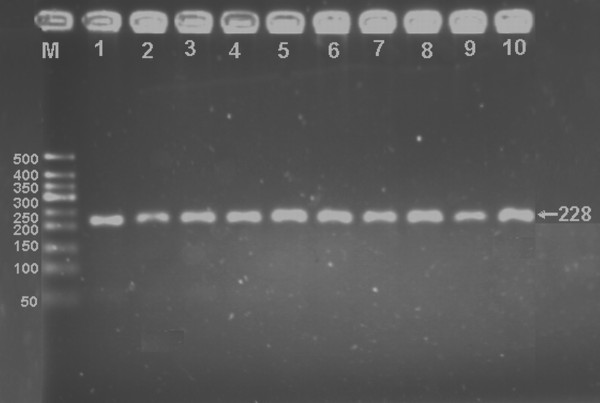
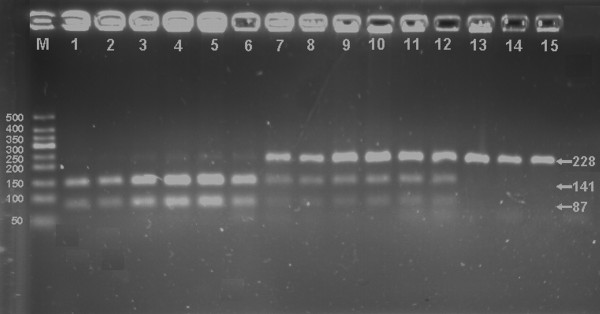
After the genomic DNA of the samples was amplified by PCR and imaged by 2.5% agarose gel electrophoresis, the PCR products of 228 bp nucleotide sequences could be found in all samples (Figure 1). The genotypes identified were named according to the presence or absence of the enzyme restriction sites. The presence of the cutting site indicates the A+ allele, while its absence indicates the A- allele (cannot be cut). Thus, A+A+ genotypes were homozygotes for the presence of the site (141- and 87-bp), A-A+ genotypes were heterozygotes for the absence and presence of the site (228-, 141- and 87-bp), and A-A- genotypes were homozygotes for the absence of the site (228-bp; Figure 2); respectively. The genotypic distribution was consistent with the Hardy-Weinberg equilibrium.
Figure 1.
Electrophoresis of PCR products of the samples. Lane M, 50 bp marker ladder; lanes 1-10, samples. The 228 bp bands are the PCR products.
Figure 2.
Genotyping of the LDL-R gene Ava Ⅱ polymorphism. Lane M, 50 bp marker ladder; lanes 1-6, A+A+ genotype (141- and 87-bp); lanes 7-12, A-A+ genotype (228-, 141- and 87-bp); and lanes 13-15, A-A- genotype (228-bp).
Genotypic and allelic frequencies
The genotypic and allelic frequencies of LDL-R gene Ava Ⅱ polymorphism in Bai Ku Yao and Han Chinese are shown in Table 2. The frequency of A- and A+ alleles was 65.5% and 34.5% in Bai Ku Yao, and 80.7% and 19.3% in Han (P < 0.001); respectively. The frequency of A-A-, A-A+ and A+A+ genotypes was 42.6%, 45.9% and 11.5% in Bai Ku Yao, and 64.9%, 31.6% and 3.5% in Han (P < 0.001); respectively. There was also significant difference in the genotypic frequencies between males and females (P < 0.05) and in the genotypic and allelic frequencies between normal LDL-C (≤ 3.20 mmol/L) and high LDL-C (>3.20 mmol/L; P < 0.05 for each) subgroups in Bai Ku Yao, and in the genotypic and allelic frequencies between males and females in Han (P < 0.05 for each).
Table 2.
Comparison of the genotypic and allelic frequencies of LDL-R gene Ava Ⅱ polymorphism in Bai Ku Yao and Han Chinese [n (%)]
| Group | n | Genotype | Allele | |||
|---|---|---|---|---|---|---|
| A-A- | A-A+ | A+A+ | A- | A+ | ||
| Bai Ku Yao | 1024 | 436 (42.6) | 470 (45.9) | 118 (11.5) | 1342 (65.5) | 706 (34.5) |
| Han Chinese | 792 | 514 (64.9) | 250 (31.6) | 28 (3.5) | 1278 (80.7) | 306 (19.3) |
| x2 | - | 101.118 | 102.054 | |||
| P | - | 0.000 | 0.000 | |||
| Bai Ku Yao | ||||||
| Male | 468 | 218(46.6) | 190(40.6) | 60(12.8) | 626(66.9) | 310(33.1) |
| Female | 556 | 220(39.6) | 278(50.0) | 58(10.4) | 718(64.6) | 394(35.4) |
| x2 | - | 9.095 | 1.204 | |||
| P | - | 0.011 | 0.272 | |||
| Normal TC | 882 | 368 (41.7) | 420 (47.6) | 94 (10.7) | 1156 (65.5) | 608 (34.5) |
| High TC | 142 | 68 (47.9) | 50 (35.2) | 24 (16.9) | 186 (65.5) | 98 (34.5) |
| x2 | - | 9.332 | 0.000 | |||
| P | - | 0.009 | 0.990 | |||
| Normal LDL-C | 902 | 392 (43.5) | 414 (45.9) | 96 (10.6) | 1198 (66.4) | 606 (33.6) |
| High LDL-C | 122 | 44 (36.1) | 56 (45.9) | 22 (18.0) | 144 (59.0) | 100 (41.0) |
| x2 | - | 6.472 | 5.199 | |||
| P | - | 0.039 | 0.023 | |||
| Han Chinese | ||||||
| Male | 330 | 230 (69.7) | 92 (27.9) | 8 (2.4) | 552 (83.6) | 108 (16.4) |
| Female | 462 | 284 (61.5) | 158 (34.2) | 20 (4.3) | 726 (78.6) | 198 (21.4) |
| x2 | - | 6.418 | 6.337 | |||
| P | - | 0.040 | 0.012 | |||
| Normal TC | 622 | 412 (66.2) | 190 (30.6) | 20 (3.2) | 1014 (81.5) | 230 (18.5) |
| High TC | 170 | 102 (60.0) | 60 (35.3) | 8 (4.7) | 264 (77.6) | 76 (22.4) |
| x2 | - | 2.593 | 2.558 | |||
| P | - | 0.274 | 0.109 | |||
| Normal LDL-C | 636 | 414 (65.1) | 202 (31.8) | 20 (3.1) | 1030 (81.0) | 242 (19.0) |
| High LDL-C | 156 | 100 (64.1) | 48 (30.8) | 8 (5.1) | 248 (79.5) | 64 (20.5) |
| x2 | - | 1.452 | 0.356 | |||
| P | - | 0.484 | 0.550 | |||
TC, total cholesterol; LDL-C, low-density lipoprotein cholesterol; Normal TC, TC ≤ 5.17 mmol/L, High TC, TC >5.17 mmol/L; Normal LDL-C, LDL-C ≤ 3.20 mmol/L, High LDL-C, LDL-C >3.20 mmol/L
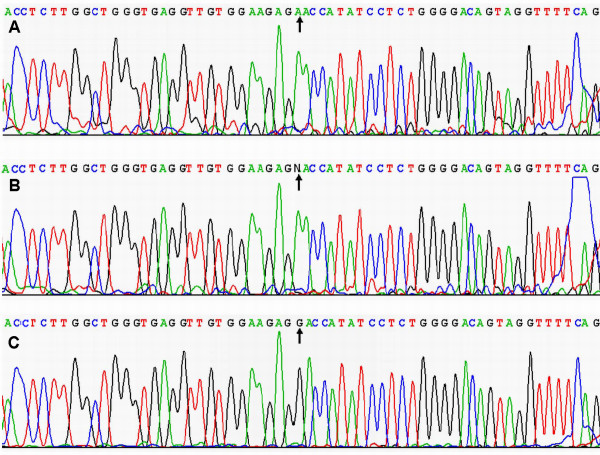
Results of sequencing
The results were shown as A-A-, A-A+ and A+A+ genotypes by PCR-RFLP, the A-A-, A-A+ and A+A+ genotypes were also confirmed by sequencing (Figure 3); respectively.
Figure 3.
A part of the nucleotide sequence of LDL-R gene Ava Ⅱ polymorphism. (A) A-A- genotype; (B) A-A+ genotype; (C) A+A+ genotype.
Genotypes and serum lipid levels
As shown in Table 3, the levels of TC and LDL-C in Bai Ku Yao were significant differences among the three genotypes (P < 0.05 and P < 0.01; respectively). The subjects with A+A+ genotype had higher serum TC levels than the subjects with A-A+ genotypes, and higher serum LDL-C levels than the subjects with A-A+ and A-A- genotypes. When the associations of genotypes and serum lipid parameters were analyzed according to sex, the levels of LDL-C in males and TC and HDL-C in females were significant differences among the three genotypes (P < 0.05 for all). The subjects with A+A+ genotype had higher serum TC, LDL-C or HDL-C levels than the subjects with A-A+ and A-A- genotypes.
Table 3.
Comparison of serum lipid levels among the genotypes of LDL-R gene Ava Ⅱ polymorphism in Bai Ku Yao and Han Chinese
| Genotype | n | TC (mmol/L) | TG (mmol/L) | HDL-C (mmol/L) | LDL-C (mmol/L) | ApoAI (g/L) | ApoB (g/L) |
|---|---|---|---|---|---|---|---|
| Bai Ku Yao | |||||||
| A-A- | 436 | 4.23 ± 0.98 | 1.26 ± 0.82 | 1.58 ± 0.42 | 2.44 ± 0.69 | 1.24 ± 0.33 | 0.82 ± 0.23 |
| A-A+ | 470 | 4.14 ± 0.81 | 1.19 ± 0.79 | 1.56 ± 0.40 | 2.45 ± 0.65 | 1.22 ± 0.31 | 0.81 ± 0.24 |
| A+A+ | 118 | 4.38 ± 0.86c | 1.27 ± 0.73 | 1.66 ± 0.49 | 2.71 ± 1.12bd | 1.28 ± 0.33 | 0.86 ± 0.24 |
| F | - | 3.673 | 1.050 | 2.676 | 6.792 | 1.724 | 2.122 |
| P | - | 0.026 | 0.350 | 0.069 | 0.001 | 0.179 | 0.120 |
| Male | |||||||
| A-A- | 218 | 4.17 ± 1.16 | 1.36 ± 1.00 | 1.57 ± 0.45 | 2.34 ± 0.80 | 1.25 ± 0.38 | 0.75 ± 0.27 |
| A-A+ | 190 | 4.05 ± 0.90 | 1.28 ± 1.00 | 1.56 ± 0.50 | 2.32 ± 0.70 | 1.23 ± 0.40 | 0.75 ± 0.23 |
| A+A+ | 60 | 4.23 ± 0.86 | 1.30 ± 0.79 | 1.62 ± 0.53 | 2.69 ± 1.45bc | 1.27 ± 0.36 | 0.82 ± 0.29 |
| F | - | 1.030 | 0.355 | 0.359 | 4.443 | 0.287 | 1.936 |
| P | - | 0.358 | 0.123 | 0.699 | 0.012 | 0.751 | 0.146 |
| Female | |||||||
| A-A- | 220 | 4.28 ± 0.77 | 1.17 ± 0.58 | 1.59 ± 0.38 | 2.55 ± 0.53 | 1.23 ± 0.28 | 0.86 ± 0.18 |
| A-A+ | 278 | 4.20 ± 0.75 | 1.13 ± 0.62 | 1.56 ± 0.31 | 2.53 ± 0.60 | 1.21 ± 0.23 | 0.85 ± 0.23 |
| A+A+ | 58 | 4.53 ± 0.84ad | 1.23 ± 0.69 | 1.70 ± 0.45ac | 2.73 ± 0.64 | 1.30 ± 0.31 | 0.90 ± 0.17 |
| F | - | 4.494 | 0.732 | 3.746 | 2.917 | 2.901 | 1.420 |
| P | - | 0.012 | 0.482 | 0.024 | 0.055 | 0.056 | 0.243 |
| Han Chinese | |||||||
| A-A- | 514 | 4.49 ± 1.02 | 1.23 ± 0.87 | 1.70 ± 0.47 | 2.63 ± 0.82 | 1.34 ± 0.34 | 0.79 ± 0.21 |
| A-A+ | 250 | 4.56 ± 0.94 | 1.31 ± 0.99 | 1.78 ± 0.50 | 2.63 ± 0.76 | 1.35 ± 0.38 | 0.79 ± 0.20 |
| A+A+ | 28 | 4.44 ± 1.08 | 1.17 ± 1.08 | 2.14 ± 0.54bd | 2.60 ± 0.77 | 1.40 ± 0.45 | 0.79 ± 0.23 |
| F | - | 0.486 | 0.759 | 12.262 | 0.018 | 0.407 | 0.004 |
| P | - | 0.615 | 0.469 | 0.000 | 0.982 | 0.666 | 0.996 |
| Male | |||||||
| A-A- | 230 | 4.45 ± 0.94 | 1.35 ± 1.13 | 1.62 ± 0.48 | 2.64 ± 0.75 | 1.30 ± 0.37 | 0.80 ± 0.21 |
| A-A+ | 92 | 4.45 ± 0.93 | 1.50 ± 1.28 | 1.73 ± 0.44 | 2.54 ± 0.82 | 1.33 ± 0.39 | 0.76 ± 0.20 |
| A+A+ | 8 | 3.80 ± 1.45 | 1.56 ± 1.78 | 1.77 ± 0.46 | 2.17 ± 0.92 | 1.02 ± 0.21 | 0.66 ± 0.21 |
| F | - | 1.823 | 0.598 | 2.059 | 1.818 | 2.549 | 2.725 |
| P | - | 0.163 | 0.550 | 0.129 | 0.164 | 0.080 | 0.067 |
| Female | |||||||
| A-A- | 284 | 4.53 ± 1.09 | 1.13 ± 0.56 | 1.76 ± 0.45 | 2.62 ± 0.87 | 1.36 ± 0.32 | 0.78 ± 0.22 |
| A-A+ | 158 | 4.62 ± 0.95 | 1.20 ± 0.76 | 1.82 ± 0.54 | 2.69 ± 0.72 | 1.37 ± 0.38 | 0.81 ± 0.20 |
| A+A+ | 20 | 4.79 ± 0.66 | 0.95 ± 0.38 | 2.35 ± 0.49 | 2.83 ± 0.61 | 1.56 ± 0.42ac | 0.87 ± 0.70 |
| F | - | 0.857 | 1.640 | 13.965 | 0.874 | 3.123 | 1.658 |
| P | - | 0.425 | 0.195 | 0.000 | 0.418 | 0.045 | 0.192 |
TC, total cholesterol; TG, triglycerides; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; ApoAI, apolipoprotein AI; ApoB, apolipoprotein B; aP < 0.05 and bP < 0.01 in comparison with A-A- genotype of same group; cP < 0.05 and dP < 0.01 in comparison with A-A+ genotype of same group.
The levels of HDL-C in Han Chinese were significant differences among the three genotypes (P < 0.001). The subjects with A+A+ genotype had higher serum HDL-C levels than the subjects with A-A+ and A-A- genotypes. When the assocoations of genotypes and serum lipid parameters were analyzed according to sex, the levels of HDL-C in males and HDL-C and ApoA1 in females were significant differences among the three genotypes (P < 0.05-0.001). The subjects with A+A+ genotype had higher serum HDL-C and ApoA1 levels than the subjects with A-A+ and A-A- genotypes.
Risk factors for the lipid parameters
Spearman rank correlation analysis showed that serum LDL-C levels in Bai Ku Yao were correlated with genotypes (P < 0.05), whereas serum HDL-C levels in Han Chinese were associated with genotypes (P < 0.01; Table 4).
Table 4.
The results of Spearman rank correlation analysis between serum lipid parameters and the genotypes of LDL-R gene Ava Ⅱ polymorphism
| Regression coefficient | Bai Ku Yao and Han | Bai Ku Yao | Han Chinese |
|---|---|---|---|
| rTC | 0.016 | 0.028 | 0.011 |
| rTG | 0.009 | 0.008 | 0.013 |
| rHDL-C | 0.062 | 0.018 | 0.116** |
| rLDL-C | 0.053 | 0.092* | 0.014 |
| rApoA1 | 0.026 | 0.020 | 0.028 |
| rApoB | 0.022 | 0.017 | 0.034 |
r, regression coefficient; TC, total cholesterol; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; ApoA1, apolipoprotein A1; ApoB, apolipoprotein B; *P < 0.05 and **P < 0.01.
Discussion
The present study showed that the levels of serum TC, HDL-C, LDL-C, ApoA1 and the ratio of ApoA1 to ApoB were lower in Bai Ku Yao than in Han. There was no significant difference in serum TG and ApoB levels between the two ethnic groups. These findings are consistent with those of our previous epidemiological studies [12-14]. It is well known that dyslipidemia is a complex trait caused by multiple environmental and genetic factors and their interactions. Differences in serum lipid profiles between the two ethnic groups might mainly result from different dietary patterns, lifestyle factors as well as genetic background. Bai Ku Yao is an isolated subgroup of the Yao minority in China. There are about 30000 people of total Bai Ku Yao population who reside in two villages, Lihu and Baxu, Nandan County. Their ancestors began their migration from Hunan and Guizhou Province about Song Dynasty (a ruling dynasty in China between 960 and 1279). Both Lihu and Baxu villages are typical infertile mountain area, usually it was called 30 percent soil with 70 percent rock. Their income mostly comes from planting corn and paddy. Thus, corn was the staple food and rice, soy, buckwheat, sweet potato, and pumpkin products were the subsidiary foods in Bai Ku Yao. Approximately 90% of the beverages were corn wine and rum. The alcohol content is about 15% (v/v). They are also accustomed to drink hempseed soup and eat hempseed products. Several experimental and clinical studies have demonstrated that the Hempseed products and Hempseed oil can decrease serum TC, TG and LDL-C levels [55-57]. In contrast, rice was the staple food and corn, broomcorn, potato, and taro products were the subsidiary foods in Han. About 90% of the beverage was rice wine. The content of alcohol is about 30% (v/v). The staple and subsidiary foods are more favorable for serum lipid profiles in Bai Ku Yao than in Han [58-62]. In addition, strict intra-ethnic marriages have been performed in this ethnic subgroup from time immemorial. Namely, only both Bai Ku Yao man and woman can intermarriage, and can not intermarry with the other subgroup of Yao minority or other ethnic groups. Therefore, this unique ethnic subgroup with a homogenous environmental and genetic background is a useful tool for genetic study.
Several previous studies have shown that the genotypic and allelic distribution of LDL-R gene Ava Ⅱ polymorphism was different in diverse populations. Ahn et al. [32] investigated the effect of LDL-R gene Ava Ⅱ polymorphism on plasma lipid profiles in 385 normolipidemic Hispanics and 543 non-Hispanic whites from the San Luis Valley, Colorado. They showed that the frequency of A+ allele was higher in Hispanics than in non-Hispanic whites (56% vs. 43%; P <0.001). Salazar et al. [38] reported that the frequency of A+A+ genotype was higher in 170 white unrelated individuals presenting a lipid profile with high risk for CHD compared to that of the 130 controls (32% vs. 16%) from São Paulo City, Brazil. Salazar et al. [40] also found that the frequency of A+ allele was higher in 50 patients with FH than in 130 normal controls (58% vs. 45%; P = 0.036). The FH patients showed a greater frequency of A+A+ homozygous genotypes when compared to the normolipidemic controls (P = 0.0292). In addtion, several studies also showed that the frequency of A+ allele was higher in patients with acute myocardial infarction [45], phlegm-dampness constitution [47], gallstone disease [48], but was lower in patients with atherosclerotic cerebral infarction [50] as compared with the corresponding normal controls; respectively. Liu et al. [43] found that the frequency of A+ allele in males but not in females was higher in hyperlipidemic (24.51%) and margin lipid patients (25.00%) than in normal controls (11.11%; P < 0.05 for each). Several studies, however, showed that there was no significant difference in the frequency of A+ allele between the CHD and control groups [44,46], and between male and female Han children [49]. In the present study, we showed that the frequency of A+ allele and A-A+ and A+A+ genotypes was higher in Bai Ku Yao than in Han. There was also significant difference in the genotypic frequencies between males and females in Bai Ku Yao, and in the genotypic and allelic frequencies between normal LDL-C (≤ 3.20 mmol/L) and high LDL-C (>3.20 mmol/L) subgroups in Bai Ku Yao and between males and females in Han. These results indicate that the prevalence of the A+ allele variants of LDL-R gene Ava Ⅱ polymorphism may have a racial/ethnic specificity.
The association of LDL-R gene Ava Ⅱ polymorphism and plasma or serum lipid levels in humans are inconsistent in several previous studies. Ahn et al. [32] reported that the LDL-R gene Ava Ⅱ polymorphism revealed a gender-specific effect on plasma lipid profiles. The gender-specific effect was significant for HDL-C (P < 0.001) and TG (P = 0.001). Ethnicity affected only the levels of TG (P = 0.02). Significant variability among Ava II genotypes was observed for TC and LDL-C in non-Hispanic women (P = 0.0015 and P = 0.014) and TC in Hispanic women (P = 0.011) and was highly suggestive for LDL-C in Hispanic women (P = 0.057). For both non-Hispanic and Hispanic women the trends seen in the cholesterol levels were similar: low in the A-A- genotype, intermediate in the A-A+ genotype, and high in the A+A+ genotype. Neither significant nor directional results were seen in men in either ethnic group. In non-Hispanic women the estimated average effect of the A+ allele was to raise LDL-C by 4.95 mg/dL, and the average effect of the A- allele was to lower LDL-C by 3.68 mg/dL; this variation explained 3.9% of the phenotypic variance in TC and 2.3% in LDL-C. In Hispanic women the estimated average effect of the A+ allele was to increase LDL-C by 2.92 mg/dL, and the A- allele lowered levels by 3.81 mg/dL; this variation explained 3.7% and 2% of the phenotypic variance in TC and LDL-C; respectively. Humphries et al. [37] showed that the individuals with one or two A- alleles (absence of cutting site) had levels of LDL-C that were lower by 8.0% and 12.4%; respectively. Variation associated with this RFLP explained 4.2% of the sample variance. Pongrapeeporn et al. [41] also revealed that the mean LDL-C levels was slightly higher in the A+A+ genotype than the A-A+ and A-A- genotypes in 54 normolipidemic Thai subjects. The difference was significant at the 5 per cent level although there were only three homozygotes with the A+A+ genotype. The average effect of the A+ allele was to increase LDL-C level by 6.75 mg/dl. A gene-dosage effect was not observed for this polymorphism. In addition, the subjects with the A+A+ genotype also tended to have high serum TC and TG and low HDL-C levels. The LDL-R gene Ava Ⅱ polymorphism was also found to be associate with the variability of plasma or serum lipid levels in subjects presenting a lipid profile with high risk for CHD [38], in normal individuals from five Brazilian Indian tribes [39], in males with hypercholesterolemia [43], in cases with acute myocardial infarction [45], in patients with CHD [46], and in population with phlegm-dampness constitution [47]. Several studies, however, failed to find a significant association between the LDL-R gene Ava Ⅱ polymorphism and plasma or serum lipid levels in normal controls [48,50] or in Han children [49]. In the present study, we showed that the levels of TC and LDL-C in Bai Ku Yao were significant differences among the three genotypes. The subjects with A+A+ genotype had higher serum TC levels than the subjects with A-A+ genotypes, and higher serum LDL-C levels than the subjects with A-A+ and A-A- genotypes. When the statistical analyses were performed according to sex, the levels of LDL-C in males and TC and HDL-C in females were significant differences among the three genotypes. The subjects with A+A+ genotype had higher serum LDL-C, TC or HDL-C levels than the subjects with A-A+ and A-A- genotypes. The levels of HDL-C in Han Chinese were significant differences among the three genotypes. The subjects with A+A+ genotype had higher serum HDL-C levels than the subjects with A-A+ and A-A- genotypes. When the analyses were performed for each gender category separately, the levels of HDL-C in males and HDL-C and ApoA1 in females were significant differences among the three genotypes. The subjects with A+A+ genotype had higher serum HDL-C and ApoA1 levels than the subjects with A-A+ and A-A- genotypes. Spearman rank correlation analysis showed that serum LDL-C levels in Bai Ku Yao were correlated with genotypes, whereas serum HDL-C levels in Han Chinese were associated with genotypes. The reason for this discrepancy between the two ethnic groups may relate to the difference in the LDL-R gene Ava Ⅱ polymorphism and/or LDL-R gene-enviromental interactions [44].
Conclusion
The present study shows that the frequency of A+ allele and A-A+ and A+A+ genotypes was higher in Bai Ku Yao than in Han. There was also significant difference in the genotypic frequencies between males and females and in the genotypic and allelic frequencies between normal LDL-C (≤ 3.20 mmol/L) and high LDL-C (>3.20 mmol/L) subgroups in Bai Ku Yao, and in the genotypic and allelic frequencies between males and females in Han. The levels of LDL-C in males and TC and HDL-C in females were different among the three genotypes in Bai Ku Yao, whereas the levels of HDL-C in males and HDL-C and ApoA1 in females were different among the three genotypes in Han. The A+A+ homozygotes had higher serum LDL-C, TC, HDL-C or ApoA1 levels than the A-A+ heterozygotes and A-A- homozygotes. The levels of LDL-C in Bai Ku Yao and HDL-C in Han were correlated with genotypes. The difference in the association of LDL-R gene Ava Ⅱ polymorphism and serum lipid levels between the two ethnic groups might partly result from different LDL-R gene Ava Ⅱ polymorphism or LDL-R gene-enviromental interactions.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
XJL participated in the design, undertook genotyping, and helped to draft the manuscript. RXY conceived the study, participated in the design, carried out the epidemiological survey, collected the samples, and drafted the manuscript. KLL, WYL, LZ, XLC, LM, DFW, LHHA and XJH collaborated to the genotyping. All authors read and approved the final manuscript.
Contributor Information
Xing-Jiang Long, Email: 511052795@qq.com.
Rui-Xing Yin, Email: yinruixing@yahoo.com.cn.
Ke-La Li, Email: likela.cn@gmail.com.
Wan-Ying Liu, Email: liuwanying1224@yahoo.com.cn.
Lin Zhang, Email: bugemiulin@sina.com.
Xiao-Li Cao, Email: maten1996@gmail.com.
Lin Miao, Email: caotianshuangmu@163.com.
Dong-Feng Wu, Email: wulove26@tom.com.
Lynn Htet Htet Aung, Email: lifeinnivana@gmail.com.
Xi-Jiang Hu, Email: huxijianght@126.com.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No: 30660061)
References
- Shekelle RB, Shryock AM, Paul O, Lepper M, Stamler J, Liu S, Raynor WJ Jr. Diet, serum cholesterol, and death from coronary heart disease. The Western Electric study. N Engl J Med. 1981;304:65–70. doi: 10.1056/NEJM198101083040201. [DOI] [PubMed] [Google Scholar]
- Expert Panel on Detection, Evaluation and Treatment of High Blood Cholesterol in Adults. Executive Summary of the third report of the National Cholesterol Education Program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) JAMA. 2001;285:2486–97. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- Satoh H, Nishino T, Tomita K, Tsutsui H. Fasting triglyceride is a significant risk factor for coronary artery disease in middle-aged Japanese men: Results from a 10-year cohort study. Circ J. 2006;70:227–31. doi: 10.1253/circj.70.227. [DOI] [PubMed] [Google Scholar]
- Jeppesen J, Hein HO, Suadicani P, Gyntelberg F. Triglyceride concentration and ischemic heart disease: an eight-year follow-up in the Copenhagen Male Study. Circulation. 1998;97:1029–36. doi: 10.1161/01.cir.97.11.1029. [DOI] [PubMed] [Google Scholar]
- Achari V, Thakur AK. Association of major modifiable risk factors among patients with coronary artery disease--a retrospective analysis. J Assoc Physicians India. 2004;52:103–8. [PubMed] [Google Scholar]
- März W, Scharnagl H, Winkler K, Tiran A, Nauck M, Boehm BO, Winkelmann BR. Low-density lipoprotein triglycerides associated with low-grade systemic inflammation, adhesion molecules, and angiographic coronary artery disease: the Ludwigshafen Risk and Cardiovascular Health study. Circulation. 2004;110:3068–74. doi: 10.1161/01.CIR.0000146898.06923.80. [DOI] [PubMed] [Google Scholar]
- Kwiterovich PO Jr, Coresh J, Smith HH, Bachorik PS, Derby CA, Pearson TA. Comparison of the plasma levels of apolipoproteins B and A-1, and other risk factors in men and women with premature coronary artery disease. Am J Cardiol. 1992;69:1015–21. doi: 10.1016/0002-9149(92)90856-T. [DOI] [PubMed] [Google Scholar]
- Durrington PN, Hunt L, Ishola M, Kane J, Stephens WP. Serum apolipoproteins AI and B and lipoproteins in middle aged men with and without previous myocardial infarction. Br Heart J. 1986;56:206–12. doi: 10.1136/hrt.56.3.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden WE. High-density lipoprotein cholesterol as an independent risk factor in cardiovascular disease: Assessing the data from Framingham to the Veterans Affairs High-Density Lipoprotein Intervention Trail. Am J Cardiol. 2000;86:19L–22L. doi: 10.1016/S0002-9149(00)01464-8. [DOI] [PubMed] [Google Scholar]
- Gordon DJ, Probstfield JL, Garrison RJ, Neaton JD, Castelli WP, Knoke JD, Jacobs DR Jr, Bangdiwala S, Tyroler HA. High-density lipoprotein cholesterol and cardiovascular disease. Four prospective American studies. Circulation. 1989;79:8–15. doi: 10.1161/01.cir.79.1.8. [DOI] [PubMed] [Google Scholar]
- Stamler J, Daviglus ML, Garside DB, Dyer AR, Greenland P, Neaton JD. Relationship of baseline serum cholesterol levels in 3 large cohorts of younger men to long-term coronary, cardiovascular, and all-cause mortality and to longevity. JAMA. 2000;284:311–8. doi: 10.1001/jama.284.3.311. [DOI] [PubMed] [Google Scholar]
- Ruixing Y, Qiming F, Dezhai Y, Shuquan L, Weixiong L, Shangling P, Hai W, Yongzhong Y, Feng H, Shuming Q. Comparison of demography, diet, lifestyle, and serum lipid levels between the Guangxi Bai Ku Yao and Han populations. J Lipid Res. 2007;48:2673–81. doi: 10.1194/jlr.M700335-JLR200. [DOI] [PubMed] [Google Scholar]
- Ruixing Y, Shangling P, Hong C, Hanjun Y, Hai W, Yuming C, Jinzhen W, Feng H, Meng L, Muyan L. Diet, alcohol consumption, and serum lipid levels of the middle-aged and elderly in the Guangxi Bai Ku Yao and Han populations. Alcohol. 2008;42:219–29. doi: 10.1016/j.alcohol.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Ruixing Y, Dezhai Y, Shuquan L, Yuming C, Hanjun Y, Qiming F, Shangling P, Weixiong L, Jing T, Yiyang L. Hyperlipidaemia and its risk factors in the Guangxi Bai Ku Yao and Han populations. Public Health Nutr. 2009;12:816–24. doi: 10.1017/S1368980008003273. [DOI] [PubMed] [Google Scholar]
- Meng L, Ruixing Y, Yiyang L, Xingjiang L, Kela L, Wanying L, Lin Z, Weixiong L, Dezhai Y, Shangling P. Association of LIPC -250G > A polymorphism and several environmental factors with serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2010;9:28. doi: 10.1186/1476-511X-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WY, Yin RX, Zhang L, Cao XL, Miao L, Wu DF, Aung LH, Hu XJ, Lin WX, Yang DZ. Association of the LIPG 584C > T polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2010;9:110. doi: 10.1186/1476-511X-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yin RX, Liu WY, Miao L, Wu DF, Aung LH, Hu XJ, Cao XL, Wu JZ, Pan SL. Association of methylenetetrahydrofolate reductase C677T polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2010;9:123. doi: 10.1186/1476-511X-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DF, Yin RX, Aung LH, Hu XJ, Cao XL, Miao L, Li Q, Yan TT, Wu JZ, Pan SL. Polymorphism of rs10449925 in the acyl-CoA:cholesterol acyltransferase-1 gene and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2010;9:139. doi: 10.1186/1476-511X-9-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, Yin RX, Wu DF, Cao XL, Li Q, Hu XJ, Yan TT, Aung LH, Yang DZ, Lin WX. Peroxisome proliferator-activated receptor delta +294T > C polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2010;9:145. doi: 10.1186/1476-511X-9-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung LH, Yin RX, Miao L, Hu XJ, Yan TT, Cao XL, Wu DF, Li Q, Pan SL, Wu JZ. The proprotein convertase subtilisin/kexin type 9 gene E670G polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2011;10:5. doi: 10.1186/1476-511X-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XL, Yin RX, Wu DF, Miao L, Aung LH, Hu XJ, Li Q, Yan TT, Lin WX, Pan SL. Genetic variant of V825I in the ATP-binding cassette transporter A1 gene and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2011;10:14. doi: 10.1186/1476-511X-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yin R, Deng Y, Li Y, Wu J. Interactions between alcohol intake and the polymorphism of rs708272 on serum high-density lipoprotein cholesterol levels in the Guangxi Hei Yi Zhuang population. Alcohol. 2008;42:583–91. doi: 10.1016/j.alcohol.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Ruixing Y, Yiyang L, Meng L, Kela L, Xingjiang L, Lin Z, Wanying L, Jinzhen W, Dezhai Y, Weixiong L. Interactions of the apolipoprotein C-III 3238C > G polymorphism and alcohol consumption on serum triglyceride levels. Lipids Health Dis. 2010;9:86. doi: 10.1186/1476-511X-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller DA, de Faire U, Pedersen NL, Dahlén G, McClearn GE. Genetic and environmental influences on serum lipid levels in twins. N Engl J Med. 1993;328:1150–6. doi: 10.1056/NEJM199304223281603. [DOI] [PubMed] [Google Scholar]
- Steinmetz J, Boerwinkle E, Gueguen R, Visvikis S, Henny J, Siest G. Multivariate genetic analysis of high density lipoprotein particles. Atherosclerosis. 1992;92:219–27. doi: 10.1016/0021-9150(92)90281-K. [DOI] [PubMed] [Google Scholar]
- Pérusse L, Rice T, Després JP, Bergeron J, Province MA, Gagnon J, Leon AS, Rao DC, Skinner JS, Wilmore JH, Bouchard C. Familial resemblance of plasma lipids, lipoproteins and postheparin lipoprotein and hepatic lipases in the HERITAGE Family Study. Arterioscler Thromb Vasc Biol. 1997;17:3263–9. doi: 10.1161/01.atv.17.11.3263. [DOI] [PubMed] [Google Scholar]
- Dammerman M, Breslow JL. Genetic basis of lipoprotein disorders. Circulation. 1995;91:505–12. doi: 10.1161/01.cir.91.2.505. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- Heber D, Koziol BJ, Henson LC. Low density lipoprotein receptor regulation and the cellular basis of atherosclerosis: implications for nutritional and pharmacologic treatment of hypercholesterolemia. Am J Cardiol. 1987;60:4G–8G. doi: 10.1016/0002-9149(87)90585-6. [DOI] [PubMed] [Google Scholar]
- Leren TP, Bakken KS, Rødningen OK, Gundersen KE, Sundvold H, Berg K, Tonstad S, Ose L. Application of gene technology in the diagnosis of familial hypercholesterolaemia. Tidsskr Nor Laegeforen. 1997;117:678–81. [PubMed] [Google Scholar]
- Vuorio AF, Ojala JP, Sarna S, Turtola H, Tikkanen MJ, Kontula K. Heterozygous familial hypercholesterolaemia: the influence of the mutation type of the low-density-lipoprotein receptor gene and PvuII polymorphism of the normal allele on serum lipid levels and response to lovastatin treatment. J Intern Med. 1995;237:43–8. doi: 10.1111/j.1365-2796.1995.tb01138.x. [DOI] [PubMed] [Google Scholar]
- Ahn YI, Kamboh MI, Aston CE, Ferrell RE, Hamman RF. Role of common genetic polymorphisms in the LDL receptor gene in affecting plasma cholesterol levels in the general population. Arterioscler Thromb. 1994;14:663–70. doi: 10.1161/01.atv.14.5.663. [DOI] [PubMed] [Google Scholar]
- Siidhof TC, Goldstein JL, Brown MS, Russell DW. The LDL receptor gene: a mosaic of exons shared with different proteins. Science. 1985;228:815–22. doi: 10.1126/science.2988123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren V, Luskey KL, Russell DW, Francke U. Human genes involved in cholesterol metabolism: chromosomal mapping of the loci for the low density lipoprotein receptor and 3-hydroxy-3-methylglutaryl-coenzyme A reductase with cDNA probes. Proc Natl Acad Sci USA. 1985;82:8567–71. doi: 10.1073/pnas.82.24.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitersdorf E, Chakravarti A, Hobbs HH. Polymorphic DNA haplotypes at the LDL receptor locus. Am J Hum Genet. 1989;44:409–21. [PMC free article] [PubMed] [Google Scholar]
- Hegele RA, Plaetke R, Lalouel J. Linkage disequilibrium between DNA markers at the low-density lipoprotein receptor gene. Genet Epidemiol. 1990;7:69–81. doi: 10.1002/gepi.1370070114. [DOI] [PubMed] [Google Scholar]
- Humphries S, Coviello DA, Masturzo P, Balestreri R, Orecchini G, Bertolini S. Variation in the low density lipoprotein receptor gene is associated with differences in plasma low density lipoprotein cholesterol levels in young and old normal individuals from Italy. Arterioscler Thromb. 1991;11:509–16. doi: 10.1161/01.atv.11.3.509. [DOI] [PubMed] [Google Scholar]
- Salazar LA, Hirata MH, Giannini SD, Forti N, Diament J, Issa JS, Hirata RD. Effects of Ava II and Hinc II polymorphisms at the LDL receptor gene on serum lipid levels of Brazilian individuals with high risk for coronary heart disease. J Clin Lab Anal. 1999;13:251–8. doi: 10.1002/(SICI)1098-2825(1999)13:6<251::AID-1>3.0.CO;2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattevi VS, Coimbra CE Jr, Santos RV, Salzano FM, Hutz MH. Association of the low-density lipoprotein receptor gene with obesity in Native American populations. Hum Genet. 2000;106:546–52. doi: 10.1007/s004390050023. [DOI] [PubMed] [Google Scholar]
- Salazar LA, Cavalli SA, Hirata MH, Diament J, Forti N, Giannini SD, Nakandakare ER, Bertolami MC, Hirata RD. Polymorphisms of the low-density lipoprotein receptor gene in Brazilian individuals with heterozygous familial hypercholesterolemia. Braz J Med Biol Res. 2000;33:1301–4. doi: 10.1590/S0100-879X2000001100006. [DOI] [PubMed] [Google Scholar]
- Pongrapeeporn KU, Pimsawat T, Likidlilid A, Poldee S, Yamwong P, Amornrattana A, Ong-Ajyooth S. Effect of Ava II and NcoI polymorphisms at the low density lipoprotein receptor gene on plasma lipid levels in a group of Thai subjects. J Med Assoc Thai. 2000;83:S74–80. [PubMed] [Google Scholar]
- Chae JJ, Kim SH, Kim UK, Hong SS, Kim YS, Namkoong Y, Park YB, Lee CC. Polymorphic DNA haplotypes at the human low-density lipoprotein receptor gene locus in Koreans. Hum Biol. 2001;73:105–19. doi: 10.1353/hub.2001.0004. [DOI] [PubMed] [Google Scholar]
- Liu AP, Zhan SY, Li LM. The relationship of low density lipoprotein receptor gene polymorphism and hyperlipidemia. Zhonghua Liu Xing Bing Xue Za Zhi. 2001;22:30–3. [PubMed] [Google Scholar]
- Wang CH, Zhou X, Zhou GD, Han DF, Zheng F. Interaction of ApoE and LDL-R gene polymorphisms and alcohol drinking and smoking on coronary heart disease. Zhonghua Yi Xue Za Zhi. 2004;84:554–8. [PubMed] [Google Scholar]
- Zang B, Guo JJ, Pan LL, Wu KG. Relationship between Ava II polymorphism of the LDL receptor gene and acute myocardial infarction. Shanxi Med J. 2004;33:882–4. [Google Scholar]
- Ouyang T, Song JN, Lin Q, Li L, Miao Y, Niu XH, Jin H, Chen B. Study on relationship between the polymorphisms of low density lipoprotein receptor gene and the constitutions in patients with coronary heart disease. Chin J Basic Med Tradit Chin Med. 2005;11:521–3. doi: 10.3736/jcim20050605. [DOI] [PubMed] [Google Scholar]
- Zhou J, Luo HM. Exploration of the relationship between phlegm-dampness constitution and polymorphism of low density lipoprotein receptor gene Pvu II and AvaⅡ. Chin J Integr Med. 2007;13:170–4. doi: 10.1007/s11655-007-0170-1. [DOI] [PubMed] [Google Scholar]
- Yu RB, Shen C, Tan YF, Yang S, Ding WL, Zhou WM, Gong WD, Zhou JY, Yao CL. Relationship between the Ava Ⅱ polymorphism of low-density lipoprotein receptor gene and gallstone disease. World Chin J Digestol. 2002;10:1412–4. [Google Scholar]
- Feng NP, Zhu WL, Wang Y, Ma J, Ye GJ. Gene polymorphism at LDL receptor Ava Ⅱ locus and its relations with serum lipids profile in 308 children of Han nationality. China Public Health. 2002;18:29–31. [Google Scholar]
- Guo Y, Guo J, Zheng D, Pan L, Li Q, Ruan G. Relationship between the Nco I, Ava II polymorphism of low density lipoprotein receptor gene and atherosclerotic cerebral infarction. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2002;19:209–12. [PubMed] [Google Scholar]
- People's Republic of China--United States Cardiovascular and Cardiopulmonary Epidemiology Research Group. An epidemiological study of cardiovascular and cardiopulmonary disease risk factors in four populations in the People's Republic of China. Baseline report from the P.R.C.-U.S.A. Collaborative Study. Circulation. 1992;85:1083–96. doi: 10.1161/01.cir.85.3.1083. [DOI] [PubMed] [Google Scholar]
- Ruixing Y, Weixiong L, Hanjun Y, Dezhai Y, Shuquan L, Shangling P, Qiming F, Jinzhen W, Jianting G, Yaju D. Diet, lifestyle, and blood pressure of the middle-aged and elderly in the Guangxi Bai Ku Yao and Han populations. Am J Hypertens. 2008;21:382–7. doi: 10.1038/ajh.2008.1. [DOI] [PubMed] [Google Scholar]
- Ruixing Y, Shangling P, Shuquan L, Dezhai Y, Weixiong L, Qiming F, Yuming C, Yaoheng H, Yijiang Z, Qinchen L. Comparison of hypertension and its risk factors between the Guangxi Bai Ku Yao and Han populations. Blood Press. 2008;17:306–16. doi: 10.1080/08037050802589593. [DOI] [PubMed] [Google Scholar]
- Cooperative Meta-analysis Group of China Obesity Task Force. Predictive values of body mass index and waist circumference to risk factors of related diseases in Chinese adult population. Zhonghua Liu Xing Bing Xue Za Zhi. 2002;23:5–10. [PubMed] [Google Scholar]
- Cen L, Qin W, Ye Y. Effect of Canabis Sativa L on serum cholesterol level in rats. J Guangxi Med Univ. 1984;1:20–2. [Google Scholar]
- Qin W, Cen L, Ye Y. The effect of some foods on serum cholesterol level in rats. Acta Nutrimenta Sinica. 1986;8:136–40. [Google Scholar]
- Schwab US, Callaway J, Erkkila AT, Gynther J, Uusitupa MI, Jarvinen T. Effects of hempseed and flaxseed oils on the profile of serum lipids, serum total and lipoprotein lipid concentrations and haemostatic factors. Eur J Nutr. 2006;45:470–7. doi: 10.1007/s00394-006-0621-z. [DOI] [PubMed] [Google Scholar]
- Shane JM, Walker PM. Corn bran supplementation of a low-fat controlled diet lowers serum lipids in men with hypercholesterolemia. J Am Diet Assoc. 1995;95:40–5. doi: 10.1016/S0002-8223(95)00011-9. [DOI] [PubMed] [Google Scholar]
- Zhan S, Ho SC. Meta-analysis of the effects of soy protein containing isoflavones on the lipid profile. Am J Clin Nutr. 2005;81:397–408. doi: 10.1093/ajcn.81.2.397. [DOI] [PubMed] [Google Scholar]
- Tomotake H, Shimaoka I, Kayashita J, Yokoyama F, Nakajoh M, Kato N. Stronger suppression of plasma cholesterol and enhancement of the fecal excretion of steroids by a buckwheat protein product than by a soy protein isolate in rats fed on a cholesterol-free diet. Biosci Biotechnol Biochem. 2001;65:1412–4. doi: 10.1271/bbb.65.1412. [DOI] [PubMed] [Google Scholar]
- Ludvik BH, Mahdjoobian K, Waldhaeusl W, Hofer A, Prager R, Kautzky-Willer A, Pacini G. The effect of Ipomoea batatas (Caiapo) on glucose metabolism and serum cholesterol in patients with type 2 diabetes: a randomized study. Diabetes Care. 2002;25:239–40. doi: 10.2337/diacare.25.1.239. [DOI] [PubMed] [Google Scholar]
- Adaramoye OA, Achem J, Akintayo OO, Fafunso MA. Hypolipidemic effect of Telfairia occidentalis (fluted pumpkin) in rats fed a cholesterol-rich diet. J Med Food. 2007;10:330–6. doi: 10.1089/jmf.2006.213. [DOI] [PubMed] [Google Scholar]