Abstract
The mitochondrial DNA (mtDNA) chloramphenicol (CAP)-resistance (CAPR) mutation has been introduced into the tissues of adult mice via female embryonic stem (ES) cells. The endogenous CAP-sensitive (CAPS) mtDNAs were eliminated by treatment of the ES cells with the lipophilic dye Rhodamine-6-G (R-6-G). The ES cells were then fused to enucleated cell cytoplasts prepared from the CAPR mouse cell line 501-1. This procedure converted the ES cell mtDNA from 100% wild-type to 100% mutant. The CAPR ES cells were then injected into blastocysts and viable chimeric mice were isolated. Molecular testing for the CAPR mutant mtDNAs revealed that the percentage of mutant mtDNAs varied from zero to approximately 50% in the tissues analyzed. The highest percentage of mutant mtDNA was found in the kidney in three of the chimeric animals tested. These data suggest that, with improved efficiency, it may be possible to transmit exogenous mtDNA mutants through the mouse germ-line.
Keywords: mouse, mitochondria, Rhodamine-6-G, cybrid, chloramphenicol, embryonic stem cell
Introduction
A variety of degenerative diseases affecting the central nervous system (CNS), heart, skeletal muscle, renal and endocrine systems can result from mutations in the mtDNA. While the importance of these diseases has been defined, there are currently no animal model systems available to investigate their pathophysiology or to perfect new therapeutic regimes.
The mammalian mtDNA is a 16 kb circular genome that encodes 13 polypeptides involved in oxidative phosphorylation (OXPHOS) as well as the small (12S) and large (16S) ribosomal RNAs and the 22 transfer RNAs required for mitochondrial protein synthesis. The first pathogenic mtDNA mutations were described in 1988 and the number of reported pathogenic mutations has since rapidly expanded (Wallace et al., 1988a; Wallace, 1992). Although the biochemical defect of several of these reported mutations has been studied in in vitro cell systems, there is still no clear understanding of how a systemic defect caused by a mtDNA mutation can result in tissue-specific degenerative disease. Hence animal models are required to understand the pathophysiology of mitochondrial disease.
Several experimental approaches have been employed for importing defined mtDNA mutations into the mouse. Isolated mitochondria have been microinjected into fertilized ova, but the resulting progeny have lacked any detectable foreign mtDNA (Ebert et al., 1989). When mitochondria were microinjected into zygotes of related species of mice, foreign mitochondria were detected in the cultured embryos up to day 4.5 of embryonic development. However, no viable mice with detectable levels of transferred mtDNA were observed (Pinkert et al., 1997). Mice heteroplasmic for mtDNAs with different naturally occurring polymorphisms have been created by fusion of oocyte cytoplasms as well as transplantation of embryonic karyoplasts and the segregation of the mtDNAs monitored along the maternal lineages (Jenuth et al., 1996, 1997; Meirelles and Smith, 1997, 1998). However, none of these experiments permitted introduction of potentially pathogenic mutations into the adult mouse tissues.
There was one previous report of an attempt to introduce the potentially deleterious CAPR mtDNA mutant into the mouse. This used the cytoplasmic hybrid (cybrid) transfer technique (Bunn et al., 1974; Wallace et al., 1975) to import the CAPR marker from B16 melanoma cells into the embryonal teratocarcinoma cell line OTT6050 (Watanabe et al., 1978). The CAPR B16 cells were enucleated, the cytoplasts fused to the CAPS OTT6050 cells, and the cybrids selected in CAP. The resulting CAPR cybrids were then injected into blastocysts and chimeric animals obtained, as confirmed by nuclear genome encoded isoenzyme analysis. However, no evidence was obtained that the CAPR mtDNAs were introduced into the mouse tissues.
One of the limitations of the cybrid transfer technique is that the resulting cybrid cells acquire a mixture of the resident CAPS and the transferred CAPR mtDNAs. Subsequent propagation of these heteroplasmic cells can result in the loss (segregation) of one or the other mitochondrial genomes. Therefore, to remove the resident CAPS mtDNAs in cybrid fusions, we pretreat the recipient cells with R-6-G prior to fusion with the CAPR donor cytoplasts. In cultured cell studies, this has resulted in the virtual complete replacement of the CAPS mtDNAs by the CAPR mtDNAs (Trounce and Wallace, 1996). The mechanism by which R-6-G inactivates the resident mitochondria is unknown, though R-6-G is a potent inhibitor of mitochondrial respiration. It was first thought that R-6-G inhibited the adenine nucleotide translocator, but other studies have suggested that R-6-G inhibits the ATPase and possibly inhibits the proton transfer of other respiratory complexes (Gear, 1974; Higuti et al., 1980; Mai and Allison, 1983; Wieker et al., 1987; Bullough et al., 1989).
In contrast to the toxic R-6-G, the related compound Rhodamine-123 (R-123) is non-toxic and can be used to stain active mitochondria (Lampidis et al., 1985; Chen, 1988). Both of these dyes have a de-localized positive charge at physiological pH which attracts them to the negatively charged mitochondrial matrix. Furthermore, they are able to freely transverse the outer mitochondrial membrane (Johnson et al., 1980; Chen, 1988; Mannella and Wang, 1989). Hence, they can be used to selectively stain actively respiring mitochondria.
With this background, we have attempted to transfer the CAPR mtDNAs from cultured mouse cells into adult mouse tissues using R-6-G treated female ES cells. We report here the first successful identification of mice harboring CAPR mtDNAs.
Materials and methods
Cell lines
The mouse 501–1 cell line was derived from the mouse HPRT-, LA9 cell line by isolation of a cytoplasmic CAPR mutation (Bunn et al., 1974). The CAP resistance of the 501–1 cell line is due to a mtDNA T-C transition at position 2433 in the 16S ribosomal RNA gene which creates a new MaeII restriction site (Blanc et al., 1981). Using this as a molecular marker, our 501-1 cell line has been found to be homoplasmic (100%) CAPR mutant. Clone 501-1 was grown in Gibco-BRL DMEM (Life Technologies, Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS) (Gibco-BRL) and 50 μg/ml CAP.
The female (40, XX by karyotype) mouse ES cell line AK11.1, derived from 129/SvJae@Sor mice, was provided by Akira Imamoto of the University of Chicago. These ES cells were grown on a mitotically inactivated feeder layer of SNL 76/7 cells, a leukemia inhibitory factor (LIF) producing STO cell line (McMahon and Bradley, 1990), in DMEM media (4.5 g/ml glucose, GIBCO) supplemented with 15% HyClone FBS (Logan, UT), 100 U/ml penicillin, 100 U/ml steptomycin, 2 mM L-glutamine, 100 μM non-essential amino acids, and 100 μM β-mercaptoethanol. When selecting CAPR ES cells, CAPR STO feeder cells were used. These were prepared by fusion of the CAPS STO feeder cells with enucleated 501-1 cell cytoplasts and selection in HAT medium plus 50 μg/ml CAP.
Biochemical analysis of CAP-R cell line 501-1
To determine if the CAPR mutation in the mtDNA 16S rRNA gene had an effect on mitochondrial OXPHOS, biochemical analysis was performed on 501-1 mitochondria (Trounce et al., 1996). Mitochondria were isolated from 501-1 cells and assayed for Complex I (NADH:ubiquinone oxidoreductase), Complex II+III (succinate:cytochrome c oxidoreductase), Complex III (ubiquinol:cytochrome c oxidoreductase), Complex IV (cytochrome c oxidase) and citrate synthase as previously described (Wallace et al., 1988b; Zheng et al., 1990; Trounce et al., 1996).
Rhodamine-6-G treatments
AK11.1 cells in the log phase of growth on CAPR feeder cells were transferred to 35 mm, 6-well plates (also with CAPR feeder cells) at a density of 5 × 105 cells. After a 48-h recovery period, six 6-well plates were treated with R-6-G (250 μg/ml stock in 3% ethanol) for 69 h. Three plates were treated at a concentration of 0.5 μg/ml and three at 0.75 μg/ml R-6-G, all in ES cell media supplemented with 1 mM pyruvate and 50 μg/ml uridine. The media was changed every 24–28 h. The R-6-G treatment was stopped by washing the cells three times with phosphate buffered saline (PBS) and adding ES cell media without R-6-G.
Enucleation and electrofusion
Enucleation of the 501-1 cells was carried out by Ficoll step gradients (Stocco, 1983). Ficoll was prepared as a 50% (w/w) stock solution by adding equal weights of Ficoll 400 (Sigma, St Louis, MO) and water and stirring at 37°C overnight. The resulting solution was filter sterilized and diluted to the appropriate concentration with DMEM media (no serum). Cytochalasin B was added to a final concentration of 20 μg/ml in all Ficoll solutions. Gradients were poured 12–16 h prior to enucleation by layering 25%, 17%, 16%, 15% and 12.5% Ficoll into sterile gradient tubes. Gradients were covered with a sterile cap and incubated at 37°C in a humid atmosphere until used. 4×107 trypsinized and washed 501-1 cells were resuspended in 12.5% Ficoll plus 20 μg/ml cytochalasin B and incubated 15 min at 37°C. The cell solution was then layered on the prepared gradients and spun at 77, 000 × G for 1 h at 31°C. The resulting cytoplast band was recovered and washed in 10 ml of media to remove any remaining Ficoll. Cytoplasts were then washed with 3 ml of 0.3 M mannitol electrofusion media, pH 7.2.
R-6-G treated ES cells were trypsinized, counted and washed with 0.3 M mannitol electrofusion medium, pH 7.2, 2–3 h after the R-6-G treatment. Aliquots of 2.5×106 and 5×105 cells R-6-G treated were then mixed with approximately 1 × 107 cytoplasts and fused by electric shock (Trounce et al., 1994). The electrofusion conditions used were: 50 V AC alignment field for 20 s followed by two 20 μs 800 V DC pulses (ECM200, BTX-Genetronics, San Diego, CA). No post-fusion AC field was used. After a 2 min recovery period, the cells were plated onto 35 mm plates containing CAPR feeder cells.
Selection of cybrids
After fusion, cells were immediately plated onto 35 mm dishes prepared with CAPR feeders using ES cell media supplemented with HAT (0.1 mM hypoxanthine, 0.4 μM aminopterin, 16 μM thymidine, 3 μM glycine). After a 24 h recovery period, the media was changed and the ES cells were placed under HAT selection with 50 μg/ml CAP. Media was changed every 24–48 h and clones were picked 7–9 days after fusion.
PCR and restriction analysis of mtDNA
ES cell clones were picked to 96-well feeder plates and expanded. At the first passage, cells were plated onto a second feeder cell plate for freezing as well as on gelatinized 96-well plates to expand the ES cells in the absence of CAPR feeder cells. After the cells were grown to confluence, total DNA was prepared by overnight digestion with proteinase K followed by phenol extraction and ethanol precipitation. A 600 bp fragment of the mtDNA was amplified by polymerase chain reaction (PCR) with the following primers flanking the CAPR mutation: (5′-3′) TTAACGGCCGCGGTATCCTG, representing heavy strand nucleotide pair (np) 1999–2018 (forward primer) and TTGTAAGGCTCTATTTC, representing np 2599–2580 (reverse primer). Amplification was achieved using 30 cycles of 94°C denaturation for 15 s followed by annealing at 51°C for 35 s and extension at 72°C for 45 s in a mixture containing 200 ng of genomic DNA, 200 μM dNTPs, and 0.5 U Taq polymerase (Boehringer Mannheim, Indianapolis, IN).
The MaeII site resulting from the CAPR T to C transition at np 2433 was detected by digesting 10 μl of PCR product with 1.5 U of MaeII enzyme (Boehringer Mannheim) in the manufacturer’s supplied buffer. Fragments were visualized by electrophoresis in a 2.5% agarose gel containing 0.5 μg/ml ethidium bromide. This PCR fragment contains one common MaeII site present at position np 2501 in the mtDNA, which can be compared to the presence or absence of the second MaeII site created by the CAPR mutation. Upon MaeII digestion of the PCR fragment, the common fragment is 98 np, while the polymorphic fragment is 502 np for wild-type and 434 np plus 68 np for CAPR mtDNAs.
Southern analysis of mouse tissue DNA
Adult mouse tissues were removed from euthanized animals, frozen in liquid nitrogen and ground to a powder with mortar and pestle. The samples were then digested with proteinase K overnight at 55°C, followed by phenol extraction and ethanol precipitation.
Three micrograms of total DNA was digested with MaeII and electrophoresed through a 1.5% agarose gel. The gel was transferred to positively charged nylon membrane (Hybond-N+, Amersham, Arlington Heights, IL) with 0.4 M NaOH and probed with a 600 np PCR fragment amplified from mtDNA positions 1999–2599. After washing, the blot was visualized by phosphorimager (Molecular Dynamics, Sunnyvale, CA) and exposed to autoradiography film. A diagnostic fragment of 1757 np is observed for wild-type mtDNA and a mutant fragment of 1688 np is observed for CAPR mutant mtDNAs. The relative levels of the mutant and normal mtDNAs were determined by quantitation of these bands in the Southern blot by phosphorimager in conjunction with the Image-Quant software supplied by the manufacturer.
ES cell Rhodamine-123 staining
Both treated and control ES cells were stained with R-123 to detect active mitochondria. Cells were washed three times with PBS and placed under fresh ES cell media containing 2.5 μg/ml R-123. After incubation at 37°C for 30 min, the cells were washed three times with PBS and destained for 15 min in ES media without R-123. Cells were then trypsinized and washed with PBS and observed under fluorescent microscopy. Images were captured directly to computer disk through the use of a charge-coupled device (CCD) camera and Adobe Photoshop software (San Jose, CA).
Results
The CAPR mutation is a well defined mtDNA mutation that also imparts a biochemical defect. To establish the nature of this defect, the OXPHOS enzyme activities for Complexes I–IV were determined for mitochondria of the homoplasmic mutant CAPR 501-1 cell line and compared to those of the parental wild-type CAPS LA9 cell line. The mitochondrial matrix enzyme citrate synthase was also assayed and the respiratory complex activities were normalized to both total mitochondrial protein and citrate synthase (Wallace et al., 1988b; Zheng et al., 1990; Trounce et al., 1996). The CAPR mitochondria were found to have a 50% reduction in Complexes I and IV specific activities and a 40% reduction in the Complexes II+III combined specific activities, as compared to wild-type CAPS mitochondria (Table 1). This biochemical signature is comparable to that seen in human cells which harbor mtDNA protein synthesis mutants. Hence, introduction of the CAPR mtDNA into the mouse has the potential for approximating the biochemical defects seen in humans.
Table 1.
OXPHOS enzymology of CAPS and CAPR cell mitochondria. Normalized values are specific activities/citrate synthase.
| Oxidative phosphor. complex | CAPR cells (501-1) | CAPS cells (LA9) | Normalized values |
|
|---|---|---|---|---|
| 501-1 | LA9 | |||
| I | 17 ± 4 | 28 ± 9 | 0.04 | 0.10 |
| II+III | 356 ± 54 | 440 ± 34 | 0.87 | 1.59 |
| III | 673 ± 86 | 754 ± 51 | 1.66 | 2.75 |
| IV | 795 ± 193 | 1196 ± 190 | 1.89 | 4.27 |
| Cit. Syntase | 432 ± 142 | 282 ± 56 | ||
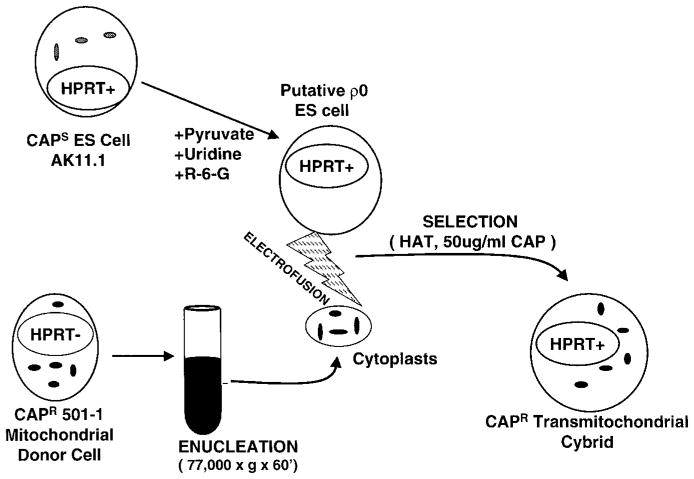
We then attempted to introduce the 501-1 CAPR mtDNA into adult mouse tissues. The 501-1 cells were enucleated and the CAPR cytoplasts fused to AK11.1 female ES cells (Figure 1). CAPR ES cells were then injected into C57BL/6 (B6) mouse blastocysts.
Figure 1.
Schematic representation of the protocol for generating CAPR AK11.1 ES cells.
The fusion of enucleated 501-1 cells to AK11.1 ES cells resulted in the efficient transfer of the CAPR mtDNAs (Figure 2A). All of the CAPR ES clones isolated contained mutant mtDNA levels of approximately 50%. No clones were either homoplasmic mutant or wild-type.
Figure 2.

PCR genotyping of CAPR ES cell clones. Lane numbers indicate the clone analyzed. Panel 2A: The PCR genotype of 13 ES cell clones derived from the fusion of AK11.1 to enucleated 501-1 without the use of the R-6-G protocol. Panel 2B: The PCR genotype of 12 of the 48 CAPR ES clones that were isolated using the R-6-G treatment protocol. Homoplasmic ES cell clones 2 and 5 and heteroplasmic ES cell clone 1 were used in blastocyst injections. The controls are PCR amplifications from the wild-type cell line LA9 (negative control) and the CAPR cell line 501-1 (positive control).
Injection of these heteroplasmic CAPR ES cells into B6 mouse blastocyts gave chimeric mice as assayed by the presence of hair pigmentation. However, none of the tissues examined from these mice had detectable levels of CAPR mtDNA.
To reduce the level of CAPS mtDNAs in the recipient ES cells, we pretreated them with R-6-G prior to fusion to the enucleated 501-1 cells. ES cells were treated with R-6-G in media supplemented with pyruvate and uridine to minimize the toxicity associated with respiratory deficiency (Trounce et al., 1996). In this medium, the growth rate of cells treated with 0.75 μg/ml R-6-G was much slower than untreated controls, with a doubling time of 48–50 h versus 18–24 h. Typically, cells double no more than twice during the 69 h treatment time. The morphology of the treated cells was more rounded, though otherwise similar to control cells.
AK11.1 ES cells, treated with 0.75 μg/ml R-6-G for 69 h, had cloning efficiencies of less than 4 × 10−7 after 9 days of culture, with no colonies being observed after plating 2.5 × 106 treated cells. When the ES cells were fused to 501-1 cytoplasts, the R-6-G treated ES cells were rescued and all ES cell cybrid clones genotyped had a high proportion of CAPR mutant mtDNAs (Figure 2B), even in fusions where CAP selection was not used (data not shown).
To better understand the physiological effect of R-6-G on ES cells, R-123 was used to stain the mitochondria of ES cells prior to R-6-G treatment, after R-6-G treatment but prior to fusion, and after fusion to enucleated 501-1 and cybrid selection (Figure 3). Prior to R-6-G treatment, the ES cells have multiple highly fluorescent and metabolically active mitochondria. Following R-6-G treatment, nearly all active mitochondria were eliminated from the ES cells, as shown by the large reduction in punctate staining. Following fusion with 501-1 cytoplasts, the ES cells were again fully repopulated with active mitochondria.
Figure 3.

Mitochondrial changes in AK11.1 ES cells during the cybrid fusion protocol as revealed by R-123 staining. Panel 1: Staining prior to R-6-G treatment. Panel 2: Staining after 69 h treatment with 0.75 μg/ml R-6-G. Panel 3: Staining after fusion of R-6-G treated ES cells with enucleated 501-1 and cybrid isolation. All images were taken by CCD camera under 400 × magnification.
Fusion of 501-1 cytoplasts with R-6-G treated ES cells resulted in colonies after 7–9 days of post-fusion culture. The cybrid frequency was 1.2 × 10−3 and 6 × 10−4 for fusion of 2.5 × 106 and 5 × 105 ES cells to 1 × 107 cytoplasts, respectively. Forty-eight individual clones were isolated, expanded and genotyped. Forty-six of the forty-eight were homoplasmic for the CAPR mutation (data not shown). Twelve of the fastest growing clones with the best morphologies were expanded and genotyped (Figure 2B). Two of these clones (clones 1 and 3) were heteroplasmic with a low level of wild-type mtDNA.
To prove that the transfer of the CAPR mtDNA also imparted resistance to chloramphenicol, the CAP resistance of one clone was assessed by testing for its ability to grow in increasing concentrations of CAP for 7 days (Figure 4). The growth of the CAPS AK11.1 ES cells was strongly inhibited by 25 μg/ml CAP while the CAPR AK11.1 ES cell cybrids were resistant to drug concentrations up to 150 μg/ml CAP, the same range of resistance seen for the CAPR 501-1 cells. All ES cell cybrids grew well in 50 μg/ml CAP, with the growth of heteroplasmic and homoplasmic clones being comparable.
Figure 4.
CAP toxicity on CAPSand CAPR ES cells as assessed by single point growth curve. 1 × 105 cells were plated into replicate 35 mm wells containing CAPR feeder cells. The ES cells were cultured for 7 days at CAP concentrations of 0, 25, 50, 75, 100, and 150 μg/ml. The cells were then removed and counted. Results are graphed as the concentration of CAP versus the percentage of cell growth relative to growth without CAP selection.
Blastocysts were then injected with the homoplasmic CAPR ES cell clones 2 and 5 and the heteroplasmic clone 1 shown in Figure 2B. A total of 77 pups were born, 20 of which were chimeric by coat color assessment (12 male and eight female). The chimeras appeared healthy without any overt phenotype.
Of the original chimeric animals, the levels of chimerism observed by coat color was low. The highest was approximately 30% in two of the females with the majority of the mice being between 10% and 20%. One female and two males had chimerism levels of less than 5%. Five representative 8–10 week male mice with 5–20% chimerism were analyzed for the presence of CAPR mtDNA by MaeII digestion of PCR amplified mtDNA or mtDNA Southern analysis. The CAPR mutation was detectable in several tissues of three of the five analyzed mice by PCR, but the accuracy of quantitating mouse mtDNA heteroplasmy by PCR has been shown to be potentially inaccurate (Jenuth et al., 1996; Jenuth et al., 1997). To more accurately assess the levels of CAPR mtDNA present in the mouse tissues, further quantitative assessments were made by Southern blot analysis (Figure 5). The ratio of mutant to wild-type mtDNAs was quantitated in the Southern blots by densitometry. The highest percentage of mutant mtDNA was found in the kidneys of the three positive mice, with percentages of mutant ranging from 20 to 50. Detectable levels of mutant mtDNA were found in the hearts of two of the mice with mutant levels of less than 15%. Finally, one sample each of brain and liver also had detectable levels of CAPR mtDNA.
Figure 5.

Molecular evidence for introduction of CAPR mtDNA into adult mouse tissues through CAPR ES cells by Southern blot. The gel shows the diagnostic CAPS (1757 np) and CAPR (1689 np) mtDNA bands detailed by Southern blot of mouse tissues. The controls are DNA from wild-type LA9 cells (negative control) and CAPR 501-1 cells (positive control). H = heart, B = brain, M = skeletal muscle, L = liver, and K = kidney.
All of the viable female chimeras were bred with B6 males and each was allowed to litter three times. However, none showed genetic evidence that the CAPR ES cells formed the germ cells.
Discussion
We report here the first successful confirmation of introduction of mtDNAs harboring a deleterious mutation into adult mouse tissues via female ES cells. The resulting chimeric animals were found to harbor both mutant and wild-type mtDNAs, with the levels of mutant mtDNAs being highest in the kidney. While heteroplasmic mice harboring naturally occurring polymorphic mtDNAs have been constructed by embryo fusion technologies or the use of microsurgical procedures (Jenuth et al., 1996, 1997; Meirelles and Smith, 1997, 1998), and mitochondria from M. spretus have been microinjected into the fertilized ova of M. m. domesticus (Pinkert et al., 1997), no mtDNA mutants which alter mitochondrial function have been introduced into the mouse. Therefore, the current introduction of CAPR mitochondria into mouse tissues is the first report of the successful transfer of a potentially pathogenic mtDNA mutation into the adult mouse.
Unfortunately, we were not able to achieve germ-line transmission of the CAPR mutation with any of the eight females that resulted from blastocyst injection of the CAPR ES cells. There are at least three possible reasons for this lack of success: the CAPR biochemical defect may be too severe, the CAPR ES cell sublines derived from the AK11.1 female ES cell line may have been developmentally impaired, or the R-6-G may have been too toxic. The biochemical defect of the CAPR mutant might be a problem since the percentage of chimerism seen in mice derived from the CAPR ES cells was low. Possibly, the inhibition of growth of the CAPR ES cells in the blastocyst limited contribution of the ES cells to the chimeric animals. On the other hand, control injections with untreated AK11.1 also yielded mice with low levels of chimerism, giving only two out of 10 mice with higher percentages of chimerism than found in the mice from CAPR ES cells. Alternatively, the AK11.1 ES cells might be developmentally impaired. Control injections using the AK11.1 ES cells have yet to yield a germ-line transmitting female mouse. Finally, the R-6-G treatment might have impaired the developmental potential of the AK11.1 ES cells. This possibility seems unlikely since the R-6-G treatment has not been found to impair function in cultured cell studies and the treated cells do not exhibit any gross morphological changes.
In conclusion, we have successfully introduced a deleterious mtDNA mutation into the tissues of adult mice. This protocol offers great promise relative to other methods for creating mouse models of mitochondrial disease since a variety of deleterious mutations have been isolated in cultured cells and can now be transferred directly into the mouse through ES cells. With the development of female ES cell lines that can provide high percentage chimerism, it is likely that this method will ultimately result in the germ-line transmission of a variety of pathogenic mtDNA mutations in the mouse. Development of mouse model systems of mitochondrial dysfunction will permit much more detailed studies of the pathophysiology of mitochondrial disease as well as provide model systems for analyzing the transmission and therapeutics of mtDNA disease.
Acknowledgments
We thank Akira Imamoto for the AK11.1 ES cell line and Allan Bradley for SNL76/7 cells. This work was supported by NIH grants NS21328, HL45572, and AG13154 awarded to D.C.W., and HD36437 awarded to G.R.M.
References
- Blanc H, Wright CT, Bibb MJ, Wallace DC, Clayton DA. Mitochondrial DNA of chloramphenicol-resistant mouse cells contains a single nucleotide change in the region encoding the 3′ end of the large ribosomal RNA. Proc Natl Acad Sci USA. 1981;78:3789–3793. doi: 10.1073/pnas.78.6.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullough DA, Ceccarelli EA, Roise D, Allison WS. Inhibition of the bovine-heart mitochondrial F1-ATPase by cationic dyes and amphipathic peptides. Biochim Biophys Acta. 1989;975:377–383. doi: 10.1016/s0005-2728(89)80346-9. [DOI] [PubMed] [Google Scholar]
- Bunn CL, Wallace DC, Eisenstadt JM. Cytoplasmic inheritance of chlormaphenicol resistance in mouse tissue culture cells. Proc Natl Acad Sci USA. 1974;71:1681–1685. doi: 10.1073/pnas.71.5.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LB. Mitochondrial membrane potential in living cells. Annu Rev Cell Biol. 1988;4:155–181. doi: 10.1146/annurev.cb.04.110188.001103. [DOI] [PubMed] [Google Scholar]
- Ebert KM, Alcivar A, Liem H, Goggins R, Hecht NB. Mouse zygotes injected with mitochondria develop normally but the exogenous mitochondria are not detectable in the progeny. Mol Reprod Dev. 1989;1:156–163. doi: 10.1002/mrd.1080010303. [DOI] [PubMed] [Google Scholar]
- Gear AR. Rhodamine 6G: A potent inhibitor of mitochondrial oxidative phosphorylation. J Biol Chem. 1974;249:3628–3637. [PubMed] [Google Scholar]
- Higuti T, Niimi S, Saito R, Nakasima S, Ohe T, Tani I, Yoshimura T. Rhodamine 6G, inhibitor of both H+−ejections from mitochondria energized with ATP and with respiratory substrates. Biochim Biophys Acta. 1980;593:463–467. doi: 10.1016/0005-2728(80)90081-x. [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA (see comments) Nat Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Shoubridge EA. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet. 1997;16:93–95. doi: 10.1038/ng0597-93. [DOI] [PubMed] [Google Scholar]
- Johnson LV, Walsh ML, Chen LB. Localization of mitochondria in living cells with rhodamine 123. Proc Natl Acad Sci USA. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampidis TJ, Munck JN, Krishan A, Tapiero H. Reversal of resistance to rhodamine 123 in adriamycin-resistant Friend leukemia cells. Cancer Res. 1985;45:2626–2631. [PubMed] [Google Scholar]
- Mai MS, Allison WS. Inhibition of an oligomycin-sensitive ATPase by cationic dyes, some of which are atypical uncouplers of intact mitochondria. Arch Biochem Biophys. 1983;221:467–476. doi: 10.1016/0003-9861(83)90165-0. [DOI] [PubMed] [Google Scholar]
- Mannella CA, Wang Q. Permeability of the mitochondrial outer membrane to organic cations. Biochim Biophys Acta. 1989;981:363–366. doi: 10.1016/0005-2736(89)90049-7. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- Meirelles FV, Smith LC. Mitochondrial genotype segregation in a mouse heteroplasmic lineage produced by embryonic karyoplast transplantation. Genetics. 1997;145:445–451. doi: 10.1093/genetics/145.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirelles FV, Smith LC. Mitochondrial genotype segregation during preimplantation development in mouse heteroplasmic embryos. Genetics. 1998;148:877–883. doi: 10.1093/genetics/148.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkert CA, Irwin MH, Johnson LW, Moffatt RJ. Mitochondria transfer into mouse ova by microinjection. Transgenic Res. 1997;6:379–383. doi: 10.1023/a:1018431316831. [DOI] [PubMed] [Google Scholar]
- Stocco DM. Rapid, quantitative isolation of mitochondria from rat liver using Ficoll gradients in vertical rotors. Anal Biochem. 1983;131:453–457. doi: 10.1016/0003-2697(83)90198-7. [DOI] [PubMed] [Google Scholar]
- Trounce I, Neill S, Wallace DC. Cytoplasmic transfer of the mtDNA nt 8993 TG (ATP6) point mutation associated with Leigh syndrome into mtDNA-less cells demonstrates cosegregation with a decrease in state III respiration and ADP/O ratio. Proc Natl Acad Sci USA. 1994;91:8334–8338. doi: 10.1073/pnas.91.18.8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trounce I, Wallace DC. Production of transmitochondrial mouse cell lines by cybrid rescue of rhodamine-6G pre-treated L-cells. Somatic Cell and Molec Gen. 1996;22:81–85. doi: 10.1007/BF02374379. [DOI] [PubMed] [Google Scholar]
- Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Meth Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Diseases of the mitochondrial DNA. Ann Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Bunn CL, Eisenstadt JM. Cytoplasmic transfer of chloramphenicol resistance in human tissue culture cells. J Cell Biol. 1975;67:174–188. doi: 10.1083/jcb.67.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ, Nikoskelainen EK. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988a;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Zheng X, Lott MT, Shoffner JM, Hodge JA, Kelley RI, Epstein CM, Hopkins LC. Familial mitochondrial encephalomyopathy (MERRF): Genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell. 1988b;55:601–610. doi: 10.1016/0092-8674(88)90218-8. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Dewey MJ, Mintz B. Teratocarcinoma cells as vehicles for introducing specific mutant mitochondrial genes into mice. Proc Natl Acad Sci USA. 1978;75:5113–5117. doi: 10.1073/pnas.75.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieker HJ, Kuschmitz D, Hess B. Inhibition of yeast mitochondrial F1-ATPase, F0F1-ATPase and submitochondrial particles by rhodamines and ethidium bromide. Biochim Biophys Acta. 1987;892:108–117. doi: 10.1016/0005-2728(87)90253-2. [DOI] [PubMed] [Google Scholar]
- Zheng XX, Shoffner JM, Voljavec AS, Wallace DC. Evaluation of procedures for assaying oxidative phosphorylation enzyme activities in mitochondrial myopathy muscle biopsies. Biochimica et Biophysica Acta. 1990;1019:1–10. doi: 10.1016/0005-2728(90)90118-n. [DOI] [PubMed] [Google Scholar]