Abstract
The majority of mitochondrial DNA (mtDNA) mutations that cause human disease are mild to moderately deleterious, yet many random mtDNA mutations would be expected to be severe. To determine the fate of the more severe mtDNA mutations, we introduced mtDNAs containing two mutations that affect oxidative phosphorylation into the female mouse germ line. The severe ND6 mutation was selectively eliminated during oogenesis within four generations, whereas the milder COI mutation was retained throughout multiple generations even though the offspring consistently developed mitochondrial myopathy and cardiomyopathy. Thus, severe mtDNA mutations appear to be selectively eliminated from the female germ line, thereby minimizing their impact on population fitness.
The maternally inherited mitochondrial DNA (mtDNA) has a high mutation rate, and mtDNA base substitution mutations have been implicated in a variety of inherited degenerative diseases including myopathy, cardiomyopathy, and neurological and endocrine disorders (1, 2). Paradoxically, the frequency of mtDNA diseases is high, estimated at 1 in 5000 (3, 4), yet only a few mtDNA mutations account for the majority of familial cases (2). Because mutations would be expected to occur randomly in the mtDNA, the paucity of the most severe mtDNA base substitutions in maternal pedigrees suggests that the severe mutations may be selectively eliminated in the female germ line.
To investigate this possibility, we have developed a mouse model in which the germline transmission of mtDNA point mutations of different severity could be tested. An antimycin A–resistant mouse LA9 cell line was cloned whose mtDNA harbored two homoplasmic (pure mutant) protein-coding gene base change mutations: one severe and the other mild. The severe mutation was a C insertion at nucleotide 13,885 (13885insC), which created a frameshift mutation in the NADH dehydrogenase subunit 6 gene (ND6). This frameshift mutation altered codon 63 and resulted in termination at codon 79 (Fig. 1A, top; fig. S1A, bottom). When homoplasmic, this mutation inactivates oxidative phosphorylation complex I (5). The mild mutation was a missense mutation at nucleotide 6589 (T6589C) in the cytochrome c oxidase subunit I gene (COI) that converted the highly conserved valine at codon 421 to alanine (V421A) (fig. S1A, top). When homoplasmic, this mutation reduces the activity of oxidative phosphorylation complex IV by 50% (6, 7).
Fig. 1.
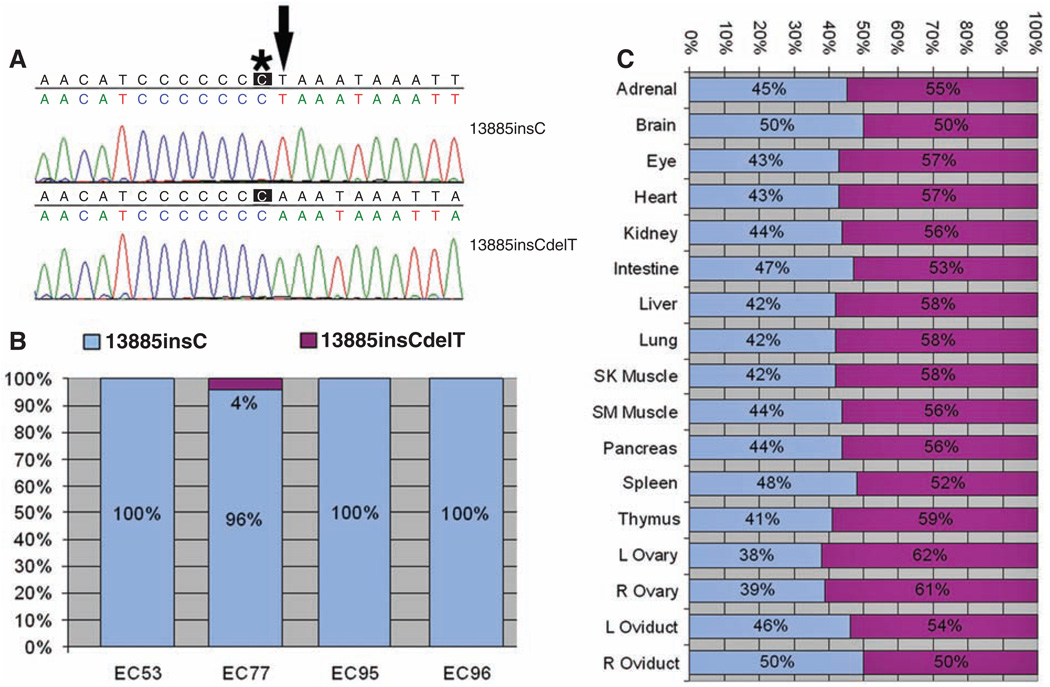
Qualitative and quantitative analysis of the ND6 frameshift (13885insC) and ND6 revertant (13885insCdelT) mutations in mouse ES cell cybrids and tissues of F1 female EC77-AG. (A) Sequence around nucleotide 13,885 of two clones of mtDNA from EC77 cells. Top (13885insC), the single C insertion causing the frameshift (asterisk); bottom (13885insCdelT), the T (arrow) deletion that restored the normal reading frame. (B) Percentages of ND6 frameshift (13885insC, blue) versus revertant (13885insCdelT, purple) in four independent mouse ES cybrids. (C) Proportions of ND6 frameshift (13885insC) and revertant (13885insCdelT) mtDNAs in the tissues of F1 female EC77-AG.
LA9 cells homoplasmic for both the ND6 frameshift and COI missense mutations were enucleated, and the mtDNAs were transferred by cytoplast fusion to the mtDNA-deficient (ρo) mouse cell line LMEB4, generating the LMJL8 transmitochondrial cybrid (8). LMJL8 mitochondria exhibited no detectable oxygen consumption when provided with NADH-linked complex I substrates (fig. S1B) and no detectable complex I enzyme activity (fig. S1C). However, the same mitochondria exhibited a 43% increase in succinate-linked respiration and a 91% increase in complex II + III activity as well as a 62% increase in complex IV activity (fig. S1, B and C), presumably as a compensatory response to the severe complex I defect (9). Relative to LM(TK−) cells, mouse L cell lines homoplasmic for the COI missense mutation also showed increased reactive oxygen species (ROS). Cells homoplasmic for both the ND6 frameshift and COI missense mutations produced fewer ROS than did the COI mutant cells. However, cells that were 50% heteroplasmic for both the ND6 frameshift and the COI missense mutations had the highest ROS production (fig. S1D).
To analyze the fates of the severe ND6 frameshift versus moderate COI missense mtDNA mutations, we introduced these mutations into the mouse germ line. LMJL8 cybrids were enucleated and the cytoplasts fused to the mouse female embryonic stem (ES) cell line CC9.3.1 that had been cured of its resident mitochondria and mtDNAs by treatment with rhodamine 6G (10–12). Of the 96 resulting ES cybrids, four (EC53, EC77, EC95, and EC96) were found to be homoplasmic for the COI missense mutation. By quantitative primer extension–denaturing high-performance liquid chromatography analysis, three of the ES cell cybrids were also found to be homoplasmic for the ND6 frameshift mutation. However, one ES cell cybrid, EC77, was heteroplasmic (mixture of mutant and normal mtDNAs); 96% of the mtDNAs harbored the ND6 frameshift mutation (13885insC), whereas 4% of the mtDNAs had sustained a secondary deletion of the adjacent T (13885insCdelT), restoring the reading frame (Fig. 1A, bottom, and Fig. 1B). This ND6 revertant mutation encodes the normal amino acid sequence but changes leucine codon 60 from TTA to TTG.
EC77 ES cells were injected into female C57BL/6NHsd blastocysts, and the chimeric embryos were transferred into pseudo-pregnant females (12). Three chimeric females were generated that contained varying proportions of three mtDNA genotypes: ND6 frameshift + COI missense, ND6 revertant + COI missense, and wild type (fig. S2). The chimeras were mated with C57BL/6J (B6) males and produced a total of 111 pups. Only one F1 agouti female pup, EC77-AG, was generated harboring the mutant mtDNAs. Analysis of tail mtDNA by primer extension and by cloning and sequencing revealed that EC77-AG was homoplasmic for the COI mutant allele but heteroplasmic for the ND6 frameshift (47%) and ND6 revertant (53%) mtDNAs (fig. S3A). Post mortem analysis at 11 months revealed that all analyzed tissues from EC77-AG had essentially the same genotype, with an average of 44 ± 3% (range 38% to 50%) of the mtDNAs harboring the ND6 frameshift plus COI missense mutations (ND6 13885insC + COI T6589C) and 56% harboring the ND6 revertant plus COI missense mutations (ND6 13885insCdelT + COI T6589C). The highest levels of the frameshift mutant mtDNA were found in the brain and right oviduct; the lowest level was found in the left ovary (Fig. 1C).
Throughout the 11 months of her life, EC77-AG displayed no overt phenotype. However, post mortem mitochondrial enzymatic assays revealed a 10% to 33% decrease of complex I activity in brain, heart, liver, and skeletal muscle (fig. S3B), a 56% and 46% decrease of complex IV activity in brain and skeletal muscle, and a 19% and 39% increase of complex IV activity in heart and liver (fig. S3C). This was associated with structures consistent with lipid droplets in the heart mitochondria by ultrastructural analysis (compare fig. S3, D and E).
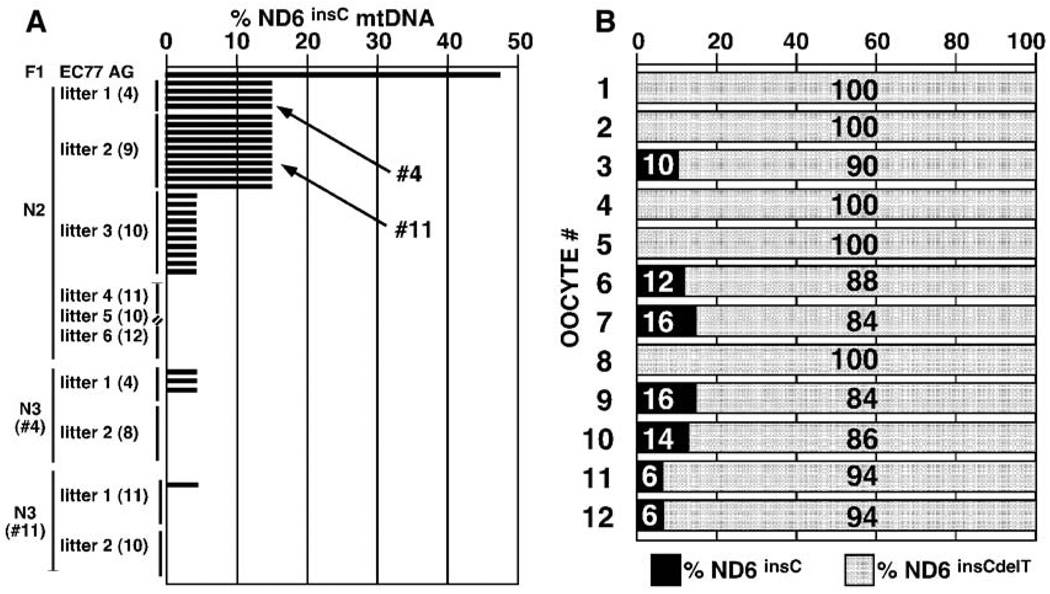
To analyze transmission of the heteroplasmic, severe, ND6 frameshift (ND6 13885insC + COI T6589C) mtDNA in successive maternal generations, we mated F1 female EC77-AG, which had 47% ND6 frameshift tail mtDNA, with B6 males. EC77-AG gave birth to six litters totaling 56 pups (N2). The proportion of ND6 frameshift tail mtDNA, as assessed by primer extension analysis, declined to 14% in the first litter of four pups (EC77 #1 to #4) and the second litter of nine pups (EC77 #5 to #13), then to 6% in the third litter of 10 pups (EC77 #14 to #23), and finally was lost (0%) in all subsequent litters (Fig. 2A).
Fig. 2.
Selective elimination of ND6 frameshift mtDNA (13885insC) from F1 female EC77-AG and her offspring. (A) Percentages of ND6 frameshift (13885insC) mtDNAs in EC77-AG and her offspring, plus the pups of her daughters EC77 #4 and #11. Each offspring was analyzed from multiple litters. Litter sizes are indicated in parentheses. (B) Percentages of the ND6 frameshift (13885insC, black) and revertant (13885insCdelT, gray) mtDNAs in 12 oocytes isolated from EC77 progeny mice containing 14% of the ND6 frameshift mutation (13885insC).
To verify the reproducibility of the progressive loss of the ND6 frameshift mtDNA, we mated N2 female EC77 #4, which had 14% ND6 frameshift mtDNA, with B6 males. EC77 #4 gave birth to two litters totaling 12 pups (N3) (Fig. 2A). Three of the four pups of the first litter had 6% ND6 frameshift mtDNA, whereas the remaining pup of the first litter and all eight pups of the second litter had lost the ND6 frameshift mtDNA (0%). We also mated N2 female EC77 #11, which had 14% ND6 frameshift mtDNA, with B6 males. EC77 #11 gave birth to two litters totaling 21 pups. One of the 11 pups of the first litter had 6% ND6 frameshift mtDNA, whereas the remaining 10 pups of the first litter and all 10 pups of the second litter had lost the frameshift mtDNA (0%). Mating of B6 males with N3 females, which had 6% ND6 frameshift mtDNA, only produced pups that lacked the ND6 frameshift mtDNA. These data suggest that the mtDNA harboring the deleterious ND6 frameshift mutation (13885insC) was selectively and directionally eliminated from the mouse female germ line within four generations.
To determine whether the ND6 frameshift plus COI missense mtDNA was eliminated from the female germ line in favor of the ND6 revertant plus COI missense mtDNA via selective loss of those fetuses with the highest percentages of ND6 frameshift mtDNA, we compared the litter sizes of females with different proportions of ND6 frameshift mtDNA. Females with higher percentages of ND6 frameshift mtDNA would be predicted to generate pups with higher proportions of the ND6 frameshift mtDNA and thus have higher fetal loss rates and smaller litter sizes. We instead observed that the percentage of ND6 frameshift mtDNA in the mother had no effect on litter size. The average litter size of F1 female EC77-AG with 47% ND6 frameshift mtDNA was 9.3 pups per litter, whereas that of two of her daughters with 14% ND6 frameshift mtDNA was 8.25 pups per litter, and that of her descendants with 6% ND6 frameshift mtDNAs was 8.75 pups per litter. Given that average litter size usually decreases slightly when backcrossing onto a B6 strain background, the average litter size appeared unaffected by the proportion of the mother’s mtDNA that harbored the ND6 frameshift mutation, thus arguing against preferential fetal loss as the segregation mechanism.
To determine whether the ND6 frameshift mtDNA was lost before fertilization or ovulation, we collected and genotyped individual oocytes from superovulated N2 females containing 14% ND6 frameshift plus COI missense and 86% ND6 revertant plus COI missense mtDNA. Of the 12 oocytes that were successfully genotyped, four retained 10 to 16% of the ND6 frameshift mtDNA, two retained 6% of the ND6 frameshift mtDNA, and five had lost the frameshift mtDNA (Fig. 2B). Previous studies on mice heteroplasmic for the normal NZB and Balb/c mtDNAs revealed that these mtDNAs segregated randomly when transmitted through the female germ line (13). If this was the case for mice that were heteroplasmic for the ND6 frameshift and ND6 revertant mtDNAs, we would expect that the percentage of frameshift versus total mtDNAs would be normally distributed around the mother’s genotype. In fact, none of the oocytes or progeny had a higher proportion of the ND6 frameshift mutant mtDNA than the mother. This indicates that proto-oocytes with the higher proportion of frameshift mutant mtDNA must have been eliminated by selection before ovulation.
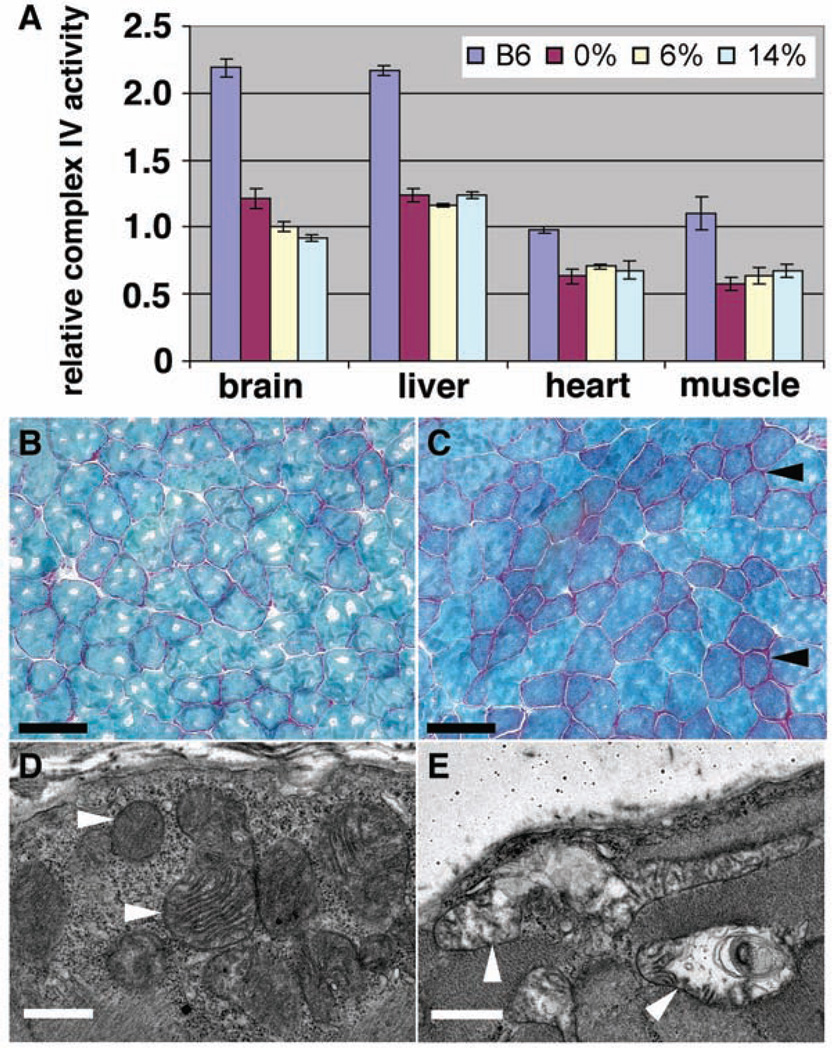
As seen with the F1 female EC77-AG, the proportion of ND6 frameshift mtDNA in the different tissues of three N2 mice containing 14% ND6 frameshift and 86% ND6 revertant mtDNA, all with the COI missense mutation, was relatively constant, ranging between 14% and 16% (fig. S4A). Ultrastructural analysis of hearts taken from the 14% ND6 frameshift plus 100% COI missense mice revealed mitochondrial proliferation, evidence of mitochondrial autophagy, and myofibrillar degeneration (fig. S4, B to D). Biochemical analysis of brain, heart, and liver of mice with 14% ND6 frameshift plus 100% COI missense mutant mtDNAs revealed little reduction in complex I activity (fig. S4E); however, complex IV activity in these tissues was reduced by 28%, 70%, and 59%, respectively (fig. S4F).
Comparison of the complex I activity in mice harboring 0%, 6%, and 14% ND6 frameshift and 100% COI missense mtDNAs confirmed that complex I was little affected, with the possible exception of a modest reduction of complex I in muscle in animals with 14% ND6 frameshift mtDNAs (fig. S5A). The complex II + III activities were also relatively stable (fig. S5B). However, complex IV was reduced about 50% in brain, liver, heart, and muscle of the 14%, 6%, and 0% ND6 frameshift plus 100% COI missense mutant mice (Fig. 3A). Hence, the predominant biochemical defect in animals with 14% or less ND6 frameshift mtDNAs can be attributed to the homoplasmic COI missense mutation.
Fig. 3.
Decreased mitochondrial complex IV activity and mitochondrial myopathy in COI mutant mice. (A) Complex IV activity was reduced to similar extents in various tissues of mutant mice harboring 0%, 6%, or 14% ND6 frameshift mutations plus 100% COI missense. Numbers of animals tested: six B6, seven 0% (0% ND6 frameshift plus 100% COI missense), three 6% (6% ND6 frameshift plus 100% COI missense), and four 14% (14% ND6 frameshift plus 100% COI missense), with three repeats performed for each test on each animal. (B and C) Gomori trichrome staining shows increased ragged red fibers (arrowheads) in skeletal muscle of 12-month-old COI mutant mice (C) compared to age-matched control (B). (D and E) Electron microscopy (EM) shows altered mitochondrial morphology (arrowheads) in skeletal muscle of 12-month-old COI mutant (E) mice compared to age-matched control (D). Scale bars, 75 µm [(B) and (C)], 1 µm [(D) and (E)].
Mice that were homoplasmic for the COI missense mutation, linked to the ND6 revertant mutation (13885insCdelT), transmitted this mtDNA to all of their offspring through multiple backcrosses to B6 males. Although the phenotype of these mice was grossly normal, muscle histology of 12-month-old animals revealed ragged red muscle fibers and abnormal mitochondria characteristic of mitochondrial myopathy (Fig. 3, B to E).
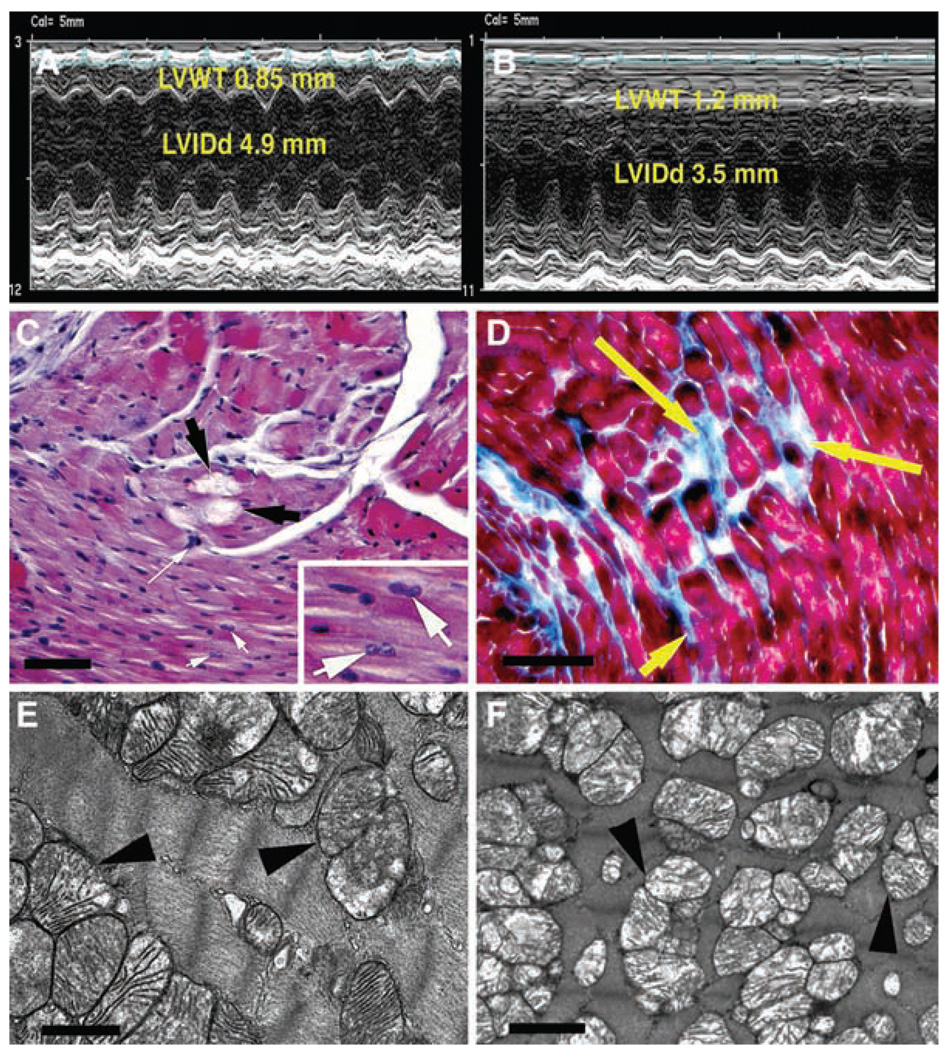
In addition to the mitochondrial myopathy, echocardiographic analysis of 12-month-old COI missense mice revealed that 100% of these animals (n = 7) had developed a striking cardiomyopathy, as compared to age-matched B6 control mice (n = 5) (Fig. 4, A and B). This cardiomyopathy was associated with a 35% increase in left ventricular wall thickness, a 23% reduction in left ventricular inner dimension at end-diastole (P < 0.001), and a 27% increase in rotation in association with a 28% reduction in circumferential strain vectors (P < 0.001) and a 42% reduction in radial stretch vectors (P < 0.001) (fig. S6, A and B). Histology of the COI missense mutant hearts revealed the presence of myocyte hypertrophy, myofibrillar lysis, binucleate cells, and interstitial fibrosis (Fig. 4, C and D). Focal inflammation, interstitial edema, and increased blood vessel number and diameter were also observed (fig. S6, C to E).However, no evidence of the myofiber disarray characteristic of hypertrophic cardiomyopathy was seen. Cardiac ultrastructural analysis revealed loss of myofilaments and mitochondrial abnormalities in mutant heart tissue, including mitochondrial proliferation, reduction in mitochondrial matrix density, and cristolysis (Fig. 4F), relative to age-matched controls (Fig. 4E). Hence, the milder mtDNA COI missense mutation was successfully transmitted through repeated maternal generations, even though it caused maternally inherited mitochondrial myopathy and cardiomyopathy.
Fig. 4.
Mitochondrial cardiomyopathy in 12-month-old mice homoplasmic for COI missense mutation. (A) Echocardiographic analysis of control heart. (B) Echocardiographic analysis of mutant heart showing increased left ventricular wall thickness (LVWT) and decreased left ventricular internal dimension in diastole (LVIDd). (C) Hematoxylin and eosin–stained mutant heart showing myofibrolysis (black arrows), myocyte hypertrophy (long white arrow), and binucleate cells (inset, white arrows). (D) Masson trichrome–stained mutant heart showing interstitial replacement fibrosis (yellow arrows). (E) EM of mitochondria (arrowheads) in normal heart. (F) EM of mutant heart showing mitochondrial proliferation, reduced matrix density, and cristolysis (arrowheads). Scale bars, 500 µm (C), 100 µm (D), 1 µm [(E) and (F)].
Our studies suggest that the female germ line has the capacity for intraovarian selection against highly deleterious mtDNA mutations such as the ND6 frameshift mutation, while permitting transmission of more moderate mtDNA mutations such as the COI missense mutation. This observation may explain why there is a dearth of severe mtDNA base substitution pedigrees in humans, yet more moderate pathogenic mtDNA mutations are repeatedly seen, such as those causing neurogenic muscle weakness, ataxia, and retinitis pigmentosa (NARP) T8993G (ATP6 L156R) and Leber hereditary optic neuropathy (LHON) G11778A (ND4 R340H) and T14484C (ND6 M64V) (2).
Although the mechanism by which the severe mtDNA mutations are recognized and eliminated remains unclear, our observation that cells heteroplasmic for both the ND6 frameshift and the COI missense mutations have the highest ROS production provides one possible explanation. It has been proposed that the primordial female germ cells have a limited number of mtDNAs permitting rapid genetic drift toward pure mutant or wild-type mtDNA during the approximately 20 female germline cell divisions (13). At birth, the ovigerous cords reorganize to form single oogonia surrounded by granulosa cells. This would lead to oogonia within fetal ovigerous cords with mtDNA genotypes symmetrically distributed around the maternal mean percent heteroplasmy. Of these oogonia, only about 30% complete meiotic maturation; the remainder undergo apoptosis (14, 15). Because apoptosis in preovulatory follicles is thought to be induced by oxidative stress (16), it is conceivable that the proto-oocytes with the highest percentage of severe mtDNA mutations produce the most ROS and thus are preferentially eliminated by apoptosis. Such a process would then lead to the progressive loss of the more deleterious mtDNA mutations over successive female generations.
Among the pathogenic missense mutations that are observed, the more severe mtDNA mutations such as NARP T8993G remain heteroplasmic through successive generations. By contrast, the milder mtDNA mutations, such as LHON G11778A and T14484C, can segregate to homoplasmic mutant (2). Because heteroplasmy would temper the biochemical defect associated with the more severe mutations, this observation supports the concept that the more severe mtDNA defects are eliminated within the maternal germ line.
The existence of a female germline filter for severely deleterious mtDNA mutations makes evolutionary sense. Assuming that mtDNA variation is pivotal to species adaptation to changing environments and that the uniparental mtDNA cannot generate diversity by recombination, then mtDNA diversity must be generated through a high mutation rate (17–19). However, a high mutation rate would generate many highly deleterious mutations that could create an excessive genetic load and endanger species fitness. This dilemma can be resolved by the addition of a graded filter in the female germ line that eliminates the most severe mutations before conception. For such a filter to succeed, multiple cell divisions resulting in a large population of proto-oocytes would be required to segregate out the new mtDNA mutations. This may explain why the mammalian female generates millions of primordial oogonia but ovulates only a few hundred mature oocytes.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/319/5865/958/DC1
Materials and Methods
Figs. S1 to S6
References
References and Notes
- 1.Wallace DC. Annu. Rev. Genet. 2005;39:359. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace DC, Lott MT, Procaccio V. In: Emery and Rimoin’s Principles and Practice of Medical Genetics. ed. 5. Rimoin DL, Connor JM, Pyeritz RE, Korf BR, editors. vol. 1. Philadelphia: Churchill Livingstone; 2007. pp. 194–298. [Google Scholar]
- 3.Schaefer AM, Taylor RW, Turnbull DM, Chinnery PF. Biochim. Biophys. Acta. 2004;1659:115. doi: 10.1016/j.bbabio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Schaefer AM, et al. Ann. Neurol. 2007 10.1002/ana.21217. [Google Scholar]
- 5.Bai Y, Attardi G. EMBO J. 1998;17:4848. doi: 10.1093/emboj/17.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acin-Perez R, et al. Hum. Mol. Genet. 2003;12:329. doi: 10.1093/hmg/ddg021. [DOI] [PubMed] [Google Scholar]
- 7.Kasahara A, et al. Hum. Mol. Genet. 2006;15:871. doi: 10.1093/hmg/ddl005. [DOI] [PubMed] [Google Scholar]
- 8.Trounce I, Wallace DC. Somat. Cell Mol. Genet. 1996;22:81. doi: 10.1007/BF02374379. [DOI] [PubMed] [Google Scholar]
- 9.Kokoszka JE, et al. Nature. 2004;427:461. doi: 10.1038/nature02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy SE, Waymire KG, Kim YL, MacGregor GR, Wallace DC. Transgen. Res. 1999;8:137. doi: 10.1023/a:1008967412955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sligh JE, et al. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14461. doi: 10.1073/pnas.250491597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacGregor GR, Fan WW, Waymire KG, Wallace DC. In: Embryonic Stem Cells. Notarianni E, Evans MJ, editors. New York: Oxford Univ. Press; 2006. pp. 72–104. [Google Scholar]
- 13.Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Nat. Genet. 1996;14:146. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- 14.Hussein MR. Hum. Reprod. Update. 2005;11:162. doi: 10.1093/humupd/dmi001. [DOI] [PubMed] [Google Scholar]
- 15.Tilly JL, Tilly KI. Endocrinology. 1995;136:242. doi: 10.1210/endo.136.1.7828537. [DOI] [PubMed] [Google Scholar]
- 16.Tsai-Turton M, Luderer U. Endocrinology. 2006;147:1224. doi: 10.1210/en.2005-1281. [DOI] [PubMed] [Google Scholar]
- 17.Wallace DC. Annu. Rev. Biochem. 2007;76:781. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Science. 2004;303:223. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz-Pesini E, Wallace DC. Hum. Mutat. 2006;27:1072. doi: 10.1002/humu.20378. [DOI] [PubMed] [Google Scholar]
- 20.Supported by California Regenerative Medicine Predoctoral Fellowship TI-00008 (W.F.), NIH grant HD45913 (G.R.M.), and NIH grants NS21328, AG13154, AG24373, DK73691, and AG16573 (D.C.W.).