Abstract
Microtubules form a cytoskeletal framework that influences cell shape and provides structural support for the cell. Microtubules in the nervous system undergo a unique post-translational modification, polyglutamylation of the C termini of their tubulin subunits. The mammalian enzymes that perform β-tubulin polyglutamylation as well as their physiological functions in the neuronal tissue remain elusive. We report identification of a mammalian polyglutamylase with specificity for β-tubulin as well as its distribution and function in neurite growth. To identify putative tubulin polyglutamylases, we searched tubulin tyrosine ligase-like (TTLL) proteins for those predominantly expressed in the nervous system. Of 13 TTLL proteins, TTLL7 was transcribed at the highest level in the nervous system. Recombinant TTLL7 catalyzed tubulin polyglutamylation with high preference to β-tubulin in vitro. When expressed in HEK293T cells, TTLL7 demonstrated specificity for β-tubulin and not for α-tubulin or nucleosome assembly protein 1. Consistent with these findings, knockdown of TTLL7 in a primary culture of superior cervical ganglion neurons caused a loss of polyglutamylated β-tubulin. Following stimulation of PC12 cells with nerve growth factor to differentiate, the level of TTLL7 increased concomitantly with polyglutamylation of β-tubulin. Short interference RNA-mediated knockdown of TTLL7 repressed nerve growth factor-stimulated MAP (microtubule-associated protein) 2-positive neurite growth in PC12 cells. Consistent with having a role in the growth of MAP2-positive neurites, TTLL7 accumulated within a MAP2-enriched somatodendritic portion of superior cervical ganglion, as did polyglutamylated β-tubulin. Anti-TTLL7 antibody revealed that TTLL7 was distributed in a somatodendritic compartment in the mouse brain. These findings indicate that TTLL7 is a β-tubulin polyglutamylase and is required for the growth of MAP2-positive neurites in PC12 cells.
Microtubules have important functions in a variety of dynamic activities within the cell including intracellular transport, cell motility, and cell division. They provide structural support for the cell and are an important component of the cytoskeletal framework that generates cell morphology. In neurons microtubules are required for formation of specialized processes, i.e. dendrites and axons. Structural microtubule-associated proteins (MAPs),2 such as MAP2 and tau, regulate the stability of microtubules in those processes.
Microtubules are predominantly composed of tubes of polymerized dimers of α- and β-tubulin. A central question in cell biology is how functional heterogeneity is imparted to different types (e.g. dendritic, axonal, and centriolar) of microtubules. Two basic mechanisms have been proposed to explain this issue. The first involves duplication and divergence of genes encoding tubulin. The second mechanism involves the addition of a variety of post-translational modifications (PTM) to microtubules that further increases the tubulin heterogeneity (1). These PTMs are associated with different microtubule states, such as the structure and its age. For example, acetylated α-tubulin (2) and detyrosinated α-tubulin (3) are enriched in highly stable, i.e. long-lived, populations of microtubules (4). Although the correlation between tubulin modifications and microtubule states has been established for more than 20 years, the physiological function of these modifications is poorly understood.
Among PTMs, polyglutamylation (5) and polyglycylation (6) are unique modifications observed in specific cell types. The great majority of tubulin in the adult mammalian brain is polyglutamylated (7), suggesting that this PTM has an important function in neural development or homeostasis. Tubulin polyglutamylation can regulate the binding affinity of MAPs, including kinesin motor proteins, to microtubules in vivo (8–11). Polyglutamylated α-tubulin is present at similar levels in the brain during development from neonate to adult. In contrast, steady-state levels of polyglutamylated β-tubulin increase gradually during this period (12), which suggests that this latter PTM may be required for maturation of the nervous system.
Janke et al. (13) have recently demonstrated that catalytic subunits of tubulin polyglutamylase belong to a family of tubulin tyrosine ligase (TTL)-like (TTLL) proteins. In Tetrahymena, TTLL1 and TTLL6A function as polyglutamylases for α- and β-tubulin, respectively (13). A mammalian polyglutamylase complex composed of TTLL1 and at least four other components, PGs1, 2, 4, and 5, has been identified (13–15). The complex biochemically purified from murine brain displays a preference for α-tubulin (13, 15, 16). Consistent with these findings, we have determined that the lack of functional PGs1 causes a gross loss of polyglutamylated α-tubulin in vivo.3 We have identified a physiological function for polyglutamylated α-tubulin in regulating kinesin motor traffic.3 In contrast to the recent exciting progress of research on α-tubulin polyglutamylation, neither the mammalian enzymes responsible for polyglutamylation of β-tubulin nor the functions for this modification have been identified. Here, we provide the first evidence that TTLL7 is a β-tubulin-specific mammalian polyglutamylase that is expressed at high levels in the nervous system. We show that TTLL7 and polyglutamylated β-tubulin accumulate in MAP2-enriched subcellular compartments and demonstrate that polyglutamylation of β-tubulin is required for MAP2-positive neurite growth in PC12 cells.
EXPERIMENTAL PROCEDURES
Antibodies
Anti-polyglutamylated tubulin monoclonal antibody, GT335 (7) was a kind gift from Dr. Bernard Eddé. Monoclonal antibodies against α-tubulin (DM1A), β-tubulin (Tub2.1), neurofilament (NN18), MAP1A (HM-1), MAP2 (HM-2), FLAG epitope (M2), and anti-actin polyclonal antibodies were purchased from Sigma-Aldrich. Monoclonal antibodies for Tau (Tau-1), glyceraldehyde-3-phosphate dehydrogenase (6C5), and anti-MAP2 polyclonal antibodies were purchased from Chemicon International (Temecula, CA). Anti-V5 monoclonal antibody was obtained from Invitrogen (Carlsbad, CA). Anti-β-tubulin monoclonal antibody (DM1B) and polyclonal antibody for α-tubulin were purchased from Lab Vision (Fremont, CA).
Anti-TTLL7 antiserum was raised in rabbits immunized with maltose-binding protein-fused TTLL7 (Δ370; corresponding to residues 371–609). The recombinant TTLL7 purified by amylose resin (New England BioLabs, Ipswich, MA) was mixed with Freund’s adjuvant (Sigma-Aldrich) and injected subcutaneously on the dorsum of rabbits. Anti-TTLL7 antibodies were purified with maltose-binding protein-TTLL7-bound column.
Plasmid Construction
cDNA containing whole coding sequences of each mouse TTLL protein and NAP1 were cloned according to the NCBI data base or were purchased from DNAFORM (Yokohama, Japan). The corresponding accession numbers of each gene TTLL are as follows: mTTL, BC018513; mTTLL1, BC010510; mTTLL3, AK080321; mTTLL6, AK077033; mTTLL7, AK014905; mTTLL13, AK029565; mNAP1, AK050375.
Mammalian expression vectors were prepared by using the GATEWAY system (Invitrogen) or by direct insertion of coding sequence. The full coding sequences of mTTLLs, except for mTTL, were amplified using primers compatible to the GATEWAY system. Amplified fragments were subcloned into the pDONR207 vector (Invitrogen). To obtain expression vector for FLAG-tagged protein, pCMV-Tag 4A vector (Stratagene, La Jolla, CA) was converted into destination vector using the GATEWAY vector conversion system (Invitrogen). GATEWAY system-compatible expression vector of EGFP-fused protein was generated by conversion of pEGFP-C1 vector (BD Biosciences, Franklin Lakes, NJ) to destination vector. Final coding sequence-harboring expression vectors were generated by performing LR reaction (Invitrogen). Expression vectors for FLAG-tagged mTTL and mNAP1 were prepared by direct ligation into pCMV-Tag 4A vector using restriction enzyme. All of the coding sequences in plasmids were verified by DNA sequencing before use. The sequences inserted to expression vectors were as follows: mTTL, 204–1334 of BC018513; mTTLL1, 337–1605 of BC010510; mTTLL3, 84–2195 of AK080321; mTTLL6, 799–2953 with a deletion of G at 951 of AK077033; mTTLL7, 360–2186 of AK014905; mTTLL13, 590–2710 of AK029565; mNAP1, 686–1858 of AK050375.
For bacterial expression, a whole coding sequence of mTTLL7 was cloned into pET vector (Novagen, Merck Biosciences, Tokyo). An enzyme-inactive form of mTTLL7 was obtained by adding a point mutation into coding sequence so that Glu at 349 was replaced with Val, which results in failure of the enzyme to capture magnesium ion (13).
Microarray Analysis Using BioExpress Data Base
We performed a data base search and statistical analysis on TTLL mRNA expression among various tissues in humans that were profiled by Affymetrix GeneChip technology and stored in the commercial Gene Logic BioExpress data base system. The expression intensities were normalized by the Affymetrix global scaling method.
In Situ Hybridization
Mouse brain sections were fixed with 4% paraformaldehyde following treatment with 1 μg/ml proteinase K in phosphate-buffered saline (PBS) containing 0.1% Tween 20. Hybridization was performed overnight at 65 °C with digoxigenin-labeled RNA probes. The hybridized probes were reacted with alkaline phosphatase-conjugated anti-digoxigenin antibody (diluted at 1:1,000; Roche Applied Science) at 4 °C overnight and then visualized by coloring reaction in the NTMT solution (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, 50 mM MgCl2, 0.1% Tween 20, and 2 mM levamisole) containing 35 mg/ml nitro blue tetrazolium (Roche Applied Science), 17.5 mg/ml 5-bromo-4-chloro-3-indolyl phosphate (Roche Applied Science), and 2 mM levamisole (Sigma).
Assay for Tubulin Polyglutamylase
Glutathione S-transferase-fused mTTLL7 synthesized in Escherichia coli Rossetta strain (DE3) was purified by chromatography on glutathione-Sepharose 4B beads (GE Healthcare Bio-Sciences, Piscataway, NJ) and then cleaved using PreScission protease at 4 °C to liberate the recombinant protein. Mouse brain tubulin was purified through two cycles of polymerization-depolymerization in a high molarity buffer as described (17). Enzyme activity was measured by incorporation of [3H]glutamate into tubulin at pH 7.0. Before the addition of recombinant TTLL7 (5 μg/ml at final concentration), L-[3H]glutamate and 2 mM ATP, the reaction mixture (50 mM Tris, pH 7.0, 8 mM MgCl2, 2.5 mM dithiothreitol, 10 μM paclitaxel, 0.2 mg/ml purified mouse brain tubulin) was warmed at 37 °C for 10 min to polymerize microtubules. After incubation of the mixture (10 μl) at 30 °C for 1 h, the reaction was subjected to SDS-PAGE using 95% purity SDS (Sigma-Aldrich) to separate α- and β-tubulins. The separated tubulins were blotted onto a nitrocellulose membrane. To visualize the migrating position of α- and β-tubulin, the membrane was stained with Ponceau S. Radioactivity incorporated into tubulins was quantified from excised bands using a Winspectral 1414 liquid scintillation counter (Wallac, Turku, Finland) for 1 min.
Cell Culture
Culture of mouse superior cervical ganglion (SCG) neurons was prepared as described (18) and cultured in minimum essential medium containing 10% fetal bovine serum (FBS). Dissociated neurons and micro-explant of SCG were seeded onto plastic plates coated with 0.1 mg/ml of poly-L-lysine- and 10 μg/ml of laminin.
Human embryonic kidney (HEK) 293T cells were grown in Dulbecco’s modified Eagle’s medium containing 10% FBS. Rat kangaroo kidney (PtK2) cells were cultured in 10% FBS-containing minimal essential medium supplemented with nonessential amino acids (Invitrogen) and sodium pyruvate. Plasmids were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
PC12 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 5% FBS and 5% horse serum. Neuronal differentiation of PC12 cells was induced by exposing cells to medium containing 100 ng/ml nerve growth factor (NGF), 0.5% FBS, and 0.5% horse serum.
RNA Interference Knockdown
Knockdown of TTLL7 was accomplished using 200 nM 21-mer short interference RNA (siRNA; from Thermo Electron, Sedansrasse, Germany) and 8 μl/ml Lipofectamine 2000 (Invitrogen). Target sequences for RNA interference were chosen using siDirect, highly effective, and target-specific siRNA design software (19, 20). The target sequences of siRNA used were 5′-UUGUUCCGAGAACGUGGAUUU-3′ (against mouse TTLL7) and 5′-UUGAUAAGAAAUGGGGAUAAA-3′ (against rat TTLL7). The sequence 5′-AAUUCUCCGAACGUGUCACGU-3′ was used as a non-specific siRNA negative control. This sequence is not found in the mouse genome. To diminish target proteins, cultures of SCG neurons were treated with siRNA twice for a total period of 1 week.
Western Blot Analysis
Proteins were separated by SDS-PAGE and blotted onto polyvinylidene fluoride membranes (Millipore, Beverly, MA). When α- and β-tubulin were separated, we used 95% purity SDS (Sigma). The proteins were labeled with primary antibodies and horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). The antibodies were used at the following dilutions: GT335, 1:20,000; DM1A, 1:10,000; Tub2.1, 1:2,000; DM1B, 1:1,000; 6C5, 1:5,000; anti-actin, 1:1,000; NN18, 1:5,000; HM-1, 1:500; HM-2, 1:2,000; tau-1, 1:5,000; M2, 1:2,000; anti-V5, 1:20,000; anti-TTLL7, 1:1,000. Signal was visualized using an ECL kit (GE Healthcare Bio-Sciences).
Immunocytochemical Analysis
The cells were fixed with 4% paraformaldehyde/PBS prewarmed to 37 °C, to prevent cold shock-induced depolymerization of microtubules. Fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min and then incubated in 5% goat serum/PBS for 1 h at room temperature. Incubation of cells in primary antibody was performed at 4 °C overnight. After rinsing in PBS, the cells were incubated with fluorescently labeled secondary antibody for 1 h at room temperature. The cells were rinsed briefly and mounted before observation with a confocal laser scanning microscope (LSM5 Pascal; Zeiss, Oberkochen, Germany; or Fluoview FV1000; Olympus, Japan).
Immunohistochemical Analysis
Adult mice were anesthetized and perfused intracardially with heparin, then with 4% paraformaldehyde, 0.1% glutaraldehyde solution in phosphate buffer, pH 7.4, followed by brief perfusions with solutions of 10, 20, and 30% sucrose. The brains and spinal cords of the mice were immersed overnight in 30% sucrose solution at 4 °C. Coronal brain sections (10 μm thick) were cut through the cerebrum using a freezing cryostat. The sections were rinsed in 0.1 M phosphate buffer (pH 7.4), treated with 50 mM glycine in phosphate buffer, and then incubated for 30 min in 3% skim milk in phosphate buffer. The sections were incubated with the primary antibody in 3% skim milk/phosphate buffer overnight at 4 °C. For the negative control, we incubated the sections with preimmune serum. The incubated sections were washed three times with PBS. The sections were processed with an Alexa Fluor fluorescent dye-conjugated secondary antibody (Invitrogen). The images were obtained with a confocal microscope (Fluoview FV1000) equipped with a 20×, 0.75 na objective lens, and appropriate fluorescence filter sets.
RESULTS
TTLL7 Is Predominantly Expressed in Neuronal Tissues and Is a Candidate for a Tubulin Polyglutamylase
TTL is a tubulin-modifying enzyme that belongs to a large superfamily of “ADP-forming” or “ATP grasp” enzymes that possesses ATP-dependent carboxylate-amine ligase activity (21, 22). TTL ligates a tyrosine to the α-carboxyl group of a C-terminal glutamate. Analysis of the mouse genome by BLAST search using a TTL identified 13 independent loci encoding “TTL domain-containing,” i.e. TTL-like (TTLL) proteins (Fig. 1, A and B, and supplemental Table S1). Recently, Janke et al. (13) have demonstrated that TTLL1 is a catalytic subunit of polyglutamylase complex biochemically purified from mouse brain. Moreover, they have identified Tetrahymena TTLL1 and TTLL6A as tubulin polyglutamylase on α-tubulin and β-tubulin, respectively (13). Given the findings by Janke et al., the 13 TTLL proteins might include mammalian β-tubulin polyglutamylases.
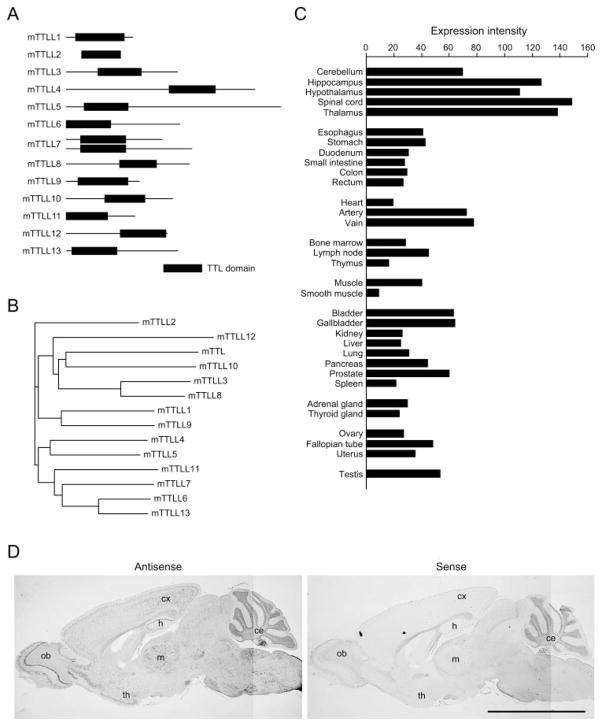
FIGURE 1. TTLL7 is predominantly expressed in neuronal tissues.
A, schematic view of mouse TTLL proteins. B, phylogenic tree of mouse TTLL proteins based on similarity of TTL domain. The phylogenic tree of mTTLL proteins was generated with NJPlot program. C, expression profile of human TTLL7. hTTLL7 is predominantly expressed in neuronal tissues, such as spinal cord, thalamus, hippocampus, and hypothalamus. D, in situ hybridization analysis of mouse TTLL7 in mouse adult brain. mTTLL7 transcripts were strongly detected in olfactory bulb (ob), hippocampus (h), thalamus (th), midbrain (m), cerebral cortex (cx), and cerebellum (ce). Scale bar, 5 mm.
Because the brain has high levels of polyglutamylated tubulin (7), we predicted that tubulin polyglutamylases are expressed in this organ. To determine whether TTLL proteins were expressed in the nervous system, we screened a human cDNA library by a microarray analysis. The results indicated that TTLL7 is highly expressed in the nervous system including spinal cord, thalamus, hippocampus, hypothalamus, and cerebellum (Table 1 and Fig. 1C). Indeed, TTLL7 transcripts were present throughout the mouse brain, especially in the hippocampus, thalamus, olfactory bulb, and cerebellum (Fig. 1D). Moreover, analysis of expressed sequence tag data bases at NCBI suggests that expression of mouse TTLL7 is dominant in whole brain.4 Thus, TTLL7 is a candidate for a tubulin polyglutamylase in the mammalian nervous system. The study by Janke et al. (13) strongly supports this conclusion because mammalian TTLL7 is a close relative to Tetrahymena TTLL6A.
TABLE 1.
Tissues dominantly expressing each TTLL protein
| TTLL proteins | Dominantly expressing tissue |
|---|---|
| TTLL1 | Nervous system, testis, fallopian tube, thyroid glanda |
| TTLL2 | Testis |
| TTLL3 | Uterus, fallopian tube, ovary, smooth muscle |
| TTLL4 | Nervous system, muscle, bone marrowa |
| TTLL5 | Testis, spinal cord |
| TTLL6 | Testis, intestine |
| TTLL7 | Nervous system |
| TTLL8 | —b |
| TTLL9 | NDc |
| TTLL10 | ND |
| TTLL11 | Muscle, bone marrowa |
| TTLL12 | Esophagus, colona |
| TTLL13 | ND |
Ubiquitously distributed to some extent (see supplemental Fig. S1).
Probe set was not present.
ND, not determined (expression intensities were under the detection threshold).
TTLL7 Is a Tubulin Polyglutamylase with Specificity for β-Tubulin
To investigate whether TTLL7 has properties of a tubulin polyglutamylase, we purified bacterially expressed recombinant mouse TTLL7 (mTTLL7) and examined it for ligase activity by measurement of its ability to incorporate [H3]glutamate into tubulin in vitro. Purified recombinant mTTLL7 catalyzed [H3]glutamate incorporation into microtubules in vitro (Fig. 2A). The preferred substrate for enzyme activity was β-tubulin rather than α-tubulin (Fig. 2A), suggesting that this protein is a β-tubulin-selective polyglutamylase in vivo. In contrast, a mutated (E349V) form of mTTLL7, which is predicted to lack the ability to bind magnesium ion co-factor (13), lost all enzyme activity (Fig. 2A).
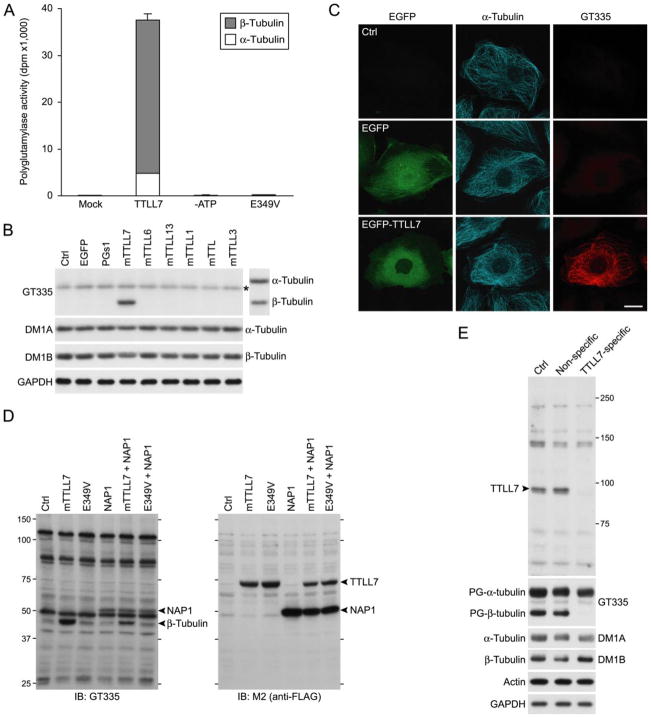
FIGURE 2. TTLL7 is a β-tubulin polyglutamylase.
A, in vitro enzyme activity of recombinant mouse TTLL7. Recombinant mTTLL7 incorporated [3H]glutamate into microtubules with a high preference to β-tubulin in vitro. These activities were lost when ATP was omitted from reaction mixture (–ATP) or when a point mutation causing failure of the enzyme to use ATP was inserted in mTTLL7 (E349V). Enzyme activities are expressed as radioactivity incorporated into microtubules (disintegration per minute; dpm). The result is representative of three independent experiments. B, in vivo enzyme activity of a subset of mouse TTLL proteins. Only mTTLL7 displayed tubulin polyglutamylase activity in HEK293T cells. Polyglutamylated tubulins were detected with a mAb, GT335. The blot was reprobed with DM1A and Tub2.1 to verify migrating positions of both tubulins (displayed on the right of GT335-labeled blot image). Note that the band marked with an asterisk, detected with GT335 in all samples, does not co-migrate with α-tubulin. Amounts of total tubulins were confirmed by reprobing with DM1A and DM1B. Amount of glyceraldehyde-3-phosphate dehydrogenase was monitored as a loading control (Ctrl). The result is representative of three independent experiments. Expression efficacies were shown in supplemental Fig. S2. C, immunocytochemical analysis of effect of overexpressed mTTLL7 in PtK2 cells. Overexpression of EGFP-mTTLL7 in PtK2 cells resulted in polyglutamylation of microtubules. Scale bar, 20 μm. D, in vivo enzyme activity of mTTLL7 on NAP1. To visualize signals other than tubulin labeled by GT335, x-ray film was overexposed. To efficiently detect polyglutamylated NAP1 signal, we overexpressed FLAG-NAP1 with or without mTTLL7. mTTLL7 did not enhance signal intensities of bands other than β-tubulin. Ectopic NAP1 also was not polyglutamylated by mTTLL7. E, knockdown of endogenous TTLL7 and loss of β-tubulin polyglutamylation in primary cultures of SCG neurons. Antiserum raised against TTLL7 detected a protein of ~90 kDa (arrowhead) that disappeared following treatment with siRNA targeted to mTTLL7. Polyglutamylated (PG-) tubulins were detected with mAb GT335. Total α-tubulin and β-tubulin were detected with mAb DM1A and DM1B, respectively.
To investigate the enzyme activity of TTLL7 in vivo, we expressed FLAG-tagged mTTLL7 in HEK293T cells and analyzed polyglutamylated tubulin by Western blotting using the GT335 monoclonal antibody that has preferential specificity for polyglutamylated tubulin (7). Consistent with the results of the in vitro analysis, expression of mTTLL7 in HEK293T cells caused a prominent increase in polyglutamylated β-tubulin but not α-tubulin (Fig. 2B). Total amounts of α- and β-tubulin were not altered by overproduction of mTTLL7 (Fig. 2B). These results indicate that TTLL7 functions as a β-tubulin-specific polyglutamylase. Consistent with the function of TTLL7 as a β-tubulin polyglutamylase, overproduction of EGFP-fused mTTLL7 lead to an increase in polyglutamylated microtubules in PtK2 cells (Fig. 2C).
Tubulins and nucleosome assembly proteins (NAPs) are currently the only reported substrates for polyglutamylation (23). To investigate the substrate specificity of TTLL7, we used the known cross-reactivity of GT335 for glutamylated forms of proteins other than tubulins, e.g. NAPs. Many additional proteins could be detected when the film was exposed for a prolonged period to the GT335-probed blot (Fig. 2D). However, other than β-tubulin, overexpression of mTTLL7 in HEK293T cells did not generate any GT335-specific band that was not also observed in untransfected negative control cells (Fig. 2D). We also investigated whether TTLL7 could glutamylate NAP1 by co-expression of both mTTLL7 and mNAP1 in HEK293T cells. GT335 reacted with exogenously overproduced mNAP1, indicating that this protein is glutamylated by endogenous polyglutamylase enzyme(s) in these cells. Overproduction of the mutant form of TTLL7, in which Glu at 349 was replaced by Val, did not decrease the NAP1 glutamylation (Fig. 2D). Because this mutant form of TTLL7 can function in a dominant negative manner, this suggests that the endogenous NAP1 glutamylase is not TTLL7. Co-expression of mTTLL7 also had no significant effect on the extent of polyglutamylation of mNAP1 (Fig. 2D). Thus, to the extent that glutamylated proteins are detected with mAb GT335, these results suggest that TTLL7 has specificity for β-tubulin in vivo in HEK293T cells.
To test this conclusion, we examined the function of endogenous TTLL7 in neuronal cells. We inhibited endogenous TTLL7 activity in primary cultures of mouse SCG neurons by siRNA-mediated knockdown of mTTLL7. A siRNA targeted to mTTLL7 effectively knocked down the endogenous TTLL7 in the primary culture of SCG neurons (Fig. 2E). This treatment resulted in a complete loss of β-tubulin polyglutamylation but had no overt effect on polyglutamylation of α-tubulin (Fig. 2E). Collectively, these results demonstrate that endogenous TTLL7 functions as a β-tubulin-specific polyglutamylase in neuronal cells.
TTLL7 and Polyglutamylated β-Tubulin Are Required for Neurite Growth
The presence of polyglutamylated β-tubulin increases progressively during neuronal maturation in vitro and in vivo (12, 24). In addition, assessment of the expression of TTLL7 by analysis of expressed sequence tags using NCBI, shows that TTLL7 is predominantly expressed in adult mice.4 This suggests that polyglutamylated β-tubulin is required for some aspect of postnatal development of the nervous system, such as growth of axodendritic processes and maturation of neural network (12, 24). Consistent with this notion, TTLL7 was expressed in neuronal PC12 cells that had been induced to differentiate by exposure to NGF but was not expressed in undifferentiated (naïve) PC12 cells (Fig. 3A). Moreover, polyglutamylated β-tubulin was detected in differentiated PC12 cells but not in undifferentiated cells (Fig. 3B). To investigate whether TTLL7-mediated β-tubulin polyglutamylation was required for neurite growth, we investigated the effect of knockdown of TTLL7 on formation and growth of neurites in PC12 cells. siRNA targeted to rTTLL7 produced significant reduction of the steady-state level of endogenous TTLL7 protein in PC12 cells (Fig. 3A), which resulted in block of β-tubulin polyglutamylation (Fig. 3B). Knockdown of TTLL7 also had a pronounced effect on reducing neurite outgrowth (Fig. 3C). Both the number and length of neurites in TTLL7 knockdown PC12 cells were reduced at 24 h following NGF stimulation (Fig. 3, D–G). These results indicate a causal relationship between TTLL7 expression and β-tubulin polyglutamylation in PC12 cells and that these activities are required for NGF-stimulated neurite growth in this cell type.
FIGURE 3. TTLL7 and polyglutamylated β-tubulin are involved in neurite growth.
A, expression of TTLL7 in neuronally differentiated PC12 cells. Neuronal PC12 cells expressed TTLL7 at 14 days following NGF stimulation. The amount of TTLL7 was diminished by rTTLL7-specific siRNA treatment. B, effect of TTLL7 knockdown on β-tubulin polyglutamylation in PC12 cells. Neuronal PC12 cells contained both polyglutamylated α- and β-tubulin. Polyglutamylated β-tubulin was lost by rTTLL7-specific siRNA treatment. C, effect of the knockdown of TTLL7 on neurite growth of PC12 cells. PC12 cells treated with siRNA for 24 h were differentiated to neuronal cells by application of NGF for 48 h. TTLL7-targeted siRNA suppressed neurite formation and extension. Scale bar, 10 μm. D and E, quantification of neurite number in PC12 cells. Neurite number was quantified at 24 h after NGF-stimulated differentiation. The data were from three independent experiments. ***, p < 0.001 with Mann-Whitney U test. F and G, quantification of neurite length in PC12 cells. Neurite length was quantified at 24 h after NGF-stimulated differentiation. The data were from three independent experiments. ***, p < 0.001 with Mann-Whitney U test. H, immunocytochemical staining of differentiated PC12 cell. Neuronally differentiated PC12 cells were stained with FITC-DM1A (green) and with anti-MAP2 antibodies (red). Scale bar, 20 μm. Ctrl, control.
TTLL7 and Polyglutamylated β-Tubulin Accumulate in MAP2-positive Somatodendritic Region of Neurons
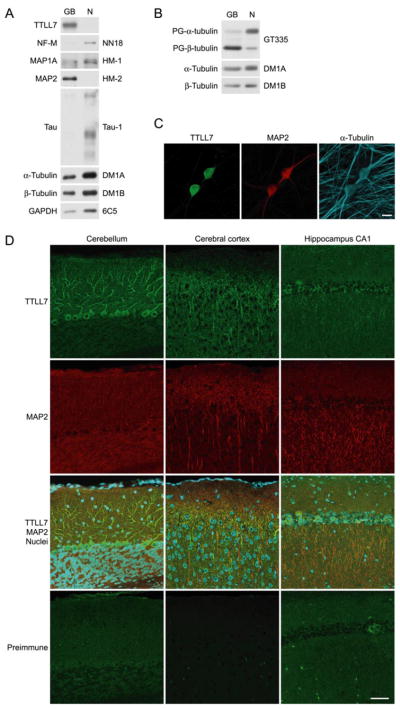
Neurites of differentiated PC12 cells contained MAP2 (Fig. 3H). This and the finding that TTLL7 is required for formation of neurites in these cells suggest that TTLL7 may normally be localized within MAP2-positive processes in neuronal cells, i.e. within the somatodendritic compartment of neurons. To address this question, we explanted SCG in culture, dissected the neurons into somatodendritic and axonal portions, and examined the protein content of each compartment using Western blot analysis (25). The results indicated that TTLL7 is present within MAP2-positive ganglion bodies, i.e. the somatodendritic compartment, but is absent from the MAP2-negative and Tau-1-positive neurite compartment, i.e. the axonal portion (Fig. 4A). Consistent with these results, almost all polyglutamylated β-tubulin was located in the ganglion body compared with the neurites of primary culture of SCG (Fig. 4B). Cytoimmunofluorescence analysis of TTLL7 intracellular distribution in cultured SCG neurons revealed that TTLL7 was exclusively localized in soma, where MAP2 was also enriched (Fig. 4C). This is consistent with the result of explant culture (Fig. 4A). We next analyzed the distribution of TTLL7 in the adult mouse CNS by immunostaining with an anti-TTLL7 antibody (Fig. 4D). As with the earlier results, intracellular distribution of TTLL7 was mainly somatodendritic; TTLL7 was predominantly localized in the soma and dendrites of Purkinje neurons in cerebellum and in apical dendrites of pyramidal neurons in cerebral cortex and hippocampal CA1 region (Fig. 4D). A subset of TTLL7 was co-localized with MAP2 (Fig. 4D). These results suggest that TTLL7 and polyglutamylated β-tubulin could also be required for the growth of MAP2-positive neurites, i.e. dendrites, in certain neurons in vivo.
FIGURE 4. TTLL7 and polyglutamylated β-tubulin are located in MAP2-positive subcellular compartments.
A, intracellular distribution of TTLL7 in explant culture of SCG neurons. TTLL7 was exclusively located in ganglion bodies (GB), where MAP2 accumulated. Equal amounts of protein were loaded in GB and neuritic portion (N). B, distribution of polyglutamylated tubulins in explant culture of SCG neurons. Polyglutamylated β-tubulin concentrated in ganglion bodies (GB), whereas polyglutamylated α-tubulin was dominant in neurite compartment (N). To obtain equal amounts of tubulin, the samples from the ganglion body contained three times the amount of protein compared with samples from neurites. C, immunocytochemical analysis of TTLL7 distribution in SCG neurons. TTLL7 was exclusively accumulated in cell bodies where MAP2 was enriched. Scale bar, 20 μm. D, distribution of TTLL7 in mouse brain. TTLL7 was concentrated in soma and dendrites of Purkinje neurons in cerebellum (left panels), in apical dendrites of pyramidal neurons in cerebral cortex (middle panels), and in apical dendrites of pyramidal neurons in hippocampal CA1 region (right panels). The signals of TTLL7 and MAP2 partially overlapped. The nuclei were stained with TOTO-3 (cyan). Staining with preimmune serum was performed as a negative control for the TTLL7 staining. Scale bar, 50 μm.
DISCUSSION
This report provides the first evidence that TTLL7 functions as a polyglutamylase with preference for β-tubulin in vitro and in vivo. TTLL proteins have recently been identified as a family of proteins possessing a TTL domain (13). Janke et al. (13) identified mammalian TTLL1 as a component of an enzyme complex having polyglutamylase activity specific for α-tubulin. They also reported that Tetrahymena TTLL1 and TTLL6A are polyglutamylases for α- and β-tubulin, respectively (13). A function of mammalian TTLL7 as a β-tubulin polyglutamylase is plausible because mammalian TTLL7 is phylogenetically related to Tetrahymena TTLL6A (13). In vitro enzyme activity assay demonstrates that pure recombinant TTLL7 performs its enzyme activity, indicating that this enzyme can function without additional proteins. This mammalian enzyme might form a homodimer as predicted in Tetrahymena TTLL6A (13).
In our experimental systems, ectopic expression of TTLL6 or TTLL13 in HEK293T cells failed to induce polyglutamylation of either α- or β-tubulin (Fig. 2B). This was unexpected because these two mammalian TTLL proteins are also close relatives of mammalian TTLL7 and Tetrahymena TTLL6A (13). For some reason, tubulin in HEK293T cells may be competent to be glutamylated by TTLL7 but not by TTLL6 or TTLL13. An alternate possibility is that these TTLL proteins may require additional co-factors to function as does TTLL1 (13). We verified our expression constructs by sequencing prior to use; thus it is unlikely that this is the cause of the failure to generate polyglutamylase activity. However, we cannot currently exclude other reasons including the possibility of existence of TTLL6/TTLL13 mRNA splice variants with different polyglutamylation activities.
In neurons, dynamic regulation of microtubule stability is required for formation and maintenance of two specialized processes, i.e. dendrites and axons, along which molecular motors transport important cargo (11) such as glutamate receptors (26, 27) and synaptic vesicles (28) to appropriate sites over long distances. Do polyglutamylated α- and β-tubulins have different roles in neurons, in particular, in such processes? We found that differentially modified α- and β-tubulins were localized in different subcellular compartments (Fig. 4B). Presumably this is due to differential subcellular localization of enzyme machinery required for the addition and removal of polyglutamylation. Moreover, polyglutamylated β-tubulin increases progressively during neuronal maturation in culture and in brain, whereas the level of polyglutamylated α-tubulin reaches a plateau at birth (12, 24). Together, this raises the possibility that polyglutamylated α- and β-tubulins could play specialized roles in neuronal maturation and function.
Polyglutamylated α-tubulin, which is enriched within axons (Fig. 4B), regulates trafficking of a subset of kinesins and is required for control of synaptic transmission.3 Mice with a mutation in PGs1, a component of α-tubulin polyglutamylase complex (13, 15) have a gross loss of polyglutamylated α-tubulin.3 Strikingly, these mutant mice do not display any obvious anatomical defect in nervous system development (29), and neurons from PGs1-deficient mice are able to form neuronal processes both in vivo and in vitro.5 However, PGs1-deficient mice do display a dramatic behavioral phenotype (29), suggesting that neuronal function may be altered in these mice. In contrast, polyglutamylated β-tubulin, which is found in MAP2-enriched subcellular compartments, was required for the growth of MAP2-positive neurites. Thus, one possibility is that polyglutamylated β-tubulin might play roles in structural development of the nervous system, whereas polyglutamylated α-tubulin might function in regulating neural activity.
Knockdown of TTLL7 in PC12 cells suppressed neurite growth (Fig. 3, C–G). In SCG neurons, which are primary culture of cells derived from the same origin as PC12 cells, both TTLL7 and the modified β-tubulin were concentrated in a MAP2-enriched compartment (Fig. 4, A and B). Suppression of MAP2 production by treatment with siRNA leads to slowdown of NGF-induced neurite extension in PC12 cells (30). MAP2 is enriched in a somatodendritic compartment (31) and regulates dendrite growth in the nervous system (32). Together, these findings imply the existence of a functional interaction between MAP2 and polyglutamylated β-tubulin within somatodendritic compartment, where TTLL7 and polyglutamylated β-tubulin could be required for the growth of dendrites. Dendrite development follows axon formation and growth during neuronal differentiation and nervous system development. This delayed development of dendrites agrees with occurrence of polyglutamylated β-tubulin during neuronal maturation in culture and in brain (12, 24).
How might polyglutamylated β-tubulin function in the neuronal development, especially in dendrite growth? MAP2, which is important for dendrite formation, binds preferentially to β-tubulin (33, 34). Interaction between MAP2 and tubulin is influenced in a biphasic manner by increase of tubulin polyglutamylation (10). The binding affinity of MAP2 with tubulin increases until the length of polyglutamyl units on the side chain of tubulin reaches three and decreases with further glutamyl chain elongation (10). The pattern of immunostaining for TTLL7 and MAP2 partially overlapped (Fig. 4D). Thus, a simple model for a function of TTLL7 in regulating neurite extension is to generate an appropriate level of polyglutamylation of β-tubulin that results in localization of MAP2 within dendrites during their formation.
Microtubule binding of other MAPs could also be influenced by TTLL7-mediated β-tubulin polyglutamylation. For example, MAP1A efficiently binds to hyperglutamylated tubulin (10). Interestingly, the staining pattern of TTLL7, especially within the cerebellum, is similar to that of MAP1A (35, 36). MAP1A levels increase in the brain during the second week postnatal (36), as does the level of polyglutamylated β-tubulin (12). Thus, transition of MAPs-tubulin interaction from MAP2-tubulin to MAP1A-tubulin during nervous system development may be dependent upon an increase in the average number of glutamyl units on side chains of β-tubulin from three to more than three. In our assays, where we used cultured neuronal cells derived from the peripheral nervous system, MAP1A is unlikely to be required for neurite growth that is dependent on β-tubulin polyglutamylation because PC12 cells express extremely low levels of MAP1A (37). Moreover, intracellular distribution of MAP1A and polyglutamylated β-tubulin was not comparable in SCG neurons (Ref. 25 and Fig. 4A), which suggests that additional mechanisms exist to direct MAPs to specific microtubule structures.
Binding of MAP1B to microtubules is also affected by the extent of tubulin polyglutamylation in a manner similar to that of MAP2 (8, 10). Moreover, MAP1B is the major MAP in differentiated PC12 cells (37–40). Thus, a decrease in binding affinity between hypoglutamylated microtubules and MAP1B could account for the failure of neurite formation in PC12 cells treated with siRNA against TTLL7. Consistent with this idea, suppression of MAP1B expression in PC12 cells by an antisense nucleotide also blocks neurite outgrowth (41). However, contribution of MAP1B could be minor in regulating the neurite growth dependent on TTLL7-mediated β-tubulin polyglutamylation. MAP1B is a juvenile MAP and decreases in parallel with neuronal development (42), whereas polyglutamylated β-tubulin gradually arises during postnatal neuronal maturation in culture and in brain (12, 24).
Tau, an Alzheimer disease-related protein, is a major MAPs localized mostly in axons in the nervous system (43). A study of tau-deficient mice revealed that this protein controls the stability of axonal microtubules rather than the axonal growth (44). PC12 cells expressing antisense RNA against tau exhibit slower neurite growth (45). Interaction between tau and tubulins is also influenced in a similar way to that between MAP2 and tubulins (8, 10). As yet we have found no convincing evidence to support the idea that interaction between tau and polyglutamylated β-tubulin accounts for neurite growth in PC12 cells and might be involved in dendrite development. SCG neurons under TTLL7-knockdown condition normally developed their neurites (data not shown), which contained tau as their major MAP (Ref. 25 and Fig. 4A).
Our findings shed light on the regulation of nervous system development by tubulin glutamylation. The identification of mammalian polyglutamylases for α-tubulin (13) and now β-tubulin offers new opportunities to investigate the physiological functions of tubulin modification. In neuropathological situations, dysfunction of the nervous system might be modified via dysregulation of the tubulin modification. Further, in addition to the roles in neuronal tissues, functions for post-translational modification of tubulin in non-neuronal cells, e.g. in cell cycle regulation (16, 46, 47), can be investigated. Such research might reveal a causal relationship between specific human pathology and the tubulin modification.
Supplementary Material
Acknowledgments
We thank Dr. Eddé for providing monoclonal antibody, GT335, and valuable suggestions. We thank Drs. Takeshita, Nakagawa, and Gaertig for constructive discussions and Drs. Nagai, Sekiya, Nagayama, and Hirokawa for generous support and valuable advice. We thank also members of the Mitsubishi Kagaku Institute of Life Sciences, especially Dr. Hatanaka, K. Hatanaka, Y. Sugiura, Y. Yasutake, and M. Takamatsu for technical assistance and advice. Finally, we thank the anonymous reviewers for valuable suggestions about our work.
Footnotes
This work was supported by a WAKATE-B grant from the Japan Society for the Promotion of Science (to K. I.), a grant from the National Institutes of Health (to G. R. M.), PRESTO and SENTAN grants from the Japan Science and Technology Agency (to M. S.), and a WAKATE-A grant from the Japan Society for the Promotion of Science (to M. S.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
The abbreviations used are: MAP, microtubule-associated protein; PTM, post-translational modifications; TTL, tubulin tyrosine ligase; TTLL, tubulin tyrosine ligase-like; PBS, phosphate-buffered saline; SCG, superior cervical ganglion; FBS, fetal bovine serum; HEK, human embryonic kidney; NGF, nerve growth factor; siRNA, short interference RNA; NAP, nucleosome assembly protein; mAb, monoclonal antibody.
K. Ikegami, M. Taruishi, R. L. Heier, H. Takagi, M. Mukai, S. Shimma, S. Taira, K. Hatanaka, N. Morone, M. Matsumoto, I. Yao, P. K. Campbell, S. Yuasa, C. Janke, G. R. MacGregor, and M. Setou, manuscript submitted.
Expression profile suggested by analysis of expressed sequence tag counts for mouse TTLL7 can be accessed through NCBI UniGene Data Base under-NCBI accession number Mm.187793.
K. Ikegami and M. Setou, unpublished observations.
References
- 1.Luduena RF. Int Rev Cytol. 1998;178:207–275. doi: 10.1016/s0074-7696(08)62138-5. [DOI] [PubMed] [Google Scholar]
- 2.L’Hernault SW, Rosenbaum JL. Biochemistry. 1985;24:473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- 3.Argarana CE, Barra HS, Caputto R. Mol Cell Biochem. 1978;19:17–21. doi: 10.1007/BF00231230. [DOI] [PubMed] [Google Scholar]
- 4.Cambray-Deakin MA, Burgoyne RD. Cell Motil Cytoskel. 1987;8:284–291. doi: 10.1002/cm.970080309. [DOI] [PubMed] [Google Scholar]
- 5.Eddé B, Rossier J, Le Caer JP, Desbruyères E, Gros F, Denoulet P. Science. 1990;247:83–85. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- 6.Redeker V, Levilliers N, Schmitter JM, Le Caer JP, Rossier J, Adoutte A, Bré MH. Science. 1994;266:1688–1691. doi: 10.1126/science.7992051. [DOI] [PubMed] [Google Scholar]
- 7.Wolff A, de Néchaud B, Chillet D, Mazarguil H, Desbruyères E, Audebert S, Eddé B, Gros F, Denoulet P. Eur J Cell Biol. 1992;59:425–532. [PubMed] [Google Scholar]
- 8.Boucher D, Larcher JC, Gros F, Denoulet P. Biochemistry. 1994;33:12471–12477. doi: 10.1021/bi00207a014. [DOI] [PubMed] [Google Scholar]
- 9.Larcher JC, Boucher D, Lazereg S, Gros F, Denoulet P. J Biol Chem. 1996;271:22117–22124. doi: 10.1074/jbc.271.36.22117. [DOI] [PubMed] [Google Scholar]
- 10.Bonnet C, Boucher D, Lazereg S, Pedrotti B, Islam K, Denoulet P, Larcher JC. J Biol Chem. 2001;276:12839–12848. doi: 10.1074/jbc.M011380200. [DOI] [PubMed] [Google Scholar]
- 11.Setou M, Hayasaka T, Yao I. J Neurobiol. 2004;58:201–206. doi: 10.1002/neu.10324. [DOI] [PubMed] [Google Scholar]
- 12.Audebert S, Koulakoff A, Berwald-Netter Y, Gros F, Denoulet P, Eddé B. J Cell Sci. 1994;107:2313–2322. doi: 10.1242/jcs.107.8.2313. [DOI] [PubMed] [Google Scholar]
- 13.Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, Temurak N, van Dijk J, Boucher D, van Dorsselaer A, Suryavanshi S, Gaertig J, Eddé B. Science. 2005;308:1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- 14.Regnard C, Audebert S, Desbruyères E, Denoulet P, Eddé B. Biochemistry. 1998;37:8395–8404. doi: 10.1021/bi9804131. [DOI] [PubMed] [Google Scholar]
- 15.Regnard C, Fesquet D, Janke C, Boucher D, Desbruyères E, Koulakoff A, Insina C, Travo P, Eddé B. J Cell Sci. 2003;116:4181–4190. doi: 10.1242/jcs.00743. [DOI] [PubMed] [Google Scholar]
- 16.Regnard C, Desbruyères E, Denoulet P, Eddé B. J Cell Sci. 1999;112:4281–4289. doi: 10.1242/jcs.112.23.4281. [DOI] [PubMed] [Google Scholar]
- 17.Castoldi M, Popov AV. Protein Expression Purif. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- 18.Ikegami K, Koike T. Neuroscience. 2003;122:617–626. doi: 10.1016/j.neuroscience.2003.08.057. [DOI] [PubMed] [Google Scholar]
- 19.Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naito Y, Yamada T, Ui-Tei K, Morishita S, Saigo K. Nucleic Acids Res. 2004;32:124–129. doi: 10.1093/nar/gkh442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galperin MY, Koonin EV. Protein Sci. 1997;6:2639–2643. doi: 10.1002/pro.5560061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dideberg O, Bertrand J. Trends Biochem Sci. 1998;23:57–58. doi: 10.1016/s0968-0004(97)01149-3. [DOI] [PubMed] [Google Scholar]
- 23.Regnard C, Desbruyères E, Huet JC, Beauvallet C, Pernollet JC, Eddé B. J Biol Chem. 2000;275:15969–15976. doi: 10.1074/jbc.M000045200. [DOI] [PubMed] [Google Scholar]
- 24.Przyborski SA, Cambray-Deakin MA. Brain Res Dev Brain Res. 1997;100:133–138. doi: 10.1016/s0165-3806(97)00031-x. [DOI] [PubMed] [Google Scholar]
- 25.Peng I, Binder LI, Black MM. J Cell Biol. 1986;102:252–262. doi: 10.1083/jcb.102.1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Setou M, Nakagawa T, Seog DH, Hirokawa N. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- 27.Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. Nature. 2002;417:83–87. doi: 10.1038/nature743. [DOI] [PubMed] [Google Scholar]
- 28.Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- 29.Campbell PK, Waymire KG, Heier RL, Sharer C, Day DE, Reimann H, Jaje JM, Friedrich GA, Burmeister M, Bartness TJ, Russell LD, Young LJ, Zimmer M, Jenne DE, MacGregor GR. Genetics. 2002;162:307–320. doi: 10.1093/genetics/162.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fontaine-Lenoir V, Chambraud B, Fellous A, David S, Duchossoy Y, Baulieu EE, Robel P. Proc Natl Acad Sci U S A. 2006;103:4711–4716. doi: 10.1073/pnas.0600113103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matus A, Bernhardt R, Hugh-Jones T. Proc Natl Acad Sci U S A. 1981;78:3010–3014. doi: 10.1073/pnas.78.5.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harada A, Teng J, Takei Y, Oguchi K, Hirokawa N. J Cell Biol. 2002;158:541–549. doi: 10.1083/jcb.200110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Littauer UZ, Giveon D, Thierauf M, Ginzburg I, Ponstingl H. Proc Natl Acad Sci U S A. 1986;83:7162–7166. doi: 10.1073/pnas.83.19.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maccioni RB, Rivas CI, Vera JC. EMBO J. 1988;7:1957–1963. doi: 10.1002/j.1460-2075.1988.tb03033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bloom SB, Schoenfeld TA, Vallee RB. J Cell Biol. 1984;98:320–330. doi: 10.1083/jcb.98.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szebenyi G, Bollati F, Bisbal M, Sheridan S, Faas L, Wray R, Haferkamp S, Nguyen S, Caceres A, Brady ST. Curr Biol. 2005;15:1820–1826. doi: 10.1016/j.cub.2005.08.069. [DOI] [PubMed] [Google Scholar]
- 37.Brugg B, Matus A. J Cell Biol. 1988;107:643–650. doi: 10.1083/jcb.107.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greene LA, Liem RKH, Shelanski ML. J Cell Biol. 1983;96:76–83. doi: 10.1083/jcb.96.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drubin DG, Feinstein SC, Shooter EM, Kirschner MW. J Cell Biol. 1985;101:1799–1807. doi: 10.1083/jcb.101.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Black MM, Aletta JM, Greene LA. J Cell Biol. 1986;103:545–557. doi: 10.1083/jcb.103.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brugg B, Reddy D, Matus A. Neuroscience. 1993;52:489–496. doi: 10.1016/0306-4522(93)90401-z. [DOI] [PubMed] [Google Scholar]
- 42.Lewis SA, Sherline P, Cowan NJ. J Cell Biol. 1986;102:2106–2114. doi: 10.1083/jcb.102.6.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Binder LI, Frankfurter A, Rebhun LI. J Cell Biol. 1985;101:1371–1378. doi: 10.1083/jcb.101.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harada A, Oguchi K, Okabe S, Kuno J, Terada S, Ohshima T, Sato-Yoshitake R, Takei Y, Noda T, Hirokawa N. Nature. 1994;369:488–491. doi: 10.1038/369488a0. [DOI] [PubMed] [Google Scholar]
- 45.Esmaeli-Azad B, McCarty JH, Feinstein SC. J Cell Sci. 1994;107:869–879. doi: 10.1242/jcs.107.4.869. [DOI] [PubMed] [Google Scholar]
- 46.Bobinnec Y, Moudjou M, Fouquet JP, Desbruyères E, Eddé B, Bornens M. Cell Motil Cytoskel. 1998;39:223–232. doi: 10.1002/(SICI)1097-0169(1998)39:3<223::AID-CM5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 47.Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Eddé B, Bornens M. J Cell Biol. 1998;143:1575–1589. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.