Abstract
It is difficult to study gene expression in mammalian embryonic germ cells as PGCs constitute only a minor proportion of the mouse embryo. We have overcome this problem by using a novel combination of established molecular and transgenic approaches. A line of mice has been generated in which the cells of the germ lineage express the β-galactosidase reporter gene during embryogenesis. Using this line, germ cells have been purified to near homogeneity from embryos at discrete stages during germline development by use of a stain for β-gal activity and a fluorescence activated cell sorter. Subsequently, cDNA libraries have been constructed from each germ cell population using a modified lone-linker PCR strategy. These combined cDNA libraries represent genes expressed in PGCs during mammalian germline development. To facilitate a molecular genetic approach to studying mammalian germline development, these cDNA libraries will be pooled to form an arrayed, addressed reference embryonic germ cell cDNA library. In parallel with large-scale cDNA sequencing efforts, genes that are differentially expressed in germ cells will be identified by screening the reference library with probes generated by subtractive hybridization. Complementary DNAs identified using this approach will be analyzed by sequencing, database comparison, genomic mapping and in situ hybridization to ascertain the potential functional importance of each gene to germline development. In addition to providing a wealth of novel information regarding patterns of gene expression during mammalian germline development, these results will form the basis for future experiments to determine the function of these genes in this process.
Keywords: germline, embryogenesis, mammalian, molecular genetic
Introduction
Development of the germline is essential to the survival of a species. The primordial germ cells (PGCs) formed during mammalian embryogenesis ultimately give rise, in the male, to spermatogonia, the only self-renewing cell type in an adult capable of making a genetic contribution to the next generation. Several important advances have recently been made in the ability to cryopreserve (Avarbock et al., 1996) and transplant (Brinster and Avarbock, 1994; Brinster and Zimmermann, 1994; Russell et al., 1996) this stem cell population to a host testis. Remarkably, apparently normal spermatogenesis can be generated following spermatogonial transplantation to testes of xenobiotic species (Clouthier et al., 1996; Russell and Brinster, 1996). Thus, contemporary mammalian germ cell research is of importance to both basic and applied biological science. However, despite this fact, our knowledge of the molecular basis for germline development during mammalian embryogenesis is relatively poor (Eddy and Hahnel, 1983; McLaren and Wylie, 1983; Snow and Monk, 1983; De Felici et al., 1992; McCarrey, 1993; Wylie and Heasman, 1993; McLaren, 1994).
In this review we give a brief summary of the biology of germline formation during mouse embryonic development including current experimental approaches to identify genes whose function is required for this process. We conclude by describing a novel approach we have developed to facilitate a systematic molecular genetic study of mammalian germline development.
Origin and development of the mammalian germ lineage
Germline formation in the mouse
Most of our current knowledge about the embryological origin and genetic basis for mammalian germline development has come from studies using the mouse. Cell transplantation or cell marking approaches have been used to assess the developmental potency of cells from the early mouse embryo. Results of these studies have provided insight about the location and time of formation of the mammalian germ lineage. Analysis of potency of cells from the pre- and early post-implantation embryo indicated that while the descendants of these cells can contribute to the germline, they do not do so exclusively (Gardner et al., 1985; Gardner and Beddington, 1988). Results of fate-mapping experiments involving labeling of individual cells from the proximal epiblast of egg-cylinder stage embryos indicated that the descendant of these cells could contribute to both somatic and germ lineages (Lawson and Hage, 1994). Significantly, distal epiblast cells of an egg-cylinder stage embryo could also contribute to the germline upon grafting to a proximal epiblast location although the converse was not true (Tam and Zhou, 1996). Collectively, these results support the theory that the murine germline is not determined before gastrulation.
One of the difficulties in studying germline development in mammals concerns the paucity of markers specific for this lineage. In the mouse, PGCs can first be distinguished from somatic cells on the basis of their relatively high level of expression of tissue non-specific alkaline phosphatase (TNAP) within the extra-embryonic mesoderm (EEM) (McKay et al., 1953; Chiquoine, 1954; Ozdzenski, 1967; Ginsburg et al., 1990; MacGregor et al., 1995). Although alkaline phosphatase is a marker of the developing germ lineage in many species, its activity is not required for development of PGCs in mouse (MacGregor et al., 1995). While within the EEM, the PGCs are not tightly clustered but appear to be interspersed with additional cells that do not express TNAP. Approximately 50 TNAP positive cells can be found in this location at this time (Ginsburg et al., 1990; MacGregor et al., 1995). This figure is very similar to an estimate for the founding population size of the mouse germline of 45 that was generated from fate-mapping studies of Lawson and Hage (Lawson and Hage, 1994). These findings are consistent with a model in which the mouse germline is specified from approximately 50 cells within the EEM shortly after the onset of gastrulation.
The EEM in which the PGCs are found is closely bordered by extra-embryonic visceral endoderm. Local embryonic environment appears to play a role in many inductive events during vertebrate embryogenesis. For example, visceral endoderm is involved in the induction of cardiac mesoderm in several species (Kelley et al., 1993; Schultheiss et al., 1995; Soudais et al., 1995; Narita et al., 1997). As with formation of hematopoietic cells within the EEM component of the yolk sac, it is interesting to speculate that the over-lying visceral endoderm may play some part in the specification of the PGCs within the EEM. Snow (1981) demonstrated that PGCs would form in fragments isolated from the extreme posterior of e7 and e7.5 embryos following their culture in vitro. As discussed (Copp et al., 1986; Ginsburg et al., 1990), these fragments were likely to have been composed of EEM with overlying extra-embryonic visceral endoderm. It would be of interest to determine if PGCs would still form in EEM in culture following removal of the associated endoderm or alternatively if heterotopic EEM could be induced to form PGCs following co-culture with visceral endoderm isolated from the region just posterior to the primitive streak.
Migration of PGCs during embryogenesis
After specification, the PGCs follow guidance cues that will ultimately lead them to the developing genital ridges. In addition, between the time of specification and arrival in the genital ridges, the PGCs must not only survive, but proliferate. Finally, during this period the PGCs must not respond to any inappropriate positional cues that would compromise their totipotency.
At around e8.0 (1–5 somite stage), the PGCs are associated with the hindgut pocket. Snow observed that pieces dissected from the posterior of early gastrulation stage embryos that were subsequently cultured in vitro could form an allantois containing PGCs (Snow, 1981). However, if the cultured portion of the embryo also formed a hindgut, all PGCs would be localized around the endoderm. This suggests that the PGCs have an affinity for the hindgut endoderm. Similar phenomena are also observed during Drosophila (Jaglarz and Howard, 1995), Xenopus (Whitington and Dixon, 1975) and avian (Fujimoto et al., 1976) embryonic development.
During the next day and a half, the growth of the hindgut appears responsible for the passive movement of the PGCs to an internal location within the developing embryo. However, at around the 5–15 somite stage (e8.5–9.0), PGCs begin an active migration from the hindgut endoderm, through the newly forming dorsal mesentery to the genital ridges which they begin to colonize at around the 20–30 somite stage (e9.5 to e10.0). Several lines of evidence suggest that the PGCs are guided to the developing gonadal ridge at least in part by chemotropic means (Kuwana et al., 1986; Godin et al., 1990; Godin and Wylie, 1991). During this migratory phase, the PGCs extend long filopodial processes that link them together in extensive arrays (Gomperts et al., 1994a). These filopodia disappear following the arrival of the PGCs in the genital ridge. The filopodia may play an adhesive role between PGCs during migration as after arrival in the genital ridges, the PGCs are found to be tightly associated (Gomperts et al., 1994b).
Control of adhesion also appears to play a role in guiding migratory PGCs to the gonadal ridge. Extra-cellular matrix glycoproteins including collagen type IV, fibronectin, laminin and tenascin-C are expressed by somatic cells found in the pathways over which PGCs migrate in birds, amphibians and mammals (Heasman et al., 1981; Fujimoto et al., 1985; Alvarez-Buylla and Merchant-Larios, 1986; England et al., 1986; De Felici and Dolci, 1989; Urven et al., 1989; Anstrom and Tucker, 1996). Moreover, migratory and post-migratory mouse PGCs are known to behave differently in culture (Donovan et al., 1986) and have been shown to modulate their adhesiveness to certain of these glycoproteins during germline formation (Ffrench-Constant et al., 1991; Garcia-Castro et al., 1997). It has been proposed that adhesion of PGCs to laminin in the early mouse embryo may involve expression of integrins and a heparin sulfate proteoglycan on the surface of the PGCs (Garcia-Castro et al., 1997). The migratory process is essentially completed by the 45 somite stage (e11.5–e12.0).
During movement from the hindgut pocket to the genital ridges, the number of PGCs increases from 100 to around 2500 (Tam and Snow, 1981; Buehr et al., 1993). Following their arrival in the genital ridges, the PGCs continue to divide until around e13.5 at which time they number approximately 25,000 (Chiquoine, 1954; Tam and Snow, 1981; Buehr et al., 1993). Results of in vitro culture and genetic studies suggest that several growth factors and other factors can influence the survival and/or proliferation of PGCs during embryogenesis. These include mast cell growth factor (MGF), leukemia inhibitory factor (LIF), tumor necrosis factor-α (TNF-α), interleukin-11 (IL-11), and oncostatin M (OSM) (Besmer et al., 1993; Wylie and Heasman, 1993; Cheng et al., 1994; De Felici and Pesce, 1994; Donovan, 1994; Kawase et al., 1994; Koshimizu et al., 1996).
Sexual differentiation of primordial germ cells
Following the onset of morphologic gonadal differentiation at around e13.5, PGCs differentiate into oogonia in the fetal ovary or M (mitotic)-prospermatogonia in the fetal testis. In females, a proportion of the oogonia enter meiotic prophase almost immediately after the mitotic proliferation at e13.5 to form oocytes while in males the prospermatogonia continue to proliferate for an additional day prior to entering mitotic arrest around e14.5. Most oogonia have formed oocytes in meiotic prophase by e15.5 while T (transition) - prospermatogonia continue to be held in G1-phase mitotic arrest until after birth (McLaren, 1988).
It is well established that the somatic component of the fetal testis produces a local factor(s) that inhibits meiosis in PGCs as well as directing the germ cells to develop as prospermatogonia. Experimental evidence to support this includes the finding that PGCs of either sex that end up in the adrenal instead of gonad enter meiosis before birth (Zamboni and Upadhyay, 1983). In addition, studies using inter-sex chimeric mice indicate that XX germ cells are subject to meiotic and mitotic arrest and differentiate into prospermatogonia in the fetal testis while XY PGCs enter meiosis in the fetal ovary and attempt to produce oocytes (McLaren, 1983,1993). Results of more recent studies involving tissue reaggregation experiments indicated that the fetal testis produces a factor at around e12 that is responsible for inhibiting entry of prospermatogonia into meiosis (McLaren and Southee, 1997).
Genetic basis for germline development in the mouse
The molecular basis for germline development is perhaps best understood in Drosophila and Caenorhabditis where sophisticated genetic screens have provided the mutants required to help elucidate this process (Ellis and Kimble, 1994; Williamson and Lehmann, 1996). Strategies to perform large-scale mutagenesis in the mouse have been greatly assisted by the exploitation of simple sequence length polymorphisms (SSLP) that facilitate rapid genetic mapping. In addition, retroviral gene trapping is an efficient approach of mutagenesis that provides information about the pattern of expression of mutated genes as well a method to clone them (Gossler et al., 1989; Friedrich and Soriano, 1991; Skarnes et al., 1992,1995; Wurst et al., 1995; Forrester et al., 1996; Baker et al., 1997; Holzschu et al., 1997). However, despite such technical advances, the cost and space required to conduct a mutagenesis screen for germline defects in the mouse is considerable as such a screen requires test breeding of F2 animals. This has forced the use of alternative experimental strategies to identify genes that have an essential function in mammalian germline development.
Molecular genetic studies of early aspects of mouse germline development are particularly problematic as the lineage constitutes a relatively small population of cells within the embryo. Without sufficient quantities of pure PGCs, it is difficult to use a molecular approach to identify the genes whose function is required for germline development. However, several different genetic approaches have been used to study genes that are expressed in PGCs during murine embryogenesis. These have included empirical approaches e.g., RT-PCR using small amounts of PGC containing material or in situ hybridization of candidate genes to germ cell containing sections; RT-PCR based approaches using degenerate oligonucleotides that have specificity for different classes of gene products; or by cloning murine homologs of genes known to be expressed in embryonic germ cells in other species. Reverse-genetic approaches (i.e., knock-outs) have also been used to provide information regarding function of known genes in germline development. Finally, in a genetic and arguably most direct approach, the cloning of genes associated with the relatively few existing mutations that are known to affect germline development can identify genes that a priori have an essential function during germline development.
Perhaps the best examples of the latter approach are mice mutant at the dominant white-spotting (W) and steel (Sl) loci which have overlapping mutant phenotypes that include drastic reductions in the number of gonadal PGCs found during embryogenesis (Besmer et al., 1993; Buehr et al., 1993; Donovan, 1994). Molecular genetic analyses have revealed that the W locus encodes the c-kit tyrosine kinase transmembrane receptor while the product of the Sl locus, mast cell growth factor (MGF) is a natural ligand for the c-kit receptor. Further analyses have shown that c-kit is expressed by PGCs during germline development while the surrounding somatic mesenchyme expresses MGF. Although soluble MGF can act through the c-kit receptor to enhance survival of PGCs in culture, it is the transmembrane form of MGF that appears to be more important for germ cell survival in vivo as Sld mice which make the secreted form of MGF but lack the transmembrane form display a severe germ cell deficiency (Donovan, 1994; Marziali et al., 1993). Hertwig’s anemia (an), atrichosis (at), Teratoma (Ter) and germ cell deficient (gcd) are four additional mutations that affect germ cell development during embryogenesis although to date none of the genes affected in these mutants has been cloned (Lyon et al., 1996).
Reverse genetic approaches have also been used to analyze the role of growth factors in PGC proliferation and survival. As described earlier, LIF, OSM, TNF-α and IL-11 have all been shown to stimulate survival and/or proliferation of PGCs in culture. However, mice deficient in LIF are not agametic (Stewart et al., 1992) and there is no overt difference in numbers of PGCs found in LIF deficient embryos compared to control animals (P. Donovan and C. Stewart, personal communication) As these cytokines appear to be both structurally and functionally related, it is possible that functional redundancy may mask the effects of loss of a single growth factor on germline development. Significantly, all of these cytokines appear to signal through receptor complexes that includes a common subunit, gp130 (Kishimoto et al., 1994). Moreover, mutation of gp130 results in a 20–60% reduction in PGC in vivo (Koshimizu et al., 1996). One interpretation of these findings is that gp130 mediated cytokine signaling may be more important in proliferation rather than survival of PGCs in vivo.
A reverse genetic approach has been used to analyze the function of additional genes in germ cell development. Examples of genes expressed in PGCs include the transcription factor Oct-3/4 (Scholer et al., 1990), TNAP (Hahnel et al., 1990), the Pem homeobox gene product (Lin et al., 1994), alpha 3, 5, 6 and beta 1 integrins (De Felici et al., 1992) and a mouse homolog (Mvh) of the Vasa gene that is required for germline development in Drosophila (Fujiwara et al., 1994). TNAP is not required for PGC development (MacGregor et al., 1995), germline development appears to be normal in Pem deficient embryos (J. Pitman and C. MacLeod, personal communication) and although analyses of α–5 integrin deficient animals has been reported, the status of PGCs in mutants was not addressed (Yang et al., 1993; Goh et al., 1997).
RT-PCR based approaches have been used to survey the expression of known genes in differentiating germ cells (Coucouvanis and Jones, 1993). In addition, degenerate oligonucleotide probes with homology to functional domains of known proteins have been used as a method to identify novel genes that are potentially important in germline development. Sky encodes a receptor tyrosine kinase that is expressed in PGCs and which can bind Gas-6 as a ligand (Matsubara et al., 1996). A similar approach was also used to identify Gek-1 which encodes a novel serine - threonine kinase expressed in PGCs (Yanagisawa et al., 1996). Gek-1 maps to proximal chromosome 11 in mouse (Yanagisawa et al., 1996). Of potential interest, the as yet uncloned gcd insertional transgenic mutation in which embryos display significant reduction in numbers of PGCs (Pellas et al., 1991) also maps to this chromosomal region (Duncan et al., 1995).
Epigenetic aspects of embryonic germ cell development
PGCs represent a unique mammalian embryonic cell lineage as they provide the source of genetic information for a subsequent generation. Several epigenetic features that regulate gene expression are modified during development of this unique lineage including genomic imprinting and X-inactivation. During germ cell development the parental imprint is erased and a new imprint established. The precise timing of erasure of the parental imprint has not yet been established for any of the imprinted genes although Szabo and Mann (Szabo and Mann, 1995) have demonstrated that inherited imprinting is either erased, or not recognized, in PGCs prior to the time of genital ridge colonization. Thus, a functional generalized neutralization of imprinting is associated with the totipotent state of this unique cell lineage. Tam and co-workers have suggested that X-inactivation occurs within the PGCs during their migration through the dorsal mesentery while reactivation may occur shortly after arrival within the genital ridge (Tam et al., 1994). The molecular basis for control of either of these processes is currently poorly understood.
Many of the aforementioned studies have provided important information about the genetic basis for germline development in the mouse. However, several important fundamental questions remain largely unanswered. These include (a) precisely when and where is the germline specified during embryogenesis and how is this achieved? (b) What protects germ cells from responding to inappropriate developmental cues during germline development? (c) How is the migration of PGCs to the endoderm and subsequently the embryonic gonad regulated? (d) How is survival and proliferation of PGCs regulated during embryogenesis? (e) When and how is genomic imprinting and X-inactivation modified? (f) Which genes are required to regulate sexual differentiation of both male and female germ cells?
A systematic molecular genetic approach to the study of mammalian germline development
Overview
Recent advances in genome analysis technology now make a large-scale systematic analysis of gene expression during development a practical endeavor. One such approach involves examination of patterns of gene expression by hybridization of complex probes to high density arrays of cDNA clones. This approach was used over 15 years ago to examine gene expression during Xenopus and Aspergillus development although at the time the expression of only around 1000 cDNAs were examined (Dworkin and Dawid, 1980; Timberlake, 1980). The concept of creating an arrayed cDNA library or a cDNA catalog as a permanent reference and using hybridization for the simultaneous monitoring of expression of many genes has been described by several groups (Ko, 1990; Lennon and Lehrach, 1991). Although originally involving the spotting of cDNA clones at high density onto nylon filters (Gress et al., 1992; Meier-Ewert et al., 1993; Harrison et al., 1995; Zhao et al., 1995), more advanced technology has recently been developed that permits spotting of cDNAs on glass plates at even greater densities (Schena et al., 1995). A similar method has also been developed to synthesize oligonucleotides in arrays on glass wafers (Fodor et al., 1993). One significant advantage of the latter technique is the dramatic increase in sensitivity of detection of low abundance mRNAs that is achieved when used in conjunction with a laser-based multi-color fluorescence detection system.
A second approach that has been used with success is high through-put, single-pass sequencing of cDNA clones (Adams et al., 1991; Hoog, 1991; Khan et al., 1992; Okubo et al., 1992). With current rates of sequence through-put and quality of automated DNA sequencers, this approach has been successful in generating more than 1 million ESTs from various cell types and organisms (Aaronson et al., 1996; Hillier et al., 1996). Furthermore, the ability of this approach to compare expression profiles has been validated for many cell types (Okubo et al., 1995). One variant of this approach is serial analysis of gene expression (SAGE) that involves sequencing of large numbers of cDNAs each of which is a concatenate of shorter cDNA fragments generated from many independent cDNA clones (Velculescu et al., 1995). Both techniques can rapidly provide large amounts of information about expression profiles of genes in different cell types.
We have devised a unique strategy to facilitate a systematic analysis of gene expression during mammalian germline development. A line of transgenic mice has been created in which the developing germline is marked by expression of a beta-galactosidase reporter gene. With these animals, PGCs can be purified from embryos at discrete stages of germline development and cDNA libraries generated from each stage of PGCs. These cDNA libraries can then be analyzed by a large-scale systematic approach using genome analysis methodology. This approach benefits from a low inherent bias in that all genes expressed in embryonic germ cells are theoretically amenable to analysis. While we are in essence constructing a large scale EST database, our approach is unique in that the library is from a single cell type – i.e., embryonic germ cells. Moreover, the reference library represents genes expressed at multiple stages during the process of germline development. In addition to generating valuable novel markers of the germ lineage and providing an addressed database of differential gene expression during germline development, this permanent resource will facilitate future experiments to determine the function of these novel genes in this fundamental biological process.
Generation of transgenic mice that express a reporter gene within the developing germline
The main barrier to a direct analysis of gene expression in the mammalian germline during embryogenesis has been in obtaining sufficient quantities of pure populations of PGCs. Several techniques have been described to purify PGCs from developing mouse embryos using antibodies that define markers of PGCs and either fluorescence activated cell sorting (FACS) (McCarrey et al., 1987) or para-magnetic beads (MACS) (Pesce and De Felici, 1995). In both cases, PGCs were identified using monoclonal antibodies (EMA-1 or TG-1 respectively) that recognize epitopes expressed by PGCs during embryonic development. By definition use of these antibodies is limited to the period during embryonic development that the target epitope is expressed by germ cells. TNAP is currently the PGC marker with the widest window of expression during germline development in the mouse (Cooke et al., 1993; Enders and May, 1994). Thus, use of this marker to purify PGCs should facilitate an analysis of gene expression in the developing germline at the greatest number of stages during mouse embryonic development. However, purification of PGCs from early post-gastrulation stage embryos using (tissue non-specific) alkaline phosphatase as a marker is confounded by the expression of an additional isoenzyme, embryonic alkaline phosphatase (Ginsburg et al., 1990; Hahnel et al., 1990). Therefore, an alternative strategy was required to purify PGCs on the basis of TNAP expression.
The bacterial lacZ (beta-galactosidase) gene has been used successfully as a reporter gene in several diverse species. Techniques have also been developed to purify mammalian cells expressing lacZ using a FACS and the purified lacZ expressing cells are viable following this procedure (Nolan et al., 1988; MacGregor et al., 1991). Consequently, a line of transgenic mice was generated that expressed lacZ under control of the TNAP locus, in the developing germ lineage. The rationale for choosing this method to facilitate purification of germ cells was that in addition to generating high purities of germ cells at a wide range of developmental stages, the purified cells could also be used for either in vitro culture experiments, or for experiments involving grafting to host embryos. The developmental potency of the purified and marked germ cells could then be assessed following culture of the embryos in vitro or after transfer of the host embryo to pseudo-pregnant female recipients for development in vivo.
Analysis of patterns of lacZ activity assessed by X-gal staining of embryos transgenic for different TNAP promoter – lacZ minigenes failed to identify a line of mice in which the developing germline was marked by lacZ expression (Escalante-Alcalde et al., 1996). The reason for this appeared to involve a requirement for several cis-acting sequences that regulate TNAP expression and which are dispersed throughout the TNAP locus (Escalante-Alcalde et al., 1996). To minimize risk of experimental failure due to absence of such cis-elements in a transgene construct, we used homologous recombination in mouse embryonic stem (ES) cells to ‘knock-in’ a lacZ gene at the TNAP (Akp2) locus (MacGregor et al., 1995). In TNAPβ-geo embryos, β-galactosidase expression in embryonic germ cells precisely follows the expression of the endogenous TNAP gene although germline development appears to be unaffected by this mutation (MacGregor et al., 1995) (Fig. 1).
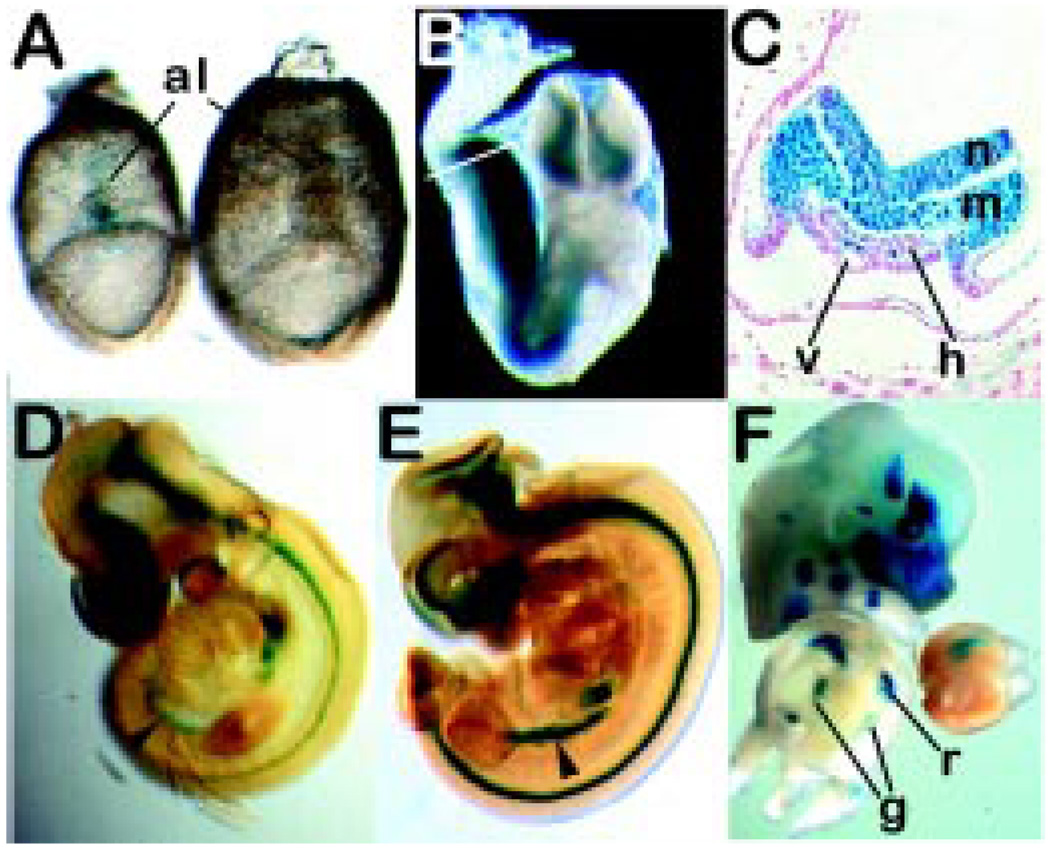
Fig. 1. Marking of the embryonic germ lineage with lacZ expression in TNAPβ-geo mouse embryos.
(A) Expression of TNAPβ-geo in PGCs at the base of the allantois (al) in e7.5 mouse embryos as indicated by X-gal staining. The left embryo is heterozygous for TNAPβ-geo, right embryo is wild-type (negative control). (B) Heterozygous TNAPβ-geo embryo at e8.5. At this stage, the PGCs are located within the hindgut endoderm [see transverse section indicated by white line, shown in (C)]. (C) Transverse section of embryo in Figure 1B. The PGCs (arrowheads) at this stage are masked by expression of TNAPβ-geo in overlying neurectoderm (n) and mesoderm (m), (h) hindgut endoderm, (v) vitelline artery. (D) PGCs at e9.5 are located in developing hindgut (arrowhead). (E) By e10.5, PGCs have arrived in genital ridge (arrowhead). (F) At e14, gonocytes continue to express TNAPβ-geo and are located in the differentiating gonad (g); (r), ribs. The embryo has been cut transversely, eviscerated and the caudal half stood upright to permit observation of the gonads.
Isolation and purification of PGCs from TNAPβgeo embryos
We have modified and optimized the use of FACS-gal (MacGregor et al., 1991; Nolan et al., 1988) to purify PGCs from TNAPβ-geo embryos (Abe et al., 1996). Gonads or gonadal ridges prior to sexual differentiation were isolated from TNAPβgeo embryos and the tissues dissociated by incubation in trypsin and trituration using a siliconized pipette until a single cell suspension was achieved (Abe et al., 1996; Buehr and McLaren, 1993; Cooke et al., 1993). After loading of a β-gal fluorescent substrate, fluorescein di-galactoside (FDG) into dissociated cells by osmotic shock and enzymatic conversion of FDG by incubating cells on ice for 1–2 h, the germ cells were purified by FACS-gal as described (Abe et al., 1996).
As seen in Figure 2A, a cell population displaying high fluorescence appeared only in samples prepared from embryos derived from a cross involving a TNAPβ-geo parent. Cell viability is routinely approximately 90% following the FACS-gal procedure and the purified PGCs can be maintained in culture (K. Abe; unpublished observations). To confirm that the fluorescein positive [F(+)] cells were PGCs, they were examined both for morphology and status for existing markers of mouse PGCs (Fig. 2B–E). Microscopically, the sorted F(+) cells were relatively large, round cells with a large nuclear-cytoplasmic ratio, each of which is a characteristic of PGCs (Eddy and Hahnel, 1983). When stained for AP activity, 96 ± 2% of the F(+) cells are routinely found to be AP positive (Abe et al., 1996) (Fig. 2B). In contrast, only a few percent of fluorescent negative cells are AP(+) (Fig. 2C). These cells are presumptive wild-type PGCs (i.e., not expressing a TNAPβ-geo allele) that would not fluoresce and therefore would not be detected by the FACS. On average we can routinely identify approximately 2000 AP positive cells in the gonads of a e12.5 embryo and approximately 3500 AP positive cells from an e13.5 stage embryo with approximately 2–5% of the total input cells being sorted on the basis of their high fluorescence intensity. This estimate for total numbers of germ cells at this stage is lower than those previously reported (Tam and Snow, 1981; Pesce and De Felici, 1995). This might be due to factors such as the procedure for counting cell numbers, the precise developmental stage of embryos used, the genetic background of the mice in each study or the stochastic nature of β-gal expression in mammalian cells (MacGregor et al., 1987; Ko et al., 1990b).
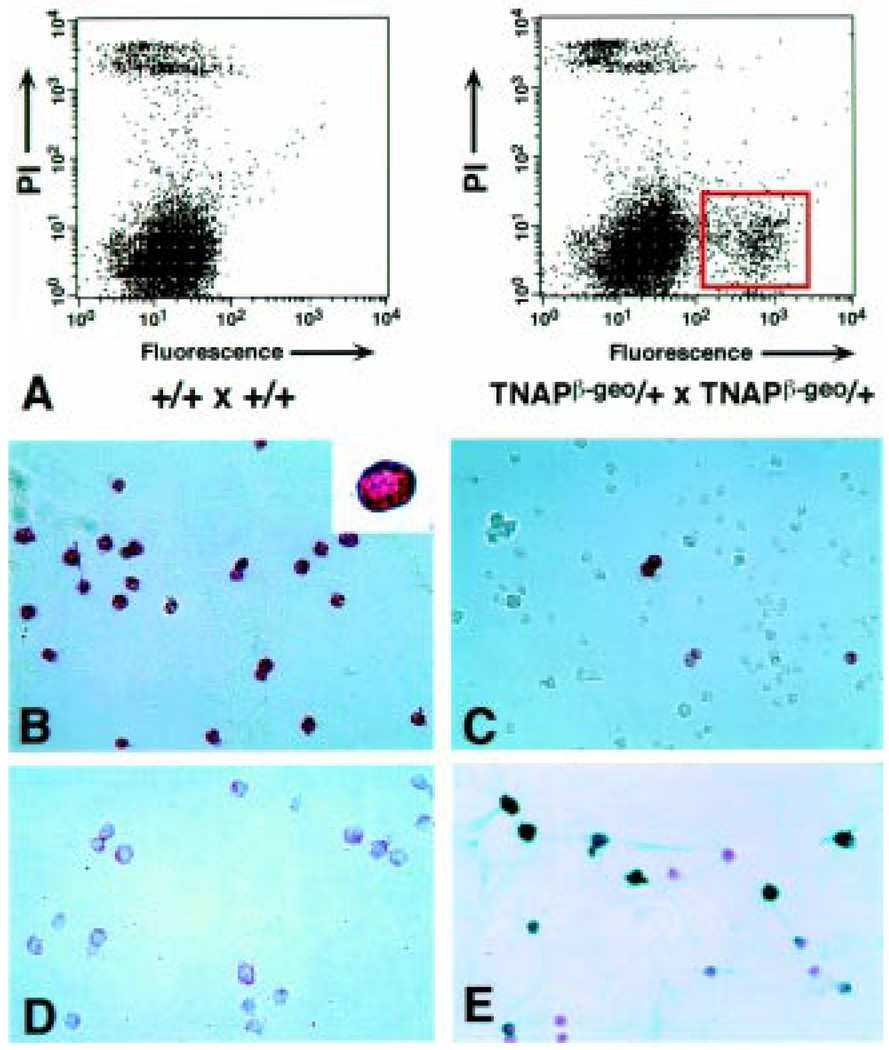
Fig. 2.
(A). Purification of PECs from TNAPβ-geo embryos. FACS profile of disaggregated gonads isolated from e12.5 wild-type embryos (left) or embryos obtained from a cross involving a TNAPβ-geo parent (right). The cells were stained with FDG to detect lacZ activity. X-axis indicates fluorescence, Y-axis indicates viability (propidium iodide exclusion). The F(+) population is boxed in red (right panel). (B) Alkaline phosphatase (AP) staining of the F(+) sorted cells from the e12.5 embryonic gonads. Inset – higher magnification of a single AP(+) cell. (C) AP staining of the F(−) sorted cells from e12.5 gonads. (D) The F(+) sorted cells of a e12.5 embryonic gonad were stained with the ACK2 antibody that defines c-kit expression using streptavidin-biotin peroxidase conjugate (Histostain SP kit, Zymed, CA). The red color indicates a positive reaction. Inset –higher magnification of c-kit positive cell. (E) Immunocytochemical staining of F(+) cells sorted from an e13.5 TNAPβ-geo gonad using the EMA-1 antibody. Cells were stained as in (D) except tetramethyl benzidine (blue color) was used to detect a positive immunoreactivity. Cells were counter-stained with nuclear fast red to reveal nuclei. Figure from Abe et al. (1996), used with permission.
An antibody against c-kit tyrosine kinase, that is expressed in PGCs reacted specifically with the e12.5 and e13.5 sorted F(+) cells at a similar rate as for AP (Fig. 2D). Interestingly, the EMA-1 epitope was expressed in only a subset of PGCs at e12.5 and even fewer at e13.5 (Fig. 2E). In agreement with previous studies (Hahnel and Eddy, 1986, 1987), this suggests that the epitope defined by the EMA-1 antibody is down-regulated shortly after PGCs have colonized the genital ridge. These data demonstrate that the FACS-gal method can isolate highly pure populations of PGCs from mouse embryos. More recently we have used FACS-gal to purify PGCs from e11.5 and e10.5 embryos and oogonia and prospermatogonia from e13.5 and e14.5 male and female gonads.
Prior to e10.5, TNAPβ-geo embryos are dissected and the hindgut or base of allantois separated from contaminating tissues by mechanical or enzymatic means. Following trypsinization of the isolated tissues, the cells are loaded with FDG. However, there are relative few PGCs present in embryos at these earlier developmental stages. Consequently, instead of purification using a FACS, these early stages of PGCs are collected manually using a pipette controlled with micro-manipulator, in conjunction with an inverted microscope equipped with a stage-cooler, a CCD-camera, Nomarski optics and epifluorescence (G.M., unpublished observations). As we routinely construct cDNA libraries from as few as 200 cells (M.S.H. Ko and K. Abe, unpublished observations) sufficient PGCs can easily be isolated from around twenty embryos using this technique.
Table one lists the embryonic stages from which germ cells will be purified. At each stage listed the germ cells are developmentally distinct from other stages. It follows that cDNA libraries generated at each stage will contain genes whose function is required during that particular step of the developmental process. For example, a proportion of cDNAs found in the male e13.5 library but not the female e13.5 library should encode products required for prospermatogonial differentiation and meiotic arrest while the complementary analysis will identify genes whose function is associated with oogonial differentiation and entry into meiosis. To facilitate identification of genes in the germ cell reference cDNA library that are not germ cell specific, cDNA libraries have also been constructed from e6.5 epiblast and from non-PGC cells isolated from e12.5 gonads.
Amplification and validation of cDNA generated from limiting amounts of germ cell RNA
FACS-gal can be used to generate pure populations of embryonic germ cells. However, it would require prohibitively large numbers of embryos to produce sufficient germ cell RNA to construct a cDNA library by conventional means. To circumvent this type of problem, the lone linker-PCR (LL-PCR) technique was developed that enables unbiased amplification of complex cDNA mixtures (Ko et al., 1990a; Abe, 1992) from small amounts of input material. This technique can be routinely used to generate sufficient cDNA for construction of plasmid based cDNA libraries from as little as 200 cells (M.S.H.Ko and K.Abe, unpublished observations). LL-PCR or similar methods have been applied to construct cDNA libraries from various RNA sources (Ko et al., 1990a; Froussard, 1993; Takahashi and Ko, 1994). Such cDNA libraries have been successfully used by several large scale cDNA sequencing projects including the HHMI-Washington University Mouse EST project and the ERATO-Wayne State University Mouse EST project.
RNA is isolated from the purified germ cells and cDNA synthesized and amplified using LL-PCR as described (Abe, 1992). The PGCs are lysed immediately after purification in Trizol (BRL) to isolate total RNA. Following DNase treatment to eliminate contaminating genomic DNA, double strand cDNA is synthesized by conventional methods from 10–100ng of total template RNA using an oligo dT primer containing a Not I site. An asymmetrical double stranded DNA adaptor having a non-palindromic protruding end and a blunt end is ligated to the ends of the double stranded cDNA such that only a single adaptor (i.e., a ‘Lone Linker’) is added to each end with high efficiency. Subsequently, the entire pool of cDNA is amplified using a single oligonucleotide primer in a PCR. With a refined ‘Long PCR protocol’, LL-PCR can amplify DNA fragments greater than 2 kb in length with unbiased efficiency, generating cDNA pools that are representative of the original population (Ko et al., 1990a) (K. Abe, unpublished).
Prior to construction of cDNA libraries, it is essential to validate the synthesized cDNA as being representative of genes expressed in germ cells during embryogenesis. Consequently, the individual cDNA reactions were analyzed using PCR to determine if each cDNA pool contained clones representing genes known to be expressed during mammalian germline development. Individual cDNA pools were synthesized using total RNA isolated using PGCs from e11.5, e12.5, e13.5 (both mixed and sex-specific) embryos as well as from both e6.5 epiblast and the somatic cells isolated as a by-product of PGC sorting from e12.5 gonads. The latter two samples were generated for use as negative controls for the validation process. In addition, they will also be used as a source of non-germ cell specific cDNA driver DNA in subtractive hybridization experiments. Each cDNA pool was examined for the presence of the following sequences –Mvh the mouse homolog of the Drosophila vasa gene that is expressed in murine germ cells (Fujiwara et al., 1994), TNAP (Hahnel et al., 1990; MacGregor et al., 1995), Oct-3/4 (Scholer et al., 1990), β-geo (in TNAPβ-geo mice) (MacGregor et al., 1995), SF-1/Ad4BP (Hatano et al., 1994), WT1 (Kreidberg et al., 1993) and Hprt. All of these genes are known to be expressed in germ cells during embryogenesis with the exception of Ad4BP and WT-1 which are expressed in the somatic component of the gonad.
Each of the genes known to be expressed in the germ cells was indeed found to be represented in the synthesized cDNA pools (data not shown). In addition, the somatically expressed genes were absent from the germ cell specific cDNA pools, being represented only in the e12.5 gonadal somatic cell samples. Thus, the synthesized cDNA appeared to being of sufficient quality to proceed with individual cDNA library construction. As the cDNA represents genes transcribed in germ cells during embryogenesis, it is in itself a valuable reagent that can be used to evaluate whether a candidate gene is expressed in the developing germline.
Construction of embryonic germ cell cDNA libraries
Having validated the individual cDNA syntheses, the amplified cDNA is digested with Not I then Sal I and is ligated to Not I -Sal I digested pBluescript prior to electro-transformation of E.coli DH10B. Results of preliminary sequencing analyses of individual cDNA clones from libraries constructed in this manner indicate that this force cloning method is efficient in generating high quality cDNA libraries from starting material of total RNA. Specifically, no inserts with homology to rRNA were found in this library in contrast to similar libraries that were constructed using cDNA digested with Sal I alone. The cDNAs for the reference library will be stored as ordered arrays, created by robotic picking of clones from the individual libraries into 384 well micro-titer trays.
Analysis of embryonic germ cell cDNA libraries
High through-put sequencing of cDNA clones
The initial analysis of the embryonic germ cell library will be conducted by large scale, single pass sequencing of cDNA clones. This is a proven strategy to rapidly obtain information about gene expression in a particular tissue or organ. In addition, relative abundance of expressed genes or ‘expression profiles’ can be monitored by compiling and analyzing the sequence information (Okubo et al., 1995; Kawamoto et al., 1996; Yokoyama et al., 1996). Furthermore, as the cDNA libraries in this case are made from purified germ cells, a priori, each gene is expressed in germ cells at some stage during germline development. Moreover, we can identify genes whose expression is germline specific by comparing this information with databases of somatic expressed genes. With the compiled data of PGC-expressed sequences, database searches can also be performed to identify homologs in other model organisms e.g., yeast and worm. This method is rapid and does not suffer from the technical difficulties that can be encountered using hybridization methods. Functional studies with homologs can then be performed in organisms with sophisticated genetics which can provide insight regarding the biochemical pathway in which the gene functions (Kuwabara, 1997). Finally, this method is less prone to bias and should enable identification of differentially expressed genes that might be overlooked by the hybridization approach.
Differential hybridization
The second method of analysis involves screening reference of cDNA library filters by differential hybridization, focusing on identification of genes that are differentially expressed during discrete stages of germline development (listed in Table 1). The rationale for doing so is that genes whose function is required during a particular period of germline development (e.g., during migration of PGCs from the hindgut to the genital ridges) should be expressed in germ cells during this stage. Two principal methods will be used to perform the analysis. Each method uses hybridization filters onto which cDNA clones have been spotted at high density in an addressed, arrayed manner. The spotted clones represent a mixture of all the individual cDNA libraries that were constructed from the discrete populations of purified germ cells. Thus the filters will contain clones that represent the majority of genes expressed in germ cells during several stages of germline development.
TABLE 1.
Discrete stages in germ line development during mouse embryogenesis
| age (sex) | location and developmental stage of PGCs or gonocytes |
|---|---|
| e7.25 | PGCs at the base of the allantois |
| e8.75 – e9.25 | PGCs mostly in hindgut endoderm, non-migrating, mitotic (12–18 somite stage) |
| e10.25 - 10.5 | PGCs mostly in hindgut mesentery, migrating, mitotic (30–35 somite stage) |
| e11.5 | All PGCs in genital ridge, non-migrating, mitotic |
| e12.5 | gonadal differentiation initiates, PGCs mitotic |
| e13.5 (M) | testis discernible, PGCs differentiate into M-prospermatogonia, mitotic |
| e13.5 (F) | ovary discernible, PGCs differentiate into oogonia, some enter meiotic prophase = oocyte |
| e15.5 (M) | T1-prospermatogonia, in mitotic arrest |
| e15.5 (F) | few oogonia; mostly oocytes in meiotic prophase |
Construction of full scale PGC reference cDNA library
The addressed embryonic germ cell reference cDNA library will be made by gridding cDNA clones on filters at high density in ordered arrays (Meier-Ewert et al., 1993). These filters will be screened with the complex cDNA probes. As all the clones on these filters have defined addresses in permanent storage, analysis of the hybridization signals using image analysis software enables the simultaneous identification of clones in addition to quantitation of their relative expression levels (Meier-Ewert et al., 1993; Nguyen et al., 1995; Zhao et al., 1995).
Based on an estimate of 100,000 expressed sequences in the mammalian genome, we estimate that a reference cDNA library composed of 2×105 independent clones, spotted on a total of seven filters will have approximately 95% probability of containing rare transcripts (Sambrook et al., 1989; Ausbel et al., 1994). The clones will be propagated, DNA purified and each plasmid cDNA clone will be spotted in duplicate (to generate consistency for treatment of individual filters) in 5×5 arrays onto hybridization membranes at a density of approx. 28,000 individual clones per 20 cm × 20 cm filter. In total, approximately 200,000 clones (seven filters) will be picked to constitute the embryonic germ cell reference cDNA library. This will be performed by a commercial company so that additional sets of filters of this library can be constructed and distributed to the international research community at minimal cost.
The number of independent cDNA clones required to give complete representation of genes expressed in the developing germ line could theoretically be reduced by construction of an ‘equalized’ cDNA library, i.e. a library with minimized redundancy (Ko, 1990,1995; Takahashi and Ko, 1994). However, in our experience, this approach is technically more complex and requires significantly greater amounts of input germ cell RNA. One also loses information about the expression level of individual genes that is provided by the frequency a cDNA is observed in a non-equalized library. For these reasons we will construct the reference library from non-equalized libraries.
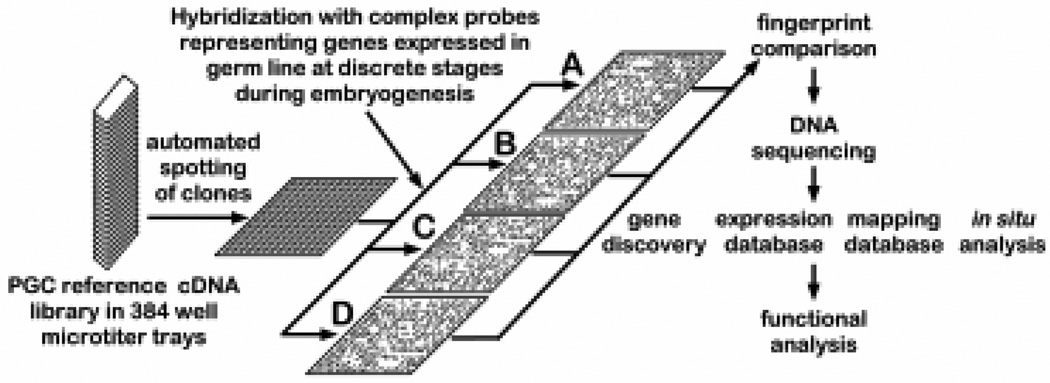
The hybridization signature approach, using high density cDNA filters and labeled complex probes can provide extremely high throughput measurement of relative gene transcriptional activity (Fig. 3). Such application of established genome analysis technology has gained rapid acceptance as a method to analyze differential gene expression during development on a large scale basis (Harrison et al., 1995; Nguyen et al., 1995; Takahashi et al., 1995; Lashkari et al., 1997). Image analysis software called AIS (Analytical Imaging System) with the capacity to locate and quantitate hybridization signals from arrayed, high density filters has been developed in collaboration with Fuji Film Co., Japan (Fig. 4). Each set of filters will be analyzed following hybridization with a particular complex probe mixture. To facilitate accumulation and retrieval of information, the data will be stored in the Wayne State University –Mouse EST database on the ORACLE relational database system that operates on a UNIX platform (M.S.H.Ko, unpublished). In addition to the AIS software, comparison of expression levels of all genes between different cDNA library will be performed using an independent analysis package –the EXPANAL, X-Window program, that is dynamically linked to the ORACLE database (M.S.H.Ko, unpublished).
Fig. 3. Hybridization signature approach to analyze gene expression.
See main text for description of details.
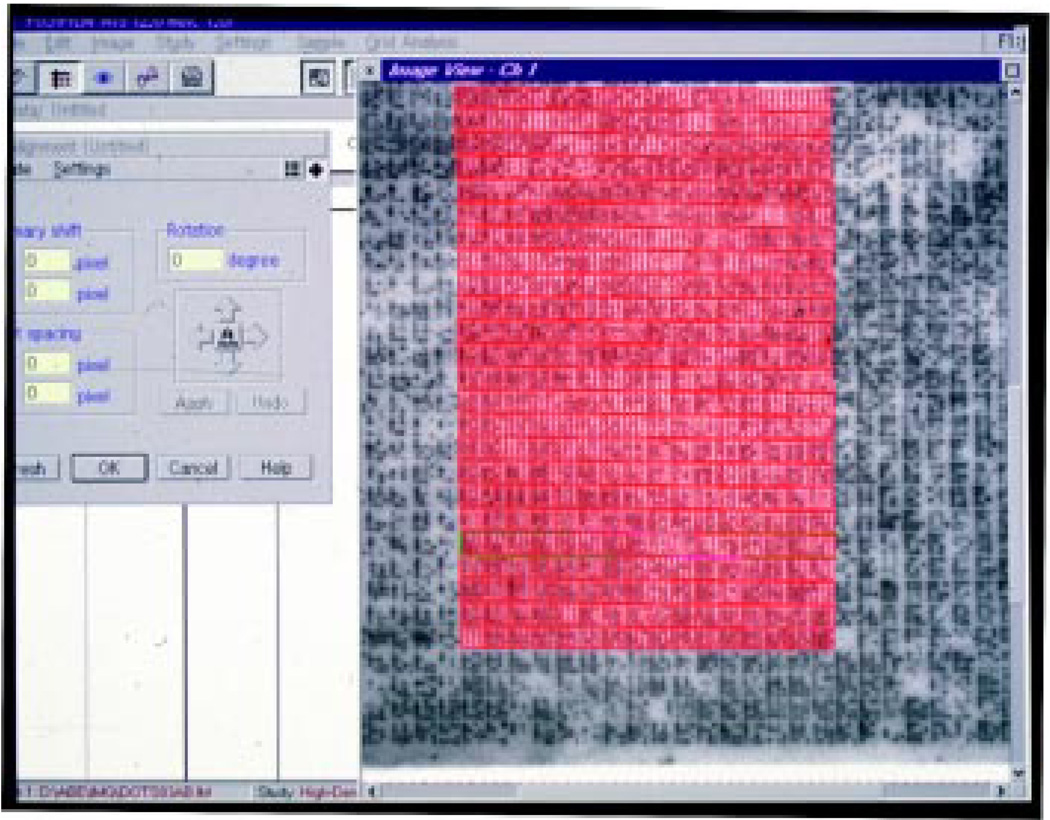
Fig. 4. Use of AIS software to quantitate relative levels of gene expression.
A 22×22cm hybridization filter containing approximately 36864 (4×4×384) individual mouse genomic DNA BAC clones spotted in 4×4 arrays was hybridized with a complex probe, washed and exposed to a phospho-storage screen. The plate was read by a phospho-imager (Fuji BAS 2000) and the image directly imported by the AIS software program. The screen-shot illustrates a grid used by the software to define individual areas that are subjected to relative quantitation. The grid represents one sixth of the entire filter. The software locates spots, quantitates the signals and converts them into numerical data that can be imported to any spread-sheet application. Signals are normalized to a standard reference set of target sequences on the filter. The AIS software can also compare one hybridization ‘signature’ with another and present the results in either a numerical or graphic format. More recently, EXPANEL software has been developed to perform relative quantitation of hybridization signatures (M.S.H.K., unpublished). The EXPANEL application provides a 2-dimensional color-graded graphical representation of differential expression of addressed cDNAs.
In the first approach, independent sets of such filters will be screened with complex probes that have been enriched for genes expressed in germ cells at specific embryonic stages. These probes will be prepared by subtraction using different cDNA libraries. Subtracted cDNA probes will be generated by enzymatic degradation subtraction (EDS) (Zeng et al., 1994).This is a highly efficient method of subtractive hybridization that involves phenolemulsion reassociation of lone linker amplified cDNA followed by exonuclease digestion of tester/driver or driver/driver hybrids. The technique can be used to identify DNA sequences that are differentially represented between two populations of highly complex molecules with great sensitivity (as low as 2.5 fold difference) and efficiency (Zeng et al., 1994). In addition, with this method the subtracted probes can be made without converting cDNA into conventional libraries. The use of subtractive hybridization to generate probes enriched for genes expressed at specific stages during germline development has two primary advantages over conventional differential screening. First, in addition to depleting common clones, the enrichment increases the sensitivity of detection, permitting identification of cDNAs representing rarer mRNAs (Harrison et al., 1995). Secondly, the use of subtraction enriched probes in combination with high-density addressed cDNA filters can reduce the required data analysis to less than 1% of that generated using non-subtracted probes (Harrison et al., 1995).
While subtractive hybridization has been successfully used by several groups to identify genes that are differentially expressed during a developmental process, this technique may not identify all transcripts whose expression levels differ subtly during development. In addition, as genes that play an important role in germline development (e.g., c-kit, Mgf) can be expressed in additional embryonic tissues or in germ cells for an extended period during germline development, genes in this category could be lost in a specific subtraction style screen.
To address this issue, the reference library will also be screened using a second, complementary experimental approach, i.e. conventional differential hybridization (Nguyen et al., 1995). This will involve hybridization of complex probes synthesized from each independent cDNA library to independent sets of reference library filters. In addition, the e6.5 epiblast and e12.5 gonadal somatic cell libraries will also be used as complex probes to screen the library. The results of the hybridization will be analyzed using the AIS and EXPANAL software packages each of which provides both quantitative and qualitative information about the expression of each clone in the reference cDNA library at specific stages during germline development. Clones displaying differential expression during germline development will be sequenced and the information analyzed to assist in determining which clones will be selected for further analysis.
Pilot experiments
High through-put automated DNA sequencing and PCR-based cDNA mapping mapping technologies are powerful genome analysis tools that can be applied to provide direct information regarding gene expression during a developmental process (Takahashi and Ko, 1993; Ko et al., 1994 and submitted). In our experience, between 400–600bp of sequence information from each end of a clone is routinely obtained by single pass sequencing. The 3’-end cDNA sequence information can be used to identify the signature of genes by performing a homology search of the current data bases. PCR primers can be made from specific genes and these and the individual cDNA libraries screened to determine if each clones expression is sex-specific in nature. The 3’-end cDNA sequence information is used to design a PCR primer pair to facilitate gene mapping (Takahashi and Ko, 1993; Ko et al., 1994) using an interspecific species backcross panel (Rowe et al, 1994). An example of the data generated is shown in Figure 5.
Fig. 5. PCR based analysis of gene expression in PGCs and somatic tissues from mouse embryos.
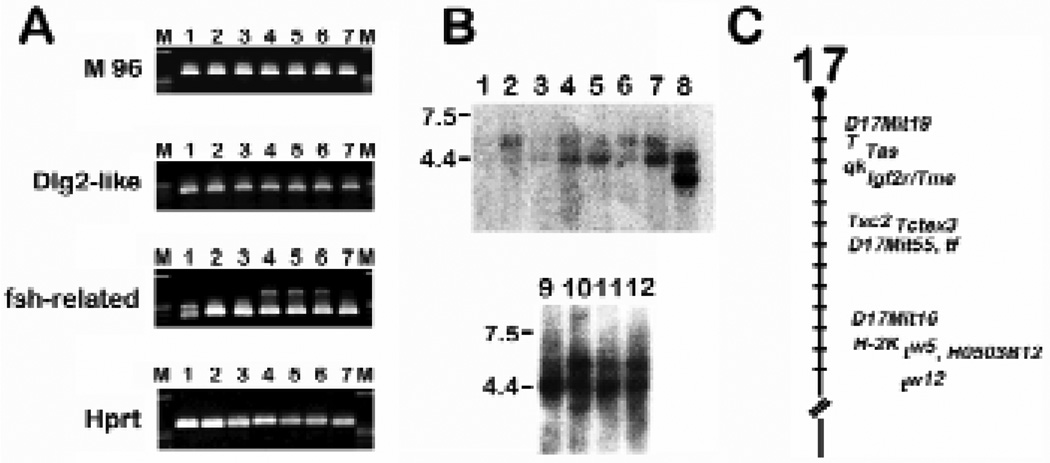
(A) Expression of M 96, Dlg2-like, fsh-related and Hprt (control) was determined by PCR analysis of the following cDNA samples-(M)-marker; (1) e6.5 epiblast; (2) e11.5 PGC; (3) e12.5 PGC; (4) e13.5 PGC; (5) e13.5 male PGC; (6) e13.5 female PGC; (7) e12.5 F(−) somatic cells. (B) Expression of fsh-related gene (H0503B12) during embryonic development and in adult mouse tissues. Northern blots containing adult (upper panel) or embryonic mouse total RNA (lower panel) were hybridized with a fsh-related cDNA. (1) heart; (2) brain; (3) spleen; (4) lung; (5) liver; (6) muscle; (7) kidney; (8) testis; (9) e7.5; (10) e11.5; (11) e15.5; (12) e17.5. Molecular weight sizes are in kb. (data courtesy of Rhonda H. Nicholson). (C) H0503B12 (fsh-related) maps to mouse chromosome 17 within the t-complex. fsh-related was mapped using an inter-specific species backcross panel (Rowe et al., 1994). (Data courtesy of Xueqian Wang).
In a preliminary test, we sequenced approximately 200 clones from e12.5 and e13.5 PGC libraries. Twenty-two independent sequences from an e13.5 mixed-sex PGC cDNA library were screened against the current genetic database by a BLAST search. Nine of the sequences appeared to be novel (Table 2) while six displayed high similarity to existing genes. The remaining seven sequences were either identical or displayed significant homology to known genes in the mouse, rat or human. PCR was used to confirm whether each of these genes was indeed expressed in PGCs and whether their expression was restricted to PGCs. Figure 5A indicates that the three genes analyzed, M96, Dlg 2 like and fsh-related are indeed represented in the cDNA synthesized from the purified germ cells although each is also expressed in somatic cells. Interestingly, the fsh-related gene product may be alternatively spliced in e6.5 epiblast and in e13.5 germ cells.
TABLE 2.
Example of blastn search results of cDNA clones from mixture of male and female e13.5 PGCs
| Clone # | Reads | Ns | Putative ID (Blastn score > 800) or Similar to (Blastn score >300) | Accession | Score | P value |
|---|---|---|---|---|---|---|
| H0501A01-3 | 648 | 7 | UNKNOWN | D10471 | 132 | 8.30E-00 |
| H0501A04-3 | 567 | 0 | UNKNOWN | X07077 | 135 | 5.60E-00 |
| H0501A12-3 | 564 | 6 | UNKNOWN | D10444 | 130 | 8.70E-00 |
| H0501B01-3 | 613 | 10 | Mouse mRNA for ubiquitin, score = 1490 | X51703 | 1490 | 0.00E+01 |
| H0501B02-3 | 490 | 6 | Rat mRNA for ribosomal protein L27, score = 1313 | X07424 | 1313 | 0.00E+01 |
| H0503A03-3 | 641 | 3 | Human peptidyl-prolyl isomerase and essential mitotic, score = 1448 | U49070 | 1448 | 0.00E+01 |
| H0503A07-3 | 723 | 87 | UNKNOWN | X63234 | 194 | 1.10E-04 |
| H0503A11-3 | 291 | 0 | UNKNOWN | M99063 | 122 | 9.90E-00 |
| H0503A12-3 | 568 | 0 | UNKNOWN, similar to Human splicing factor SRp55-1 (SRp-55) mRNA, complete cds. | U30883 | 390 | 0.00E+01 |
| H0503B01-3 | 670 | 86 | UNKNOWN | 0 | 0.00E+01 | |
| H0503B03-3 | 282 | 22 | UNKNOWN, similar to M.musculus mRNA for 17 beta-hydroxysteroid dehydrogenase | X89998 | 331 | 0.00E+01 |
| H0503B04-3 | 614 | 26 | Mouse single stranded DNA binding protein p9 mRNA, complete, score = 1394 | J03750 | 1394 | 0.00E+01 |
| H0503B07-3 | 522 | 2 | UNKNOWN | U12235 | 144 | 1.30E-00 |
| H0503B12-3 | 595 | 46 | UNKNOWN, similar to H.sapiens mRNA for Drosophila female sterile homeotic (FSH) | X62083 | 487 | 0.00E+01 |
| H0504A07-3 | 162 | 0 | UNKNOWN, similar to Rat matrin 3 mRNA. | M63485 | 471 | 0.00E+01 |
| H0504A09-3 | 677 | 71 | UNKNOWN, similar to R.rattus mRNA for ribosomal protein L23a | X65228 | 519 | 0.00E+01 |
| H0504A10-3 | 758 | 38 | UNKNOWN, similar to H.sapiens mRNA for DLG2 | X82895 | 594 | 0.00E+01 |
| H0504A12-3 | 702 | 18 | M96 = metal response element DNA-binding protein | S78454 | 1938 | 0.00E+01 |
| H0505A03-3 | 604 | 19 | UNKNOWN | 0 | 0.00E+01 | |
| H0505A05-3 | 421 | 36 | UNKNOWN | 0 | 0.00E+01 | |
| H0505A06-3 | 530 | 18 | Rat mRNA for ribosomal protein S13, score = 2015 | X53378 | 2015 | 0.00E+01 |
| H0505A09-3 | 722 | 76 | Mouse 24.6 kda protein mRNA, complete cds., score = 1521 | M93980 | 1521 | 0.00E+01 |
All three genes are homologs of genes known to be required for Drosophila development. M96 is a homolog of Polycomb-like while Dlg2 is a homolog of Disc large. The fsh (female sterile homeotic) gene product is a positive regulator of the bithorax complex in Drosophila (Haynes et al., 1989) and a human homolog (Ring3) has also been identified (Beck et al., 1992). Ring3 is known to function as nuclear kinase (Denis and Green, 1996). To determine the potential association of the fsh-related mouse homolog of fsh/Ring3 with existing mutations known to affect germline development, the expression pattern of fsh-related in embryos and adult tissues was analyzed (Fig. 5B) and the gene mapped (Fig. 5C). In the tissues analyzed, the gene appears to be expressed at highest levels in the testis (Rhonda H. Nicholson and M.S.H. Ko, unpublished results). Fsh-related is also expressed at different stages of embryogenesis. The fsh-related gene maps to chromosome 17 close to the H-2 locus which is consistent with the conserved chromosomal location of its human homolog Ring3 within the human MHC class II locus (Beck et al., 1992). Interestingly, this gene maps to the t-complex tw5 developmental mutation (Xueqian Wang and M.S.H. Ko) that is associated with a cell proliferation defect specific to e6.5 embryonic ectoderm (Bennett and Dunn, 1958; Bennett, 1975) (K. Abe, unpublished observations).
This brief example serves to illustrate how this approach will provide enormous amounts of novel information regarding genes expressed in the germline during embryonic development. In essence, this data will represent a mammalian germline EST database. Importantly however, in contrast to many current EST projects, the embryonic germ cell reference cDNA library is unique as the database represents genes that are expressed in a single cell type during its development. Once identified, specific germ cell expressed genes will be mapped to identify regions of germline-specific or stage-specific gene clustering on the mouse genome. Such clustering within the mouse genome, has been discovered for genes expressed within extra-embryonic tissues (Ko et al., submitted). Lastly, the combined sequencing and mapping information will be used to screen for candidate cDNAs for the genes affected by the an, at, Ter, and gcd mutations.
More recently, we have completed sequencing of 4000 clones from independent male and female e13.5 PGC libraries. A total of 1600 sequences from each library have been compared to the existing database by BLAST analysis. While many sequences were found with homology to known genes, approximately one-third of the sequences appeared to be novel (M.S.H. Ko and K. Abe.; unpublished observations). Significantly, a preliminary analysis of the known sequences indicated the presence of transcripts whose expression is sex-specific.
Criteria for selection of cDNA clones from the PGC reference library for in depth characterization
One of the major problems that arises from the generation of such large amounts of primary data is the choice of criteria that will be used to define a subset of cDNA clones for subsequent in depth molecular analyses. The criteria we have adopted are (a) the requirement for a sequence motif that implies functional importance (e.g. known DNA/RNA binding motif such as zinc finger, DEAD, Y-box, bHLH domains or motif related to cell signaling such as tyrosine kinase domains; (b) sequence homology to previously characterized genes whose function is relevant to germline development in other species; (c) a germ cell specific expression pattern as revealed by whole mount in situ analysis or high through-put in situ analysis of embryonic tissue sections mounted in 96 well plates (Komiya et al., 1997) or (d) a chromosomal location supportive of candidacy for an existing mutation known to affect germline development. In these cases, full length cDNAs will be derived, sequenced and the cognate genes will be analyzed in the appropriate mutant strains. These results will lay the foundation for future gene disruption experiments to ascertain the function of these genes during germline development.
Future perspectives
The major barrier to studying the molecular basis for mammalian germline development has been the inability to obtain sufficient quantities of pure embryonic germ cells with which to conduct molecular analyses. We have developed and validated an experimental strategy that has successfully overcome this problem. This technology will be used to generate an arrayed addressed reference cDNA library of genes expressed in the germline during mammalian development. By combining several novel techniques, we will be able to provide a unique and valuable resource with which to perform a systematic molecular genetic analysis of mammalian germline development. By making this library available to the international research community, any research group with basic molecular biology skills will be able to take advantage of this resource and in turn contribute to the study of the molecular basis for mammalian germline development. The permanent addressed nature of the library will minimize redundant analyses and facilitate long term accumulation of information regarding each clone in the library.
The reference library will also facilitate identification of genes that are genetically downstream of the loci affected in an, gcd, Ter, at and any additional mutants that may be identified in the future in which germline development is affected. This can be accomplished by crossing the TNAPβ-geo marker into each of these mutant backgrounds and using PGCs purified from the mutant animals as a source of complex cDNA probes to ‘fingerprint’ the reference library. Downstream loci affected by these mutations would then be identified by differences in the ‘fingerprints’ between normal and mutant complex probes.
Collectively, these three approaches will generate unprecedented amounts of novel information regarding the molecular basis for mammalian germline development. Most importantly, the approach is a reasonable one as its feasibility to isolate novel genes expressed during mouse gastrulation has demonstrated (Harrison et al., 1995). Moreover, each step of the entire procedure has now been validated by our group. This study will generate, at the very least an extensive battery of valuable molecular markers and significant amount of sequence information on embryonic germ cell specific expressed genes. This information is in itself an important resource for future functional studies to analyze the molecular characteristics of embryonic germ cells as well as understanding the biological process of germ cell development. Finally, this study epitomizes the growing utility of ‘genome analysis’ technology to study developmental processes at the molecular level (Harrison et al., 1995; Bernard et al., 1996; Granjeaud et al., 1996; Heller et al., 1997; Lashkari et al., 1997; Wodicka et al., 1997; Zhang et al., 1997).
Summary
Development of the germ lineage is essential for maintenance of a species. Despite this importance, relatively little is known about the molecular basis for mammalian germline development. The paucity of reports concerning gene expression in mammalian embryonic germ cells is in part the result of technical considerations as PGCs constitute only a minor proportion of the mouse embryo. Thus, in the absence of a suitable long term in vitro culture system, it has been difficult to isolate sufficient quantities of pure germ cells with which to conduct molecular genetic studies. We have overcome this problem by using a novel combination of established molecular and transgenic approaches. A line of mice has been generated in which the cells of the germ lineage express the β-galactosidase reporter gene during embryogenesis. Using this line, germ cells have been purified to near homogeneity from embryos at discrete stages during germline development by use of a viable stain for b-gal activity and a fluorescence activated cell sorter (FACS). Subsequently, cDNA libraries have been constructed from each germ cell population using a modified lone-linker PCR strategy. These combined cDNA libraries represent genes that are expressed in PGCs during mammalian germline development. To facilitate a molecular genetic approach to studying mammalian germline development, these cDNA libraries will be pooled to form an arrayed, addressed reference embryonic germ cell cDNA library. In parallel with large-scale cDNA sequencing efforts, genes that are differentially expressed in germ cells either prior to gonadal differentiation or in a sexually dimorphic manner following gonadal differentiation will be identified by screening the reference library with probes generated by subtractive hybridization. Complementary DNAs identified using this approach will be analyzed by sequencing, database comparison, genomic mapping and in situ hybridization to ascertain the potential functional importance of each gene to germline development. In addition to providing a wealth of novel information regarding patterns of gene expression during germline development, these results will form the basis for future experiments to determine the function of these genes in this process. The addressed reference embryonic germ cell cDNA library will constitute a valuable, permanent shared resource that will enable the entire international research community to study the molecular basis for germline development during mammalian embryogenesis.
Acknowledgments
We thank members of Ko laboratory for providing preliminary results, especially Rhonda H. Nicholson for the northern analyses in Figure 5, Marija J. Grahovac and Meng K. Lim for the large scale cDNA sequencing analyses, and Xueqian Wang for the mapping of Fsh-related gene. We also thank Colin Stewart, Peter Donovan, Jeff Pitman and Carol MacLeod for communicating results prior to publication. The work described is supported in part by grants to K.A. from the Science and Technology Agency of Japan, with Special Coordinating Funds for Promoting Science and Technology and by grants from the Ministry of Education, Science and Culture of Japan, and to M.S.H.K. (R01HD32243) and G.R.M. (R01HD36437), from the NIH, USA.
Abbreviations used in this paper
- AIS
analytical imaging software
- cDNA
complementary DNA
- PGC
primordial germ cell
- EEM
extra-embryonic mesoderm
- MGF
mast cell growth factor
- TNF-α
tumour necrosis factor - alpha
- OSM
oncostatin M
- LIF
leukemia inhibitory factor
- IL-11
interleukin-11
- RT-PCR
reverse transcriptase polymerase chain reaction
- TNAP
tissue non-specific alkaline phosphatase
- FACS
fluorescence activated cell sorter
- MACS
magnetic antibody cell sorting
- EST
expressed sequence tag
- e7.5
embryonic day 7.5
- FDG
fluorescein di-galactoside
- MHC
major histocompatibility complex
- LL-PCR
lone-linker - PCR
- CCD
charge coupled device
References
- Aaronson JS, Eckman B, Blevins RA, Borkowski JA, Myerson J, Imran S, Elliston KO. Toward the development of a gene index to the human genome: an assessment of the nature of high-throughput EST sequence data. Genome Res. 1996;6:829–845. doi: 10.1101/gr.6.9.829. [DOI] [PubMed] [Google Scholar]
- Abe K. Rapid isolation of desired sequences from lone linker PCR amplified cDNA mixtures: Application to identification and recovery of expressed sequences in cloned genomic DNA. Mammal. Genome. 1992;2:252–259. doi: 10.1007/BF00355435. [DOI] [PubMed] [Google Scholar]
- Abe K, Hashiyama M, MacGregor G, Yamamura K-I, Abe K. Purification of primordial germ cells from TNAPβ-geo mouse embryos using FACS-gal. Dev. Biol. 1996;180:468–472. doi: 10.1006/dbio.1996.0320. [DOI] [PubMed] [Google Scholar]
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Merchant-Larios H. Mouse primordial germ cells use fibronectin as a substrate for migration. Exp. Cell Res. 1986;165:362–368. doi: 10.1016/0014-4827(86)90590-2. [DOI] [PubMed] [Google Scholar]
- Anstrom KK, Tucker RP. Tenascin-C lines the migratory pathways of avian primordial germ cells and hematopoietic progenitor cells. Dev. Dynamics. 1996;206:437–446. doi: 10.1002/(SICI)1097-0177(199608)206:4<437::AID-AJA9>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Ausbel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Brooklyn, N Y: Wiley Interscience; 1994. [Google Scholar]
- Avarbock MR, Brinster CJ, Brinster RL. Reconstitution of spermatogenesis from frozen spermatogonial stem cells. Nature Med. 1996;2:693–696. doi: 10.1038/nm0696-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RK, Haendel MA, Swanson BJ, Shambaugh JC, Micales BK, Lyons GE. In vitro preselection of gene trapped ES cell clones for characterizing novel developmentally regulated genes in the mouse. Dev. Biol. 1997;185:201–214. doi: 10.1006/dbio.1997.8541. [DOI] [PubMed] [Google Scholar]
- Beck S, Hanson I, Kelley A, Pappin DJC, Trowsdale J. A homolog of the Drosophila female sterile homeotic (fsh) gene in the class II region of the human MHC. J. DNA Seq. Map. 1992;2:203–210. doi: 10.3109/10425179209020804. [DOI] [PubMed] [Google Scholar]
- Bennett D. The T-locus of the mouse. Cell. 1975;6:441–454. [Google Scholar]
- Bennett D, Dunn LC. Effects on embryonic development of a group of genetically similar lethal alleles derived from different populations of wild housed mice. J. Morphol. 1958;103:135–158. [Google Scholar]
- Bernard K, Auphan N, Granjeaud S, Victorero G, Schmitt-Verhulst AM, Jordan BR, Nguyen C. Multiplex messenger assay: simultaneous, quantitative measurement of expression of many genes in the context of T cell activation. Nucleic Acids Res. 1996;24:1435–1442. doi: 10.1093/nar/24.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besmer P, Manova K, Duttlinger R, Huang EJ, Packer A, Gyssler C, Bachvarova RF. The kit-ligand (steel factor) and its receptor c-kit/W: pleiotropic roles in gametogenesis and melanogenesis. Development. 1993 Suppl.:125–137. [PubMed] [Google Scholar]
- Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proc. Natl. Acad. Sci. USA. 1994;91:11303–11307. doi: 10.1073/pnas.91.24.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc. Natl. Acad. Sci. USA. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr M, McLaren A. Isolation and culture of primordial germ cells. In: Wassarman PM, DePamphilis ML, editors. Methods in Enzymology: Guide to techniques in mouse development. Vol. 225. San Diego: Academic Press; 1993. pp. 58–77. [DOI] [PubMed] [Google Scholar]
- Buehr M, McLaren A, Bartley A, Darling S. Proliferation and migration of primordial germ cells in We/We mouse embryos. Dev. Dynamics. 1993;198:182–189. doi: 10.1002/aja.1001980304. [DOI] [PubMed] [Google Scholar]
- Cheng L, Gearing DP, White LS, Compton DL, Schooley K, Donovan PJ. Role of leukemia inhibitory factor and its receptor in mouse primordial germ cell growth. Development. 1994;120:3145–3153. doi: 10.1242/dev.120.11.3145. [DOI] [PubMed] [Google Scholar]
- Chiquoine AD. The Identification, origin, and migration of the primordial germ cells in the mouse embryo. Anat. Rec. 1954;118:135–146. doi: 10.1002/ar.1091180202. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Avarbock MR, Maika SD, Hammer RE, Brinster RL. Rat spermatogenesis in mouse testis. Nature. 1996;381:418–421. doi: 10.1038/381418a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke JE, Godin I, Ffrench-Constant C, Heasman J, Wylie CC. Culture and manipulation of primordial germ cells. In: Wassarman PM, DePamphilis ML, editors. Methods in Enzymology: Guide to techniques in mouse development. Vol. 225. San Diego: Academic Press; 1993. pp. 37–57. [DOI] [PubMed] [Google Scholar]
- Copp AJ, Roberts HM, Polani PE. Chimerism of primordial germ cells in the early post-implantation mouse embryo following microsurgical grafting of posterior primitive streak cells in vitro. J. Embryol. Exp. Morphol. 1986;95:95–115. [PubMed] [Google Scholar]
- Coucouvanis EC, Jones PP. Changes in protooncogene expression correlated with general and sex-specific differentiation in murine primordial germ cells. Mech. Dev. 1993;42:49–58. doi: 10.1016/0925-4773(93)90097-h. [DOI] [PubMed] [Google Scholar]
- De Felici M, Dolci S. In vitro adhesion of mouse fetal germ cells to extracellular matrix components. Cell Differ. Dev. 1989;26:87–96. doi: 10.1016/0922-3371(89)90011-7. [DOI] [PubMed] [Google Scholar]
- De Felici M, Pesce M. Growth factors in in mouse primordial germ cell migration and proliferation. Prog. Growth Factor Res. 1994;5:135–143. doi: 10.1016/0955-2235(94)90001-9. [DOI] [PubMed] [Google Scholar]
- De Felici M, Dolci S, Pesce M. Cellular and molecular aspects of mouse primordial germ cell migration and proliferation in culture. Int. J. Dev. Biol. 1992;36:205–213. [PubMed] [Google Scholar]
- Denis GV, Green MR. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev. 1996;10:261–271. doi: 10.1101/gad.10.3.261. [DOI] [PubMed] [Google Scholar]
- Donovan PJ. Growth factor regulation of mouse primordial germ cell development. Curr. Top. Dev. Biol. 1994;29:189–225. doi: 10.1016/s0070-2153(08)60551-7. [DOI] [PubMed] [Google Scholar]
- Donovan PJ, Stott D, Cairns LA, Heasman J, Wylie CC. Migratory and postmigratory mouse primordial germ cells behave differently in culture. Cell. 1986;44:831–838. doi: 10.1016/0092-8674(86)90005-x. [DOI] [PubMed] [Google Scholar]
- Duncan MK, Lieman J, Chada KK. The germ cell deficient locus maps to mouse chromosome 11A2-3. Mammal. Genome. 1995;6:697–699. doi: 10.1007/BF00354290. [DOI] [PubMed] [Google Scholar]
- Dworkin MB, Dawid IB. Use of a cloned library for the study of abundant poly(A)+RNA during Xenopus laevis development. Dev. Biol. 1980;76:449–464. doi: 10.1016/0012-1606(80)90393-0. [DOI] [PubMed] [Google Scholar]
- Eddy EM, Hahnel AC. Establishment of the germ cell line in mammals. In: McLaren A, Wylie CC, editors. Current problems in germ cell differentiation. Cambridge: Cambridge University Press; 1983. pp. 41–69. [Google Scholar]
- Ellis RE, Kimble J. Control of germ cell differentiation in Caenorhabditis elegans. Ciba Found Symp. 1994;182:179–188. doi: 10.1002/9780470514573.ch10. discussion 189-92. [DOI] [PubMed] [Google Scholar]
- Enders GC, May JJ. Developmentally regulated expression of a mouse germ cell nuclear antigen examined from embryonic day 11 to adult in male and female. Dev. Biol. 1994;163:331–340. doi: 10.1006/dbio.1994.1152. [DOI] [PubMed] [Google Scholar]
- England MA, Swan AP, Dane P. The migration of amphibian primordial germ cells in the chick embryo. Scan. Electron Microsc. 1986;Pt3:1175–1182. [PubMed] [Google Scholar]
- Escalante-Alcalde D, Recillas-Targa F, Hernandez-Garcia D, Castro-Obregon S, Terao M, Garattini E, Covarrubias L. Retinoic acid and methylation cis-regulatory elements control the mouse tissue non-specific alkaline phosphatase gene expression. Mech. Dev. 1996;57:21–32. doi: 10.1016/0925-4773(96)00524-2. [DOI] [PubMed] [Google Scholar]
- Ffrench-Constant C, Hollingsworth A, Heasman J, Wylie CC. Response to fibronectin of mouse primordial germ cells before, during and after migration. Development. 1991;113:1365–1373. doi: 10.1242/dev.113.4.1365. [DOI] [PubMed] [Google Scholar]
- Fodor SP, Rava RP, Huang XC, Pease AC, Holmes CP, Adams CL. Multiplexed biochemical assays with biological chips. Nature. 1993;364:555–556. doi: 10.1038/364555a0. [DOI] [PubMed] [Google Scholar]
- Forrester LM, Nagy A, Sam M, Watt A, Stevenson L, Bernstein A, Joyner AL, Wurst W. An induction gene trap screen in embryonic stem cells: Identification of genes that respond to retinoic acid in vitro. Proc. Natl. Acad. Sci. USA. 1996;93:1677–1682. doi: 10.1073/pnas.93.4.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Froussard P. rPCR: a powerful tool for random amplification of whole RNA sequences. PCR Methods Appl. 1993;2:185–190. doi: 10.1101/gr.2.3.185. [DOI] [PubMed] [Google Scholar]
- Fujimoto T, Ukeshima A, Kiyofuji R. The origin, migration and morphology of the primordial germ cells in the chick embryo. Anat. Rec. 1976;185:139–145. doi: 10.1002/ar.1091850203. [DOI] [PubMed] [Google Scholar]
- Fujimoto T, Yoshinaga K, Kono I. Distribution of fibronectin on the migratory pathway of primordial germ cells in mice. Anat. Rec. 1985;211:271–278. doi: 10.1002/ar.1092110307. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Komiya T, Kawabata H, Sato M, Fujimoto H, Furusawa M, Noce T. Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc. Natl. Acad. Sci. USA. 1994;91:12258–12262. doi: 10.1073/pnas.91.25.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Castro MI, Anderson R, Heasman J, Wylie C. Interactions between germ cells and extracellular matrix glycoproteins during migration and gonad assembly in the mouse embryo. J. Cell Biol. 1997;138:471–480. doi: 10.1083/jcb.138.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RL, Beddington RSP. Multi-lineage ‘stem’ cells in the mammalian embryo. J. Cell Sci. 1988;10 Suppl.:11–27. doi: 10.1242/jcs.1988.supplement_10.2. [DOI] [PubMed] [Google Scholar]
- Gardner RL, Lyon MF, Evans EP, Burtenshaw MD. Clonal analysis of X-chromosome inactivation and the origin of the germ line in the mouse embryo. J. Embryol. Exp. Morphol. 1985;88:349–363. [PubMed] [Google Scholar]
- Ginsburg M, Snow MHL, McLaren A. Primordial germ cells in the mouse during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- Godin I, Wylie CC. TGF beta 1 inhibits proliferation and has a chemotropic effect on mouse primordial germ cells in culture. Development. 1991;113:1451–1457. doi: 10.1242/dev.113.4.1451. [DOI] [PubMed] [Google Scholar]
- Godin I, Wylie CC, Heasman J. Genital ridges exert long range effects on lmouse primordial germ cell numbers and direction of migration in culture. Development. 1990;108:357–363. doi: 10.1242/dev.108.2.357. [DOI] [PubMed] [Google Scholar]
- Goh KL, Yang JT, Hynes RO. Mesodermal defects and cranial neural crest apoptosis in a5 integrin-null embryos. Development. 1997;124:4309–4319. doi: 10.1242/dev.124.21.4309. [DOI] [PubMed] [Google Scholar]
- Gomperts M, Garcia-Castro M, Wylie C, Heasman J. Interactions between primordial germ cells play a role in their migration in mouse embryos. Development. 1994a;120:135–141. doi: 10.1242/dev.120.1.135. [DOI] [PubMed] [Google Scholar]
- Gomperts M, Wylie C, Heasman J. Primordial germ cell migration. Ciba Found. Symp. 1994b;182:121–134. doi: 10.1002/9780470514573.ch7. discussion 134-9. [DOI] [PubMed] [Google Scholar]
- Gossler A, Joyner AL, Rossant J, Skarnes WC. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science. 1989;244:463–465. doi: 10.1126/science.2497519. [DOI] [PubMed] [Google Scholar]
- Granjeaud S, Nguyen C, Rocha D, Luton R, Jordan BR. From hybridization image to numerical values: a practical, high throughput quantification system for high density filter hybridizations. Genet Anal. 1996;12:151–162. doi: 10.1016/1050-3862(95)00128-x. [DOI] [PubMed] [Google Scholar]
- Gress TM, Hoheisel JD, Lennon GG, Zehetner G, Lehrach H. Hybridization fingerprinting of high-density cDNA-library arrays with cDNA pools derived from whole tissues. Mammal. Genome. 1992;3:609–619. doi: 10.1007/BF00352477. [DOI] [PubMed] [Google Scholar]
- Hahnel AC, Eddy EM. Cell surface markers of mouse promordial germ cells defined by two monoclonal antibodies. Gamete Res. 1986;15:25–34. [Google Scholar]
- Hahnel AC, Eddy EM. The distribution of two cell surface determinants of mouse embryonal carcinoma and early embryonic cells. J Reprod. Immunol. 1987;10:89–110. doi: 10.1016/0165-0378(87)90069-6. [DOI] [PubMed] [Google Scholar]
- Hahnel AC, Rappolee DA, Millán JL, Manes T, Ziomek CA, Theodosiou NG, Werb Z, Pederson RA, Schultz GA. Two alkaline phosphatase genes are expressed during early development in the mouse embryo. Development. 1990;110:555–564. doi: 10.1242/dev.110.2.555. [DOI] [PubMed] [Google Scholar]
- Harrison SM, Dunwoodie SL, Arkell RM, Lehrach H, Beddington RSP. Isolation of novel tissue-specific genes from cDNA libraries representing the individual tissue constituents of the gastrulating mouse embryo. Development. 1995;121:2479–2489. doi: 10.1242/dev.121.8.2479. [DOI] [PubMed] [Google Scholar]
- Hatano O, Takayama K, Imai T, Waterman MR, Takakusu A, Omura T, Morohashi K. Sex-dependent expression of a transcription factor, Ad4BP, regulating steroidogenic P-450 genes in the gonads during prenatal and postnatal rat development. Development. 1994;120:2787–2797. doi: 10.1242/dev.120.10.2787. [DOI] [PubMed] [Google Scholar]
- Haynes SR, Mozer BA, Bhatia-Dey N, Dawid IB. The Drosophila fsh locus, a maternal effect homeotic gene encodes apparent membrane proteins. Dev. Biol. 1989;134:246–257. doi: 10.1016/0012-1606(89)90094-8. [DOI] [PubMed] [Google Scholar]
- Heasman J, Hynes RO, Swan AP, Thomas V, Wylie CC. Primordial germ cells of Xenopus embryos: the role of fibronectin in their adhesion during migration. Cell. 1981;27:437–447. doi: 10.1016/0092-8674(81)90385-8. [DOI] [PubMed] [Google Scholar]
- Heller RA, Schena M, Chai A, Shalon D, Bedilion T, Gilmore J, Woolley DE, Davis RW. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc. Natl. Acad. Sci. USA. 1997;94:2150–2155. doi: 10.1073/pnas.94.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LD, Lennon G, Becker M, Bonaldo MF, Chiapelli B, Chissoe S, Dietrich N, Dubuque T, Favello A, Gish W, Hawkins M, Hultman M, Kucaba T, Lacy M, Le M, Le N, Mardis E, Moore B, Morris M, Parsons J, Prange C, Rifkin L, Rohlfing T, Schellenberg, Bento Soares M, Tan F, Thierry-Meg J, Trevaskis E, Underwood K, Wohldman P, Waterston R, Wilson R, Marra M. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- Holzschu D, Lapierre L, Neubaum D, Mark WH. A molecular strategy designed for the rapid screening of gene traps based on sequence identity and gene expression pattern in adult mice. Transgenic Res. 1997;6:97–106. doi: 10.1023/a:1018465402294. [DOI] [PubMed] [Google Scholar]
- Hoog C. Isolation of a large number of novel mammalian genes by a differential cDNA library screening strategy. Nucleic Acids Res. 1991;19:6123–6127. doi: 10.1093/nar/19.22.6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglarz MK, Howard KR. The active migration of Drosophila primordial germ cells. Development. 1995;121:3495–3503. doi: 10.1242/dev.121.11.3495. [DOI] [PubMed] [Google Scholar]
- Kawamoto S, Matsumoto Y, Mizuno K, Okubo K, Matsubara K. Expression profiles of active genes in human and mouse livers. Gene. 1996;174:151–158. doi: 10.1016/0378-1119(96)00512-4. [DOI] [PubMed] [Google Scholar]
- Kawase E, Yamamoto H, Hashimoto K, Nakatsuji N. Tumor necrosis factor-alpha (TNF-alpha) stimulates proliferation of mouse primordial germ cells in culture. Dev. Biol. 1994;161:91–95. doi: 10.1006/dbio.1994.1011. [DOI] [PubMed] [Google Scholar]
- Kelley C, Blumberg H, Zon LI, Evans T. GATA-4 is a novel transcription factor expressed in endocardium of the developing heart. Development. 1993;118:817–827. doi: 10.1242/dev.118.3.817. [DOI] [PubMed] [Google Scholar]
- Khan AS, Wilcox AS, Polymeropoulos MH, Hopkins JA, Stevens TJ, Robinson M, Orpana AK, Sikela JM. Single pass sequencing and physical and genetic mapping of human brain cDNAs. Nature Genet. 1992;2:180–185. doi: 10.1038/ng1192-180. [DOI] [PubMed] [Google Scholar]
- Kishimoto T, Taga T, Akira S. Cytokine signal transduction. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- Ko MSH. An ‘equalized cDNA library’ by the reassociation of short double-stranded cDNAs. Nucleic Acids Res. 1990;18:5705–5711. doi: 10.1093/nar/18.19.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MSH. Equalized cDNA libraries. In: Larrick JW, Siebert PD, editors. Reverse Transcriptase PCR. London: Ellis Horwood; 1995. pp. 245–264. [Google Scholar]
- Ko MSH, Ko SBH, Takahashi N, Nishiguichi K, Abe K. Unbiased amplification of a highly complex mixture of DNA fragments by “lone linker”-tagged PCR. Nucleic Acids Res. 1990a;18:4293–4294. doi: 10.1093/nar/18.14.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MSH, Nakauchi H, Takahashi N. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J. 1990b;9:2835–2842. doi: 10.1002/j.1460-2075.1990.tb07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MSH, Wang X, Horton JH, Hagen MD, Takahashi N, Maezaki Y, Nadeau JH. Genetic mapping of 40 cDNA clones on the mouse genome by PCR. Mammal. Genome. 1994;5:349–355. doi: 10.1007/BF00356553. [DOI] [PubMed] [Google Scholar]
- Komiya T, Tanigawa Y, Hirohashi S. A large scale in situ hybridization with an equalized cDNA library. Anal. Biochem. 1997;254:23–30. doi: 10.1006/abio.1997.2399. [DOI] [PubMed] [Google Scholar]
- Koshimizu U, Taga T, Watanabe M, Saito M, Shirayoshi Y, Kishimoto T, Nakatsuji N. Functional requirement of gp130-mediated signaling for growth and survival of mouse primordial germ cells in vitro and derivation of embryonic germ (EG) cells. Development. 1996;122:1235–1242. doi: 10.1242/dev.122.4.1235. [DOI] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- Kuwabara PE. Worming your way through the genome. Trends Genet. 1997;13:455–460. doi: 10.1016/s0168-9525(97)01253-5. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Maeda-Suga H, Fujimoto T. Attraction of chick primordial germ cells by gonadal anlage in vitro. Anat. Rec. 1986;215:403–406. doi: 10.1002/ar.1092150411. [DOI] [PubMed] [Google Scholar]
- Lashkari DA, Derisi JL, McCusker JH, Namath AF, Gentile C, Hwang SY, Brown PO, Davis RW. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl. Acad. Sci. USA. 1997;94:13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson K, Hage W. Germline development. Vol. 182. Wiley, Chichester: Ciba Found. Symp.; 1994. Clonal analysis of the origin of primordial germ cells in the mouse; pp. 68–91. [DOI] [PubMed] [Google Scholar]
- Lennon GG, Lehrach H. Hybridization analyses of arrayed cDNA libraries. Trends Genet. 1991;7:314–317. doi: 10.1016/0168-9525(91)90420-u. [DOI] [PubMed] [Google Scholar]
- Lin TP, Labosky PA, Grabel LB, Kozak CA, Pitman JL, Kleeman J, MacLeod CL. The Pem homeobox gene is X-linked and exclusively expressed in extraembryonic tissues during early murine development. Dev. Biol. 1994;166:170–179. doi: 10.1006/dbio.1994.1305. [DOI] [PubMed] [Google Scholar]
- Lyon MF, Rastan S, Brown SDM. Genetic variants and strains of the laboratory mouse. Oxford: Oxford University Press; 1996. [Google Scholar]
- MacGregor GR, Mogg AE, Burke JF, Caskey CT. Histochemical staining of clonal mammalian cell lines expressing E.coli beta-galactosidase indicates heterogeneous expression of the bacterial gene. Somatic Cell Mol. Genet. 1987;13:253–265. doi: 10.1007/BF01535207. [DOI] [PubMed] [Google Scholar]
- MacGregor GR, Nolan GP, Fiering S, Roederer M, Herzenberg LA. Use of E.coli lacZ (beta-galactosidase) as a reporter gene. In: Murray EJ, editor. Gene Transfer and Expression Protocols. Vol. 7. Clifton: Humana Press; 1991. [Google Scholar]
- MacGregor GR, Zambrowicz BP, Soriano P. Tissue nonspecific alkaline phosphatase is expressed in both embryonic and extra-embryonic lineages during mouse embryogenesis but is not required for migration of primordial germ cells. Development. 1995;121:1487–1496. doi: 10.1242/dev.121.5.1487. [DOI] [PubMed] [Google Scholar]
- Marziali G, Lazzaro D, Sorrentino V. Binding of germ cells to mutant Sld Sertoli cells is defective and is rescued by expression of the transmembrane form of the c-kit ligand. Dev. Biol. 1993;157:182–190. doi: 10.1006/dbio.1993.1122. [DOI] [PubMed] [Google Scholar]
- Matsubara N, Takahashi Y, Nishina Y, Mukouyama Y, Yanagisawa M, Watanabe T, Nakano T, Nomura K, Arita H, Nishimune Y, Obinata M, Matsui Y. A receptor tyrosine kinase, Sky, and its ligand Gas 6 are expressed in gonads and support primordial germ cell growth or survival in culture. Dev. Biol. 1996;180:499–510. doi: 10.1006/dbio.1996.0323. [DOI] [PubMed] [Google Scholar]
- McCarrey JR. Development of the Germ Cell. In: Desjardins C, Ewing LL, editors. Cell and Molecular Biology of the Testis. Oxford: Oxford University Press; 1993. pp. 58–89. [Google Scholar]
- McCarrey JR, Hsu KC, Eddy EM, Klevecz RR, Bolen JL. Isolation of viable mouse primordial germ cells by antibody directed flow sorting. J. Exp. Zool. 1987;242:107–111. doi: 10.1002/jez.1402420116. [DOI] [PubMed] [Google Scholar]
- McKay RG, A.T H, Adams EC, Danziger S. Histochemical observations on the germ cells of human embryos. Anat. Rec. 1953;117:201–219. doi: 10.1002/ar.1091170206. [DOI] [PubMed] [Google Scholar]
- McLaren A. Studies on mouse germ cells inside and outside the gonad. J. Exp. Zool. 1983;228:161–171. doi: 10.1002/jez.1402280203. [DOI] [PubMed] [Google Scholar]
- McLaren A. The developmental history of female germ cells in mammals. Oxford Rev. Reprod. Biol. 1988;10:162–179. [PubMed] [Google Scholar]
- McLaren A. Germ cell sex determination. Semin. Dev. Biol. 1993;4:171–177. [Google Scholar]
- McLaren A. Development of primordial germ cells in the mouse. Andrologia. 1994;24:243–247. doi: 10.1111/j.1439-0272.1992.tb02647.x. [DOI] [PubMed] [Google Scholar]
- McLaren A, Southee D. Entry of mouse embryonic germ cells into meiosis. Dev. Biol. 1997;187:107–113. doi: 10.1006/dbio.1997.8584. [DOI] [PubMed] [Google Scholar]
- McLaren A, Wylie CC. Current problems in germ cell differentiation. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Meier-Ewert S, Maier E, Ahmadi A, Curtis J, Lehrach H. An automated approach to generating expressed sequence catalogues. Nature. 1993;361:375–376. doi: 10.1038/361375a0. [DOI] [PubMed] [Google Scholar]
- Narita N, Bielinska M, Wilson DB. Wild-type endoderm abrogates the ventral developmental defects associated with GATA-4 deficiency in the mouse. Dev. Biol. 1997;189:270–274. doi: 10.1006/dbio.1997.8684. [DOI] [PubMed] [Google Scholar]
- Nguyen C, Rocha D, Granjeaud S, Baldit M, Bernard K, Naquet P, Jordan BR. Differential gene expression in the murine thymus assayed by quantitative hybridization of arrayed cDNA clones. Genomics. 1995;29:207–216. doi: 10.1006/geno.1995.1233. [DOI] [PubMed] [Google Scholar]
- Nolan GP, Fiering S, Nicolas JF, Herzenberg LA. Fluorescence-activated cell analysis and sorting of viable mammalian cells based on beta-D-galactosidase activity after transduction of Escherichia coli lacZ. Proc Natl Acad Sci USA. 1988;85:2603–2607. doi: 10.1073/pnas.85.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo K, Hori N, Matoba R, Niiyama T, Fukushima A, Kojima Y, Matsubara K. Large scale cDNA sequencing for analysis of quantitative and qualitative aspects of gene expression. Nature Genet. 1992;2:173–179. doi: 10.1038/ng1192-173. [DOI] [PubMed] [Google Scholar]
- Okubo K, Itoh K, Fukushima A, Yoshii J, Matsubara K. Monitoring cell physiology by expression profiles and discovering cell type-specific genes by compiled expression profiles. Genomics. 1995;30:178–186. doi: 10.1006/geno.1995.9887. [DOI] [PubMed] [Google Scholar]
- Ozdzenski W. Observations on the origin of primordial germ cells in the mouse. Zool. Pol. 1967;17:367–379. [Google Scholar]
- Pellas TC, Ramachandran B, Duncan M, Pan SS, Marone M, Chada K. Germ-cell deficient (gcd), an insertional mutation manifested as infertility in transgenic mice. Proc. Natl. Acad. Sci. USA. 1991;88:8787–8791. doi: 10.1073/pnas.88.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce M, De Felici M. Purification of mouse primordial germ cells by miniMACS magnetic separation system. Dev. Biol. 1995;170:722–725. doi: 10.1006/dbio.1995.1250. [DOI] [PubMed] [Google Scholar]
- Rowe LB, Nadeau JH, Turner R, Frankel WN, Letts VA, Eppig JT, Ko MSH, Thurston SJ, Birkenmeier EH. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mammal. Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- Russell LD, Brinster RL. Ultrastructural observations of spermatogenesis following transplantation of rat testis cells into mouse seminiferous tubules. J. Androl. 1996;17:615–627. [PubMed] [Google Scholar]
- Russell LD, Franca LR, Brinster RL. Ultrastructural observations of spermatogenesis in mice resulting from transplantation of mouse spermatogonia. J. Androl. 1996;17:603–614. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. Cold Spring Harbor: CSHL Press; 1989. [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Scholer H, Ruppert S, Suzuki N, Chowdury K, Gruss P. New type of POU domain in germ-line specific protein. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- Schultheiss TM, Xydas S, Lassar AB. Induction of avian cardiac myogenesis by anterior endoderm. Development. 1995;121:4203–4214. doi: 10.1242/dev.121.12.4203. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Auerbach BA, Joyner AL. A gene trap approach in mouse embryonic stem cells: the lacZ reported is activated by splicing, reflects endogenous gene expression, and is mutagenic in mice. Genes Dev. 1992;6:903–918. doi: 10.1101/gad.6.6.903. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Moss JE, Hurtley SM, Beddington RS. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc. Natl. Acad. Sci. USA. 1995;92:6592–6596. doi: 10.1073/pnas.92.14.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow MHL. Autonomous development of parts isolaed from primitive-streak-stage mouse embryos. Is development clonal? J. Embryol. Exp. Morphol. 1981;65:269–287. [PubMed] [Google Scholar]
- Snow MHL, Monk M. Emergence and migration of mouse primordial germ cells. In: McLaren A, Wylie CC, editors. Current problems in germ cell differentiation. Cambridge: Cambridge University Press; 1983. pp. 115–135. [Google Scholar]
- Soudais C, Bielinska M, Heikinheimo M, Macarthur CA, Narita N, Saffitz JE, Simon MC, Leiden JM, Wilson DB. Targeted mutagenesis of the transcription factor GATA-4 gene in mouse embryonic stem cells disrupts visceral endoderm differentiation in vitro. Development. 1995;121:3877–3888. doi: 10.1242/dev.121.11.3877. [DOI] [PubMed] [Google Scholar]
- Stewart CL, Kaspar P, Brunet LJ, Bhatt H, Gadi I, Kontgen F, Abbondanzo SJ. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- Szabo PE, Mann JR. Biallelic expression of imprinted genes in the mouse germ line: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev. 1995;9:1857–1868. doi: 10.1101/gad.9.15.1857. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Ko MSH. The short 3'-end region of complementary DNAs as PCR-based polymorphic markers for an expression map of the mouse genome. Genomics. 1993;16:161–168. doi: 10.1006/geno.1993.1153. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Ko MSH. Toward a whole cDNA catalog: construction of an equalized cDNA library from mouse embryos. Genomics. 1994;23:202–210. doi: 10.1006/geno.1994.1478. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Hashida H, Zhao N, Misumi Y, Sakaki Y. High-density cDNA filter analysis of the expression profiles of the genes preferentially expressed in human brain. Gene. 1995;164:219–227. doi: 10.1016/0378-1119(95)00396-n. [DOI] [PubMed] [Google Scholar]
- Tam PP, Zhou SX. The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev. Biol. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- Tam PP, Zhou SX, Tan SS. X-chromosome activity of the mouse primordial germ cells revealed by the expression of an X-linked lacZ transgene. Development. 1994;120:2925–2932. doi: 10.1242/dev.120.10.2925. [DOI] [PubMed] [Google Scholar]
- Tam PPL, Snow MHL. Proliferation and migration of primordial germ cells during compensatory growth in mouse embryos. J. Embryol. Exp. Morphol. 1981;64:133–147. [PubMed] [Google Scholar]
- Timberlake WE. Developmental gene regulation in Aspergillus nidulans. Dev. Biol. 1980;78:497–510. doi: 10.1016/0012-1606(80)90349-8. [DOI] [PubMed] [Google Scholar]
- Urven LE, Abbott UK, Erickson CA. Distribution of extracellular matrix in the migratory pathway of avian primordial germ cells. Anat. Rec. 1989;224:14–21. doi: 10.1002/ar.1092240104. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Whitington PM, Dixon KE. Quantitative studies of germ plasm and germ cells during early embryogenesis of Xenopus laevis. J. Embryol. Exp. Morphol. 1975;33:57–74. [PubMed] [Google Scholar]
- Williamson A, Lehmann R. Germ cell development in Drosophila. Annu. Rev.Cell Dev. Biol. 1996;12:365–391. doi: 10.1146/annurev.cellbio.12.1.365. [DOI] [PubMed] [Google Scholar]
- Wodicka L, Dong H, Mittmann M, Ho M-S, Lockhart DJ. Genome-wide exprssion monitoring in Saccharomyces cerevisiae. Nature Biotech. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- Wurst W, Rossant J, Prideaux V, Kownacka M, Joyner A, Hill DP, Guillemot F, Gasca S, Cado D, Auerbach A. A large-scale gene-trap screen for insertional mutations in developmentally regulated genes in mice. Genetics. 1995;139:889–899. doi: 10.1093/genetics/139.2.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie CC, Heasman J. Migration, proliferation and potency of primordial germ cells. Semin. Dev. Biol. 1993;4:161–170. [Google Scholar]
- Yanagisawa M, Mukouyama Y, Watanabe T, Obinata M, Matsui Y. A novel serine/threonine kinase gene, Gek1, is expressed in meiotic testicular germ cells and primordial germ cells. Mol. Reprod. Dev. 1996;45:411–420. doi: 10.1002/(SICI)1098-2795(199612)45:4<411::AID-MRD2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Yang JT, Rayburn H, Hynes RO. Embryonic mesodermal defects in α5 integrin deficient mice. Development. 1993;119:1093–1105. doi: 10.1242/dev.119.4.1093. [DOI] [PubMed] [Google Scholar]
- Yokoyama M, Nishi Y, Yoshii J, Okubo K, Matsubara K. Identification and cloning of neuroblastoma-specific and nerve tissue-specific genes through compiled expression profiles. DNA Res. 1996;3:311–320. doi: 10.1093/dnares/3.5.311. [DOI] [PubMed] [Google Scholar]
- Zamboni L, Upadhyay S. Germ cell differentiation in mouse adrenal glands. J. Exp. Zool. 1983;228:178–193. doi: 10.1002/jez.1402280204. [DOI] [PubMed] [Google Scholar]
- Zeng J, Gorski RA, Hamer D. Differential cDNA cloning by enzymatic degrading subtraction (EDS) Nucleic Acids Res. 1994;22:4381–4385. doi: 10.1093/nar/22.21.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- Zhao N, Hashida H, Takahashi N, Misumi Y, Sakaki Y. Highdensity cDNA filter analysis: a novel approach for large-scale, quantitative analysis of gene expression. Gene. 1995;156:207–213. doi: 10.1016/0378-1119(95)00023-y. [DOI] [PubMed] [Google Scholar]