Abstract
Maintaining genome integrity during cell division requires regulated interactions between chromosomes and spindle microtubules. To ensure that daughter cells inherit the correct chromosomes, the sister kinetochores must attach to opposite spindle poles. Tension across the centromere stabilizes correct attachments, while phosphorylation of kinetochore substrates by the conserved Ipl1/Aurora B kinase selectively eliminates incorrect attachments. Here, we review our current understanding of how mechanical forces acting on the kinetochore are linked to biochemical changes to control chromosome segregation. We discuss models for tension sensing and regulation of kinetochore function downstream of Aurora B, and mechanisms that specify Aurora B localization to the inner centromere and determine its interactions with substrates at distinct locations.
Introduction
The accurate segregation of chromosomes during cell division is essential to maintain genomic stability. In eukaryotic cells, the microtubule-based mitotic spindle generates forces to align the sister chromatids at the metaphase plate, and then to pull the sister chromatids in opposite directions to segregate them to the two daughter cells. The kinetochore assembles at the centromere of each chromosome to mediate interactions with spindle microtubules. Kinetochores can initially bind to microtubules in any configuration, but accurate chromosome segregation requires that each pair of sister kinetochores ultimately attach to microtubules from opposite spindle poles (bi-orientation). Although there is a bias towards bi-orientation due to geometric constraints imposed by chromosome structure [1, 2], frequent errors in kinetochore-microtubule attachments do occur [3, 4] and would lead to unequal segregation if left uncorrected. Therefore, kinetochore-microtubule attachments must be carefully regulated: incorrect attachments are destabilized, while correct attachments are stabilized. In this way, all kinetochores eventually reach the correct attachment state in a trial-and-error process, with destabilization providing a fresh opportunity to bi-orient (reviewed in [5]). Defining the mechanism that selectively stabilizes only correct attachments is critical to understanding proper chromosome segregation. Here, we review recent work to understand the molecular mechanisms by which erroneous attachments are detected and corrected, focusing on the role of Aurora B kinase in this process. We discuss the processes that act upstream to control the activity of Aurora B and its phosphorylation of kinetochore substrates, and the downstream consequences of Aurora B phosphorylation for kinetochore activity and function.
Regulating attachments: reconciling mechanical and molecular mechanisms
Classic experiments by Bruce Nicklas using micromanipulation in insect spermatocyes provided direct experimental evidence that attachments are stabilized through tension across the centromere. In cells, this tension is established as spindle microtubules pull bi-oriented kinetochores in opposite directions. Experimentally induced tension, applied with a glass microneedle, stabilizes unipolar attachments that are otherwise unstable [6, 7]. These experiments laid the foundation for a model to explain the general principle of how bi-orientation can be achieved before any molecular details of this regulation had been defined.
One of the first pieces to the molecular puzzle of tension-dependent regulation was the identification of the Ipl1 kinase in budding yeast in a screen for mutants that display an increase-in-ploidy (ipl) phenotype [8]. Ipl1 was subsequently shown to be required for accurate chromosome segregation and to phosphorylate kinetochore substrates regulating microtubule binding [9–11]. Furthermore, Ipl1 promotes the turnover of attachments in the absence of tension [12], suggesting that it might function in the pathway described by Nicklas. Parallel work in Drosophila, C. elegans, and vertebrates identified Aurora kinases, the Ipl1 homologues, as key regulators of cell division (reviewed in [13]). The functional homolog of Ipl1 is Aurora B, which localizes to the inner centromere as the enzymatic component of the chromosome passenger complex (CPC), which also includes the inner centromere protein (INCENP), Survivin, and Borealin (also known as Dasra or CSC-1) (reviewed in [14]). The CPC remains at the inner centromere until anaphase onset and then redistributes to the midzone of the anaphase spindle and the equatorial cell cortex. Although we will focus here on the role of Aurora B and the CPC in kinetochore function, the CPC also regulates cytokinesis. In vertebrates, Aurora B inhibition using small molecules or inhibitory antibodies leads to stabilization of incorrect attachments, for example with both sister kinetochores attached to a single spindle pole [15–17]. Activation of Aurora B by removing an inhibitor leads to correction of these attachment errors by selectively destabilizing incorrect attachments [18]. Together, these studies demonstrate that Ipl1/Aurora B phosphorylates kinetochore substrates in the absence of tension to destabilize incorrect attachments and allow re-orientation.
Aurora B belongs to a family of serine/threonine protein kinases that includes Aurora A, but also has strong structural similarity to Protein Kinase A (PKA; cAMP-dependent protein kinase). Thus, while the cellular localizations and functions of Aurora A, Aurora B, and PKA are distinct, their substrate preference is extremely similar. The preferred phosphorylation consensus sequence for each of these kinases is [RK]×[TS][ILV] [10, 19]. This consensus site provides a good approximation for those sequences targeted by Aurora B, although some established substrates lack the downstream hydrophobic residue. In addition, extra upstream positively charged residues appear to increase the probability of phosphorylation. Aurora B substrates often contain multiple, closely clustered phosphorylation sites. These multiple sites may allow a switch-like behavior for the regulation of a given substrate, as described for the CDK target Sic1 [20], or may be required to generate charge effects to alter certain biochemical functions. Unlike kinases such as Polo-like kinase (Plk1), additional targeting regions in Aurora B substrates have not been defined outside of the consensus phosphorylation site. Thus, a primary determinant of whether a potential substrate will be targeted by Aurora B is its proximity to the kinase. Aurora B localizes to the inner centromere, close to the kinetochore and thus proximal to the site where regulation is required to ensure that improper attachments are destabilized.
How is tension sensed?
A key question for understanding the error correction process is how correct and incorrect attachments are distinguished. Based on the tension hypothesis established by the Nicklas micromanipulation experiments, phosphorylation of Aurora B substrates should respond to tension. A priori, there are multiple ways in which this regulation could be accomplished. Mechanical stretching can lead to various functional responses, for example by exposing binding sites or phosphorylation sites, accelerating the disassociation of non-covalent bonds, or gating ion channels through tension in the lipid bilayer [21, 22]. One model for tension sensing at centromeres is that Aurora B activity might be directly regulated by mechanically induced structural changes in a component of the CPC. INCENP is particularly important for kinase activation [23] and binds directly to microtubules in vitro [24, 25]. Furthermore, work in budding yeast demonstrated that a complex of Bir1 and Sli15 (Survivin/INCENP) can link centromeres to microtubules in vitro, which might reflect the ability to regulate Ipl1 (Aurora B) activity through a tension-dependent change in the complex [26].
Several other mechanisms that regulate Aurora B activity have been described, such as interactions with the protein TD-60, binding of the CPC to microtubules, activation by other kinases such as Chk1, Tousled-like kinase (TLK-1), and Mps1, or inactivation by phosphatases including PP1 and PP2A (reviewed in [27]). Mps1 is particularly intriguing because, like Aurora B, it is required for the correction of attachment errors in both budding yeast and mammalian cells [28, 29]. Mps1 phosphorylates Borealin, but the effect of this phosphorylation on Aurora B activity is controversial [29–35]. Overall, it is not clear whether Aurora B kinase activity is regulated on the short time scales associated with the error correction process. Instead, these upstream regulators appear to act primarily to generally “license” Aurora B activity during mitosis, rather than to modulate its function at a specific kinetochore. Any factor that is required for full Aurora B activity will also likely be required for proper regulation of kinetochore-microtubule attachments. Attachments can form when Aurora B activity is low, but they are unlikely to fully achieve correct bi-orientation.
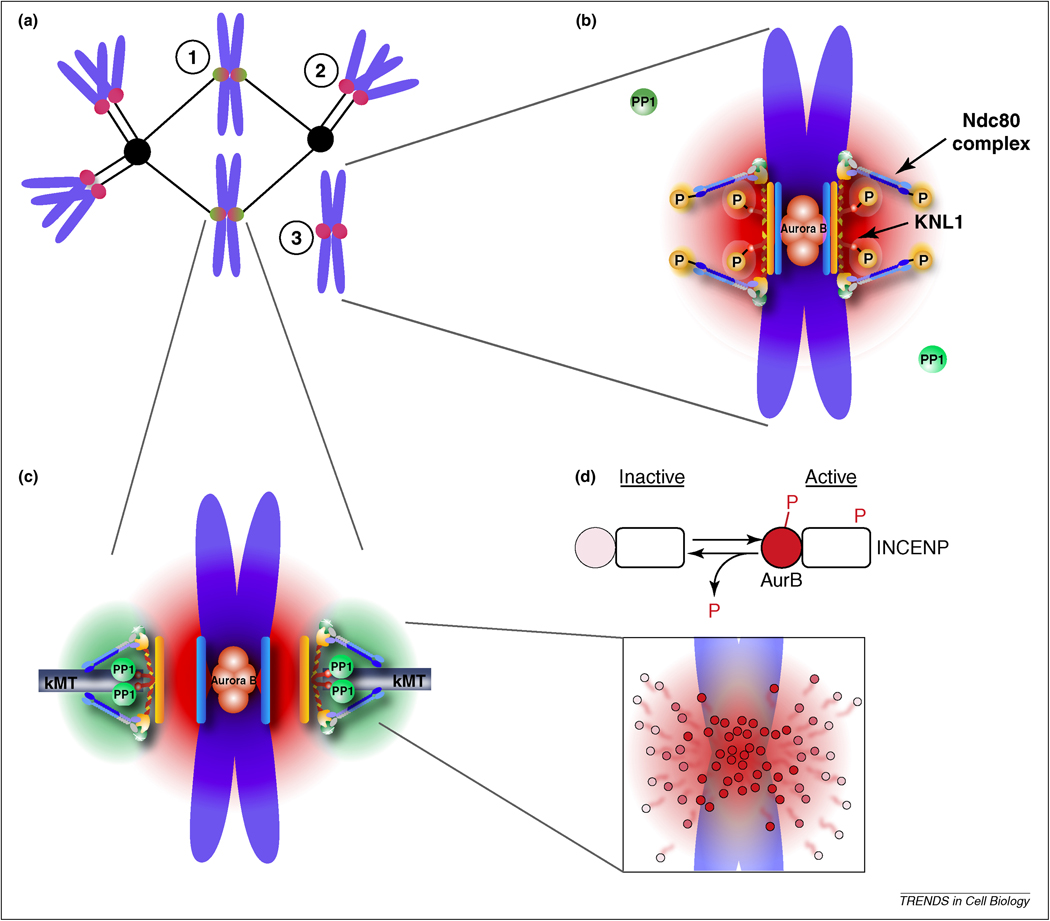
Rather than changes in intrinsic kinase activity, recent work has suggested that the ability to sense tension depends on the localization of Aurora B relative to its substrates at the outer kinetochore. Under this hypothesis, the force exerted on bi-oriented kinetochores separates the kinase at the inner centromere from its outer kinetochore substrates, making Aurora B less able to access these substrates [12, 36, 37]. Evidence for this “spatial separation” model was provided in two ways. First, the phosphorylation level of an Aurora B substrate is strongly related to its position within the kinetochore. When positioned close to the kinase, a FRET-based biosensor that reports on phosphorylation by Aurora B is constitutively phosphorylated, independent of tension, suggesting that kinase activity itself is not force-dependent. However, if the same sensor is positioned at the outer kinetochore, more distant from the kinase, it is dephosphorylated when tension is high [38]. Moreover, placing this sensor at increasing distances from the inner centromere leads to progressively greater dephosphorylation in response to tension [39]. Importantly, endogenous outer kinetochore substrates behave similarly to this FRET-based reporter and are also dephosphorylated when centromeres bi-orient [39, 40]. Second, it is possible to manipulate this spatial tension sensing mechanism by altering the position of the kinase. If Aurora B is artificially targeted to the outer kinetochore, the outer kinetochore is constitutively phosphorylated, independent of tension, and microtubule attachments cannot be stabilized [38]. Together, these findings support the model that the tension-sensing mechanism first demonstrated by Nicklas depends on the distance of Aurora B from its substrates at the outer kinetochore (Figure 1A–C).
Figure 1. Model for tension sensing by spatial separation of Aurora B from kinetochore substrates.
(a) Cartoon depiction of a spindle with correctly bi-oriented chromosomes (1) and incorrect kinetochore-microtubule attachments including a syntelically attached chromosome (2) or an unattached chromosome (3). (b,c) A phosphorylation gradient is generated by concentration of Aurora B at the inner centromere. Aurora B sites within the KMN network are phosphorylated due to their position within this gradient (red) at incorrect attachments, where tension is low, which destabilizes kinetochore microtubules (b). These substrates are dephosphorylated at correct attachments, where tension is high, because they are positioned farther from the kinase (c). Recruitment of PP1 to the outer kinetochore provides a counteracting gradient of dephosphorylation (green). (d) Model for how a phosphorylation gradient might be generated. Aurora B is activated (dark red circles) at the inner centromere by autophosphorylation, both of Aurora B and INCENP, followed by release and inactivation (lighter circles) by dephosphorylation as Aurora B diffuses away from the inner centromere.
The spatial separation model explains how differences in tension can be sensed to distinguish sister chromatids that are attached to opposite spindle poles (bi-oriented) from those that are attached to the same pole (syntelic attachment). This model also suggests a pathway for correction of merotelic errors, in which a single kinetochore is attached to both poles simultaneously. Such defects make a significant contribution to the total number of chromosome segregation errors [4]. Merotelic kinetochores are deformed and stretched compared to bi-oriented kinetochores [41], which may position the incorrect attachment sites closer to the inner centromere and locally increase phosphorylation of these sites to selectively destabilize the incorrect attachment [36]. There is also an intriguing observation that the CPC is enriched at merotelic attachments [42], which could also contribute to an error correction mechanism, but how this enrichment occurs is unclear. Consistent with the idea that Aurora B contributes to correcting merotelic errors, Aurora B inhibition leads to an increased frequency of these errors [36, 42].
Connecting Aurora B activity to changes in kinetochore function
As described above, the position of Aurora B relative to its substrates (Table 1) is a key factor in controlling the phosphorylation of its downstream targets. As defined substrates for Aurora B have distinct localizations within the kinetochore, this position has important implications for their relative phosphorylation. Those substrates that are located proximally to Aurora B are likely to be constitutively phosphorylated at the times when Aurora B localizes to centromeres (prophase until anaphase onset). In contrast, substrates positioned at the outer kinetochore will display tension sensitive phosphorylation with higher phosphorylation on misaligned kinetochores and lower phosphorylation on aligned kinetochores. Recent work has demonstrated that there is a spectrum of behaviors even within the kinetochore, with the exact spatial position of a protein providing a critical determinant for its phosphorylation by Aurora B. For example, substrates located at the periphery of the outer kinetochore, such as Ndc80, exhibit differential phosphorylation compared to substrates that are closer to the inner kinetochore, such as the Mis12 complex subunit Dsn1 [39].
Table 1.
Examples of Aurora B substrates
| Protein | Organism | Localization | Reference |
|---|---|---|---|
| Histone H3 | Conserved | Chromosomes | [92] |
| Aurora B (autophosphorylation) | Conserved | Inner centromere | [108] |
| INCENP (CPC subunit) | Conserved | Inner centromere | [87, 88] |
| Ndc80 | Conserved | Outer kinetochore | [45, 49] |
| KNL1 | Conserved | Outer kinetochore | [39] |
| Dsn1 | Conserved | Outer kinetochore | [39] |
| MCAK | Metazoa | Inner centromere | [37, 51, 52] |
| CENP-A | Human | Inner Kinetochore | [109] |
| CENP-E | Vertebrates | Outer kinetochore | [43] |
| Dam1 Complex | Fungi | Outer kinetochore | [10] |
| EB1 | Fungi | Midzone | [110] |
| MKLP1 | Metazoa | Midzone | [111] |
| MgcRacGAP | Metazoa | Midzone | [112] |
| Op18/Stathmin | Xenopus | Cytoplasm | [89, 91] |
The same principle of position-dependent phosphorylation applies to Aurora kinase substrates at other sites in the cell. Since the consensus phosphorylation site for Aurora B is identical to that of Aurora A, it is possible for a substrate to be targeted by both kinases. For example, the kinetochore motor kinesin CENP-E is phosphorylated by both Aurora A and Aurora B to regulate its affinity for microtubules [43]. In addition, the dynactin subunit p150glued, which localizes to both the spindle and kinetochores, is regulated by Aurora A at spindle poles where this kinase is concentrated [44]. Although not yet tested directly, it is possible that Aurora B would target and regulate p150glued at kinetochores.
Several of the substrates listed in Table 1 have multiple phosphorylation sites targeted by Aurora B, and phosphorylation of a single site often does not have a strong effect on protein activity or behavior [10]. However, due to the presence of multiple substrates at distinct spatial positions within a kinetochore, the combined action of Aurora B on kinetochore function can act in a graded manner [39]. In addition, the consequences of Aurora B phosphorylation can vary dramatically between different substrates. A primary function for Aurora B is to destabilize improper kinetochore-microtubule interactions, so Aurora B should have a net negative effect on the microtubule binding activity of the kinetochore, particularly the key proteins that contribute to the kinetochore-microtubule interface. Indeed, phosphorylation of the kinetochore proteins Ndc80/Hec1 and KNL1 strongly reduces their microtubule binding activity, likely by introducing negative charges that prevent interactions with the negatively charged microtubule polymers [39, 45–49]. Phosphorylation of the Mis12 complex subunit Dsn1 also has a negative effect on the microtubule binding activity of the kinetochore [39]. However, in this case regulation appears to occur through an allosteric change in Dsn1 that modulates the conformation by which KNL1 and Ndc80 interact with microtubules, as the Mis12 complex itself does not display a direct microtubule binding activity. Phosphorylation by Aurora B also has a negative effect on the microtubule depolymerase activity of the kinesin-13 proteins MCAK and Kif2a and also regulates MCAK localization, and both effects may be important for regulating kinetochore-microtubules [37, 50–53]. Finally, Aurora B also phosphorylates the fungal Dam1 complex, an important player in generating processive interactions with microtubules. Although Ipl1/Aurora B phosphorylation has been proposed to alter the microtubule binding activity of the Dam1 complex [54], the tested phosphorylation site in this case does not reside within the microtubule binding domain of Dam1. Instead, phosphorylation of Dam1 appears to alter its interaction with the Ndc80 complex [55], disrupting the connection between these two co-functional components of the outer kinetochore [56, 57].
In the cases described above, Aurora B phosphorylation disrupts the activity of specific kinetochore proteins by targeting critical regions of those proteins required for function. Aurora B phosphorylation therefore acts as a switch that can be reversed to restore protein function, as long as the protein itself remains at kinetochores. However, in addition to altering protein activity, Aurora B has been implicated in controlling the recruitment of proteins to kinetochores. As with the control of protein activity, this effect could either be constitutive if the protein is located close to Aurora B, or could occur in a tension-dependent manner. For constitutive control of protein localization to kinetochores, Aurora B phosphorylation may “license” the recruitment of that protein. This mechanism has been proposed for the recruitment of the KMN (KNL1-Mis12-Ndc80) network to kinetochores in Xenopus extracts [58], although a similarly strong effect is not observed in human cells [39]. In addition, Aurora B and the CPC are required to recruit Shugoshin family proteins to centromeres [59–64].
In contrast, Aurora B-dependent phosphorylation of outer kinetochore substrates could act as a switch to control kinetochore composition. Perhaps the best understood example of controlled kinetochore localization downstream of Aurora B is Protein Phosphatase 1 (PP1), which localizes to kinetochores and opposes Aurora B (reviewed in [65]). A major PP1 targeting factor at kinetochores is the outer kinetochore protein KNL1 [66]. PP1 binds to a conserved RVSF motif within KNL1, which is an example of the RVxF motifs commonly found in PP1 interacting proteins [67, 68]. Aurora B directly phosphorylates the RVSF motif of KNL1, which disrupts the interaction between KNL1 and PP1 [66]. Thus, phosphorylation of the outer kinetochore by Aurora B prevents the recruitment of PP1, the opposing phosphatase, to kinetochores. In addition to generating a switch-like behavior for PP1 recruitment to kinetochores, it also provides an elegant feedback mechanism between phosphorylation derived from the inner centromere-localized Aurora B and dephosphorylation generated by outer kinetochore PP1. A similar mechanism has been suggested for another outer kinetochore protein, CENP-E, in which phosphorylation of a conserved motif by Aurora kinases regulates PP1 binding [43]. Phosphorylation of this residue clearly regulates CENP-E function, but the significance of this binding site for PP1 recruitment to kinetochores is unclear, as CENP-E depletion or mutation of the site were not shown to affect kinetochore PP1 levels. Finally, recent work suggests that the PP1-associated protein Sds22 may also contribute to controlling the levels of Aurora B phosphorylation at kinetochores [69].
Thus, in addition to its role in error correction, Aurora B phosphorylation can specify kinetochore composition to provide different activities for prometaphase kinetochores, which are in the process of spindle attachment and congression, and metaphase kinetochores that are bi-oriented and must be stabilized and prepared for anaphase. There are two distinct groups of proteins that participate in this switch in kinetochore composition. The first group requires high levels of Aurora B phosphorylation to bind kinetochores and thus localizes preferentially to prometaphase kinetochores, including Kif2b, the RZZ complex, Spindly, dynein (and its associated proteins), and BubR1 [17, 70–72]. The localization of the second group of proteins, which includes PP1 and the Astrin/SKAP complex [66, 73, 74], is prevented by high levels of Aurora B phosphorylation and thus preferentially localizes to metaphase bi-oriented kinetochores. Thus a tension-dependent switch from a phosphorylated to a dephosphorylated state may serve as a general mechanism to regulate the composition of the outer kinetochore.
How does Aurora B touch its substrates?
The spatial separation model explains how chromosome bi-orientation can be sensed through changes in tension across the centromere, which is a critical component for any model of error correction. As discussed above, this model is based on the idea that local Aurora B activity at the outer kinetochore depends on the distance from the inner centromere, where the kinase is localized. As these localizations are clearly distinct from each other, possibly separated by as much as 100 nm, it is unclear how Aurora B contacts its substrates at a molecular level, even when tension is low. One proposed model is that only kinase molecules at the outer edges of the inner centromere are able to contact substrates at the outer kinetochore, while the majority of kinase molecules would have no role in phosphorylating these substrates [33, 75]. INCENP may act as a flexible extended linker, or “leash,” that determines the distance that Aurora B can reach from binding sites at the inner centromere. Perturbations that affect the length of the leash should disrupt the error correction process according to this model. However, deletion of the INCENP coiled-coil region, which would reduce the “reach” of this protein, does not inhibit the error correction process [76], arguing against this domain of INCENP acting as a leash to control kinetochore-microtubule attachments. In contrast, the coiled-coil domain is required for targeting the CPC to spindle microtubules [25, 77, 78]. To further assess the leash model, the exact positions of the CPC binding sites at the inner centromere should be determined, as how these positions change relative to the outer kinetochore in response to centromere tension is crucial. The positions of outer kinetochore components relative to each other along the kinetochore-kinetochore axis have been measured with nanometer accuracy by calculating the centroids of spots of labeled kinetochore proteins [79, 80]. Unfortunately, this method may not be applicable to the CPC if the outer edge of the distribution is important rather that the centroid. Thus, we currently do not have a high resolution picture of precisely where the CPC localizes relative to kinetochore substrates. Furthermore, the distance from Aurora B to its substrates will vary significantly even over short time scales because inter-kinetochore distance and intra-kinetochore stretch both change rapidly during normal chromosome oscillations [81–83]. According to the spatial separation model, the level of substrate phosphorylation may reflect a time average over these oscillations, which would depend on the rates of phosphorylation and dephosphorylation relative to the spatial changes. However, phosphorylation changes have not yet been measured on these time scales, and the significance of the oscillations remains unclear. Whether Aurora B can directly contact outer kinetochore substrates while tethered to the inner centromere is an open question.
An alternative model for how Aurora B reaches its outer kinetochore substrates is that active kinase molecules diffuse from the inner centromere. This model draws on several established aspects of Aurora B function. Although Aurora B localizes to the inner centromere, both Aurora B and INCENP turn over with a t1/2 of ~50 sec [84, 85]. It is unknown whether the CPC turns over as a complex, or whether individual components or sub-complexes function independently. Aurora B activation depends on phosphorylation of the INCENP C-terminus, which is predicted to occur in trans [86–88]. This activation occurs by locally concentrating Aurora B and is opposed by cytoplasmic phosphatases that suppress kinase activity [89]. Based on these findings, Aurora B should be activated by its concentration at centromeres, with active Aurora B being released to diffuse away from the centromere, and subsequently inactivated by phosphatases. The combination of these factors would generate a diffusion-based gradient of kinase activity (Figure 1D). This diffusion model is conceptually similar to one proposed to explain a gradient of phosphorylation by Aurora B observed in anaphase [90], in which the CPC is concentrated on the spindle midzone instead of at centromeres. Aurora B substrates on chromatin (histone H3), in the cytoplasm (Op18/Stathmin), and on spindle microtubules are phosphorylated in mitosis [78, 89, 91, 92], suggesting that there is indeed soluble active kinase. However, phosphorylation gradients have not been observed prior to anaphase, so whether a diffusible gradient of active kinase is produced at centromeres remains an open question. A key constraint of the diffusion model is that differences in substrate positions at the kinetochore are <100 nm [39, 80], and whether a gradient of soluble Aurora B could act on these length scales is unclear. It may be possible to fine tune the production of active Aurora B at centromeres because both the concentration of CPC components at centromeres and their rates of turnover are regulated by post-translational modifications, including phosphorylation, ubiquitination, and sumoylation [85, 93–98]. By modulating both kinase concentration and turnover, the properties of a diffusion-based gradient of active kinase could be set to achieve a tightly spatially defined gradient. Local phosphatase activity may also contribute to defining a phosphorylation gradient, both by inactiving Aurora B and by directly dephosphorylating substrates.
An important question for any model of Aurora B function at kinetochores is how the kinase localization is specified. In a human cell line carrying a neocentromere on chromosome 4, in which the normal centromere has been silenced, Aurora B and INCENP localize to the neocentromere, which lacks repetitive α-satellite DNA, but not to the silenced centromere [99]. CPC targeting is therefore determined by an epigenetic property of the centromere rather than a specific DNA sequence. Recent findings suggest an appealing model for CPC targeting based on two distinct histone phosphorylations. The mitotic kinase Bub1 localizes to kinetochores and phosphorylates histone H2A on centromeric heterochromatin, and this phospho-mark creates a binding site for Shugoshin family proteins [100]. Shugoshin acts as a centromeric adaptor that binds to the CPC after CPC components are phosphorylated by Cdk1 [101]. In addition, the mitotic kinase Haspin phosphorylates histone H3, which creates a binding site for Survivin [102–104]. Point mutations in the phospho-H3 binding site of Survivin prevent centromere localization [103, 105]. Curiously, CENP-A is still phosphorylated when Aurora B fails to fully concentrate at centromeres in haspin-depleted cells [103]. Thus, there may be some residual kinase activity in this case, or the control of substrate phosphorylation may depend on factors in addition to the spatial proximity of the kinase. Unlike Bub1, Haspin localizes to heterochromatin, but not specifically to centromeres, in a cohesin dependent process. The combination of the Bub1-dependent H2A phosphorylation and the Haspin-dependent H3 phosphorylation specifies CPC localization to centromeres [104]. There is some additional complexity, as work in multiple organisms has shown that localization of Shugoshin also depends on the CPC [59–64], suggesting that a complex between Shugoshin and the CPC might contribute to the centromere targeting of both.
Concluding remarks
In conclusion, recent work has provided important insights that explain how mechanical forces at kinetochores are translated into chemical signals to ensure correct kinetochore-microtubule attachments, as envisioned by Nicklas more than 40 years ago. This review has focused on Aurora B in the context of inter-kinetochore tension, or stretching of the centromere in response to sister kinetochore bi-orientation. However, recent evidence indicates that stretching within a single kinetochore, which increases the distance between inner and outer kinetochore components, is a crucial component of mitotic checkpoint signaling [82, 83]. Remarkably, inter-kinetochore stretch and intra-kinetochore stretch can change independently of each other. An appealing model was recently proposed in which intra-kinetochore stretch and checkpoint signaling depend on dynamic microtubule attachments at a single kinetochore, while inter-kinetochore stretching depends on bi-orientation of sister kinetochores [106]. The two types of deformation are likely related because inter-kinetochore tension determines local Aurora B activity, which regulates microtubule dynamics at kinetochores. In addition, intra-kinetochore stretch may affect the positions of Aurora substrates within a phosphorylation gradient [107]. The relationship between intra- and inter-kinetochore stretch, Aurora B, and the spindle checkpoint remains unresolved and will likely be an active area of future investigation.
Acknowledgments
Research in our labs is supported by grants from the US National Institutes of Health (GM083988 to M.A. Lampson and GM088313 to I.M. Cheeseman) and the Searle Scholars Program. We thank the (mostly) anonymous reviewers for their helpful, thoughtful, and constructive comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Indjeian VB, Murray AW. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr Biol. 2007;17:1837–1846. doi: 10.1016/j.cub.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 2.Loncarek J, et al. The centromere geometry essential for keeping mitosis error free is controlled by spindle forces. Nature. 2007;450:745–749. doi: 10.1038/nature06344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ault JG, Rieder CL. Chromosome mal-orientation and reorientation during mitosis. Cell motility and the cytoskeleton. 1992;22:155–159. doi: 10.1002/cm.970220302. [DOI] [PubMed] [Google Scholar]
- 4.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. The Journal of cell biology. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicklas RB. How cells get the right chromosomes. Science (New York, N.Y.) 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- 6.Nicklas RB, Koch CA. Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. The Journal of cell biology. 1969;43:40–50. doi: 10.1083/jcb.43.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicklas RB, Ward SC. Elements of error correction in mitosis: microtubule capture, release, and tension. The Journal of cell biology. 1994;126:1241–1253. doi: 10.1083/jcb.126.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan CS, Botstein D. Isolation and characterization of chromosome-gain and increase-inploidy mutants in yeast. Genetics. 1993;135:677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biggins S, et al. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes & development. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheeseman IM, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 11.Francisco L, et al. Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Molecular and cellular biology. 1994;14:4731–4740. doi: 10.1128/mcb.14.7.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka TU, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 13.Giet R, Prigent C. Aurora/Ipl1p-related kinases, a new oncogenic family of mitotic serine-threonine kinases. Journal of cell science. 1999;112(Pt 21):3591–3601. doi: 10.1242/jcs.112.21.3591. [DOI] [PubMed] [Google Scholar]
- 14.Ruchaud S, et al. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 15.Hauf S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. The Journal of cell biology. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kallio MJ, et al. Inhibition of Aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr Biol. 2002;12:900–905. doi: 10.1016/s0960-9822(02)00887-4. [DOI] [PubMed] [Google Scholar]
- 17.Ditchfield C, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. The Journal of cell biology. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lampson MA, et al. Correcting improper chromosome-spindle attachments during cell division. Nature cell biology. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 19.Feramisco JR, et al. Optimal spatial requirements for the location of basic residues in peptide substrates for the cyclic AMP-dependent protein kinase. The Journal of biological chemistry. 1980;255:4240–4245. [PubMed] [Google Scholar]
- 20.Nash P, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 21.Vogel V, Sheetz MP. Cell fate regulation by coupling mechanical cycles to biochemical signaling pathways. Current opinion in cell biology. 2009;21:38–46. doi: 10.1016/j.ceb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janmey PA, McCulloch CA. Cell mechanics: integrating cell responses to mechanical stimuli. Annual review of biomedical engineering. 2007;9:1–34. doi: 10.1146/annurev.bioeng.9.060906.151927. [DOI] [PubMed] [Google Scholar]
- 23.Carmena M, et al. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Current opinion in cell biology. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheatley SP, et al. INCENP binds directly to tubulin and requires dynamic microtubules to target to the cleavage furrow. Experimental cell research. 2001;262:122–127. doi: 10.1006/excr.2000.5088. [DOI] [PubMed] [Google Scholar]
- 25.Kang J, et al. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. The Journal of cell biology. 2001;155:763–774. doi: 10.1083/jcb.200105029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandall S, et al. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 2006;127:1179–1191. doi: 10.1016/j.cell.2006.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly AE, Funabiki H. Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Current opinion in cell biology. 2009;21:51–58. doi: 10.1016/j.ceb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maure JF, et al. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr Biol. 2007;17:2175–2182. doi: 10.1016/j.cub.2007.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jelluma N, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132:233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 30.Sliedrecht T, et al. Chemical genetic inhibition of Mps1 in stable human cell lines reveals novel aspects of Mps1 function in mitosis. PloS one. 2010;5:e10251. doi: 10.1371/journal.pone.0010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwiatkowski N, et al. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nature chemical biology. 2010;6:359–368. doi: 10.1038/nchembio.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maciejowski J, et al. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. The Journal of cell biology. 2010;190:89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santaguida S, et al. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. The Journal of cell biology. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hewitt L, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. The Journal of cell biology. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourhis E, et al. Phosphorylation of a borealin dimerization domain is required for proper chromosome segregation. Biochemistry. 2009;48:6783–6793. doi: 10.1021/bi900530v. [DOI] [PubMed] [Google Scholar]
- 36.Cimini D, et al. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 37.Andrews PD, et al. Aurora B regulates MCAK at the mitotic centromere. Developmental cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 38.Liu D, et al. Sensing chromosome bi-orientation by spatial separation of Aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welburn JP, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Molecular cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keating P, et al. Ipl1-dependent phosphorylation of Dam1 is reduced by tension applied on kinetochores. Journal of cell science. 2009;122:4375–4382. doi: 10.1242/jcs.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cimini D, et al. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 42.Knowlton AL, et al. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 43.Kim Y, et al. Aurora Kinases and Protein Phosphatase 1 Mediate Chromosome Congression through Regulation of CENP-E. Cell. 2010;142:444–455. doi: 10.1016/j.cell.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rome P, et al. Aurora A contributes to p150(glued) phosphorylation and function during mitosis. The Journal of cell biology. 2010;189:651–659. doi: 10.1083/jcb.201001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheeseman IM, et al. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 46.Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guimaraes GJ, et al. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller SA, et al. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Curr Biol. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X, et al. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Molecular biology of the cell. 2007;18:3264–3276. doi: 10.1091/mbc.E07-01-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lan W, et al. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 52.Ohi R, et al. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Molecular biology of the cell. 2004;15:2895–2906. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Knowlton AL, et al. ICIS and Aurora B coregulate the microtubule depolymerase Kif2a. Curr Biol. 2009;19:758–763. doi: 10.1016/j.cub.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gestaut DR, et al. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nature cell biology. 2008;10:407–414. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shang C, et al. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Molecular biology of the cell. 2003;14:3342–3355. doi: 10.1091/mbc.E02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tien JF, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by Aurora B. The Journal of cell biology. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lampert F, et al. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. The Journal of cell biology. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emanuele MJ, et al. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. The Journal of cell biology. 2008;181:241–254. doi: 10.1083/jcb.200710019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanoosthuyse V, et al. Shugoshin 2 regulates localization of the chromosomal passenger proteins in fission yeast mitosis. Molecular biology of the cell. 2007;18:1657–1669. doi: 10.1091/mbc.E06-10-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawashima SA, et al. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes & development. 2007;21:420–435. doi: 10.1101/gad.1497307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pouwels J, et al. Shugoshin 1 plays a central role in kinetochore assembly and is required for kinetochore targeting of Plk1. Cell cycle. 2007;6:1579–1585. doi: 10.4161/cc.6.13.4442. [DOI] [PubMed] [Google Scholar]
- 62.Huang H, et al. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. The Journal of cell biology. 2007;177:413–424. doi: 10.1083/jcb.200701122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Resnick TD, et al. INCENP and Aurora B promote meiotic sister chromatid cohesion through localization of the Shugoshin MEI-S332 in Drosophila. Developmental cell. 2006;11:57–68. doi: 10.1016/j.devcel.2006.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dai J, et al. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Developmental cell. 2006;11:741–750. doi: 10.1016/j.devcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 65.De Wulf P, et al. Protein phosphatases take the mitotic stage. Current opinion in cell biology. 2009;21:806–815. doi: 10.1016/j.ceb.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 66.Liu D, et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. The Journal of cell biology. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Egloff MP, et al. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. The EMBO journal. 1997;16:1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hendrickx A, et al. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chemistry & biology. 2009;16:365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 69.Posch M, et al. Sds22 regulates Aurora B activity and microtubule-kinetochore interactions at mitosis. The Journal of cell biology. 2010;191:61–74. doi: 10.1083/jcb.200912046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bakhoum SF, et al. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nature cell biology. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Famulski JK, Chan GK. Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores. Curr Biol. 2007;17:2143–2149. doi: 10.1016/j.cub.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 72.Chan YW, et al. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. The Journal of cell biology. 2009;185:859–874. doi: 10.1083/jcb.200812167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manning AL, et al. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. The EMBO journal. 2010;29:3531–3543. doi: 10.1038/emboj.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmidt JC, et al. Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores. The Journal of cell biology. 2010;191:269–280. doi: 10.1083/jcb.201006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Santaguida S, Musacchio A. The life and miracles of kinetochores. The EMBO journal. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vader G, et al. The chromosomal passenger complex controls spindle checkpoint function independent from its role in correcting microtubule kinetochore interactions. Molecular biology of the cell. 2007;18:4553–4564. doi: 10.1091/mbc.E07-04-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mackay AM, et al. Molecular analysis of the INCENPs (inner centromere proteins): separate domains are required for association with microtubules during interphase and with the central spindle during anaphase. The Journal of cell biology. 1993;123:373–385. doi: 10.1083/jcb.123.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tseng BS, et al. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Developmental cell. 18:903–912. doi: 10.1016/j.devcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Joglekar AP, et al. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Waters JC, et al. Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push. Journal of cell science. 1996;109:2823–2831. doi: 10.1242/jcs.109.12.2823. [DOI] [PubMed] [Google Scholar]
- 82.Uchida KS, et al. Kinetochore stretching inactivates the spindle assembly checkpoint. The Journal of cell biology. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. The Journal of cell biology. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ahonen LJ, et al. Perturbation of INCENP function impedes anaphase chromatid movements and chromosomal passenger protein flux at centromeres. Chromosoma. 2009;118:71–84. doi: 10.1007/s00412-008-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murata-Hori M, Wang YL. Both midzone and astral microtubules are involved in the delivery of cytokinesis signals: insights from the mobility of Aurora B. The Journal of cell biology. 2002;159:45–53. doi: 10.1083/jcb.200207014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sessa F, et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Molecular cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 87.Bishop JD, Schumacher JM. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. The Journal of biological chemistry. 2002;277:27577–27580. doi: 10.1074/jbc.C200307200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Honda R, et al. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Molecular biology of the cell. 2003;14:3325–3341. doi: 10.1091/mbc.E02-11-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kelly AE, et al. Chromosomal enrichment and activation of the Aurora B pathway are coupled to spatially regulate spindle assembly. Developmental cell. 2007;12:31–43. doi: 10.1016/j.devcel.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fuller BG, et al. Midzone activation of Aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gadea BB, Ruderman JV. Aurora B is required for mitotic chromatin-induced phosphorylation of Op18/Stathmin. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4493–4498. doi: 10.1073/pnas.0600702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hsu JY, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/Aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 93.Wheatley SP, et al. Phosphorylation by Aurora-B negatively regulates survivin function during mitosis. Cell cycle. 2007;6:1220–1230. doi: 10.4161/cc.6.10.4179. [DOI] [PubMed] [Google Scholar]
- 94.Delacour-Larose M, et al. Role of survivin phosphorylation by Aurora B in mitosis. Cell cycle. 2007;6:1878–1885. doi: 10.4161/cc.6.15.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Beardmore VA, et al. Survivin dynamics increases at centromeres during G2/M phase transition and is regulated by microtubule-attachment and Aurora B kinase activity. Journal of cell science. 2004;117:4033–4042. doi: 10.1242/jcs.01242. [DOI] [PubMed] [Google Scholar]
- 96.Vong QP, et al. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 2005;310:1499–1504. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- 97.Sumara I, et al. A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Developmental cell. 2007;12:887–900. doi: 10.1016/j.devcel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 98.Fernandez-Miranda G, et al. SUMOylation modulates the function of Aurora-B kinase. Journal of cell science. 123:2823–2833. doi: 10.1242/jcs.065565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bassett EA, et al. Epigenetic centromere specification directs Aurora B accumulation but is insufficient to efficiently correct mitotic errors. J. Cell Biol. 190:177–185. doi: 10.1083/jcb.201001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kawashima SA, et al. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 101.Tsukahara T, et al. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature. 467:719–723. doi: 10.1038/nature09390. [DOI] [PubMed] [Google Scholar]
- 102.Kelly AE, et al. Survivin Reads Phosphorylated Histone H3 Threonine 3 to Activate the Mitotic Kinase Aurora B. Science. 330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang F, et al. Histone H3 Thr-3 Phosphorylation by Haspin Positions Aurora B at Centromeres in Mitosis. Science. 330:231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yamagishi Y, et al. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 330:239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- 105.Yue Z, et al. Deconstructing Survivin: comprehensive genetic analysis of Survivin function by conditional knockout in a vertebrate cell line. J. Cell Biol. 2008;183:279–296. doi: 10.1083/jcb.200806118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Khodjakov A, Pines J. Centromere tension: a divisive issue. Nat. Cell Biol. 12:919–923. doi: 10.1038/ncb1010-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maresca TJ, Salmon ED. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. J. Cell Sci. 123:825–835. doi: 10.1242/jcs.064790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yasui Y, et al. Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J. Biol. Chem. 2004;279:12997–13003. doi: 10.1074/jbc.M311128200. [DOI] [PubMed] [Google Scholar]
- 109.Zeitlin SG, et al. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 2001;155:1147–1157. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zimniak T, et al. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 2009;186:379–391. doi: 10.1083/jcb.200901036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guse A, et al. Phosphorylation of ZEN-4/MKLP1 by Aurora B regulates completion of cytokinesis. Curr Biol. 2005;15:778–786. doi: 10.1016/j.cub.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 112.Minoshima Y, et al. Phosphorylation by Aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Developmental cell. 2003;4:549–560. doi: 10.1016/s1534-5807(03)00089-3. [DOI] [PubMed] [Google Scholar]