Abstract
Gain-of-function mutations in leucine-rich repeat kinase 2 (LRRK2) cause familial as well as sporadic Parkinson’s disease (PD) characterized by age-dependent dopaminergic neuron (DN) degeneration1, 2. The molecular mechanism of LRRK2 action is not known. Here we show that LRRK2 interacts with the microRNA (miRNA) pathway to regulate protein synthesis. Drosophila e2f1 and dp mRNAs are translationally repressed by let-7 and miR-184*, respectively. Pathogenic LRRK2 antagonizes these miRNAs, leading to overproduction of E2F1/DP previously implicated in cell cycle and survival control3, and shown here to be critical for LRRK2 pathogenesis. Genetic deletion of let-7, antagomir-mediated blockage of let-7 and miR-184* action, transgenic expression of dp target protector, or replacing endogenous dp with a dp transgene non-responsive to let-7 all had similar toxic effects as pathogenic LRRK2. Conversely, increasing let-7 or miR-184* level attenuates pathogenic LRRK2 effects. LRRK2 associates with Drosophila Argonaute-1 (dAgo1) or human Argonaute-2 (hAgo2) of the RNA-induced silencing complex (RISC). In aged fly brain, dAgo1 protein level is negatively regulated by LRRK2. Further, pathogenic LRRK2 promotes the association of phospho-4E-BP1 with hAgo2. Our results implicate deregulated synthesis of E2F1/DP caused by miRNA pathway impairment as a key event in LRRK2 pathogenesis and suggest novel miRNA-based therapeutic strategies.
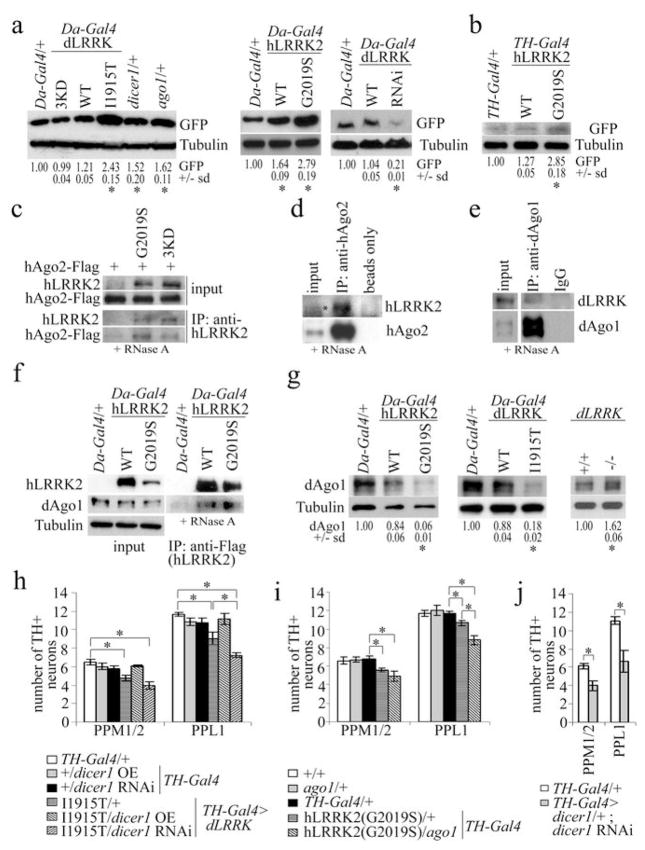
Analyses of Dicer knockout mice have implicated the miRNA pathway in maintaining post-mitotic neurons4,5. To test whether LRRK2 might affect miRNA function, we generated an in vivo EGFP reporter with let-7-binding sites in the 3′UTR (EGFP-let-7-3′UTR). Loss of one copy of Drosophila dicer-1 or argonaute 1 (ago1) genes of the miRNA pathway enhanced reporter expression (Fig.1a), suggesting that this reporter faithfully monitors endogenous miRNA activity. Da-Gal4-directed ubiquitous or TH-Gal4-directed DN-specific co-expression of pathogenic dLRRK(I1915T) or hLRRK2(G2019S) all stimulated reporter expression, more so than WT proteins (Fig.1a, 1b). This effect correlated with differential toxicity6, rather than expression levels of the proteins, since the pathogenic proteins were actually expressed at lower levels (Fig.2a). dLRRK RNAi led to decreased reporter expression, indicating that endogenous dLRRK also impacts miRNA function (Fig.1a). The kinase-dead dLRRK(3KD) did not affect reporter expression (Fig.1a, s1a). This result, together with the observation that I1915T and G2019S mutations augmented kinase activity7,8, suggests that the effect of LRRK2 is kinase-dependent. gfp mRNA levels were not affected (Fig.s1b,c). Moreover, the expression of an EGFP transgene without let-7-binding sites in the 3′UTR was not affected by dLRRK(I1915T) (Fig.s3a). LRRK2 thus antagonizes let-7 function at the translational level.
Figure 1. Pathogenic LRRK2 suppresses let-7 function and interacts with RISC component Argonaute.
a, effects of various genotypes on the expression of EGFP in Da-Gal4>EGFP-let-7-3′UTR flies. Da-Gal4/+ serves as control (*P<0.05, n=3). b, EGFP level in TH-Gal4>EGFP-let-7-3′UTR flies co-expressing hLRRK2(WT) or hLRRK2(G2019S), compared to TH-Gal4/+ control (*P<0.05, n=3). c-f, co-IP experiments between transfected hLRRK2(G2019S) or hLRRK2(3KD) and hAgo2 culture (c), endogenous hAgo2 and hLRRK2 in culture (d), endogenous dAgo1 and dLRRK in fly head extracts (e), or transgenic hLRRK2 and endogenous dAgo1 in fly head extracts (f). g, endogenous dAgo1 level in fly head extracts of the indicated genotypes (*P<0.05, n=3, 65 days). h, i, effects of genetic interaction between dLRRK(I1915T) and Dicer1 (h, *P<0.05, n=8) or hLRRK2(G2019S) and dAgo1 (i, *P<0.05, n=10) on DN number. j, effects of Dicer1 knockdown in DNs (*P<0.0001, n=10).
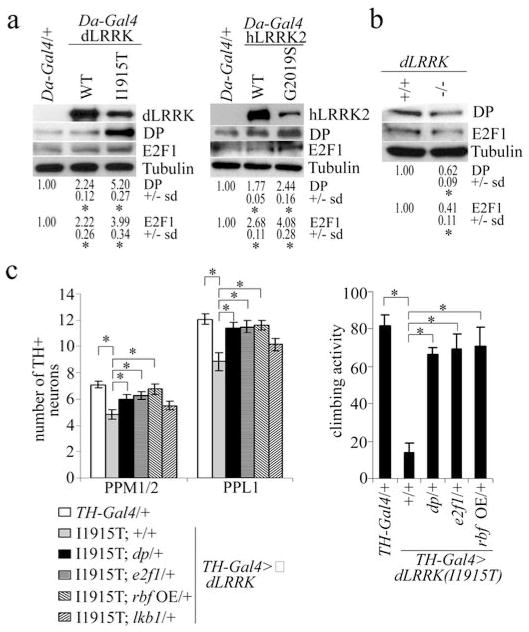
Figure 2. Identification of E2F1 and DP as key translational targets of pathogenic LRRK2.
a, WB analysis of E2F1 and DP expression in control (Da-Gal4/+), dLRRK transgenic, and hLRRK2 transgenic flies. Values represent EGFP levels in the transgenic crosses relative to the control from three independent experiments (*P<0.05, n=3). Tubulin serves as a loading control. Fly head extracts prepared from 35-day-old flies raised at 29°C were used. b, WB analysis of E2F1 and DP levels (*P<0.05, n=3) in dLRRK mutant and wild-type animals. c, effects of e2f1, dp, or lkb1 heterozygosity, or overexpression of rbf on DN number (left, n=20) and climbing activity (right, n=20) in TH-Gal4>dLRRK(I1915T) flies (*P<0.05).
We further tested the effect of pathogenic LRRK2 on miRNA-regulated translation in HEK293T cells, using a luciferase reporter harboring let-7-binding sites in the 3′UTR9. hLRRK2(I2020T) or hLRRK2(G2019S) efficiently suppressed the inhibitory effect of let-7 on reporter expression (Fig.s2a,s2b). Introduction of kinase-inactivating mutations (3KD) into hLRRK2-G2019S or -I2020T abolished the effects of these proteins on reporter expression (Fig.s2b), supporting kinase activity-dependency of pathogenic LRRK2 action. Mutant hLRRK2 carrying another pathogenic mutation R1441G had no effect on let-7 miRNA function (Fig.s2b), and transgenic flies expressing dLRRK carrying the equivalent R1069G mutation showed no DN loss (Fig.s2c), suggesting that this mutation act through a different mechanism.
We next investigated how LRRK2 regulates miRNA function. No significant change of let-7 level in hLRRK2(G2019S) or dLRRK(I1915T) transgenics was detected (Fig.s4a). We asked whether LRRK2 impairs miRNA function through RISC components10. In fly DNs, hLRRK2 and dAgo1 were both cytoplasmic and partially co-localized (Fig.s5a). In co-immunoprecipitation (co-IP) assays, transfected hLRRK2 associated with hAgo2 in HEK293T cells (Fig.1c), transgenic hLRRK2 associated with dAgo1 in fly head extracts (Fig.1f), and endogenous hLRRK2 or dLRRK associated with hAgo2 or dAgo1, respectively (Fig.1d, 1e, s6b). Since similar co-IP results were obtained with (Fig.1c,f) or without (Fig.s6a,c) RNase A treatment of extracts, the interaction was likely direct and not mediated by RNA.
We examined the physiological consequence of the LRRK2-dAgo1 interaction. While no significant change in dAgo1 level was observed in young flies (Fig.s7a), in aged flies dAgo1 protein was significantly reduced by pathogenic hLRRK2 (Fig.1g, s5b). Conversely, dAgo1 level was increased in dLRRK mutant (Fig.1g, s7a). Consistent with a negative regulation of dAgo1 by LRRK2, ago1 heterozygosity exacerbated the climbing defect (Fig.s8b) and DN loss phenotypes (Fig.1i) in pathogenic dLRRK or hLRRK2 transgenics. Aged ago1 heterozygote showed ~26% reduction of dAgo1 protein compared to control and did not exhibit obvious phenotypes (Fig.1i, s7b). We also examined genetic interaction between LRRK2 and Dicer1. In TH-Gal4>dLRRK(I1915T) background, Dicer1 overexpression suppressed, whereas Dicer1 RNAi exacerbated, dLRRK(I1915T) toxicity, although similar manipulations in a wild type background had no obvious effect (Fig.1h, s8a). However, stronger inhibition of Dicer1 by Dicer1 RNAi in dicer1 heterozygous background did result in DN loss and climbing defects (Fig.1j, s8c), which were partially reduced by dLRRK RNAi (Fig.s8d, s19a). Since reducing bulk translation through overexpression of active forms of 4E-BP did not rescue the toxicity of Dicer1 knockdown (data not shown), the rescue by dLRRK RNAi may not be attributable to reduced bulk translation. These results support a specific link between LRRK2 and the miRNA pathway.
Since the miRNA-antagonizing function of LRRK2 is kinase-dependent, we investigated possible interaction between the miRNA pathway and LRRK2 substrate 4E-BP6. 4E-BP1 associated with hAgo2 in HEK293T cells (Fig.s10a). dLRRK(I1915T) and hLRRK2(G2019S) more effectively promoted this interaction than WT proteins. This presumably resulted from increased phosphorylation of 4E-BP1 by pathogenic LRRK26, and preferential binding of phospho-4E-BP1 to hAgo2 (Fig.s10b, s10c). Moreover, the phospho-mimetic 4E-BP(TE) was more effective than 4E-BP(WT) or 4E-BP(TA) in attenuating let-7 effect in vivo or in 4E-BP1(−/−) MEF cells (Fig.s10d, s10e), and overexpression of 4E-BP(TE) was toxic to DNs (Fig.s10f, s10g). Since no change in dAgo1 level was detected in aged Da-Gal4>d4E-BP(TE) animals (Fig.s10h), and loss of dAgo2 had no effect on DN number (Fig.s10i), phospho-4E-BP effect is unlikely through regulating dAgo1 or dAgo2. Pathogenic hLRRK2 with hyperactive kinase activity thus promotes the association of phospho-4E-BP with hAgo2, relieving miRNA-mediated translational repression. Consistently, non-phosphorylatable 4E-BP(TA) blocked the effect of hLRRK2(G2019S) in antagonizing let-7 (Fig.s10j).
We next sought to identify mRNA targets whose translation is upregulated by pathogenic LRRK2. Da-Gal4>dLRRK(WT) or Da-Gal4>dLRRK(I1915T) fly heads were subjected to mRNA translational profiling11. From a list of candidate genes showing differential polysome association (Table s1), we chose e2f1 and dp for further analysis, since previous studies have implicated E2F deregulation in PD12. Puromycin treatment, which specifically releases actively translating polysomes, showed that most e2f1 and dp mRNAs were associated with active polysomes (Fig.s11b). Western blot analysis confirmed upregulation of E2F1 and DP by dLRRK(I1915T) or hLRRK2(G2019S) (Fig.2a, s12a,b); conversely, both proteins were downregulated in dLRRK(−/−) mutant (Fig.2b), with mRNA levels not significantly changed in both conditions (Fig.s12c,d). Significantly, removing one copy of e2f1 or dp, or overexpressing RBF, a negative regulator of E2F1/DP13, suppressed the DN phenotypes in TH-Gal4>dLRRK(I1915T) animals (Fig.2c). Removing one copy of lkb1, another potential target identified by translational profiling, showed no effect (Fig.2c). Removing one copy of e2f1 or dp, but not lkb1, also attenuated oxidative stress sensitivity of Da-Gal4>dLRRK(I1915T) transgenics (Fig.s13).
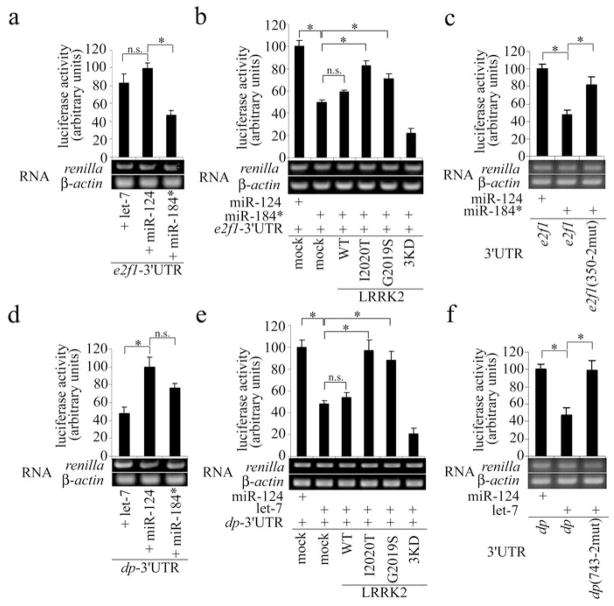
To test whether dLRRK and hLRRK2 affect the translation of E2F1 and DP through the miRNA pathway, we used the RNAhybrid algorithm to predict potential miRNA-binding sites within e2f1- and dp-3′UTRs. One candidate miR-184*-binding site in e2f1-3′UTR responded to miR-184* activity in reporter assays (Fig.3a). Mutating two central nucleotides in the seed sequence completely abolished this miR-184* effect (Fig.3c, s16a). Similarly, we identified one functional let-7-binding site within dp-3′UTR (Fig.3d, 3f, s16b). Co-transfection of pathogenic hLRRK2 strongly relieved the repressive effects of let-7 and miR-184* on the expression of dp- or e2f1-3′UTR reporters, respectively (Fig.3b, e).
Figure 3. Control of E2F1 and DP expression by miR-184* and let-7 and their regulation by pathogenic hLRRK2.
a, d, reporter assays showing the response of the 3′UTRs of Drosophila e2f1 (a) and dp (d) to exogenous let-7 and miR-184*, with miR-124 serving as a control (*P<0.05, n=3). RT-PCR measures the level of luciferase mRNA, with β-actin serving as a control. b, e, reporter assays showing the effects of WT, 3KD, and pathogenic forms of hLRRK2 on miRNA-mediated regulation of e2f1- (b) and dp-3′UTR (e) (*P<0.05, n=3). c, f, reporter assays using e2f1- and dp-3′UTRs containing mutagenized seed sequences in let-7- or miR-184*-binding sites (*P<0.05, n=3). Luciferase activity was normalized with renilla mRNA level in all assays.
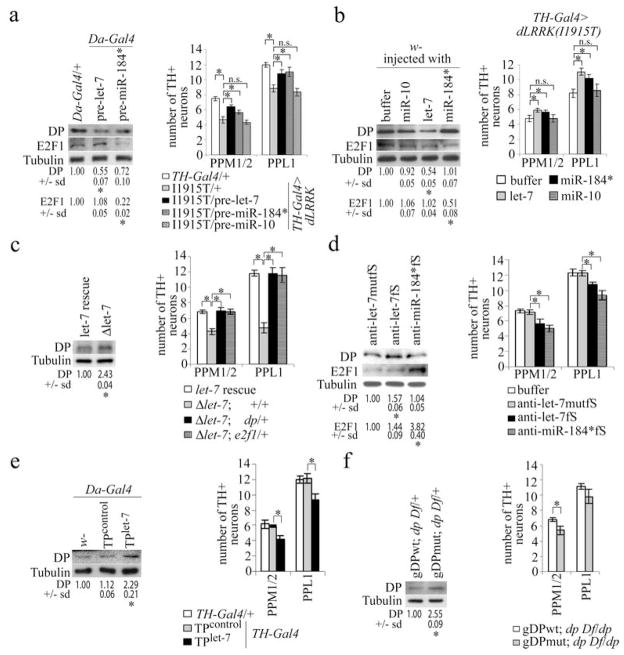
To test whether let-7 and miR-184* regulate endogenous e2f1 and dp translation, we overexpressed precursor (pre)-miRNAs or injected synthetic mature miRNAs into wild-type fly heads. Ubiquitous overexpression of pre-let-7 (Fig.s4b), or injection of synthetic let-7 (Fig.s3b) significantly reduced DP level in head extracts (Fig. 4a, 4b). Overexpressed pre-miR-184* or injected miR-184* reduced E2F1 level (Fig. 4a, 4b), validating E2F1 and DP as in vivo targets of miR-184* and let-7. Importantly, DN-specific pre-let-7 or pre-miR-184* overexpression or mature miRNA injection partially rescued TH-Gal4>dLRRK(I1915T) mutant phenotypes, whereas buffer or control miR-10 injection had no effect (Fig. 4a, 4b, s8e). Injection of miRNAs into wild-type fly heads also had no effect (Fig.s8g). Regulation of E2F1 and DP by miRNA is therefore causally involved in dLRRK(I1915T) pathogenesis.
Figure 4. Regulated E2F1 and DP translation by let-7 and miR-184* impacts DN maintenance and function in vivo.
a, effects of pre-let-7 and pre-miR-184* on DP and E2F1 level (left, *P<0.05, n=3) and DN number (right, *P<0.05, n=6). b, effects of injected mature let-7 and miR-184* miRNAs on DP and E2F1 level (left, *P<0.05, n=3) and DN number (right, *P<0.05, n=16). c, DP level (left, *P<0.05, n=3) and DN number (right, *P<0.05, n=6) in the indicated genotypes. d, effects of let-7 and miR-184* antagomir injection on E2F1 and DP level (left, *P<0.05, n=3) and DN number (right, *P<0.05, n=6) in wild-type flies. Anti-let-7mutfS serves as control. e, effect of ubiquitous dp-TPcontrol or dp-TPlet-7 expression on DP level (left, *P<0.05, n=3), or DN-specific expression on DN number (right, *P<0.004, n=15). f, effects of gDPwt and gDPmut transgene expression on DP level in dp Df/+ heterozygous background (left, *P<0.05, n=3) and DN number (right, *P<0.015, n=5). Flies were raised at 29°C for 33 days.
We further tested whether inhibition of let-7 or miR-184* function in wild-type animals is sufficient to phenocopy pathogenic LRRK2. We first verified that let-7 is expressed in DNs (Fig.s14). Homozygous let-7 mutant (Δlet-7) showed reduced locomotor activity and a specific reduction of DNs, which could be rescued by a let-7 transgene (Fig.4c, s8f, s15c, d). Consistent with let-7 regulating DP expression in vivo, DP level was increased in Δlet-7 animals (Fig.4c). Removing one copy of dp or e2f1 suppressed Δlet-7 mutant phenotypes (Fig.4c, s8f). These results support that DP overproduction contributes significantly to Δlet-7 phenotypes. Alternatively, we injected antagomirs into fly heads to block miRNA function. Injected anti-let-7fS attenuated endogenous let-7 function in DNs (Fig.s3b). Anti-let-7fS injection into wild-type flies increased DP level in head extracts, whereas a control antagomir (anti-let-7mutfS) had no effect. Similarly, anti-miR-184*fS increased E2F1 level (Fig.4d). Anti-let-7fS and anti-miR-184*fS injection resulted in specific DN loss (Fig.4d, s15a, b), and reduced climbing activity (Fig.s9c). In comparison, anti-let-7fS injection into dLRRK mutant failed to produce such effects (Fig.s9a), suggesting that the miRNA pathway is more robust when dLRRK is absent. Significantly, the toxic effects of anti-miR-184*fS and anti-let-7fS were attenuated by e2f1/+ or dp/+ heterozygosity (Fig.s9b,c), supporting E2F1/DP as important mediators of antagomir toxicity. Consistently, E2F1 overexpression is sufficient to cause DN degeneration and reduced locomoter activity (Fig.s9f, s15e).
To more conclusively prove the critical role of the specific miRNA-mRNA interactions in DNs, we employed two approaches. First, we generated transgenics expressing a dp target protector (dp-TPlet-7, Fig.s16d), which is expected to specifically interferes with let-7-miRNA/dp mRNA 3′UTR interaction14. dp-TPlet-7 is functional in a reporter assay (Fig.s18a). Ubiquitous expression of dp-TPlet-7 using Da-Gal4 resulted in DP overproduction without affecting dp mRNA level (Fig.4e, s18b), and TH-Gal4-driven dp-TPlet-7 expression led to DN loss and reduced climbing activity (Fig.4e, s9d, s19b). Second, we generated genomic transgenes expressing a mutant form of dp (gDPmut) with potential let-7-binding sites rendered non-functional (Fig.s16b, c). Compared to gDPwt, gDPmut caused DP accumulation in a dp Df/+ background (Fig.4f). Although both gDPwt and gDPmut rescued the viability of Df/dpR217H transheterozygote, gDPmut-rescued animals exhibited loss of DNs and locomotor defects (Fig.4f, s9e, s19c). Together, these in vivo studies establish the importance of LRRK2-regulated let-7-miRNA/dp mRNA interaction in DNs. Interestingly, GMR-Gal4-driven dLRRK(I1915T) expression in the eye did not affect the activities of bantam and miR-14 miRNAs (Fig.s17a, b), which protect against MJD.tr-Q78 and Reaper-induced neurodegeneration, respectively15,16. Thus pathogenic dLRRK may regulate miRNA function in a miRNA- and/or tissue-specific manner. The recent identification of let-7 miRNA-specific ribonucleoprotein complexes supports miRNA-specific regulation17.
We have identified let-7 and miR-184* and their targets E2F1 and DP as critical mediators of pathogenic LRRK2 in DNs. E2F1 and DP are better known for their roles in driving the cell cycle. In post-mitotic neurons, their upregulation may lead to abortive cell division and cell death, as shown in DNs of PD patients and animal models12. Mechanistically, pathogenic LRRK2 associate with and regulate dAgo1 stability, and promote the association of phospho-4E-BP with dAgo1. The exact mechanism of phospho-4E-BP action in this process and the potential involvement of other LRRK2 targets await further investigation. Intriguingly, 4E-BP overexpression was first shown to be neuroprotective in the LRRK2 model6, and later extended to other PD models18, supporting a general role of deregulated protein translation in Parkinsonism. The fact that the effects of LRRK2 only manifest in aged animals suggests that LRRK2 is particularly important for miRNA regulation under stress conditions, where the function of 4E-BP is also most pronounced19. This may have important implications for understanding PD pathogenesis.
Methods Summary
Counting of TH+ and FMRFamide+ neurons was performed by whole-mount immunostaining of brain samples from adult flies raised at 25°C for 65 days unless otherwise indicated. For co-immunoprecipitation in HEK293 cells, cells were co-transfected with hLRRK2 constructs and Flag-tagged human Ago221. For Western blotting, the following primary antibodies were used: Drosophila DP22 (Yun3, Dyson lab), Drosophila E2F123 (Wharton lab), Drosophila β-Tubulin (E7, University of Iowa Hybridoma Bank), EGFP (11E5 and 3E6, Qbiogene, Inc), FMRFamide (Chaoyang Zeng), Flag (M2, Sigma), dAgo1 (Abcam), hLRRK2 (Novus), 4E-BP1 (Cell Signaling), phospho-4E-BP1(Thr37/46) (Cell Signaling), hAgo2 (11A9, Meister lab24).
UAS-dLRRK, UAS-4E-BP, UAS-4E-BP(TA), UAS-dLRRK RNAi and dLRRK mutant lines were used as described6. The UAS-eIF4E RNAi, UAS-dicer1 RNAi and UAS-dicer1 flies were obtained from the Vienna Drosophila RNAi Center (VDRC) and Dr. B. Dickson, respectively, UAS-rbf and UAS-e2f1 were requested from Dr. B. Edgar13, dcr-1Q1147X from Dr. R.W. Carthew25, lkb14A from Dr. D. St. Johnston26, UAS-4E-BP and UAS-4E-BP(LL)w from Dr. N. Sonenberg27, UAS-hLRRK2 from Dr. W. Smith28, ago2(414) from Dr. H. Siomi10, GMR-miR-14 from Dr. B. Hay16, and UAS-banD from Dr. S. Cohen29. The Δlet-7 mutant, Δlet-7 mutant rescued with a let-7 transgene, and let-7-C-Gal4 flies were described20. All other stocks are from Bloomington Drosophila Stock Center. 4E-BP1 depleted mouse embryonic fibroblast (MEF) line was obtained from Dr. N. Sonenberg30.
The oxidative stress assay and climbing activity assay were performed essentially as described6. To measure climbing activity, we used flies aged at 25°C for 65 days or otherwise indicated. Each measurement was repeated using three independent fly cohorts per genotype.
Unpaired Student’s t-tests were used for statistical analysis between two groups and one-way ANOVA analysis for statistical analysis involving more than two experimental groups. The RNAhybrid algorithm available at [http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html] was used to predict potential miRNA target sites.
Methods
Isolation of polysomal mRNA, cDNA Synthesis, and DNA microarray analysis
Four hundred fly heads (50% males and 50% females of 25-day-old flies) were dissected in ice-cold dissection buffer [137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4, 0.1 mg/ml cycloheximide (Sigma), 0.05% Triton X-100]. Fly heads were transferred into extraction buffer [125 mM Sucrose, 25 mM HEPES, pH 6.9, 100 mM KCl, 5 mM MgCl2, 1 mM DTT supplemented with RNase inhibitor and Complete protease inhibitor cocktail] and subjected to 10 strokes of homogenization in a pre-chilled homogenizer. The extract was centrifuged at 10,000 rpm for 5 minutes at 4°C and loaded onto the top of 10–50% sucrose gradients prepared in 300 mM NaCl, 15 mM MgCl2, 15 mM Tris-HCl, pH 7.5, 0.1 mg/ml cycloheximide. After centrifugation at 35,000 rpm for 3 hours at 4°C using SW40Ti rotor (Beckman), fractions were harvested from the bottom. The RNA from each polysomal fraction (fractions 2 to 7) was extracted using Guanidine-HCl and precipitated with 100% ethanol. Equal amounts of mRNA were reverse transcribed into cDNA and amplified using SMARTTM PCR cDNA Synthesis Kit (Clontech) according to manufacturer’s recommendation. The Klenow fragment was used to incorporate biotin-16-dUTP (Roche) into the cDNA. After a subsequent treatment with DNase I, the fragmented and biotinylated cDNA was hybridized onto the Drosophila Genome 2.0 Array (Affymetrix) and data analyzed according to manufacturer’s instruction.
Quantitative real-time PCR, RT-PCR and Northern blot analysis of gene expression
Total RNA was extracted from fly heads using an Rneasy® Mini kit (Qiagen). Quantitative real-time PCR was performed using an ABI PRISM 7700 (Applied Biosystems) and Power SYBR Green PCR Master Mix (Applied Biosystems) as one-step reaction mixture supplemented with Superscript Reverse Transcriptase (LifeTechnologies) and sequence specific primers, according to the manufacturer’s protocol. Data were generated from three independent qPCR reactions and normalized with rp49 mRNA level as control.
For RT-PCR analysis, total RNA was extracted from fly heads using an RNeasy® Mini kit (Qiagen) and one-step RT-PCR was performed using an RT-PCR kit (Qiagen). For RT-PCR analysis of polysomal mRNAs, we prepared extracts from 150 fly heads and incubated the extracts in the presence of RNase inhibitor (protectRNA, Sigma) and 0.1 mg/ml cycloheximide or 1mM puromycin for 2 hours at 37°C and subsequently fractionated on 10–50% sucrose gradients. Fractions 2 to 7 from the bottom were pooled, RNA extracted, and one-step RT-PCR performed using the RT-PCR kit (Qiagen).
The primer sequences are: rp49 internal control 5′ primer: 5′ CCAAGGACTTCATCCGCCACC-3′ and 3′ primer: 5′-GCGGGTGCG CTTGTTCGATCC-3′; β-actin internal control 5′ primer: 5′-GCGGGAAATCGTGCG TGACATT-3′ and 3′ primer: 5′-GATGGAGTTT GAAGG TAGTTTCGTG-3′; Drosophila dp 5′ primer: 5′-GAGCTGATGGTGCCGCCG-3′ and 3′ primer: 5′-GGGC ATGTGGTTTCGCGG-3′; Drosophila e2f1 5′ primer: 5′-CAACACAAAATTGCC GCG-3′ and 3′ primer: 5′-GGCTGGGACTGCTTGGCG G-3′; renilla luciferase 5′ primer: 5′-ATGACTTCGAAAGTTTATGTCC-3′ and 3′ primer: 5′-CTCGAAG CGGCCGCTCTCTAG-3′; egfp 5′ primer: 5′-GGGCATCG ACTTCAAGGAGG-3′ and 3′ primer: 5′-TCGCGCTTCTCGTTGGGG-3′. Northern blot analysis was performed essentially as described elsewhere20.
Immunohistochemistry, Immunoprecipitation and Western blot analysis
Fly brains were dissected and fixed at 4°C for 2 hours in phosphate buffered saline (pH 7.2) containing 4% paraformaldehyde and 0.3 % Triton X-100. After three washing steps, primary antibodies against TH (1:1000, Immunostar) and FMRFamide (1:2000, gift of Chaoyang Zeng) were incubated at 4°C overnight in the presence of 5% normal goat serum. As secondary antibodies, we used Alexa Fluor® 568-conjugated goat anti-mouse and Alexa Fluor® 488-conjugated goat anti-rabbit (1:500, Molecular Probes). Immunofluorescence analysis was performed using a Carl Zeiss laser-scanning confocal microscope. A Nikon Eclipse 3000 fluorescence microscope was used for live imaging.
For immunoprecipitation, cell extracts of HEK293T and fly heads were homogenized in lysis buffer [50 mM Tris-HCl, pH7.4, 150 mM NaCl, 5 mM EDTA, 10% glycerol, 1% Triton X-100, 0.5 mM DTT, 60 mM β-glycerolphosphate, 1 mM sodium vanadate, 20 mM NaF, and complete inhibitor cocktail (Roche)]. After centrifugation at 16,000 g for 10 min, the supernatant was subjected to immunoprecipitation for 4 hours at 4°C using the indicated antibodies. Immunocomplexes were washed 3 times for 5 min each at 4°C. For immunoprecipitation of endogenous fly proteins, fly heads were homogenized in lysis buffer and fractionated using 10–50% sucrose gradients. Fractions 9 and 10 from the top were pooled and co-immunoprecipitation of dAgo1 and dLRRK performed. For co-immunoprecipitation in HEK293 cells, cells were co-transfected with the indicated plasmids. Forty-eight hours after transfection, cells were harvested in lysis buffer and subjected to immunoprecipitation as described above.
To prepare head extracts for Western blotting, 4 fly heads were directly homogenized in 10 μl/head of SDS sample buffer using a hand-held motorized homogenizer. After centrifugation at 16,000 g for 10 min, the extract was separated on 10% SDS-PAGE gel, transferred to Hybond P membrane, and detected with an Enhanced Chemiluminescence Plus Western Blotting Detection kit (Amersham). The following primary antibodies were used: Drosophila DP22 (Yun3, 1:4, mouse monoclonal supernatant, Dyson lab), Drosophila E2F123 (1:200, rabbit polyclonal, Wharton lab), Drosophila β-Tubulin (E7, mouse monoclonal, 1:20,000, University of Iowa Hybridoma Bank), EGFP (11E5, 1:1000, mouse monoclonal, Qbiogene, Inc), EGFP (3E6, 1:1000, mouse monoclonal, Qbiogene, Inc), FMRFamide (1:2000, rabbit polyclonal, Chaoyang Zeng), Flag (M2, WB: 1:3000, IHC: 1:500, Sigma), Drosophila Ago1 (WB: 1:500, IHC: 1:150, rabbit polyclonal, Abcam), human LRRK2 (1:3000, rabbit polyclonal, Novus), 4E-BP1 (1:1000, rabbit polyclonal, Cell Signaling), phospho-4E-BP1(Thr37/46) (1:1000, rabbit monoclonal, Cell Signaling), human Ago2 (11A9, rat monoclonal, Meister lab24).
DNA cloning, Transfection and Reporter Assays
Human LRRK2 cDNA was purchased from Origene and FLAG-tag sequence was added at the C-terminus. dLRRK cDNA was also inserted into the pcDNA5/FRT vector. Introduction of desired point mutations was performed using QuikChange II XL Site-directed mutagenesis kit (Stratagene). Kinase-dead forms of dLRRK/LRRK2 (3KD) were generated by introduction of triple mutations (K1781M, D1882A and D1912A in dLRRK; K1906M, D1994A and D2017A in hLRRK2). To make UAS-d4E-BP(TE) transgenic lines, the T37/46/86E mutant form of d4E-BP generated by site-directed mutagenesis was subcloned into the pUAST vector. To generate mammalian expression vectors encoding d4E-BP(WT), d4E-BP(TA), and d4E-BP(TE), the appropriate cDNA fragments were cleaved with EcoRI/XhoI from the pUAST vectors used to generate the corresponding transgenic animals and subcloned into the same restriction sites into the pcDNA3-Nmyc vector.
The cloning of 3′UTRs was performed by RT-PCR on total RNAs. The primers for amplifying Drosophila e2f1 3′UTR were 5′ primer: 5′-GCTCTAGAGAGGAGA CGTCCACGAACAC-3′; 3′ primer: 5′-ATAAGAATGCGGCCGCCTAATTTACAGA CAGTTCTAGCCC-3′. For amplifying dp 3′UTR, 5′ primer: 5′-ATAAGAATGCGGC CGCTTAAATTCTCTGTTTATCATATTTAC-3′ and 3′ primer: 5′-GCTCTAGAGCT GATGGTGCCGCCGTCATTGTGCC-3′ were used. The 3′UTRs were cloned into the XbaI and NotI restriction sites of pRL-TKlet7A (Addgene). Mutations at miRNA-binding sites were introduced by site-directed mutagenesis. To generate pUAST-EGFP-let-7-3′UTR, we first subcloned EGFP cDNA from pAc5.1A/GFP into pUAST, using the EcoRI and XbaI sites. Subsequently, we used pRL-TKlet7A as template and amplified the existing two incomplete let-7 binding sites as XbaI-NotI and NotI-XbaI fragments, with restriction sites added to the PCR primers to facilitate cloning. The two fragments were ligated and simultaneously inserted into the XbaI site of pUAST-EGFP. For the generation of pre-miRNA transgenic flies, genomic DNA containing the precursor sequences were amplified and subcloned into the appropriate restrictions sites of pUAST. The following primers were used for amplifying Drosophila pre-miRNAs: for pre-let-7, 5′ primer: 5′ GAAGATCTGCATT TTTAATATGATTTCT CCG-3′ and 3′ primer: 5′-GCTCTAGAGGTTGCATTTACAT ACTTTGGC-3′; for pre-miR-184, 5′ primer: 5′-GAAGATCTGTTGTTCAACACCTT CCCC-3′ and 3′ primer: 5′-GCTCT AGAGATTTTGCACACTGAGCAGCC-3′; and for pre-miR-10, 5′ primer: 5′-GAAG ATCTGACTAGAGGAAACTGCTA GCC-3′ and 3′ primer: 5′-CCGCTCGAGTTTC AGCTCCTA TATGAGGG-3′.
To generate Drosophila dp genomic transgenes, we performed a triple ligation of dp 5′UTR as a HindIII-SacI fragment and dp CDS as a SacI-XhoI fragment into the HindIII-XhoI sites of pcDNA3.1/myc-His. Mutations at nt82 of the 5′UTR and nt891 of the CDS (see Fig.s14a for the mutations made) were introduced using QuikChange II XL Site-directed mutagenesis kit (Stratagene). At the same time we amplified the previously cloned wild type and mutant dp 3′UTRs from pRL-TK-dp3′UTRwt and pRL-TK-dp3′UTR743 as XhoI-XbaI fragments (see Fig.3h for the mutations introduced into dp3′UTR). The dp promoter was amplified from genomic DNA of wild-type animals as BglII-HindIII fragment. The dp promoter (BglII-HindIII) and the dp 5′UTR-CDS (HindIII-XhoI) were ligated into the BamHI-XhoI sites of P{W30}. The dp 3′UTR was subsequently ligated into the XhoI-XbaI sites of P{W30}/dp promoter-5′UTR-CDS to generate P{W30}/dp promoter-5′UTR-CDS-3′UTR constructs containing the wild-type sequences (gDPwt) or with three mutations (Fig.3h, s14a) at potential let-7 miRNA binding sites (gDPmut). The following primers for Drosophila dp were used: dp promoter, 5′ primer: 5′-GAAGATCTCGAAATGTTTTCC GAAGGCCCCGGC-3′ and 3′ primer: 5′-CCCAAGCTTAGTCGCTATCGCACA CTGGTCGCGG-3′; dp 5′UTR, 5′ primer: 5′-CCCAAGCTTGTCG CACACAGCTGCTTACAACACTGCG- 3′ and the 3′primer: 5′-AGTCCGAGCTC TTTTCGGCCGCCGGAGATCAAGTG-3′; dp CDS, 5′primer: 5′-AGTCCGAGCTCATGGCGCATTCGACGGGCGGTACGG-3′ and 3′ primer: 5′-CCGCTCGAGTCAATCAATGTCGTCGTCCAGCTCG-3′; dp 3′UTR, 5′ primer: 5′-CCGCTCGAGGCTGATGGTGCCGCCGTCATTGTGCC-3′ and 3′ primer: 5′-GCTCTAGAACAGTTTTGGTATATTATATTATTAAGAGACAATTA CCC-3′. For the mutagenesis of dp 5′UTR and CDS, we used the following primers: for dp 5′UTR(nt82), 5′ primer: 5′-CAGCCTGCTTTGAATTTCCCACATGAAAACG-3′ and 3′ primer: 5′-CGTTTTCATGTGGGAAATTCAAAGCAGGCTG-3′; for CDS(nt891), 5′ primer: 5′-GCCTTCCCCGAATGAATCGATCCAGCTACCG-3′ and 3′ primer: 5′-CGGTAGCTGGATCGATTCATTCGGGGAAGGC-3′. All PCR reactions were performed with Pfu DNA polymerase (Stratagene) and PCR products confirmed by DNA sequencing.
To generate dp-TPcontrol and dp-TPlet-7 (Fig.s14b) constructs, we ligated the following annealed and restricted primer pairs as an EcoRI-XhoI fragment into the appropriate sites of pUAST vector. The primers used were: for dp-TPcontrol, 5′ primer: 5′-GGAATTCGGGTCGGATTGGGTGTGGAGTGGACTCGAGCGG-3′ and 3′ primer: 5′-CCGCTCGAGTCCACTCCACACCCAATCCGACCCGAATTCC-3′; for dp-TPlet-7, 5′ primer: 5′-GGAATTCTT GTTATTACTAATTAGGTAGGTTGCT CGAGCGG-3′ and 3′ primer: 5′-CCG CTCGAGCAACCTACCTAATTAGTAATAACAAGAA TTCC-3′. All insertions were confirmed by DNA sequencing.
For dual-luciferase reporter assays, HEK293T cells were seeded onto 12-well plates (3.7cm2/well) and for each well a mixture of 100 ng of RL reporter, 50 nM final concentration of siRNA-like duplex, and 20 ng of pGL3 control vector (Promega) was transfected using Lipofectamine (Invitrogen) following manufacturer’s instructions. Six hours after transfection, luciferase activities were determined using the Dual-Glo Luciferase Assay system (Promega). For reporter assays testing the effect of hLRRK2, we transfected 1.2 μg (per 3.7cm2 well) of the indicated hLRRK2 construct into HEK293T cells using Lipofectamine Plus (Invitrogen) and followed the protocol provided by the manufacture. Twenty-four hours after transfection, we prepared a second transfection of reporters and siRNA-like duplexes and measured luciferase activities as described above.
For renilla luciferase-let-7-3′UTR reporter assay, we transfected 0.75 μg/well (12 well plate) of empty pcDNA3.1(+) as control or plasmid DNA encoding d4E-BP(WT), d4E-BP(TA), and d4E-BP(TE) into m4E-BP1 depleted 4E-BP(−/−) MEF cells. 24 hours after the first transfection we performed a second transfection using 120 ng/well (12 well plate) m7GpppG-capped renilla-let-7(8x) (Addgene) mRNA plus 100 nM of indicated miRNAs or 120 ng/well (12 well plate) m7GpppG-capped renilla-let-7(8x) mRNA plus 0.5 μM of indicated antagomirs. All transfections were prepared with lipofectamine according to manufacturer suggested protocol. Renilla luciferase activities were measured 20 hours after the second transfection.
siRNA-like duplex and antagomir injection, in vitro transcription
The siRNA-like duplexes were formed by annealing the following sense and anti-sense oligonucleotides: let-7 sense 5′-UGAGGUAGUAGGUUGUAUAGU-3′; let-7 antisense 5′-UAUACAAC CUACUACCUUAUU-3′; dme-mir-124 sense 5′-UAAGGCACGCGGUGAAUGCC AAG-3′; dme-mir-124 antisense 5′-UGGCAUUCACCGCGUGCCUCAUU-3′; dme-miR-184* sense 5′-CCUUAUCAUUCUCUCGCCCCG-3′; dme-miR-184* antisense 5′-GGGCGAGAGAAUGAUAAUGUU-3′; dme-miR-10 sense 5′-ACCCUGUAGAUCC GAAUUUGU-3′; dme-miR-10 antisense 5′-AAAUUCGGAUCUACAGGAUUU-3′. The siRNA-like duplexes were dissolved in 5 mM KCl, 0.5 mM sodium phosphate (pH 7.5) buffer at a final concentration of 2.5 μM or 100 μM as indicated, and injected dorso-laterally into the cavity between the head cuticle and central brain complex. Five injections over an 11-day period were performed. Anti-let-7fS, anti-let-7mutfS, and anti-miR-184*fS were synthesized at Dharmacon. They contain the following sequences: anti-let-7fS 5′-ascsusasusascsasascscsusascsusascscsuscsa-3′, anti-let-7mutfS 5′-usgsasasususgs asasgsgsu sasgsasascscsgsgsu-3′ (introduced mutations are highlighted in bold), and anti-miR-184*fS 5′-csgsgsgs gscsgsasgsasgsasasusgsasusasasgsg-3′. The lower case letters represent 2′-OMe-modified nucleotides; subscript ‘s’ represents a phosphorothioate linkage. All antagomirs were injected five times over a 12-day period at the indicated concentration.
The renilla let-7 (8x) reporter mRNA was transcribed using the AmpliScribeTM T7-Flash transcription kit (Epicentre Biotechnologies). Briefly, psiCHECK2-let-7 8x [renilla-let-7(8x)] reporter plasmid DNA (Addgene) was linearized with BamHI, purified and transcribed. The manufacturer recommended protocol was modified by reducing GTP to 10 mM final concentration and adding 10 mM m7GpppG.
Drosophila genetics
To generate UAS-EGFP-let-7-3′UTR, UAS-pre-let-7, UAS-pre-miR-184*, UAS-pre-miR-10, UAS-dp-TPControl, UAS-dp-TPlet-7, gDPwt, gDPmut, UAS-dLRRK(R1069G), and UAS-4E-BP(TE) transgenic flies, the corresponding plasmids were injected into w− embryos following standard P-element mediated transformation protocol. P[PZ]e2f107172, Df(2R)BSC272, dp49Fk-1, P[lacW]ago1K08121, UAS-GFP, UAS-Hsap\MJD.tr-Q78, and GMR-reaper were obtained from the Bloomington Drosophila stock center. The UAS-eIF4E RNAi, UAS-dicer1 RNAi and UAS-dicer1 flies were obtained from the Vienna Drosophila RNAi Center (VDRC) and Dr. B. Dickson, respectively, UAS-rbf and UAS-e2f1 were requested from Dr. B. Edgar13, dcr-1Q1147X from Dr. R.W. Carthew25, lkb14A from Dr. D. St. Johnston26, UAS-4E-BP and UAS-4E-BP(LL)w from Dr. N. Sonenberg27, UAS-hLRRK2 from Dr. W. Smith28, ago2(414) from Dr. H. Siomi10, GMR-miR-14 from Dr. B. Hay16, and UAS-banD from Dr. S. Cohen29. The Δlet-7 mutant, Δlet-7 mutant rescued with a let-7 transgene, and let-7-C-Gal4 flies were described before20. All other general fly lines were obtained from Bloomington Drosophila stock center. 4E-BP1 depleted mouse embryonic fibroblast (MEF) line was obtained from Dr. N. Sonenberg30.
Oxidative stress and climbing activity assays
The survival rate of 10-day-old male flies (n=15–20) kept in a vial containing a tissue paper socked with 2 mM paraquat prepared in Schneider’s insect medium was measured as previously described6. To measure climbing activity, we aged flies at 25°C for 65 days unless otherwise indicated, transferred 5 to 23 flies into empty plastic vials and counted the number of flies climbing up to the top quarter of vertically standing vials within a 10-second period, after being bumped to the bottom. We repeated each measurement using three independent fly cohorts per genotype.
Supplementary Material
Acknowledgments
We are grateful to Drs. R.W. Carthew, B. Dickson, B. Edgar, D. St. Johnston, N. Sonenberg, W. Smith, Z. Zhang, H. Siomi, B. Hay, S. Cohen, the Vienna Drosophila RNAi Center and the Bloomington Drosophila Stock Center for fly stocks and cell line, and Bestgene Inc. for help with making gDPwt and gDPmut transgenics; Drs. N. J. Dyson, R. P. Wharton, C. Zeng, G. Meister and Y. Liu for antibodies; Drs. T. Tuschl, P. A. Sharp, and Y. Tomari for plasmids; Drs. P. Sarnow and R. Cevallos for advice on polysome preparation; Drs. S. Guo and Andy Fire for reading the manuscript. Special thanks go to W. Lee and G. Silverio for technical supports and members of the Lu lab for discussions. Supported by the NIH (R01AR054926, R01MH080378, and R21NS056878), McKnight, Beckman, and Sloan Foundations (B.L.) and Program for Young Researchers from Special Coordination Funds for Promoting Science and Technology commissioned by MEXT in Japan and Asahi Glass Foundation Research Grant (Y.I).
Footnotes
Author Contributions: S.G. designed and performed the experiments and wrote the manuscript; Y.I. performed the experiments; N.S. provided key reagents and advice; B.L. designed the experiments, wrote the paper, and provided funding.
References
- 1.Zimprich A, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44 (4):601. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Paisan-Ruiz C, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44 (4):595. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 3.Girling R, et al. A new component of the transcription factor DRTF1/E2F. Nature. 1993;362 (6415):83. doi: 10.1038/362083a0. [DOI] [PubMed] [Google Scholar]
- 4.Kim J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317 (5842):1220. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaefer A, et al. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204 (7):1553. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imai Y, et al. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27 (18):2432. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gloeckner CJ, et al. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum Mol Genet. 2006;15 (2):223. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- 8.West AB, et al. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102 (46):16842. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309 (5740):1573. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 10.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18 (14):1655. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johannes G, et al. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci U S A. 1999;96 (23):13118. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoglinger GU, et al. The pRb/E2F cell-cycle pathway mediates cell death in Parkinson’s disease. Proc Natl Acad Sci U S A. 2007;104 (9):3585. doi: 10.1073/pnas.0611671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93 (7):1183. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- 14.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318 (5848):271. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 15.Bilen J, et al. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol Cell. 2006;24 (1):157. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 16.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13 (9):790. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 17.Chan SP, Ramaswamy G, Choi EY, Slack FJ. Identification of specific let-7 microRNA binding complexes in Caenorhabditis elegans. RNA. 2008;14 (10):2104. doi: 10.1261/rna.551208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tain LS, et al. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci. 2009;12 (9):1129. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tettweiler G, et al. Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes Dev. 2005;19 (16):1840. doi: 10.1101/gad.1311805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22 (12):1591. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15 (2):185. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Du W, Vidal M, Xie JE, Dyson N. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 1996;10 (10):1206. doi: 10.1101/gad.10.10.1206. [DOI] [PubMed] [Google Scholar]
- 23.Asano M, Nevins JR, Wharton RP. Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev. 1996;10 (11):1422. doi: 10.1101/gad.10.11.1422. [DOI] [PubMed] [Google Scholar]
- 24.Rudel S, et al. A multifunctional human Argonaute2-specific monoclonal antibody. RNA. 2008;14 (6):1244. doi: 10.1261/rna.973808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117 (1):69. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 26.Martin SG, St Johnston D. A role for Drosophila LKB1 in anterior-posterior axis formation and epithelial polarity. Nature. 2003;421 (6921):379. doi: 10.1038/nature01296. [DOI] [PubMed] [Google Scholar]
- 27.Miron M, et al. The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat Cell Biol. 2001;3 (6):596. doi: 10.1038/35078571. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, et al. A Drosophila model for LRRK2-linked parkinsonism. Proc Natl Acad Sci U S A. 2008;105 (7):2693. doi: 10.1073/pnas.0708452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennecke J, et al. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113 (1):25. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 30.Le Bacquer O, et al. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest. 2007;117 (2):387. doi: 10.1172/JCI29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.