Abstract
Pharmacogenetic studies in DNA repair pathway have consistently demonstrated correlations between the XRCC1 Arg399Gln, XPD Lys751Gln and XPD Asp312Gln genotypes, previously associated with suboptimal DNA repair, and differential cancer treatment outcomes. We evaluated these polymorphisms and XPD haplotypes in adult de novo (n=214) and secondary (n=79) acute myeloid leukemia (AML) patients treated with cytarabine and anthracycline chemotherapy. Genotyping was performed by MALDI-TOF mass spectrometry. Logistic and proportional hazards regression models were used to evaluate relationships. Differential responses were observed in secondary, but not de novo, AML. Among secondary AML patients, the odds of achieving complete remission (CR) were higher for the XPD 312Asn/Asn (OR= 11.23; 95% CI, 2.23-56.63) and XPD 751Gln/Gln (OR= 7.07; 95% CI, 1.42-35.18) genotypes. The XPD diplotypes were coded as the combination of two of the following haplotypes: haplotype A=(Lys)751A/(Asp) 312G; B=(Gln)751C/(Asn)312A; C=(Lys)751A/(Asn)312A; and D=(Gln)751C/(Asp)312G. The BB diplotype was associated with CR attainment [OR=18.31; 95% CI: 2.08-283.57] and longer survival [HR=0.31; 95% CI: 0.14-0.73] compared to the referent AA diplotype. The XPD 751 CC, 312GA, 312AA genotypes and the XPD DC diplotype were also associated with longer overall survival (OS).Thus, XPD codon 312 and 751 variant genotypes and haplotypes containing at least one variant allele may predict better treatment responses. If validated, these findings could support stratification of chemotherapy in secondary AML.
Keywords: Acute Myeloid Leukemia (AML), secondary AML, pharmacogenetics/pharmacogenomics, DNA repair gene polymorphisms
Introduction
Numerous studies have shown that anti-neoplastic drug effects are subject to significant inter-individual variability. Based on an abundance of accumulated literature from twin studies, pre-clinical experimental data, clinical trials, biomarker-driven and epidemiologic studies, it is widely accepted that this variability is largely due to inherited genetic variation [1-3]. The aim of pharmacogenetics/genomics is to identify genetic predictors of treatment outcomes that would allow individualization of therapy for cancer patients.
Identification of pharmacogenomic determinants is especially needed for cancers with poor treatment outcomes. Acute myeloid leukemia (AML) in adult populations is associated with poor (approximately 20%) overall five-year relative survival (OS) rates. Despite continuing efforts to stratify AML patients according to prognostic host and tumor characteristics, including crucial cytogenetic and molecular markers, and despite new pharmaceutical agents, outcome remains suboptimal [4]. Overall prognosis for patients with secondary AML (sAML), including both AML following an antecedent hematologic disorder (AHD) and therapy-related acute mye-loid leukemia (t-AML), is even worse, with an average life expectancy of eight to ten months from diagnosis [5,6]. Cytosine arabinoside (ARA-C), an a nti metabolite, anthracyclines (daunorubicin, idarubicin) and etoposide are the most commonly used drugs in AML treatment. They have different mechanisms of action, including inhibition of DNA synthesis, formation of irreversible complexes with topoisomerases, generation of free radicals and intracellular oxi-dative damage, leading to accumulation of single- and double-srand DNA breaks.
Among the major DNA repair pathways, base excision repair (BER) is used for minor base alterations and non-bulky DNA adducts, while nucleotide excision repair (NER) is involved in fixing more complex DNA lesions such as bulky DNA adducts. The double-strand break repair (DSB) pathway repairs broken DNA strands by either homologous recombination (HR) or non-homologous end joining (NHEJ) [7,8]. It has been noted previously that acquired resistance to alkylating agents, anthracyclines and epipodophyllotoxins may develop as a result of enhanced DNA repair, NER repair in particular [9].
Overall, more than 130 genes have been associated with human DNA repair, and numerous studies have investigated associations between single nucleotide polymorphisms (SNPs) in nearly all DNA repair pathways and clinical outcomes [10]. Variant alleles associated with less proficient DNA repair activity may lead to decreased repair of DNA damage in malignant cells, and thereby improve response to chemotherapy. However, compromised repair activity may also lead to accumulation of DNA damage in normal cells, leading to more profound treatment-related toxicities in normal tissues, and may predispose towards secondary cancers.
Germline variations in DNA repair activity of genes including those encoding the X-ray cross-complementing group 1 (XRCC1) protein in the BER pathway and the XPD protein in the NER pathway, may have a significant impact on AML patients' response to treatment, including their induction chemotherapy outcomes, toxicity profiles and survival [10-13]. The XRCC1 codon 399Gln homozygous variant genotype has been found to decrease risk of both de novo and t-AML, and has been associated with both poor and favorable survival outcomes for other cancers [14-17]. The variant XPD allele Lys751Gln SNP was associated with increased risk of therapy-related AML (t-AML) as well as shorter disease-free survival (DFS) and OS for AML patients [18]. Additionally, AML chemotherapy is associated with a wide range of toxicities, the most serious of which are hematologic (myelo-suppression), hematologic (myelosuppression), gastrointestinal, infectious, dermatologic [9,19], and there are few data on impact of DNA repair gene polymorphisms on treatment-related toxicities [20-23].
We previously reported on associations between AML and variant XPD genotypes in a cohort of elderly AML patients in a Southwest Oncology Group (SWOG) clinical trial. Those with the XPD 751Gln/312Asp haplotype had significantly better odds of complete remission (CR), as well as significantly reduced risk of resistant disease [20]. Here we evaluated relationships between functional SNPs in XRCC1 Arg399Gln, ERCC2/(XPD) Lys751Gln and ERCC2/(XPD) Asp312Asn and patients' responses to induction chemotherapy, chemotherapy-induced toxicities and OS in adult AML patients treated at Roswell Park Cancer Institute (RPCI). The latter patient population was larger and included a larger number of sAML patients, and thus therapeutic outcomes in sAML were investigated in this work.
Materials and methods
Patients
The study population included 293 adult patients with de novo AML or sAML with all French-American-British subtypes (FAB) other than M3 or acute promyelocytic leukemia who were treated with ARA-C and anthracycline-based chemotherapy at RPCI between 1994 and 2006 and had cryopreserved vials of pre-treatment marrow or peripheral blood cells available in the RPCI Leukemia Tissue Bank. Among 79 (27%) patients with sAML, 57 had an AHD, 32 had t-AML, and 10 had both. Patients were predominantly Caucasian (90%). For remission induction, 45% received standard-dose ARA-C, daunorubicin and etoposide, 23% received high-dose ARA-C and idarubicin, and 32% were enrolled in clinical trials and received experimental agents, including BCL-2 inhibitor, P-glycoprotein (P-gp) modulators or arsenic trioxide in addition to ARA-C and an anthracycline. Sixty-six patients (23%) underwent hematopoietic stem cell transplantation (HSCT), either allogeneic (n=41) or autologous (n=25), as part of their AML therapy. Clinical information was obtained from the RPCI Leukemia Section Database and from patients' medical records. The study was approved by the RPCI Institutional Review Board.
Data collection and analysis
Data on patient and disease characteristics, toxicities of induction chemotherapy, and treatment outcomes were collected and evaluated according to standard procedures. De novo AML and sAML patient subgroups were defined by absence or presence of AHD prior to AML diagnosis or t-AML. Complete remission (CR) was defined by normal peripheral blood cell counts, including absolute neutrophil count of 1×109/L or more, platelet count of 100×109/L or more, and less than 5% blasts in the peripheral blood or bone marrow (BM). Relapse was defined as reappearance of more than 5 % leukemic blasts in the bone marrow not attributable to any other cause. Overall Survival (OS) was measured from the date of treatment until death from any cause, with observation censored on the date the patient was last known to be alive. Relapse-free survival (RFS) was measured from the date CR was achieved until the date of the first relapse of AML or death from any cause, with observation censored on the date of last contact for patients without report of relapse. Clinical toxicity data during remission induction chemotherapy were abstracted from medical records. Severity of toxicities was graded 1 (mild) through 5 (fatal) according to the NCI Common Toxicity Criteria, version 3.0 (http://ctep.cancer.gov/protocolDevelopment/electronic_applications/docs/ctcaev3.pdf). For the analyses described here, induction chemotherapy toxicities were combined into the following categories: genitourinary, gastrointestinal (including but not limited to: nausea/vomiting, diarrhea, oral mucositis, esophagitis), hepatic, pulmonary, cardiovascular, hemorrhage, dermatologic, neurologic, metabolic, hypersensitivity reactions and infectious complications. For each patient, the grade of toxicity assigned to an organ group was the maximum grade of all the specific toxicities within that group.
Gen otyping
DNA was extracted from cryopreserved bone marrow samples using Gentra PureGene DNA extraction kits (Minneapolis, MN) and geno-typed for SNPs in XRCC1 (rs25487) and XPD/ERCC2 (codon 751, rsl3181 and codon 312, rsl799793) by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS) [24]. Polymerase chain reaction amplification was performed using SNP-specific primers (XPD312: F: 5′- ACGTTGGAT-GACGACGC CCACCTGGCCAA - 3′ and R: 5′ -CGTTGGATGGG AGGCGGGAAAGGGACT - 3′; for XPD751: F: 5′- ACGTTGGATGAGCAGCTAGAATCA-GAGGAG - 3′ and R: 5′- ACGTTGGATGCACCAG-GAACCGTTTATG GC -3′ and for XRCC1: F: 5′-ACGTTGGAT GCAGGATAAGGAGCAGGGTTG - 3′ and R: 5′- ACG TTGGATGTAAGAGTGGGTGCTGG ACT -3′). Duplicate aliquots for approximately 10% of the samples were randomly distributed throughout the plates for quality control purposes. Controls for genotype and two ‘no template’ controls were also included on each plate. All genotyping results were reviewed manually for quality control.
Statistical analyses
Chi-Square test statistics with 1 degree of freedom were used to test for deviation from Hardy-Weinberg equilibrium for each polymorphism. The Estimation Haplotype (EH) genetic linkage utility program was used to evaluate possible linkage disequilibrium (LD) for the XPD SNPs located on chromosome 19q 13.2-13.3. XPD haplotypes were coded as previously reported 20: haplotype A=(Lys)751A/(Asp)312G; B=(Gln) 751C/(Asn)312A; C=(Lys)751A/(Asn)312A; and D=(Gln)751C/(Asp)312G. Data analysis was performed using SAS 9.1 software (SAS Institute, Cary, NC). Categorical distributions of patient, disease and outcome characteristics (age, sex, race, de novo vs. secondary AML onset, FAB type, WBC (white blood cell) count, treatment outcomes, toxicity grades) were compared among genotypes and haplotypes using Chi-square analysis, and Fisher's exact tests when appropriate, with calculation of Monte-Carlo estimates for the exact p-vlaues across nine haplotype groups.
Distributions of OS and RFS were estimated by Kaplan-Meier (KM) method and compared between genotypes/haplotypes by log-rank tests. Crude and adjusted unconditional logistic regression (LR) models were run for the assessment of relationships between CR, resistant disease (RD) and genotype/haplotypes. Odds ratios (OR) and their 95% confidence intervals (CI) for the associations between DNA repair genotypes/haplotypes and CR were adjusted for age and WBC count (continuous), as well as cytogenetic group and HSCT treatment. The prognostic cytogenetic groups were determined as favorable, intermediate, unfavorable as described by Byrd J. et al. from Cancer and Leukemia Group B results [25]; a few patients with not known karyotype belonged to the ‘unknown’ cytogenetic prognostic group. The OR and their 95% CI for the associations between DNA repair genoypes/haplotypes and CR were additionally adjusted for the type of induction chemotherapy when RD outcome was modeled. Hazard ratios (HR) and their 95% confidence intervals (CI) for the associations between DNA repair genotypes/haplotypes and OS were adjusted for age and WBC count (continuous), as well as cytogenetic group and HSCT in the Cox proportional hazards regression analysis. Analyses of treatment outcomes (CR, RD, OS) were stratified by de novo vs. secondary AML, except that statistical analysis between ungrouped XPD diplotypes and treatment outcomes was not stratified by AML onset due to small numbers. In order to make our current findings comparable to the ones from the previous study, in the logistic regression and Cox proportional hazards models the grouping of XPD haplotypes into diplotype groups was performed in the same manner [20]. We also grouped the haplotypes by the number of variant alleles in order to observe any variant allele dose response relationship and stratified the analysis by AML onset. Patients who underwent HSCT (n=41) were censored at the time of HSCT for all RFS analyses. Unconditional and polytomous logistic regression analysis of toxicities in relation to genotype/ha plotype group was performed. For the unconditional regression analysis, toxicity outcomes were dichotomized (grades 3-5 versus 0-2). For each gene and each organ group, poly-tomous logistic regression analyses were performed to test whether the distributions of highest toxicity grade varied among genotypes (each of these analyses excluded patients with only toxicities of unknown grade). These analyses treated each patient's maximum grade of a given type of toxicity as an ordinal response variable.
Results
Patient and disease characteristics are shown in Table 1. Median age was 65 years, with approximately equal numbers of men and women. Seventy-nine (27%) patients had sAML. Outcomes of remission induction therapy in the 293 patients included CR in 157 (53%), RD in 83 (28%), death in 50 (17%) and unknown due to lack of BM evaluation in three. Most patients (256, or 87%) experienced grade 3-5 toxicities of one or more organ systems during remission induction therapy. The most common grade 3-5 toxicities were infectious in 151 (52%) patients, pulmonary in 30%, cardiac in 27%, metabolic in 26%, dermatologic in 23%, neurologic in 17%, genitourinary in 16%, diarrhea in 15% and mucositis in 14%. Outcomes correlated with previously established adverse prognostic factors for AML, including unfavorable cytogenetic group, secondary AML and age 60 years and greater 26-28 (log rank p<0.0001). OS and RFS did not differ by race (White non-Hispanic vs. others), gender, or FAB types (data not shown), but OS differed significantly for standard versus high-dose cytarabine induction regimens with or without inclusion of investigational drugs (p<0.0001) and HSCT (p<0.0001).
Table 1.
Summary of selected patient and disease characteristics
| Characteristic | Description | No. | % |
|---|---|---|---|
| Sex | Female | 135 | 46 |
| Male | 158 | 54 | |
| Race/Ethnicity | Asian | 3 | 1 |
| Black | 15 | 5 | |
| Hispanic, NOS | 6 | 2 | |
| White, non-Hispanic | 264 | 90 | |
| Other/unknown | 5 | 2 | |
| AML Onset | de novo | 214 | 73 |
| Secondary | 79 | 27 | |
| Cytogenetic | Favorable | 20 | 7 |
| Risk Group | Intermediate | 149 | 51 |
| Unfavorable | 96 | 33 | |
| Unknown | 28 | 10 | |
| FAB Type | M0 | 17 | 6 |
| M1 | 65 | 22 | |
| M2 | 125 | 43 | |
| M4 | 51 | 17 | |
| M5 | 13 | 4 | |
| M6 | 12 | 4 | |
| M7 | 2 | 1 | |
| Other AML | 8 | 3 | |
| Median | Min – Max | ||
| Age, yrs | 65 | 20–85 | |
| Marrow Blasts (%) | 62 | 0–98 | |
| WBC (X1000/μL) | 19.03 | 0.43–555 | |
Final sample sizes were 289 for the XRCCl SNP, 288 for the XPD codon 751 SNP, 277 for the XPD codon 312 SNP and 273 patients for XPD haplotypes due to DNA amplification failure. All genotypes were in Hardy Weinberg equilibrium. DNA repair genotype/haplotypes did not differ significantly by age, WBC count or cytogenetic risk groups, remission induction outcomes or toxicity grades (data not shown). Significant differences were observed for XPD312 genotypes by AML onset (p=0.02), and for XPD312 and XPD751 distributions by race (p=0.01 and p=0.003, respectively), though the latter observations may be due to small numbers of non-Caucasian patients.
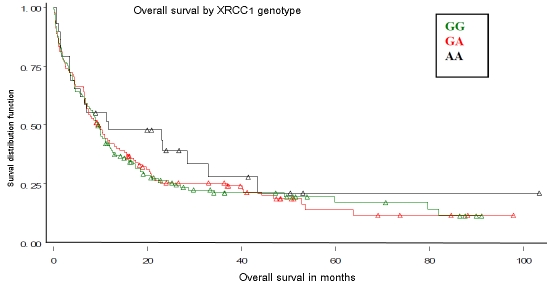
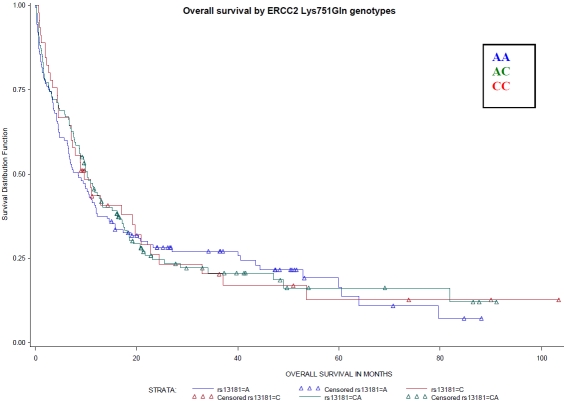
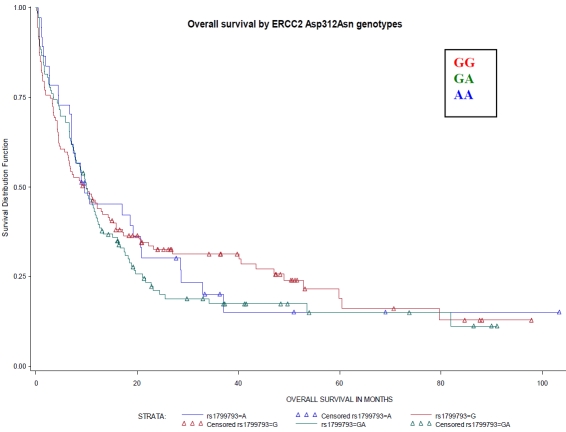
There were no significant associations between XRCC1 genotypes or XPD diplotypes and OS (Figures 1 and 2) or RFS (data not shown). Though we previously found that the DA XPD diplotype was associated with better treatment outcomes, OS did not differ among XPD haplotype group categories including DA as a separate diplotype group (Figure 3).
Figure 1.
Overall survival by DNA repair XRCC1 Arg399Gln genotypes (rs25487). Referent genotype – GG, variant genotype – AA. No differences by XRCC1 genotypes were observed (Log-Rank P-value=0.58).
Figure 2.
Overall survival by ERCC2 Lys751Gln genotypes (rs13181). Referent genotype – AA, variant genotype – CC. No differences by XPD, codon 751, genotypes were observed (Log-Rank P-value=0.93).
Figure 3.
Overall survival by ERCC2 Asp312Asn genotypes (rsl799793). Referent genotype – GG, variant genotype – AA. No differences by XPD, codon 312, genotypes were observed (Log-Rank P-value=0.74).
Table 2 displays associations between CR and OS and XRCC1 and XPD genotypes. Significantly increased odds of CR were associated with one or both variant XPD751 genotypes (OR 2.05; 95% CI, 1.20-3.52 for AC+CC). Patients with sAML and any of the variant genotypes had highly significant greater odds of achieving CR (OR=8.42 [2.08-34.01] for AC+CC). Similar trends were observed for the XPD312 genotypes: in the overall AML RPCI cohort the odds of achieving CR in homozygous variant AA genotype carriers were 4 times higher than in those with the common GG genotype (OR 4.34, [1.80-10.48], and within the sAML subgroup, odds were 11-fold higher (OR 11.23, 95% CI, 2.23-56.63). Variant XPD312 (GA, AA, GA+AA) and XPD751 (CC, AA+CC) genotypes predicted significantly lower hazards of death among patients with sAML, with OR 0.39 [0.20-0.79] and OR 0.46 [0.23-0.88] for the XPD312 Asn/Asn (or AA) genotype and XPD751 Gln/Gln (or CC) genotypes, respectively. In the sAML subgroup there were inverse associations between risk of RD and the AC XPD751 gene polymorphism (OR=0.28 [95% CI, 0.08-0.88]), but no other significant associations with RD were seen (data not shown).
Table 2.
Associations between Complete Remission, Overall Survival and DNA Repair Gene Polymorphisms
| Complete Remission (CR) * | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | CR, Overall | CR in de novo AML | CR in Secondary AML | ||||||||
| Polymorphism | N | OR | CI | N | OR | CI | N | OR | CI | ||
| XRCC1 | |||||||||||
| Arg399Gln: | GG** | 137 | 73 | 1.00 | – | 106/64 | 1.00 | – | 31/9 | 1.00 | – |
| GA | 123 | 66 | 1.05 | 0.60–1.83 | 84/53 | 1.01 | 0.53–1.95 | 39/13 | 1.10 | 0.35–3.46 | |
| AA | 29 | 15 | 1.03 | 0.42– 2.50 | 20/10 | 0.65 | 0.23–1.86 | 9/5 | 2.56 | 0.44–14.79 | |
| GA or AA | 152 | 81 | 1.05 | 0.62–1.77 | 104/63 | 0.92 | 0.50–1.70 | 48/18 | 1.29 | 0.43–3.86 | |
| Total | 289 | 154 | |||||||||
| XPD | |||||||||||
| Lys751Gln: | AA** | 125 | 57 | 1.00 | – | 93/52 | 1.00 | – | 32/5 | 1.00 | – |
| AC | 118 | 68 | 2.00 | 1.12–3.56 | 90/55 | 1.39 | 0.72–2.67 | 28/13 | 9.52 | 2.10–43.12 | |
| CC | 45 | 27 | 2.22 | 1.00– 4.91 | 27/19 | 1.68 | 0.61–4.60 | 18/8 | 7.07 | 1.42–35.18 | |
| AC or CC | 163 | 95 | 2.05 | 1.20–3.52 | 117/74 | 1.44 | 0.78–2.68 | 46/21 | 8.42 | 2.08–34.01 | |
| Total | 288 | 152 | |||||||||
| XPD | |||||||||||
| Asp312Asn: | GG** | 127 | 60 | 1.00 | – | 93/53 | 1.00 | – | 34/7 | 1.00 | – |
| GA | 113 | 64 | 1.54 | 0.86–2.75 | 90/54 | 1.24 | 0.64–2.40 | 23/10 | 3.39 | 0.89–12.85 | |
| AA | 37 | 26 | 4.34 | 1.80–10.48 | 21/16 | 2.97 | 0.95–9.23 | 16/10 | 11.23 | 2.23–56.63 | |
| GA or AA | 150 | 90 | 1.98 | 1.15–3.40 | 111/70 | 1.47 | 0.79–2.75 | 39/20 | 5.19 | 1.54–17.51 | |
| Total | 277 | 150 | |||||||||
| Overall Survival (OS) | |||||||||||
| Total | OS | OS in de novo AML | OS in Secondary AML | ||||||||
| Polymorphism | N | Deaths | HR | CI | Deaths | HR | CI | Deaths | HR | CI | |
| XRCC1 | |||||||||||
| Arg399Gln: | GG | 137 | 107 | 1.00 | – | 78 | 1.00 | – | 29 | 1.00 | – |
| GA | 123 | 97 | 0.96 | 0.72–1.27 | 62 | 1.05 | 0.75–1.48 | 35 | 0.84 | 0.48–1.46 | |
| AA | 29 | 20 | 0.77 | 0.48–1.25 | 12 | 0.89 | 0.48–1.64 | 8 | 0.56 | 0.23–1.35 | |
| GA or AA | 152 | 117 | 0.92 | 0.70–1.20 | 74 | 1.02 | 0.74–1.40 | 43 | 0.78 | 0.46–1.33 | |
| Total | 289 | 224 | 152 | 72 | |||||||
| XPD | |||||||||||
| Lys751Gln: | AA | 125 | 99 | 1.00 | – | 69 | 1.00 | – | 30 | 1.00 | – |
| AC | 118 | 91 | 0.86 | 0.65–1.15 | 65 | 1.00 | 0.71–1.41 | 26 | 0.59 | 0.34–1.04 | |
| CC | 45 | 35 | 0.81 | 0.54–1.21 | 20 | 1.15 | 0.69–1.92 | 15 | 0.46 | 0.24–0.88 | |
| AC or CC | 163 | 126 | 0.85 | 0.65–1.11 | 85 | 1.03 | 0.75–1.42 | 41 | 0.53 | 0.32–0.88 | |
| Total | 288 | 225 | 154 | 71 | |||||||
| XPD | |||||||||||
| Asp312Asn: | GG | 127 | 94 | 1.00 | – | 62 | 1.00 | – | 32 | 1.00 | – |
| GA | 113 | 91 | 1.01 | 0.76–1.36 | 71 | 1.32 | 0.93–1.86 | 20 | 0.48 | 0.26–0.86 | |
| AA | 37 | 29 | 0.77 | 0.50–1.19 | 15 | 1.09 | 0.61–1.93 | 14 | 0.39 | 0.20–0.79 | |
| GA or AA | 150 | 120 | 0.95 | 0.72–1.25 | 86 | 1.27 | 0.91–1.77 | 34 | 0.44 | 0.26–0.75 | |
| Total | 277 | 214 | 148 | 66 | |||||||
CR and OS estimates are adjusted for the following covariates: age, cytogenetic group, white blood cell count, HSCT
Referent genotype
Associations between CR, RD, OS and XPD haplotypes are presented in Table 3. Because of the limited number of CRs in each of the seven ungrouped XPD diplotypes (and CC and DD were excluded due to their small numbers), these analyses were not stratified by AML onset. The AA diplotype (with all common genotype alleles for both XPD polymorphisms) was used as the referent category. The BB diplotype (containing all variant alleles of both XPD genotypes) was the best predictor of CR achievement (OR=4.53, 95% CI, 1.60-12.87). When a joint analysis was performed grouping haplotypes by number of variant alleles, no clear effect was observed. However, the DC diplotype with two variant alleles was associated with a 2-fold increase in CR odds (OR=2.00, 95% CI 1.04-3.83), and patients with the BB diplotype (with four variant alleles) had the highest CR odds, as mentioned above. Signficantly higher CR odds were not observed in patients with the DA diplotype and/or the D haplotype-containing diplotype group (DA, DB, DC, DD), and XPD haplotypes were not associated with RD.
Table 3.
Associations between Treatment Outcomes and XPD Haplotypes*
| Complete Response | Resistant Disease | Overall Survival | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| XPD Haplotype | Totla N | |||||||||
| N | OR | CI | N | OR | CI | N of Deaths | HR | CI | ||
| AA** | 105 | 48 | 1.00 | – | 31 | 1.00 | – | 80 | 1.00 | – |
| AC | 11 | 4 | 0.84 | 0.19–3.63 | 6 | 2.35 | 0.61–9.09 | 10 | 1.21 | 0.62–2.36 |
| BB | 24 | 17 | 4.53 | 1.60–12.87 | 6 | 0.66 | 0.23–1.88 | 19 | 0.71 | 0.42–1.19 |
| BC | 10 | 6 | 3.58 | 0.81–15.81 | 2 | 0.40 | 0.08–2.09 | 8 | 0.92 | 0.44–1.92 |
| DA | 18 | 10 | 2.05 | 0.64–6.54 | 4 | 0.63 | 0.19–2.15 | 12 | 0.74 | 0.40–1.36 |
| DB | 16 | 9 | 0.82 | 0.21–3.17 | 4 | 1.14 | 0.31–4.14 | 12 | 1.15 | 0.62–2.14 |
| DC† | 85 | 50 | 2.00 | 1.04–3.83 | 22 | 0.73 | 0.37–1.45 | 68 | 0.90 | 0.65–1.25 |
| AA (‘0’ variant alleles) | 105 | 48 | 1.00 | – | 31 | 1.00 | – | 80 | 1.00 | – |
| AC/DA (1 variant allele) | 29 | 14 | 1.45 | 0.57–3.70 | 10 | 1.09 | 0.43–2.73 | 22 | 0.90 | 0.56–1.45 |
| CC/DD/DC (2 variant alleles) | 89 | 52 | 2.00 | 1.05–3.79 | 24 | 0.75 | 0.38–1.48 | 71 | 0.89 | 0.65–1.24 |
| BC/DB (3 variant alleles) | 26 | 15 | 1.62 | 0.57–4.59 | 6 | 0.71 | 0.25–2.05 | 20 | 1.04 | 0.63–1.72 |
| BB (4 variant alleles) | 24 | 17 | 4.56 | 1.60–12.95 | 6 | 0.65 | 0.23–1.86 | 19 | 0.71 | 0.42–1.18 |
| AA** | 105 | 48 | 1.00 | – | 31 | 1.00 | – | 80 | 1.00 | – |
| DA | 18 | 10 | 2.05 | 0.64–6.53 | 4 | 0.63 | 0.19–2.16 | 12 | 0.74 | 0.40–1.36 |
| DC | 85 | 50 | 2.00 | 1.04–3.83 | 22 | 0.72 | 0.36–1.44 | 68 | 0.90 | 0.65–1.25 |
| Other | 65 | 38 | 2.20 | 1.08–4.48 | 20 | 0.91 | 0.45–1.86 | 52 | 0.90 | 0.63–1.30 |
| AA/AC/BB/BC/CC | 151 | 76 | 1.00 | – | 45 | 1.00 | – | 118 | 1.00 | – |
| DA/DB/DC/DD | 122 | 70 | 1.27 | 0.74–2.16 | 32 | 0.84 | 0.48–1.49 | 94 | 0.95 | 0.72–1.25 |
| Total | 273 | 146 | 77 | 212 | ||||||
Haplotypes: A=Lys751A/Asp312G; B=Gln751C/Asn312A; C=Lys751A/Asn312A; D=Gln751C/Asp312G; Haplotype frequencies: A=0.424; B=0.121; C=0.211; D=0.242
CR and OS estimates are adjusted for the following covariates: age (continuous), AML onset (de novo vs. secondary), cytogenetic group (favorable, intermediate, unfavorable, unknown), peripheral white cell count (continuous), type of HSCT (allogenic, autologous, none); RD estimates are adjusted for: age, AML onset, cytogenetic group, induction therapy, HSCT
Referent haplotype or haplotype category OR = estimated odds ratio, HR = estimated hazard ratio, CI = 95% confidence interval
Haplotypes CC (n=1) and DD (n=3) were excluded from this analysis because of small numbers OR = estimated odds ratio, HR = estimated hazard ratio, CI = 95% confidence interval.
Table 4 presents results for XPD diplotype groups, stratified by AML onset. In these analyses, the DC and BB diplotypes emerged as predictors of better CR and OS: The AA group served as the referent and consisted of all common XPD alleles. As mentioned above, there was a 4.56-fold increase in odds of CR in the BB group, likely associated with deficient DNA repair. The associations were not significant in de novo AML, but among sAML patients an 18.31-fold higher CR rate [OR=18.31, 95% CI, 2.08-283.57] was observed in the BB group; due to small numbers (7 patients with CR in BB diplotype category) the exact conditional probabilities were calculated in the logistic regression model for these estimates. Overall Survival in the BB group was also longer among sAML patients only [HRO.31, 95% CI, 0.14-0.73]. Compared to the AA diplotype, a significantly higher CR rate was also observed in the DC//CC/DD group, but results were only significant in sAML (OR=6.35, 95% CI, 1.14-47.16). Since there were only one patient with CC diplotype (who had de novo AML and achieved CR) and three patients with DD diplotype (two with sAML who failed to achieve CR and one with de novo onset who achieved CR), the DC diplotype most likely contributes to these findings. OS in the DC/CC/DD group was also longer among sAML patients only [HR=0.44, 95% CI, 0.23-0.87]. No evident associations were noted for RD (results not shown).
Table 4.
Associations between treatment outcomes and XPD hapiotypes, effect of AML onset
| CR in de novo AML | CR in secondary AML | ||||||
|---|---|---|---|---|---|---|---|
| Haplotype group | Total N | ||||||
| N1* | OR/HR | CI | N1* | OR/HR | CI | ||
| AA/AC/BB/BC/CC | 151 | 62 | 1.00 | – | 14 | 1.00 | – |
| DA/DB/DC/DD | 122 | 58 | 1.08 | 0.58–2.02 | 12 | 1.92 | 0.64–5.75 |
| Total | 273 | 120 | 26 | ||||
| AA** | 105 | 43 | 1.00 | – | 5† | 1.00 | – |
| DA | 18 | 8 | 1.25 | 0.34–4.64 | 2† | 10.55 | 0.45–287.48 |
| DC | 85 | 41 | 1.42 | 0.68–2.45 | 9†† | 7.73 | 1.36–59.24 |
| Other | 65 | 28 | 1.79 | 0.75–4.26 | 10†† | 5.05 | 0.98–34.50 |
| AA (‘0’ variant alleles) | 105 | 43 | 1.00 | – | 5† | 1.00 | – |
| AC/DA (1 variant allele) | 29 | 12 | 1.13 | 0.39–3.29 | 2† | 4.32 | 0.24–72.29 |
| CC/DD/DC (2 variant alleles) | 89 | 43 | 1.52 | 0.73–3.13 | 9†† | 6.35 | 1.14–47.16 |
| BC/DB (3 variant alleles) | 26 | 12 | 1.28 | 0.34–4.78 | 3† | 2.99 | 0.31–28.12 |
| BB (4 variant alleles) | 24 | 10 | 2.54 | 0.70–9.21 | 7†† | 18 31 | 2 08–283 57 |
| OS in de novo AML | OS in secondary AML | ||||||
| AA/AC/BB/BC/CC | 151 | 77 | 1.00 | – | 41 | 1.00 | – |
| DA/DB/DC/DD | 122 | 70 | 1.06 | 0.77–1.47 | 24 | 0.70 | 0.41–1.21 |
| Total | 273 | 147 | 65 | ||||
| AA** | 105 | 55 | 1.00 | – | 25 | 1.00 | – |
| DA | 18 | 7 | 0.69 | 0.31–1.52 | 5 | 0.62 | 0.21–1.83 |
| DC | 85 | 54 | 1.15 | 0.79–1.68 | 14 | 0.41 | 0.20–0.83 |
| Other | 65 | 31 | 1.33 | 0.79–2.23 | 21 | 0.61 | 0.30–1.23 |
| AA (‘0’ variant alleles) | 105 | 55 | 1.00 | – | 25 | 1.00 | – |
| AC/DA (1 variant allele) | 29 | 14 | 0.94 | 0.52–1.69 | 8 | 0.60 | 0.25–1.45 |
| CC/DD/DC (2 variant alleles) | 89 | 55 | 1.13 | 0.78–1.65 | 16 | 0.44 | 0.23–0.87 |
| BC/DB (3 variant alleles) | 26 | 12 | 1.40 | 0.73–2.68 | 8 | 0.58 | 0.25–1.35 |
| BB (4 variant alleles) | 24 | 11 | 1.09 | 0.56–2.11 | 8 | 0.31 | 0.14–0.73 |
N1=number of CR/deaths with certain haplotype
models adjusted for age, cytogenetic group, AML onset (if not stratified), HSCT, WBC count
Exact OR and CR are calculated
p-value=0.02 (DC), 0.05 (Other), 0.03 (CC/DD/DC), 0.004 (BB) accordingly
Tables 5 and 6 present polytomous regression results of toxicity categories in relation to XRCC1 and XPD genotypes/haplotypes. Analyses of toxicities by haplotypes were limited to combined haplotype categories due to small numbers. As shown in Table 5, the homozygote variant XPD genotypes C751C and A312A were both associated with significantly reduced risks of nausea/vomiting [OR=0.47 (.23-0.94) and OR=38 (0.17-0.81), respectively]. The BB diplotype was associated with a three-fold reduction in risk of nausea/vomiting (OR 0.31; 95% CI, 0.11-0.79), and the heterozygote XPD genotypes A751C and G312A were both associated with increased risk of infectious complications [OR=1.71 (1.05-2.78) and OR=1.68 (1.03-2.76), respectively]. The DC diplotype group was also associated with increased risk of infectious complications (OR=1.77, 95%CI: 1.02-3.11). The XRCC1 variant genotypes were associated with metabolic and pulmonary toxicities (Table 6).
Table 5.
Results of polytomous logistic regression (LR) analysis on DNA repair genotypes/haplotypes and selected toxicity categories†
| Gene | Genotype | Patients | Nausea Vomiting | Oral Mucositis | Diarrhea | GI toxicity* | Skin Toxicity | Infections | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | % | OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | ||
| XRCC1 | GG | 137 | 46.8 | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – |
| GA | 123 | 41.9 | 0.90 | 0.56–1.44 | 0.82 | 0.52–1.27 | 0.88 | 0.56–1.38 | 0.88 | 0.56–1.37 | 0.90 | 0.57–1.43 | 0.91 | 0.58–1.44 | |
| AA | 29 | 9.9 | 0.97 | 0.45–2.04 | 1.18 | 0.57–2.44 | 0.85 | 0.41–1.76 | 0.96 | 0.47–1.93 | 0.83 | 0.39–1.71 | 0.76 | 0.35–1.62 | |
| GA or AA | 152 | 0.91 | 0.58–1.43 | 0.89 | 0.58–1.35 | 0.87 | 0.57–1.34 | 0.89 | 0.59–1.36 | 0.89 | 0.58–1.37 | 0.88 | 0.57–1.36 | ||
| XPD | AA** | 125 | 42.7 | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – |
| Lys751Gln | AC | 118 | 40.3 | 0.92 | 0.56–1.49 | 0.77 | 0.48–1.22 | 0.85 | 0.53–1.35 | 0.77 | 0.49–1.23 | 0.98 | 0.60–1.58 | 1.71 | 1.05–2.78 |
| CC | 45 | 15.4 | 0.47 | 0.23–0.94 | 1.20 | 0.64–2.24 | 0.73 | 0.37–1.41 | 0.94 | 0.50–1.77 | 1.33 | 0.70–2.50 | 1.22 | 0.64–2.30 | |
| AC or CC | 163 | 0.77 | 0.49–1.21 | 0.87 | 0.57–1.34 | 0.82 | 0.53–1.26 | 0.82 | 0.53–1.25 | 1.07 | 0.69–1.67 | 1.54 | 0.99–2.43 | ||
| XPD | GG** | 127 | 43.3 | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – |
| Asp312Asn | GA | 113 | 38.6 | 1.04 | 0.63–1.72 | 1.09 | 0.68–1.75 | 1.01 | 0.63–1.62 | 1.11 | 0.70–1.78 | 0.91 | 0.56–1.49 | 1.68 | 1.03–2.76 |
| AA | 37 | 12.6 | 0.38 | 0.17–0.81 | 0.99 | 0.51–1.90 | 0.64 | 0.31–1.33 | 0.83 | 0.42–1.63 | 1.57 | 0.79–3.12 | 1.39 | 0.70–2.76 | |
| GA or AA | 150 | 0.81 | 0.51–1.29 | 1.06 | 0.69–1.64 | 0.91 | 0.58–1.41 | 1.03 | 0.67–1.59 | 1.05 | 0.67–1.65 | 1.60 | 1.02–2.53 | ||
| XPD haplotype | All other | 151 | 55.3 | 1 | – | 1 | – | ||||||||
| D-group | 122 | 44.7 | 1.23 | 0.76–1.97 | 0.87 | 0.56–1.76 | 0.97 | 0.61–1.52 | 0.92 | 0.59–1.43 | 0.86 | 0.54–1.35 | 1.26 | 0.80–2.00 | |
| AA | 105 | 38.5 | 1 | – | 1 | – | |||||||||
| DA | 18 | 6.6 | 0.87 | 0.33–2.26 | 0.86 | 0.34–2.12 | 0.79 | 0.31–1.93 | 0.86 | 0.36–2.11 | 0.59 | 0.22–1.48 | 0.71 | 0.26–1.86 | |
| DC | 85 | 31.1 | 1.08 | 0.61–1.89 | 0.86 | 0.50–1.46 | 0.92 | 0.54–1.57 | 0.91 | 0.54–1.54 | 0.99 | 0.57–1.72 | 1.77 | 1.02–3.11 | |
| Other | 65 | 23.8 | 0.49 | 0.26–0.91 | 1.26 | 0.72–2.21 | 0.79 | 0.43–1.42 | 1.00 | 0.56–1.77 | 1.20 | 0.67–2.15 | 1.38 | 0.77–2.47 | |
| AA | 105 | 38.5 | 1 | – | 1 | – | |||||||||
| DA/AC | 29 | 10.6 | 0.85 | 0.39–1.86 | 1.24 | 0.60–2.59 | 0.99 | 0.46–2.08 | 1.21 | 0.58–2.53 | 0.64 | 0.29–1.39 | 0.94 | 0.42–2.04 | |
| DD/CC/DC | 89 | 32.6 | 1.06 | 0.61–1.85 | 0.82 | 0.48–1.40 | 0.91 | 0.54–1.55 | 0.85 | 0.50–1.43 | 1.08 | 0.63–1.86 | 1.73 | 1.00–3.01 | |
| DB/BC | 26 | 9.5 | 0.57 | 0.24–1.31 | 1.19 | 0.52–2.72 | 0.90 | 0.39–2.05 | 0.97 | 0.42–2.24 | 0.82 | 0.36–1.83 | 1.50 | 0.66–3.39 | |
| BB | 24 | 8.8 | 0.31 | 0.11–0.79 | 1.21 | 0.56–2.61 | 0.53 | 0.21–1.27 | 0.92 | 0.42–2.07 | 1.60 | 0.69–3.68 | 1.30 | 0.57–2.93 | |
ORs and 95% CI from polytomous logistic regression models are adjusted for induction treatment regimen and age
GI toxicity includes: nausea/vomiting, oral mucositis, diarrhea, esophagitis/dysphagia, other
referent genotype
Table 6.
Results of polytomous LR analysis on DNA repair genotypes/haplotypes and selected toxicity categories (cont.)†
| Gene | Genotype | Patients | Cardiac | Pulmonary | Genitourinary | Neurologic | Metabolic | Overal Toxicity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | % | OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | ||||
| XRCC1 | GG | 137 | 46.8 | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – |
| GA | 123 | 41.9 | 1.42 | 0.88–2.28 | 1.78 | 1.09–2.94 | 1.20 | 0.67–2.14 | 1.13 | 0.67–1.89 | 1.11 | 0.71–1.73 | 1.24 | 0.79–1.97 | |
| AA | 29 | 9.9 | 1.47 | 0.69–3.05 | 1.71 | 0.78–3.69 | 0.41 | 0.09–1.31 | 1.02 | 0.42–2.30 | 2.38 | 1.15–4.96 | 1.00 | 0.46–2.15 | |
| GA or AA | 152 | 1.43 | 0.92–2.24 | 1.77 | 1.11–2.85 | 1.03 | 0.59–1.81 | 1.10 | 0.68–1.80 | 1.28 | 0.84–1.95 | 1.19 | 0.77–1.85 | ||
| XPD | AA** | 125 | 42.7 | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – |
| Lys751Gln | AC | 118 | 40.3 | 1.24 | 0.76–2.02 | 1.05 | 0.64–1.75 | 0.94 | 0.51–1.73 | 1.20 | 0.70–2.07 | 0.93 | 0.58–1.48 | 1.03 | 0.63–1.67 |
| CC | 45 | 15.4 | 1.04 | 0.53–1.99 | 1.01 | 0.49–2.02 | 0.76 | 0.30–1.77 | 1.21 | 0.58–2.47 | 0.69 | 0.37–1.27 | 0.56 | 0.29–1.09 | |
| AC or CC | 163 | 1.18 | 0.75–1.85 | 1.04 | 0.65–1.67 | 0.89 | 0.50–1.58 | 1.20 | 0.73–1.99 | 0.85 | 0.55–1.31 | 0.87 | 0.56–1.37 | ||
| XPD | GG** | 127 | 43.3 | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – |
| Asp312Asn | GA | 113 | 38.6 | 1.08 | 0.65–1.78 | 0.91 | 0.55–1.52 | 0.73 | 0.37–1.40 | 1.02 | 0.59–1.76 | 0.88 | 0.55–1.42 | 1.05 | 0.64–1.71 |
| AA | 37 | 12.6 | 0.95 | 0.46–1.90 | 0.79 | 0.36–1.68 | 1.06 | 0.43–2.43 | 1.25 | 0.58–2.62 | 0.86 | 0.45–1.65 | 0.70 | 0.35–1.39 | |
| GA or AA | 150 | 1.04 | 0.66–1.65 | 0.88 | 0.55–1.42 | 0.81 | 0.45–1.47 | 1.07 | 0.65–1.78 | 0.88 | 0.57–1.35 | 0.94 | 0.60–1.49 | ||
| XPD haplotype | All other | 151 | 55.3 | ||||||||||||
| D-group | 122 | 44.7 | 1.08 | 0.68–1.73 | 1.01 | 0.62–1.63 | 0.76 | 0.41–1.39 | 0.94 | 0.56–1.57 | 0.79 | 0.50–1.24 | 0.95 | 0.60–1.51 | |
| AA | 105 | 38.5 | |||||||||||||
| DA | 18 | 6.6 | 1.12 | 0.44–2.77 | 0.68 | 0.24–1.78 | 1.56 | 0.50–4.44 | 1.08 | 0.37–2.93 | 1.26 | 0.50–3.15 | 0.68 | 0.26–1.78 | |
| DC | 85 | 31.1 | 1.20 | 0.68–2.12 | 0.97 | 0.55–1.71 | 0.64 | 0.30–1.36 | 1.09 | 0.59–2.01 | 0.83 | 0.48–1.42 | 1.01 | 0.59–1.77 | |
| Other | 65 | 23.8 | 0.99 | 0.54–1.80 | 0.82 | 0.43–1.55 | 1.13 | 0.52–2.37 | 1.06 | 0.54–2.03 | 0.90 | 0.52–1.58 | 0.73 | 0.40–1.32 | |
| AA | 105 | 38.5 | |||||||||||||
| DA/AC | 29 | 10.6 | 1.22 | 0.57–2.59 | 0.62 | 0.26–1.39 | 1.50 | 0.58–3.68 | 1.02 | 0.41–2.37 | 1.55 | 0.74–3.27 | 0.88 | 0.40–1.94 | |
| DD/CC/DC | 89 | 32.6 | 1.23 | 0.70–2.15 | 0.96 | 0.55–1.69 | 0.67 | 0.31–1.40 | 1.06 | 0.58–1.95 | 0.79 | 0.46–1.34 | 0.96 | 0.55–1.66 | |
| DB/BC | 26 | 9.5 | 0.58 | 0.21–1.43 | 0.95 | 0.35–2.37 | 1.75 | 0.61–4.74 | 0.69 | 0.23–1.78 | 0.90 | 0.42–1.94 | 0.76 | 0.32–1.77 | |
| BB | 24 | 8.8 | 1.20 | 0.51–2.72 | 0.88 | 0.34–2.37 | 0.64 | 0.17–1.92 | 1.71 | 0.70–4.09 | 0.74 | 0.33–1.63 | 0.64 | 0.28–1.47 | |
ORs and 95% CI from polytomous logistic regression models are adjusted for induction treatment regimen and age
referentgenot
Discussion
In this study, we evaluated the role of XPD and XRCC1 gene polymorphic variation in response to induction chemotherapy, toxicities and survival in a population of 293 predominantly Caucasian adult patients treated for AML. We found that the strongest predictors of CR were Asn/ Asn codon 312 XPD genotypes and BB diplotype (751Gln/312Asn-751Gln/312Asn).
Variant DNA sequence in ERCC2/XPD gene codons 312 and 751 is associated with impaired DNA repair activity 29-31. In accordance, Morvan et al.32 showed that ERCC2 expression in the NCI-60 tumor cell line panel was associated with reduced DNA NER activity, and enhanced drug cytotoxicity. If variant alleles of NER-related polymorphisms are associated with less proficient DNA repair activity, they may lead to decreased repair of DNA damage in malignant cells, facilitating their apoptosis. Hence, less resistance to chemotherapy and better odds of achieving CR can be anticipated, as well as longer survival of patients. Furthermore, compromised repair activity may lead to accumulation of more DNA damage, leading to more profound treatment-related toxicities in normal tissues, and may predispose towards secondary cancers.
Our findings at least partially confirm the above hypothesis. First, we observed significantly higher CR rates after induction chemotherapy for AML in association with variant codon 312 and codon 751 XPD gene polymorphisms, as well as diplotypes containing variant alleles (BB, DC). Highly significant increased CR odds were observed among sAML patients with variant allele genotypes/haplotypes in the two studied XPD polymorphisms. In particular, in the BB diplotype group, patients had 18.31 times higher chance of achieving CR (95% CI, 2.0-283.57) compared to the AA diplotype category. In regard to OS, lower hazards of dying were associated with variant XPD genotypes or XPD haplotype group, containing variant alleles, in the sAML subgroup.
The reasons for the association of variant XPD genotypes and haplotypes and better treatment outcomes in sAML, but not de novo AML, are not fully understood. These results could reflect germ-line XPD alleles that predispose toward development of MDS or t-AML by compromised DNA repair activity and resultant accumulation of mutations. The same alleles may then predict better reponse to chemotherapy by virtue of less proficient repair of cytotoxic damage. It is unclear whether the association described is direct or indirect. In a direct association, the SNPs studied here would directly govern the responses to agents in the AML cells. In a direct association, the SNPs should play a similar role in de novo and secondary AML cells, but this was not the case, suggesting that other modifier elements likley had an impact. Zijno et al. [33] demonstrated that healthy volunteers with XPD 751 Gln/GIn genotype had a 4.55-fold higher rate of sister chromatid exhanges per cell than individuals with Lys/Lys genotype. In addition, Allan et al. [18], had previously reported that the Gln/GIn genotype XPD751 polymorphism was associated with a 2.22-fold increased risk of t-AML development among patients treated with chemotherapy only [18]. In that study, however, Lys/Lys de novo patients had a better survival, which we cannot confirm. In another UK study by Seedhouse et al. [14], sAML was not associated with XPD genotypes, but carrying at least one variant XRCC1 399Gln allele predicted a lower risk of t-AML. Investigators from the Children's Oncology Group, however, did not observe any associations of XPD with outcome in children with AML [34]. In our previous analysis of the same XRCC1 and XPD genotypes in relation to clinical outcomes of a smaller-sized AML patient population from SWOG clinical trials (n=200), we did not observe significant associations with XPD variant genotypes. However, the ‘D’ (751Gln/312Asp) haplotype was associated with 4-fold higher odds of CR. In that study, we analysed patients with de novo and secondary AML together and did not stratify analyses by AML onset. Similarly to those observations, in the present study we did not observe any significant associations between XRCC1 genotypes and any treatment outcome [20], but we were unable to replicate the previous results with respect to significantly higher CR rates for the DA diplotype and D-haplotype overall.
Our findings suggest that in AML, variation in the XPD gene may be associated with suboptimal DNA repair activity and may thus predispose to sAML development. However at the same time, suboptimal DNA repair capacity may contribute to better sensitivity to chemotherapy and better treatment outcomes in AML. Since the multiplicative interaction between AML onset and genotypes/haplotypes was not observed and the numbers for the sAML analysis were limited, leading to wide confidence intervals, it cannot be clearly concluded from our data whether the genotype effect on treatment outcomes is limited to the patients with sAML.
The impact of DNA repair pathway polymorphisms has also been studied in other malignancies. A 1.3-fold longer time to treatment failure was associated with ERCC2 (XPD) codon 751 SNP, compared to the Lys/Lys common homozygote genotype in patients with multiple myeloma treated with autologous stem cell transplantation [35]. The authors attributed better reponse to suboptimal DNA repair associated with the Gin allele, which is in accordance with our hypothesis and observations. Numerous studies were also previously conducted to investigate relationships between the XRCC1 Arg399Gln, XPD codon 751 and 312 SNPs and therapy outcome of solid tumors treated with platinum analogs. Quintela et al. [15] observed significantly longer median OS in carriers of the XPD 751Gln, XPD 312Asn and XRCC1 399Gln variant alleles receiving cisplatin for head and neck squamous cell carcinoma in a small European population (n=103). In contrast to our findings and others mentioned above, variant alleles of either of these polymorphisms were strong predictors of treatment failure in patients with advanced colorectal cancer [36-38], eso-phageal cancer [39] and lung cancer [40]. Considering the amount of literature on BER and NER pharmacogenetics, it becomes clearer that modest genetic effects can be only partly, if at all, explained by single polymorphisms, and that effects are tissue- and treatment-specific.
It is not understood how DNA repair, and NER and BER mechanisms in particular, could play a role in our finding that decreased nausea/vomiting toxicity and increased risk of infectious complications are significantly associated with genetic variation in XPD. These associations could be due to LD with other true causative alleles or may be due to chance because of small numbers of events. Even though we previously reported associations between XRCC3 (double-strand break repair pathway) 241Met allele and codon 751 XPD and liver toxicities, XPD and genitourinary and gastrointestinal toxicities, and ERCC1 (NER) and lung, metabolic toxicities in AML patients on SWOG trials [20], these associations might be random due to small numbers of events. Previously, several other studies also investigated the role of DNA repair gene polymorphisms in toxicity outcomes, but only a few were conducted in AML patient populations. In a recent Chinese report [41] on treatment outcomes in adult AML, the risks of neutropenia, nausea and vomiting, and alopecia were significantly higher among patients with variant XRCC3 Thr241Met genotypes. The risk of hematuria was also significantly higher in patients with the variant, compared to the wild-type, genotype. More than two-fold increased risk of grade 3-4 gastrointestinal toxicity was reported in cisplatin-treated lung cancer patients with variant ERCC1 C8092A and XRCC1 Arg399Gln alleles in North American and Chinese studies respectively [21,22]. A recent exploratory study [23] investigated associations between toxicities and treatment outcomes in 107 Caucasian advanced colorectal patients treated with irinotecan-based regimens, and XRCC1 haplotypes. Although no genetic variants were associated with diarrhea or grade 3/4 hematologic toxicity, none of the patients homozygous for XRCC1 GGCC-G haplotype experienced severe neutropenia. Additional data are needed to interpret the associations between NER genetic variation and toxicity outcomes.
A limitation of this study is that application of the candidate-gene approach with a small number of candidate genes and SNPs does not account for genomic multi-genetic effects. This study was initiated several years ago, before the availability of current opportunities for genome-wide association studies. However, the significance of XRCC1 Arg399Gln, XPD Lys751Gln and Asp312Asn SNPs in modifying DNA repair functional capacity and cancer outcomes is well established [42]. As recently noted, cancer genetic lesions tend to cluster around certain pathways after exposure to therapeutic agents, rather than representing activation of single oncogenes [43,44]. Multiple gene variants within pathways more than likely interact with each other during repair processes [45,46]. Thus, the biological pathway approach may provide more in-depth analysis of genetic variation responsible for clinical outcomes, as well as provide clues for the follow-up functional studies. Due to explorative nature of this analysis and small number of selected genotypes the adjustment for multiple comparisons has not been performed. This could increase the odds of false positive significant findings. However, the consistency of our results across both XPD genotypes and diplotypes suggest that XPD variant alleles are associated with suboptimal repair, with relevant biological plausibility for differential clinical outcomes.
To our knowledge, this is the first report of associations between XPD haplotypes and therapy outcomes in sAML, as well as chemotherapy-induced toxicities. If confirmed in larger studies, genotyping for XPD SNPs could potentially lead to modifications of treatment strategies in sAML by incorporating stratification based on pharmacogenetic data.
Acknowledgments
The study was funded by an NIH grant R03 - CA108353.
References
- 1.Evans WE, Johnson JA. Pharmacogenomics: the inherited basis for interindividual differences in drug response. Annu Rev Genomics Hum Genet. 2001;2:9–39. doi: 10.1146/annurev.genom.2.1.9. [DOI] [PubMed] [Google Scholar]
- 2.Evans WE, McLeod HL. Pharmacogenomics-drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 3.McLeod HL, Evans WE. Pharmacogenomics: unlocking the human genome for better drug therapy. Annu Rev Pharmacol Toxicol. 2001;41:101–121. doi: 10.1146/annurev.pharmtox.41.1.101. [DOI] [PubMed] [Google Scholar]
- 4.emal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 5.Smith SM, Le Beau MM, Huo D, Karrison T, Sobecks RM, Anastasi J, Vardiman JW, Rowley JD, Larson RA. Clinical-cytogenetic associations in 306 patients with therapy-related myelodysplasia and myeloid leukemia: the University of Chicago series. Blood. 2003;102:43–52. doi: 10.1182/blood-2002-11-3343. [DOI] [PubMed] [Google Scholar]
- 6.Schoch C, Kern W, Kohlmann A, Hiddemann W, Schnittger S, Haferlach T. Acute myeloid leukemia with a complex aberrant karyotype is a distinct biological entity characterized by genomic imbalances and a specific gene expression profile. Genes Chromosomes Cancer. 2005;43:227–238. doi: 10.1002/gcc.20193. [DOI] [PubMed] [Google Scholar]
- 7.Friedberg EC. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 8.Apian PD. “You break it, you fix it”. Blood. 2005;105:1843–1844. doi: 10.1182/blood-2004-12-4760. [DOI] [PubMed] [Google Scholar]
- 9.Chabner Bruce A, Amrein Philip C, Druker Brian J, Michaelson MD, Mitsiades Constantine S, Goss Paul E, Ryan David P, Ramachandra Sumant, Richardson Paul G, Supko Jeffrey G. “Chapter 51. Antineoplastic Agents” (Chapter). Brunton LL, Lazo JS, Parker KL: Goodman & Gilman's The Pharmacological Basis of Therapeutics, lie: http://www.accessmedicine.com/contentaspx?alD=957513.
- 10.Gossage L, Madhusudan S. Cancer pharmacogenomics: role of DNA repair genetic polymorphisms in individualizing cancer therapy. Mol Diagn Ther. 2007;11:361–380. doi: 10.1007/BF03256260. [DOI] [PubMed] [Google Scholar]
- 11.Ansari M, Krajinovic M. Pharmacogenomics of acute leukemia. Pharmacogenomics. 2007;8:817–834. doi: 10.2217/14622416.8.7.817. [DOI] [PubMed] [Google Scholar]
- 12.Kelly KM, Perentesis JP. Polymorphisms of drug metabolizing enzymes and markers of genotoxic-ity to identify patients with Hodgkin's lymphoma at risk of treatment-related complications. Ann Oncol. 2002;13(Suppl 1):34–39. doi: 10.1093/annonc/13.s1.34. [DOI] [PubMed] [Google Scholar]
- 13.Seedhouse C, Russell N. Advances in the understanding of susceptibility to treatment-related acute myeloid leukaemia. Br J Haematol. 2007;137:513–529. doi: 10.1111/j.1365-2141.2007.06613.x. [DOI] [PubMed] [Google Scholar]
- 14.Seedhouse C, Bainton R, Lewis M, Harding A, Russell N, Das-Gupta E. The genotype distribution of the XRCC1 gene indicates a role for base excision repair in the development of therapy-related acute myeloblastic leukemia. Blood. 2002;100:3761–3766. doi: 10.1182/blood-2002-04-1152. [DOI] [PubMed] [Google Scholar]
- 15.Quintela-Fandino M, Hitt R, Medina PP, Gamarra S, Manso L, Cortes-Funes H, Sanchez-Cespedes M. DNA-repair gene polymorphisms predict favorable clinical outcome among patients with advanced squamous cell carcinoma of the head and neck treated with cisplatin-based induction chemotherapy. J Clin Oncol. 2006;24:4333–4339. doi: 10.1200/JCO.2006.05.8768. [DOI] [PubMed] [Google Scholar]
- 16.Giachino DF, Ghio P, Regazzoni S, Mandrile G, Novello S, Selvaggi G, Gregori D, DeMarchi M, Scagliotti GV. Prospective assessment of XPD Lys751Gln and XRCC1 Arg399Gln single nucleotide polymorphisms in lung cancer. Clin Cancer Res. 2007;13:2876–2881. doi: 10.1158/1078-0432.CCR-06-2543. [DOI] [PubMed] [Google Scholar]
- 17.Bewick MA, Conlon MS, Lafrenie RM. Polymorphisms in XRCC1, XRCC3, and CCND1 and survival after treatment for metastatic breast cancer. J Clin Oncol. 2006;24:5645–5651. doi: 10.1200/JCO.2006.05.9923. [DOI] [PubMed] [Google Scholar]
- 18.Allan JM, Smith AG, Wheatley K, Hills RK, Travis LB, Hill DA, Swirsky DM, Morgan GJ, Wild CP. Genetic variation in XPD predicts treatment outcome and risk of acute myeloid leukemia following chemotherapy. Blood. 2004;104:3872–3877. doi: 10.1182/blood-2004-06-2161. [DOI] [PubMed] [Google Scholar]
- 19.Whitlock JA, Wells RJ, Hord JD, Janco RL, Greer JP, Gay JC, Edwards JR, McCurley TL, Lukens JN. High-dose cytosine arabinoside and etoposide: an effective regimen without anthracyclines for refractory childhood acute non-lymphocytic leukemia. Leukemia. 1997;11:185–189. doi: 10.1038/sj.leu.2400572. [DOI] [PubMed] [Google Scholar]
- 20.Kuptsova N, Kopecky KJ, Godwin J, Anderson J, Hoque A, Willman CL, Slovak ML, Ambrosone CB. Polymorphisms in DNA repair genes and therapeutic outcomes of AML patients from SWOG clinical trials. Blood. 2007;109:3936–3944. doi: 10.1182/blood-2006-05-022111. [DOI] [PubMed] [Google Scholar]
- 21.Suk R, Gurubhagavatula S, Park S, Zhou W, Su L, Lynch TJ, Wain JC, Neuberg D, Liu G, Christiani DC. Polymorphisms in ERCC1 and grade 3 or 4 toxicity in non-small cell lung cancer patients. Clin Cancer Res. 2005;11:1534–1538. doi: 10.1158/1078-0432.CCR-04-1953. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Xu B, Lin D, Tan W, Leaw S, Hong X, Hu X. XRCC1 polymorphisms and severe toxicity in lung cancer patients treated with cisplatin-based chemotherapy in Chinese population. Lung Cancer. 2008 doi: 10.1016/j.lungcan.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Hoskins JM, Marcuello E, Altes A, Marsh S, Maxwell T, Van Booven DJ, Pare L, Culverhouse R, McLeod HL, Baiget M. Irinotecan pharmacogenetics: influence of pharmacodynamic genes. Clin Cancer Res. 2008;14:1788–1796. doi: 10.1158/1078-0432.CCR-07-1472. [DOI] [PubMed] [Google Scholar]
- 24.Fannon W. 2002. p. 43. Single nucleotide polymorphism (SNP) analysis of a variety of non-ideal DNA sample types by SEQUENOM MassARRAY matrix assisted laser desoption/ionization time of flight (MALDI-TOF)
- 25.Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS, Koduru PR, Moore JO, Stone RM, Mayer RJ, Feldman EJ, Davey FR, Schiffer CA, Larson RA, Bloomfield CD. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 26.Leith CP, Kopecky KJ, Godwin J, McConnell T, Slovak ML, Chen IM, Head DR, Appelbaum FR, Willman CL. Acute myeloid leukemia in the elderly: assessment of multidrug resistance (MDR1) and cytogenetics distinguishes biologic subgroups with remarkably distinct responses to standard chemotherapy. A Southwest Oncology Group study. Blood. 1997;89:3323–3329. [PubMed] [Google Scholar]
- 27.Godwin JE, Kopecky KJ, Head DR, Willman CL, Leith CP, Hynes HE, Balcerzak SP, Appelbaum FR. A double-blind placebo-controlled trial of granulocyte colony-stimulating factor in elderly patients with previously untreated acute myeloid leukemia: a Southwest oncology group study (9031) Blood. 1998;91:3607–3615. [PubMed] [Google Scholar]
- 28.Anderson JE, Kopecky KJ, Willman CL, Head D, O'Donnell MR, Luthardt FW, Norwood TH, Chen IM, Balcerzak SP, Johnson DB, Appelbaum FR. Outcome after induction chemotherapy for older patients with acute myeloid leukemia is not improved with mitoxantrone and etoposide compared to cytarabine and daunorubica Southwest Oncology Group study. Blood. 2002;100:3869–3876. doi: 10.1182/blood-2001-12-0354. [DOI] [PubMed] [Google Scholar]
- 29.Spitz MR, Wu X, Wang Y, Wang LE, Shete S, Amos CI, Guo Z, Lei L, Mohrenweiser H, Wei Q. Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res. 2001;61:1354–1357. [PubMed] [Google Scholar]
- 30.Robert J, Morvan VL, Smith D, Pourquier P, Bonnet J. Predicting drug response and toxicity based on gene polymorphisms. Crit Rev Oncol Hematol. 2005;54:171–196. doi: 10.1016/j.critrevonc.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–608. [PubMed] [Google Scholar]
- 32.Le Morvan V, Bellott R, Moisan F, Mathoulin-Pelissier S, Bonnet J, Robert J. Relationships between genetic polymorphisms and anticancer drugcytotoxicity vis-a-vis the NCI-60 panel. Pharmacogenomics. 2006;7:843–852. doi: 10.2217/14622416.7.6.843. [DOI] [PubMed] [Google Scholar]
- 33.Zijno A, Verdina A, Galati R, Leopardi P, Marcon F, Andreoli C, Rossi S, Crebelli R. Influence of DNA repair polymorphisms on biomarkers of genotoxic damage in peripheral lymphocytes of healthy subjects. Mutat Res. 2006;600:184–192. doi: 10.1016/j.mrfmmm.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Mehta PA, Alonzo TA, Gerbing RB, Elliott JS, Wilke TA, Kennedy RJ, Ross JA, Perentesis JP, Lange BJ, Davies SM. XPD Lys751Gln polymorphism in the etiology and outcome of childhood acute myeloid leukemia: a Children's Oncology Group report. Blood. 2006;107:39–45. doi: 10.1182/blood-2005-06-2305. [DOI] [PubMed] [Google Scholar]
- 35.Vangsted A, Gimsing P, Klausen TW, Nexo BA, Wallin H, Andersen P, Hokland P, Lillevang ST, Vogel U. Polymorphisms in the genes ERCC2, XRCC3 and CD3EAP influence treatment outcome in multiple myeloma patients undergoing autologous bone marrow transplantation. Int J Cancer. 2007;120:1036–1045. doi: 10.1002/ijc.22411. [DOI] [PubMed] [Google Scholar]
- 36.Stoehlmacher J, Ghaderi V, lobal S, Groshen S, Tsao-Wei D, Park D, Lenz HJ. A polymorphism of the XRCC1 gene predicts for response to platinum based treatment in advanced colorectal cancer. Anticancer Res. 2001;21:3075–3079. [PubMed] [Google Scholar]
- 37.Le Morvan V, Smith D, Laurand A, Brouste V, Bellott R, Soubeyran I, Mathoulin-Pelissier S, Robert J. Determination of ERCC2 Lys751Gln and GSTP1 Ile105Val gene polymorphisms in colorectal cancer patients: relationships with treatment outcome. Pharmacogenomics. 2007;8:1693–1703. doi: 10.2217/14622416.8.12.1693. [DOI] [PubMed] [Google Scholar]
- 38.Park DJ, Stoehlmacher J, Zhang W, Tsao-Wei DD, Groshen S, Lenz HJ. A Xeroderma pigmentosum group D gene polymorphism predicts clinical outcome to platinum-based chemotherapy in patients with advanced colorectal cancer. Cancer Res. 2001;61:8654–8658. [PubMed] [Google Scholar]
- 39.Wu X, Gu J, Wu TT, Swisher SG, Liao Z, Correa AM, Liu J, Etzel CJ, Amos CI, Huang M, Chiang SS, Milas L, Hittelman WN, Ajani JA. Genetic variations in radiation and chemotherapy drug action pathways predict clinical outcomes in esophageal cancer. J Clin Oncol. 2006;24:3789–3798. doi: 10.1200/JCO.2005.03.6640. [DOI] [PubMed] [Google Scholar]
- 40.Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, Lynch TJ, Neuberg DS, Christiani DC. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol. 2004;22:2594–2601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 41.Liu L, Yang L, Zhang Y, Xu ZF, Yu MH, Wang JX, Xiao ZJ. [Polymorphisms of RAD51(G135C) and XRCC3(C241T) genes and correlations thereof with prognosis and clinical outcomes of acute myeloid leukemia] Zhonghua Yi Xue Za Zhi. 2008;88:378–382. [PubMed] [Google Scholar]
- 42.Xu Z, Chen ZP, Malapetsa A, Alaoui-Jamali M, Bergeron J, Monks A, Myers TG, Mohr G, Sausville EA, Scudiero DA, Aloyz R, Panasci LC. DNA repair protein levels vis-a-vis anticancer drug resistance in the human tumor cell lines of the National Cancer Institute drug screening program. Anticancer Drugs. 2002;13:511–519. doi: 10.1097/00001813-200206000-00010. [DOI] [PubMed] [Google Scholar]
- 43.Ulrich CM, Robien K, McLeod HL. Cancer pharmacogenetics: polymorphisms, pathways and beyond. Nat Rev Cancer. 2003;3:912–920. doi: 10.1038/nrc1233. [DOI] [PubMed] [Google Scholar]
- 44.Tonon G. From oncogene to network addiction: the new frontier of cancer genomics and therapeutics. Future Oncol. 2008;4:569–577. doi: 10.2217/14796694.4.4.569. [DOI] [PubMed] [Google Scholar]
- 45.Cistulli C, Lavrik OI, Prasad R, Hou E, Wilson SH. AP endonuclease and poly(ADP-ribose) polymerase-1 interact with the same base excision repair intermediate. DNA Repair (Amst). 2004;3:581–591. doi: 10.1016/j.dnarep.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 46.Sossou M, Flohr-Beckhaus C, Schulz I, Daboussi F, Epe B, Radicella JP. APE1 overexpression in XRCCl-deficient cells complements the defective repair of oxidative single strand breaks but increases genomic instability. Nucleic Acids Res. 2005;33:298–306. doi: 10.1093/nar/gki173. [DOI] [PMC free article] [PubMed] [Google Scholar]