Abstract
Large intervening noncoding RNAs (lincRNAs) are pervasively transcribed in the genome1, 2, 3 yet their potential involvement in human disease is not well understood4,5. Recent studies of dosage compensation, imprinting, and homeotic gene expression suggest that individual lincRNAs can function as the interface between DNA and specific chromatin remodeling activities6,7,8. Here we show that lincRNAs in the HOX loci become systematically dysregulated during breast cancer progression. The lincRNA termed HOTAIR is increased in expression in primary breast tumors and metastases, and HOTAIR expression level in primary tumors is a powerful predictor of eventual metastasis and death. Enforced expression of HOTAIR in epithelial cancer cells induced genome-wide re-targeting of Polycomb Repressive Complex 2 (PRC2) to an occupancy pattern more resembling embryonic fibroblasts, leading to altered histone H3 lysine 27 methylation, gene expression, and increased cancer invasiveness and metastasis in a manner dependent on PRC2. Conversely, loss of HOTAIR can inhibit cancer invasiveness, particularly in cells that possess excessive PRC2 activity. These findings suggest that lincRNAs play active roles in modulating the cancer epigenome and may be important targets for cancer diagnosis and therapy.
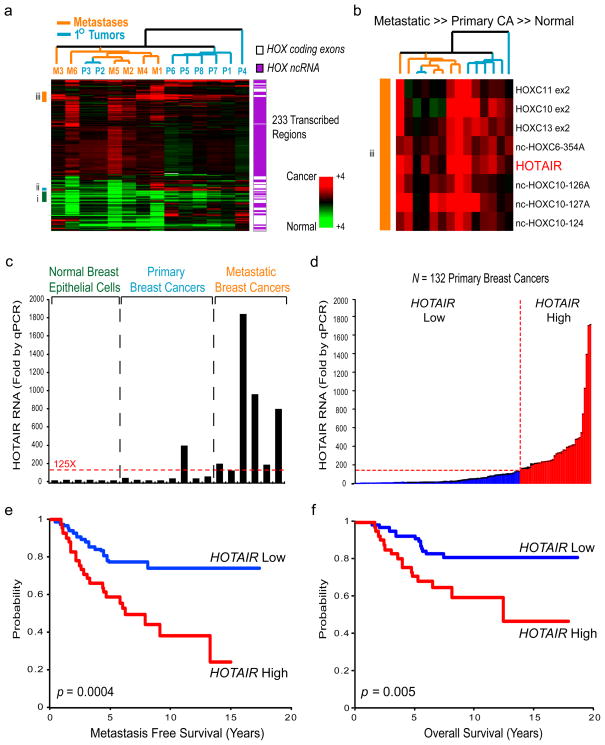
We hybridized RNA derived from normal human breast epithelia, primary breast carcinomas, and distant metastases to ultra-dense HOX tiling arrays7 (Fig. 1a, b). We found that 233 transcribed regions in the HOX loci, comprising 170 ncRNAs and 63 HOX exons, were differentially expressed (Fig. 1a). Unsupervised hierarchical clustering showed systematic variation in the expression of HOX lincRNAs among normal breast epithelia, primary tumor, and metastases. HOXA5, a known breast tumor suppressor11 along with dozens of HOX lincRNAs, are expressed in normal breast but with reduced expression in all cancer samples (Supplementary Fig. 1). A set of HOX lincRNAs and mRNAs, including the known oncogene HOXB712, are frequently expressed in primary tumors but not in metastases (Supplementary Fig. 1). A distinct set of HOX lincRNAs are sometimes overexpressed in primary tumors, and very frequently overexpressed in metastases (Fig. 1b). Notably, one such metastasis-associated lincRNA is HOTAIR (Fig. 1b), which has a unique association with patient prognosis (Supplementary Figs. 1, 2, Supplementary Table 1). HOTAIR is a lincRNA in the mammalian HOXC locus that binds to and targets the PRC2 complex to the HOXD locus, located on a different chromosome7. PRC2 is a histone H3 lysine 27 methylase involved in developmental gene silencing and cancer progression9,10. We hypothesized that altered HOTAIR expression may be involved in human cancer by promoting genomic relocalization of Polycomb complex and H3K27 trimethylation.
Figure 1. HOX lincRNAs are systematically dysregulated in breast carcinoma and have prognostic value for metastasis and survival.
a, Heat map representing unsupervised hierarchical clustering of expression values of a panel of primary and metastatic breast cancers relative to normal breast epithelial cells (pooled from 5 breast organoids). An ultra-high density HOX tiling array7 was interrogated with either normal breast organoid RNA (Cy3 channel) or RNA derived from primary or metastatic breast tumors (Cy5 channel). Each column represents the indicated clinical sample. Each row indicates a transcribed region, either a HOX coding exon or HOX ncRNA. Expression values are depicted as a ratio relative to pooled normal and represented as a red-green color scale. b, Higher resolution of subset iii identifying transcripts that show higher relative expression in metastatic as compared to primary tumors and normal epithelia. HOTAIR is one such transcript (p= 0.03, Student t-test). c, Quantitative reverse transcription (qRT)-PCR validation of the expression tiling array results measuring HOTAIR abundance in a panel of normal breast epithelial-enriched organoids, primary breast tumors, and metastatic breast tumors. Metastatic tumors had at a minimum 125-fold higher level of HOTAIR than normal breast epithelia. Error bars represent s.d. (n=3). d, qRT-PCR analysis of HOTAIR in 132 primary breast tumors (stage I or II). Approximately one third of primary breast tumors had >125 fold overexpression of HOTAIR over normal (HOTAIR high, indicated in red), while roughly two third of tumors did not (HOTAIR low, indicated in blue). Error bars represent s.d. (n=3). e and f, Kaplan-Meier curves for (e) metastasis free survival or (f) overall survival of the same 132 primary breast tumors measured in panel (d).
Quantitative PCR showed that HOTAIR is overexpressed from hundreds to nearly two thousand- fold in breast cancer metastases, and HOTAIR level is sometimes high but heterogeneous among primary tumors (Fig. 1c). We next measured HOTAIR level in an independent panel of 132 primary breast tumors (stage I and II) with extensive clinical follow-up13. Indeed, nearly one third of primary breast tumors overexpress HOTAIR by over 125-fold over normal breast epithelia, the minimum level of HOTAIR overexpression observed in bona fide metastases (Fig. 1d), and high HOTAIR level is a significant predictor of subsequent metastasis and death (p=0.0004 and p=0.005 for metastasis and death, respectively, Fig. 1e, f). Multivariate analysis showed that prognostic stratification of metastasis and death by HOTAIR is independent of known clinical risk factors such as tumor size, stage, and hormone receptor status (Supplementary Table 2).
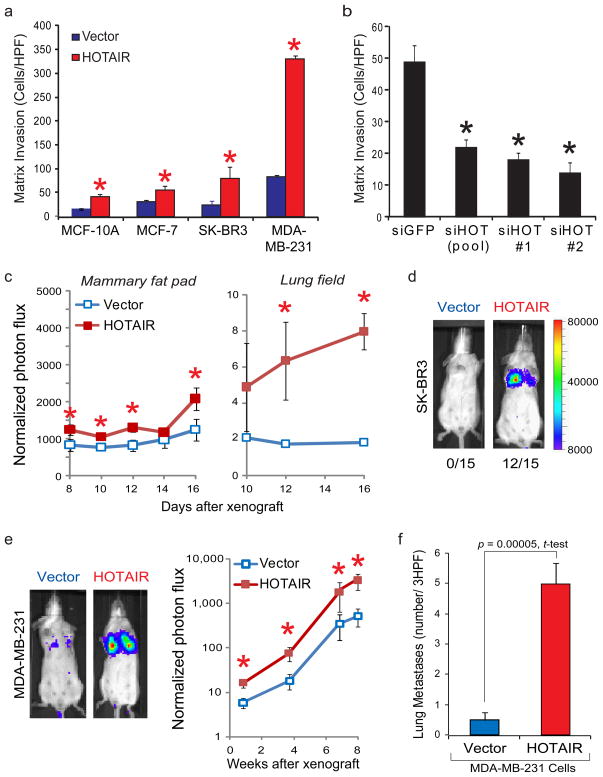
We next examined the effects of manipulating HOTAIR level in several breast cancer cell lines. HOTAIR levels in cell lines are significantly lower than those seen in primary or metastatic breast tumors (Supplementary Fig. 3, 4). Retroviral transduction allowed stable overexpression of HOTAIR to several hundred fold over vector-transduced cells, which are comparable to levels observed in patients (Supplementary Fig. 4). HOTAIR overexpression promoted colony growth in soft agar (Supplementary Fig. 5). In addition, enforced expression of HOTAIR in four different breast cancer cell lines increased cancer cell invasion through Matrigel, a basement-membrane like extracellular matrix, (Fig. 2a). Conversely, depletion of HOTAIR by small inferring RNAs (siRNAs) in MCF7, a cell line that expresses endogenous HOTAIR, decreased its matrix invasiveness (Fig. 2b and Supplementary Fig. 6). To probe the effects of HOTAIR on cancer cell dynamics in vivo, we labeled control and HOTAIR-expressing cells with firefly luciferase, enabling in vivo bioluminescence imaging. When MDA-MB-231 cells expressing vector or HOTAIR were orthotopically grafted into mammary fat pads, serial imaging showed that HOTAIR expression modestly increased the rate of primary tumor growth (Fig. 2C, left panel). Importantly, in the same animals, we observed significantly increased foci of luciferase signal in the lung fields of mice bearing HOTAIR+ primary tumors (Fig. 2C, right panel), which suggests that HOTAIR promotes lung metastasis.
Figure 2. HOTAIR promotes invasion of breast carcinoma cells.
a, Relative fold increase in matrix invasion in three breast carcinoma cell lines after enforced expression of HOTAIR. Mean±s.d. are shown (n=3). b. Matrix invasion in the MCF-7 breast carcinoma cell line transfected with individual or pooled siRNAs targeting HOTAIR (error bars = s.d., n=3). c, HOTAIR expression in MDA-MB-231 cells enhances orthotopic growth in mammary fat pads and metastasis to lung (n=5 per arm; error bars = s.e.m.). d, HOTAIR promotes transient lung colonization of SK-BR3 after tail vein xenograft (n=15 per arm). e, HOTAIR promotes lung colonization of MDA-MB-231 cells after tail vein xenograft. f, Histologic analysis of the number of lung metastasis in vector- or HOTAIR- expressing MDA-MB-231 cells 8–9 weeks after tail vein xenograft (n=18 per arm, error bars = s.e.m.). Similar results were obtained using luciferase marked cells. *, p<0.05 between control cells and cells manipulated for HOTAIR.
To further quantify metastatic potential in vivo, we performed tail vein xenografts and compared the rates of lung colonization. Vector expression in the non-metastatic cell line SK-BR3 never showed lung colonization after tail vein xenograft (0 of 15 mice), but HOTAIR expression allowed SK-BR3 cells to colonize the lung in 80% of animals (12 of 15 mice, Fig. 2C). SK-BR3 cells apparently lack additional genetic elements required to persist in the lung, because HOTAIR-transduced SK-BR3 cells in the lung disappeared after approximately one week. In contrast, HOTAIR expression in MDA-MB-231 cells resulted in approximately eight-to ten- fold more cells to engraft the lung after tail vein xenograft (Fig. 2D). These differences persisted until the end of the experiment, resulting in ten-fold more lung metastases as verified by histology (p= 0.00005, Fig. 2E). The tumors retained HOTAIR expression for the length of the experiment (Supplementary Fig. 7).
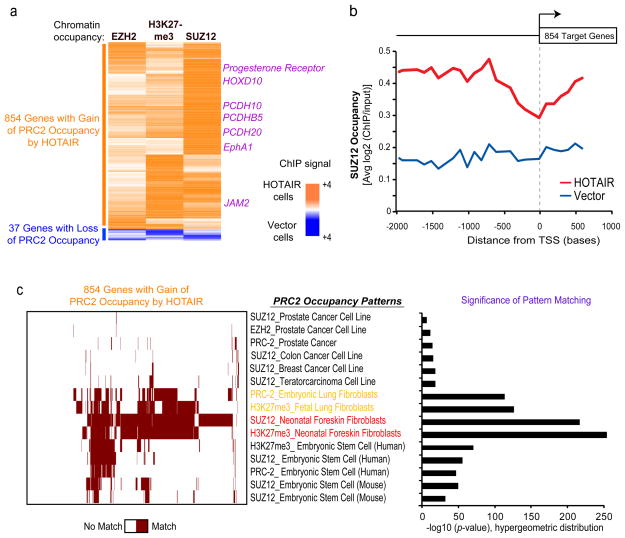
We next tested if HOTAIR overexpression affected the pattern of PRC2 occupancy. We mapped PRC2 occupancy genome-wide by chromatin immunoprecipitation followed by hybridization to tiling microarrays interrogating all human promoters (ChIP-chip, Fig. 3). Compared to vector expressing cells, HOTAIR overexpression induced localization of H3K27me3 and PRC2 subunits SUZ12, EZH2, on 854 new genes while concomitantly losing PRC2 occupancy and H3K27me3 on 37 genes (Fig. 3a). A significant fraction of these 854 genes also showed consequent changes in gene expression after HOTAIR over-expression (39% observed vs. 7.0% expected by chance alone, p = 2.5 ×10−209, hypergeometric distribution). The majority of PRC2 occupancy sites on promoters genome-wide showed little change (data not shown), and HOTAIR overexpression did not change the levels of PRC2 subunits (Fig. 4a, lane 1 vs. lane 4). A number of the genes with HOTAIR-induced PRC2 occupancy are implicated in inhibiting breast cancer progression, including transcription factors HOXD1014 and PRG1, encoding progesterone receptor (a classic favorable prognostic factor); cell adhesion molecules of the protocadherin (PCDH) gene family15 and JAM216; and EPHA117,18, encoding an ephrin receptor involved in tumor angiogenesis. Gene Ontology19 analysis suggested a majority of the 854 genes are involved in pathways related to cell-cell signaling and development (Supplementary Fig. 8). HOTAIR-induced PRC2 occupancy tended to spread over promoters, and to a lesser extent, gene bodies (Fig. 3b). HOTAIR may also induce PRC2 localization to other intergenic regions not present on our tiling arrays. ChIP followed by quantitative PCR confirmed that HOTAIR substantially increased PRC2 occupancy and H3K27me3 of all target genes examined (Supplementary Fig. 9). Notably, like HOTAIR itself, the 854 HOTAIR-PRC2 target genes are coordinately down regulated in aggressive breast tumors that tend to cause death (p<0.0003, Supplementary Fig. 10).
Figure 3. HOTAIR promotes selective re-targeting of PRC2 and H3K27me3 genome-wide.
a, Heat map representing genes with a significant relative change in chromatin occupancy of EZH2, SUZ12, and H3K27 following HOTAIR expression. MDA-MB-231 vector or HOTAIR cells were subjected to ChIP using anti-EZH2, H3K27me3, and SUZ12 antibodies followed by interrogation on a genome-wide promoter array. Values are depicted as relative ratio of HOTAIR over vector cells and represented as an orange-blue scale. b, Average SUZ12 occupancy of >800 PRC-2 target genes in HOTAIR or vector expressing cells across the length of gene promoter and gene body. All target genes are aligned by their transcriptional start sites (TSS). c, Module map20 of the 854 genes with a gain in PRC2 occupancy following HOTAR overexpression. (Left panel) Heat map of genes (column) showing a gain in PRC2 occupancy following HOTAR expression in breast carcinoma cells [see panel (a)] compared with PRC-2 occupancy patterns from the indicated cell or tissue type (rows). Binary scale is brown (match) or white (no match). (Right panel) Quantification of significance of pattern matching between gene sets.
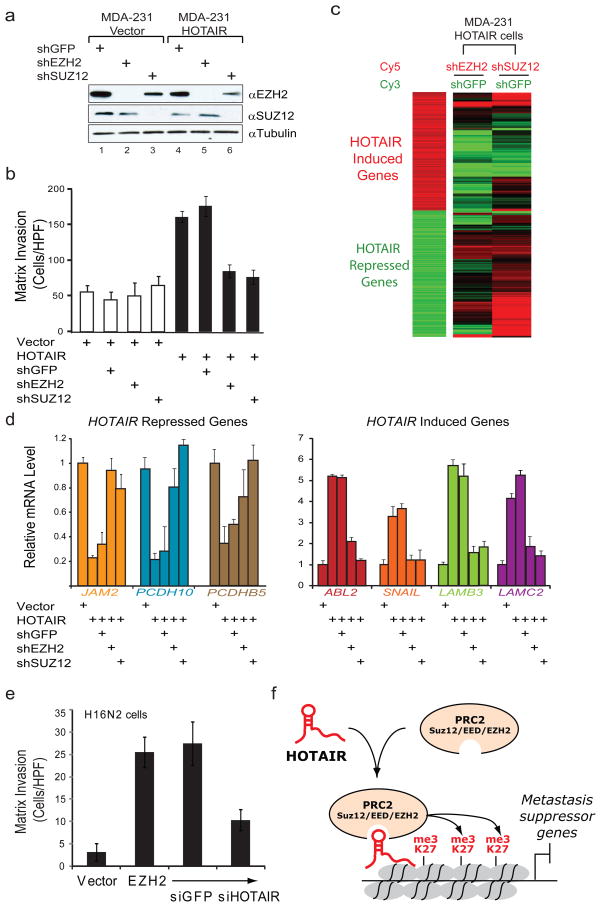
Figure 4. HOTAIR-induced matrix invasion and global gene expression changes requires PRC2.
a, Immunoblot of SUZ12 and EZH2 protein levels following transduction of MDA-MB-231 vector or HOTAIR cells with retrovirus expressing a shRNA targeting either GFP, EZH2, or SUZ12. b, Matrix invasion in vector or HOTAIR cells expressing the indicated shRNA. Mean±s.d. are shown (n=3). c, (left panel) Heat map of gene with significant induction (red) or repression (green) following HOTAIR expression in the MDA-MB-231 cells. (right panel) The relative expression of the same gene list in MDA-MB-231 HOTAIR cells expressing shEZH2 or shSUZ12 (expressed as a ratio to HOTAIR cells expressing shGFP). d, qRT-PCR of a representative panel of genes in MDA-MB-231 vector or HOTAIR cells also expressing the indicated shRNA (error bars = s.d., n=3). e, Matrix invasion in the immortalized H16N2 breast epithelial line expressing vector or EZH2 as well as EZH2-expressing cells transfected with siRNAs targeting GFP or HOTAIR (error bars = s.d., n=3). f, Working model of the role of HOTAIR in breast cancer progression. Selection for increased HOTAIR expression in a subset of breast primary tumors leads to a genome-wide retargeting of the PRC2 and H3K27me3 patterns, resulting in gene expression changes that promote tumor metastasis.
We next compared the 854 genes with HOTAIR-induced PRC2 occupancy in MDA-MB-231 cells with a compendium of published PRC2 occupancy profiles in diverse cell types (Fig. 3c). PRC2 occupancy patterns from different cancer, fibroblastic, and embryonic stem cell lines were annotated from existing databases (Supplementary Table 3). Using a pattern matching algorithm20, we found that the HOTAIR-induced PRC2 occupancy pattern in breast cancer cells most resembled the endogenous PRC2 occupancy pattern in embryonic and neonatal fibroblasts, especially fibroblasts derived from posterior and distal anatomic sites where endogenous HOTAIR is expressed7 (p<10−50 for each comparison, FDR≪0.05, Fig. 3c). These 854 genes are also significantly enriched for genes in primary fibroblasts that are bound by PRC2 in a HOTAIR dependent manner (32% overlap observed vs. 9.9% expected by chance alone, p = 8.5 ×10−93, hypergeometric distribution, M.C.T., unpublished data). These results suggest that elevated HOTAIR expression in breast cancer cells appears to reprogram the Polycomb binding profile of a breast epithelial cell to that of an embryonic fibroblast.
Finally, we addressed whether the ability of HOTAIR to induce breast cancer invasiveness required an intact PRC2 complex. We transduced vector- or HOTAIR-expressing MDA-MB-231 cells with short hairpin RNAs (shRNAs) targeting PRC2 subunits EZH2 or SUZ12. Immunoblot analyses confirmed efficient depletion of the targeted proteins (Fig. 4a). Depletion of either SUZ12 or EZH2 had little impact on the invasiveness of control cells, but completely reversed the ability of HOTAIR to promote matrix invasion (Fig. 4b). Depletion of EZH2 also inhibited HOTAIR-driven lung colonization after tail vein xenograft by approximately 50% (p < 0.05). These results suggest that PRC2 is specifically required for HOTAIR to promote cellular invasiveness. Global gene expression analysis revealed hundreds of genes that were induced or repressed as a consequence of HOTAIR overexpression (Fig. 4c, left panel). Importantly, concomitant depletion of PRC2 in large part reversed the global gene expression pattern to that of cells not overexpressing HOTAIR (Fig. 4c, right panel). Quantitative RT-PCR confirmed that HOTAIR-induced PRC2 target genes, such as JAM2, PCDH10, PCDHB5, were transcriptionally repressed upon HOTAIR expression and de-repressed upon concomitant PRC2 depletion (Fig. 4d). HOTAIR-induced genes were also reversed upon PRC2 depletion (Fig. 4d). Of note, many of the genes induced by HOTAIR are known positive regulators of cancer metastasis, including ABL221, SNAIL22, and laminins23. Conversely, overexpression of EZH2 in H16N2 breast cells is known to promote matrix invasion10, but concomitant depletion of endogenous HOTAIR in large measure inhibited the ability of EZH2 to induce matrix invasion (Fig. 4e and Supplementary Figure 6). Together, these results demonstrate a functional inter-dependency between HOTAIR and PRC2 in promoting cancer invasiveness.
In summary, the cancer transcriptome is more complex than previously believed. In addition to protein coding genes and microRNAs, dysregulated expression of lincRNAs is likely pervasive in human cancers and can drive cancer development and progression. Notably, the lincRNA HOTAIR regulates metastatic progression. HOTAIR recruits PRC2 complex to specific targets genes genome-wide, leading to H3K27 trimethylation and epigenetic silencing of metastasis suppressor genes (Fig. 4f). The concept of epigenomic reprogramming by lincRNAs may also be applicable to many other human disease states characterized by aberrant lincRNA expression and chromatin states. HOTAIR is normally involved in specifying the chromatin state associated with fibroblasts from anatomically posterior and distal sites. Within the context of cancer cells, ectopic expression of HOTAIR appear to re-impose that chromatin state, thereby enabling gene expression programs that are conducive to cell motility and matrix invasion.
The interdependence between HOTAIR and PRC2 has therapeutic implications. High levels of HOTAIR may identify tumors that are sensitive to small molecules inhibitors of PRC224. Conversely, tumors that overexpress Polycomb proteins may be sensitive to therapeutic strategies that target endogenous HOTAIR or inhibit HOTAIR-PRC2 interactions. Understanding the precise molecular mechanisms by which HOTAIR regulates PRC2 will be a critical first step in exploring these potential new avenues in cancer therapy.
METHODS SUMMARY
Human materials were obtained from Johns Hopkins Hospital and the Netherlands Cancer Institute. Expression of HOX transcripts was determined using ultra-high density HOX tiling arrays7 and qRT-PCR. Kaplan-Meier analyses of breast cancer patients were as described13. We used retroviral transduction to overexpress HOTAIR and luciferase, and used siRNA or shRNA to deplete the indicated transcripts. Matrix invasion was measured by the transwell Matrigel assay. We implanted cells in the mammary fat pad of SCID mice, and monitored primary tumor growth and lung metastasis by bioluminescence. Cells were injected into the tail vein of nude mice, and lungs were analyzed at 9 weeks to quantify lung colonization in vivo. ChIP-chip was performed as described7 using human whole genome promoter tiling arrays (Roche Nimblegen, Wisconsin). Module map and GO enrichment analyses were done using Genomica20.
METHODS
Reagents
The MDA-MB-231, SK-BR-3, MCF-10A, MCF-7, HCC1954, T47D, and MDA-MB-453 cell lines were obtained from ATCC. The H16N2 cell line was a gift from V. Band (University of Nebraska medical center). pLZRS, pLZRS-luciferase and pSuper Retro –shGFP, -shSUZ12, and –shEZH225 were obtained from P. Khavari (Stanford University). pLZRS-HOTAIR and pLZRS-EZH2-Flag were constructed by subcloning the full-length human HOTAIR7 or Flag-EZH2-ER fusion protein [representing amino acids 1–751 of EZH2 fused with the murine estrogen receptor (amino acids 281–599)] into pLZRS using the gateway cloning system (Invitrogen).
Human Materials
Normal Breast organoid RNA was prepared as reported26. Briefly, tissues from reduction mammoplasties performed at Johns Hopkins Hospital were mechanically macerated then digested overnight with hyaluronic acid and collagenase. The terminal ductal units are placed into suspension by this method; they were then isolated by serial filtration. Samples were treated with TRIzol and RNA extracted.
Fresh frozen primary breast tumor specimens were obtained from the Department of Pathology breast tumor bank; specimens were all from patients 45–55 years of age, with estrogen receptor expression by immunohistochemistry as performed during routine tumor staging at diagnosis, for uniformity of samples.
Metastatic breast carcinoma samples were obtained from the Rapid Autopsy Program at Johns Hopkins Hospital27. All specimens were snap frozen at time of autopsy and stored at −80 degrees. Twenty 20-micron sections were obtained from metastasis to the liver (for uniformity of samples) and embedded in OCT. These slices were macerated by use of the BioMasher centrifugal sample preparation device (Cartagen), with 350 uL of lysis buffer from the Qiagen RNEasy Mini Extraction kit. RNA extraction was completed with the flow-through from the BioMasher, as per the commercial protocol.
HOTAIR Expression and Survival/Metastasis Analysis of Primary Breast Tumors
The database of 295 breast cancer patients from the Netherlands Cancer Institute with detailed clinical and gene expression data was used 13. Clinical data are available at http://microarray-pubs.stanford.edu/wound_NKI, www.rii.com/publications, or http://microarrays.nki.nl. RNA from 132 primary breast tumors from the NKI 295 cohort was isolated along with RNA from normal breast organoid cultures (n=6). HOTAIR and GAPDH were measured by qRT-PCR. HOTAIR values were normalized to GAPDH and expressed relative to pooled normal HOTAIR RNA levels. For both univariate and multivariate analysis, the expression of HOTAIR was treated as a binary variable divided into “high” and “low” HOTAIR expression. To determine the criteria for “high” HOTAIR expression, the minimum relative level of HOTAIR seen in six metastatic breast cancer samples (see Fig. 1c and accompanying methods) was determined (≥ 125 above normal). By this criteria, 44 of 132 primary breast tumors were categorized as “high” and 88 of 132 tumors were labeled as “low”. For statistical analysis, overall survival was defined by death from any cause. Distant metastasis-free probability was defined by a distant metastasis as the first recurrence event. Kaplan-Meier survival curves were compared by the Cox-Mantel log-rank test in Winstat (R. Fitch Software). Multivariate analysis by the Cox proportional hazard method was done using SPSS 15.0 (SPSS, Inc.)
RNA expression analysis
qRT-PCR
Total RNA from cells was extracted using TRIzol and the RNeasy mini kit (Qiagen). RNA levels (starting with 50–100 ng per reaction) for a specific gene (primer set sequences listed in Supplementary Table S4) was measured using the Brilliant SYBR Green II qRT-PCR kit (Strategene) according to manufacturer instructions. All samples were normalized to GAPDH.
HOX tiling array
RNA samples (Primary or Metastatic breast carcinoma in channel in Cy5 channel and normal breast organoid RNA representing a pool of six unique samples in Cy3 channel) were labeled and hybridized to a custom human HOX tiling array with 50 base pair resolution (Roche Nimblegen) as described7. For each sample, RMA normalized intensity values for previously defined peaks encoding HOX coding gene exons (as defined in version HG17) and HOX lincRNAs (as defined by Rinn et al.7) were determined relative to normal. Unsupervised hierarchical clustering was performed by CLUSTER28.
Microarray
Total RNA from cells was extracted using TRIzol and the RNeasy mini kit (Qiagen) and hybridized to Stanford human oligonucleotide (HEEBO) arrays as described29. Data analysis was done using CLUSTER28.
Gene Transfer Experiments
Retrovirus was generated using amphotrophic phoenix cells and used to infect target cells as described30. For LZRS vector, HOTAIR, and EZH2-ER, and firefly luciferase no further selection was done post-infection. For pRetro-Super -shGFP, -shSUZ12, and shEZH2, target cells were selected using puromycin (0.5 μg/ml). Many of the epigenetic changes due to HOTAIR expression were only seen after several cell passages; thus all experiments post-HOTAIR transduction were done after passage 10.
Nonradioactive ISH of Paraffin Sections
Digoxigenin (DIG)-labeled sense and antisense RNA probes were generated by PCR amplification of T7 promoter incorporated into the primers. In vitro transcription was performed with DIG RNA labeling kit and T7 polymerase according to the manufacturer’s protocol (Roche Diagnostics, Indianapolis, IN, USA). Sections (5 um thick) were cut from the paraffin blocks, deparaffinized in xylene, and hydrated in graded concentrations of ethanol for 5 min each. Sections were incubated with 1% hydrogen peroxide, followed by digestion in 10 ug/ml of proteinase K at 37C for 30 min. Sections were hybridized overnight at 55C with either sense or antisense riboprobes at 200 ng/ml dilution in mRNA hybridization buffer (Chemicon). The following day, sections were washed in 2xSSC and incubated with 1:35 dilution of RNase A cocktail (Ambion, Austin, TX, USA) in 2xSSC for 30 min at 37C. Next, sections were stringently washed in 2xSSC/50% formamide twice, followed by one wash at 0.08xSSC at 55C. Biotin-blocking reagents (Dako) were applied to the section to block the endogenous biotin. For signal amplification, a horseradish peroxidase (HRP)-conjugated sheep anti-DIG antibody (Roche) was used to catalyze the deposition of biotinyl-tyramide, followed by secondary streptavidin complex (GenPoint kit; Dako). The final signal was developed with DAB(GenPoint kit; Dako), and the tissues were counterstained in hematoxylin for 30s.
RNA Interference
RNA interference for HOTAIR was done as described7. Briefly, cells were transfected with 50 nM of siRNAs targeting HOTAIR (siHOTAIR #1 GAACGGGAGUACAGAGAGAUU; #2 CCACAUGAACGCCCAGAGAUU; #3 UAACAAGACCAGAGAGCUGUU) or siGFP (CUACAACAGCCACAACGUCdTdT) using Lipofectamine 2000 (Invitrogen) per the manufacturer’s direction. Total RNA was harvested for total RNA 72 hr later for qRT-PCR analysis.
RNA interference of EZH2 and SUZ12 was done by infecting target cells with retrovirus expressing shEZH2, shSUZ12, and shGFP as described25. To confirm knock-down, protein lysates were resolved on 10% SDS-PAGE followed by immunoblot analysis as described30 using SUZ12 (Abcam), anti-EZH2 (Upstate), and anti-tubulin (Santa Cruz).
Matrigel Invasion Assay and Cell Proliferation Assay
The matrigel invasion assay was done using the Biocoat Matrigel Invasion Chamber from Becton Dickson according to manufacturer protocol. Briefly, 5 × 104 cells were plated in the upper chamber in serum free media. The bottom chamber contained DMEM media with 10% FBS. Following 24–48 hrs, the bottom of the chamber insert was fixed and stained with Diff-Quick stain. Cells on the stained membrane were counted under a dissecting microscope. Each membrane was divided into four quadrants and an average from all four quadrants was calculated. Each matrigel invasion assay was at least done in biological triplicates. For invasion assays in the H16N2 cell line using EZH2-ER, all experiments (both vector and with EZH2-ER) were done in the presence of 500 nM estradiol.
For cell proliferation assays, 1×103 cells were plated in quadruplicate in 96 well plates and cell number was calculated using the MTT assay (Roche).
Soft Agar Colony Formation Assay
Soft Agar Assays were constructed in 6-well plates. The base layer of each well consisted of 2mL with final concentrations of 1x media (RPMI (HCC1954), McCoy’s Media (SKBR3), or DMEM (MDA-MB-231) + 10% or 2% Heat-Inactivated FBS (Invitrogen)) and 0.6% low melting point agarose. Plates were chilled at 4°C until solid. Upon this, a 1 ml growth agar layer was poured, consisting of 1 × 104 cells (either infected with LZRS-HOTAIR or LZRS vector as described above) suspended in 1X media and 0.3% low melting point agarose. Plates were again chilled at 4°C until the growth layer congealed. An additional 1 mL of 1X media without agarose was added on top of the growth layer on day 0 and again on day 14 of growth. Cells were allowed to grow at 37°C for 1 month and total colonies counted (>200 micron in diameter for MDA-MB-231; >50 micron in diameter for HCC1954 and SKBR3). Assays were repeated a total of 3 times. Results were statistically analyzed by paired T-test using the PRISM Graphpad program.
Mammary Fat Pad Xenografts
Six week old female SCID Beige mice were purchased from Charles River laboratories (Wilmington, MA), housed at the animal care facility at Stanford University Medical Center (Stanford, CA) and kept under standard temperature, humidity, and timed lighting conditions and provided mouse chow and water ad libitum. MDA-MB-231-Luc or MDA-MB-231-Luc tumor cells transduced with HOTAIR were injected directly into the mammary fat pad of the mice semi-orthotopically (n=10 each) in 0.05mL of sterile DMEM (2,500,000 cells/animal).
Mouse Tail-Vein Assay
Female athymic nude mice were used. 2.5 × 106 MDA-MB-231 HOTAIR-luciferase or VECTOR-luciferase cells in 0.2 mL PBS were injected via the tail vein into individual mice (18 for each cell line). Mice were observed generally for signs of illness weekly for the length of the experiment. The lungs were excised and weighed fresh, then bisected. Half was fixed in formalin overnight then embedded in paraffin, from which sections were made and H&E stained by our pathology consultation service. These slides were examined for the presence of micrometastases, which were counted in 3 low power (5x) fields per specimen. The other half of the tumor was fast-frozen into OCT and stored at -80°C. RNA was extracted by the TRIzol protocol from 10 sections, 20 microns thick each, obtained from the frozen sections. RT-PCR confirmed expression of HOTAIR RNA in lungs bearing micrometastases of MDA-MB-231 HOTAIR cells at the end of the experiment.
Bioluminescence Imaging
Mice received luciferin (300 mg/kg, 10 minutes prior to imaging) and were anesthetized (3% isoflurane) and imaged in an IVIS spectrum imaging system (Xenogen, part of Caliper Life Sciences). Images were analyzed with Living Image software (Xenogen, part of Caliper Life Sciences). Bioluminescent flux (Photons/sec/sr/cm2) was determined for the primary tumors or lungs (upper abdomen region of interest).
ChIP-chip
ChIP-chip experiments were done as previously described7. Each experiment was done in biological triplicate. The following antibodies were used: anti-H3K27me3 (Abcam), anti-SUZ12 (Abcam) and anti-EZH2 (Upstate). Immunoprecipitated DNA was amplified using the Whole Genome Amplification kit (Sigma) based on the manufacturers protocol. Amplified and labeled DNA was hybridized to the HG18 whole genome two array promoter set from Roche Nimblegen. Probe labeling, hybridization, and data extraction and analysis was performed using Roche Nimblegen protocols. The relative ratio of HOTAIR over vector was calculated for each promoter peak by extracting the normalized (over input) intensity values for promoter peaks showing peaks with an FDR score ≤0.2 in either vector or HOTAIR cells. These values were weighted to determine the significance of the relative ratio: using Cluster 28, only those promoters with a consistent relative ratio (HOTAIR/vector) ≥1.5 fold or ≤0.5 fold in two out of the three ChIP were selected and displayed in TreeView. Selected ChIP –chip results were confirmed by PCR using the Lightcycler 480 SYBR Green I kit (see Supplementary Table S5 for primer sequences).
TaqMan® real-time PCR assays
A panel of 96 TaqMan® real-time PCR HOX assays (Supplementary Table 6) was developed targeting 43 HOX lincRNAs and 39 HOX transcription factors across the four HOX loci. Two housekeeping genes (ACTB and PPIA) were also included in this panel in triplicates as endogenous controls for normalization between samples. The transcript specificity and genome specificity of all TaqMan assays were verified using a position specific alignment matrix to predict potential cross-reactivity between designed assays and genome-wide non-target transcripts or genomic sequences. Using this HOX assay panel we profiled 88 total RNA samples from a cohort of five normal breast organoids, seventy-eight primary breast tumors (from the NKI 295 cohort) and five metastatic breast tumors. cDNAs were generated from 30ng of total RNA using the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Foster City, CA). The resulting cDNA was subjected to a 14-cycle PCR amplification followed by real-time PCR reaction using the manufacturer’s TaqMan® PreAmp Master Mix Kit Protocol (Life Technologies, Foster City, CA). Four replicates were run for each gene for each sample in a 384-well format plate on 7900HT Fast Real-Time PCR System (Life Technologies, Foster City, CA). Between the two measured endogenous control genes (PPIA and ACTB), we chose PPIA for normalization across different samples based on the fact that this gene showed the most relatively constant expression in different breast carcinomas (data not shown).
Gene Set Analysis
For gene set enrichment analysis, gene sets from fifteen different H3K27, SUZ12, or EZH2 global occupancy lists from the indicated cell lineages were procured (see Supplementary Table S3 for references and platforms). Pattern matching between the 854 gene set with increased PRC2 occupancy (Supplementary Table S7) and these fifteen gene sets were visualized using CLUSTER and TreeView. The significance of enrichment between these gene sets was calculated using module map analysis implemented in Genomica20 (corrected for multiple hypotheses using FDR).
Supplementary Material
Acknowledgments
We thank Y. Chen-Tsai, M. Guttman, G. Sen, T. Ridky, P. Khavari, V. Band, and Y. Kang for advice and reagents. Supported by NIH, Emerald Foundation, and American Cancer Society (H.Y.C.), Dermatology Foundation (R.A.G., K.C.W., D.J.W.), Susan Komen Foundation (M.C.T.), NSF (T.H.), and Department of Defense BCRP (S.S). H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
R.A.G. measured lincRNAs in cancer samples, performed all gene transfer and knockdown experiments. R.A.G. and N.S. performed cell growth, invasion, and in vivo xenograft assays. R.A.G., K.C.W., M.C.T., T.H. performed ChIP-chip studies and analyses. R.A.G., J.L.R., D.J.W. performed bioinformatic analyses. J.K. performed in vivo bioluminescence studies. H.M.H., P.A., M.J. vd V. procured and analyzed human tumor samples. Y.W., P.B., B.K. designed lincRNA Taqman probes and analyzed tumor RNAs by qRT-PCR. R.L. and R.B.W. performed in situ hybridization studies. R.A.G., N.S., S.S., and H.Y.C. designed the experiments and interpreted the results. R.A.G. and H.Y.C. wrote the paper.
AUTHOR INFORMATION
Microarray data are deposited in Gene Expression Omnibus (accession number GSE20435). Reprints and permission information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calin GA, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 5.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nature reviews. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 10.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raman V, et al. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 12.Wu X, et al. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer research. 2006;66:9527–9534. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- 13.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. The New England journal of medicine. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 14.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 15.Novak P, et al. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer research. 2008;68:8616–8625. doi: 10.1158/0008-5472.CAN-08-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naik MU, Naik TU, Suckow AT, Duncan MK, Naik UP. Attenuation of junctional adhesion molecule-A is a contributing factor for breast cancer cell invasion. Cancer research. 2008;68:2194–2203. doi: 10.1158/0008-5472.CAN-07-3057. [DOI] [PubMed] [Google Scholar]
- 17.Fox BP, Kandpal RP. Invasiveness of breast carcinoma cells and transcript profile: Eph receptors and ephrin ligands as molecular markers of potential diagnostic and prognostic application. Biochemical and biophysical research communications. 2004;318:882–892. doi: 10.1016/j.bbrc.2004.04.102. [DOI] [PubMed] [Google Scholar]
- 18.Herath NI, Doecke J, Spanevello MD, Leggett BA, Boyd AW. Epigenetic silencing of EphA1 expression in colorectal cancer is correlated with poor survival. British journal of cancer. 2009;100:1095–1102. doi: 10.1038/sj.bjc.6604970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nature genetics. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 21.Srinivasan D, Plattner R. Activation of Abl tyrosine kinases promotes invasion of aggressive breast cancer cells. Cancer research. 2006;66:5648–5655. doi: 10.1158/0008-5472.CAN-06-0734. [DOI] [PubMed] [Google Scholar]
- 22.Olmeda D, et al. SNAI1 is required for tumor growth and lymph node metastasis of human breast carcinoma MDA-MB-231 cells. Cancer research. 2007;67:11721–11731. doi: 10.1158/0008-5472.CAN-07-2318. [DOI] [PubMed] [Google Scholar]
- 23.Marinkovich MP. Tumour microenvironment: laminin 332 in squamous-cell carcinoma. Nature reviews. 2007;7:370–380. doi: 10.1038/nrc2089. [DOI] [PubMed] [Google Scholar]
- 24.Tan J, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes & development. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
ADDITIONAL REFERENCES (FOR FULL METHODS SECTION)
- 25.Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes & development. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergstraesser LM, Weitzman SA. Culture of normal and malignant primary human mammary epithelial cells in a physiological manner simulates in vivo growth patterns and allows discrimination of cell type. Cancer research. 1993;53:2644–2654. [PubMed] [Google Scholar]
- 27.Wu JM, et al. Heterogeneity of breast cancer metastases: comparison of therapeutic target expression and promoter methylation between primary tumors and their multifocal metastases. Clin Cancer Res. 2008;14:1938–1946. doi: 10.1158/1078-0432.CCR-07-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS genetics. 2006;2:e119. doi: 10.1371/journal.pgen.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adler AS, et al. Genetic regulators of large-scale transcriptional signatures in cancer. Nature genetics. 2006;38:421–430. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.