SUMMARY
Dynamic range compression (DRC) by hexapeptide libraries increases MS/MS-based identification of lower-abundance proteins in complex mixtures. However, two unanswered questions impede fully realizing DRC’s potential in shotgun proteomics. First, does DRC enhance identification of post-translationally modified proteins? Second, can DRC be incorporated into a workflow enabling relative protein abundance profiling? We sought to answer both questions analyzing human whole saliva. Addressing question one, we coupled DRC with covalent glycopeptide enrichment and MS/MS. With DRC we identified ~2 times more N-linked glycoproteins and their glycosylation sites than without DRC, dramatically increasing the known salivary glycoprotein catalog. Addressing question two, we compared differentially stable isotope-labeled saliva samples pooled from healthy and metastatic breast cancer women using a multidimensional peptide fractionation-based workflow, analyzing in parallel one sample portion with DRC and one portion without. Our workflow categorizes proteins with higher absolute abundance, whose relative abundance ratios are altered by DRC, from proteins of lower absolute abundance detected only after DRC. Within each of these salivary protein categories we identified novel abundance changes putatively associated with breast cancer, demonstrating feasibility and benefits of DRC for relative abundance profiling. Collectively, our results bring us closer to realizing the full potential of DRC for proteomic studies.
INTRODUCTION
Combinatorial chemistry derived hexapeptide libraries, ProteoMiner (PM) and its carboxylated derivative, Library-2 (LIB2) have been used to effectively compress the dynamic range of protein abundances in complex protein mixtures, thus enabling increased mass spectrometric detection of lower abundance proteins1–4. In this dynamic range compression (DRC) method, millions of hexapeptide sequences act as affinity “baits”, with each hexapeptide putatively having high binding affinity for one/few related proteins within the complex protein mixture. Most proteins present at lower-abundance levels are effectively fully bound by their hexapeptide baits. Simultaneously, very high abundance proteins quickly saturate their hexapeptide baits, such that a significant proportion of these proteins does not bind (i.e., is depleted) and removed in the flow-through. Upon elution of bound proteins from the hexapeptide bead library, the resultant complex mixture is compressed for dynamic range owing to the “partial depletion” of higher abundance proteins and “enrichment” of lower-abundance proteins. After DRC, the complex sample can be further resolved at the protein level5 or integrated with advanced peptide fractionation schemes6, to result in significantly increased protein identifications by mass spectrometry.
Despite the demonstrated effectiveness of DRC in increasing protein identifications within complex protein mixtures, important questions remain about its effectiveness in other contexts important to proteomic studies. One question pertains to the potential of DRC for enhancing identification of protein post-translational modifications (PTMs)- information that is often critical to understanding a protein’s functional state or potential role in disease. It remains unknown whether the presence of PTMs might affect overall protein capture efficiencies by hexapeptide libraries as PTMs can significantly modify physicochemical properties of targeted proteins, or if the existing diversity of hexapeptides within libraries is able to account for these potential modifications. Glycosylation is a prominent candidate PTM, wherein relatively large and chemically complex carbohydrate structures are added to modified proteins7–9.
A second question pertains to the incorporation of DRC in quantitative proteomic studies measuring relative protein abundance levels between samples via stable isotope labeling. Incorporation of DRC into quantitative proteomic studies has the potential to expand the sensitivity of such analyses. However, this is not without challenges. It has been shown2 that proteins of higher absolute abundance do not retain their quantitative accuracy subsequent to hexapeptide library treatment; these saturate their peptide binding partners, resulting in a loss of the unbound portion of the protein. In contrast, proteins present at lower absolute abundance retain their quantitative accuracy subsequent to treatment with hexapeptide libraries, since these do not saturate their peptide binding partners and are fully captured2, 5, 10, 11. These studies all relied either on “spike-in” of single proteins or whole proteomes at known molar amounts in order to determine the absolute concentrations within which proteins retained their quantitative accuracy subsequent to treatment with hexapeptide libraries.
However, in the practical application of DRC (e.g., differential analysis of clinical samples or cellular lysates), distinguishing a priori proteins of higher absolute abundance from proteins of lower absolute abundance is not possible. Thus, it is also not possible to discriminate those relative abundance ratios which are most likely accurate (derived from proteins with lower absolute abundance) and those that are most likely altered from DRC treatment (derived mostly from proteins with higher absolute abundance). This information is critical when prioritizing proteins based on relative abundance differences between samples (e.g., healthy versus disease states or cellular functional states) for further validation. Therefore, a workflow that facilitates such discrimination is needed for incorporating the enhanced sensitivity offered by DRC in quantitative proteomic studies.
In this work, we sought to answer the two questions above using the proteome of human whole saliva as a model sample. Whole saliva, with its non-invasive and easy collection, is gaining appreciation as a fluid rich in protein content6 for the potential clinical detection of systemic disease12–14. In addressing the first question, we targeted protein glycosylation, known to be a key PTM for salivary protein function15–19, and providing a suitable example to test the effects of DRC on PTM identification. We found that DRC provides a significant increase in the number of N-linked salivary glycoproteins identified, greatly expanding the known catalog of N-glycosylated proteins in this fluid. In addressing the second question, we evaluated a workflow that incorporated DRC into relative protein abundance profiling of pooled saliva samples from healthy women and women with metastatic breast cancer. Our results suggest that a workflow using multidimensional peptide fractionation and parallel sample analysis with and without DRC treatment allows for categorization of proteins with putatively higher absolute abundance (i.e. detected without the need for DRC treatment) from those with putatively lower absolute abundance (i.e. only detected with DRC treatment). Importantly this workflow retains the enhanced sensitivity offered by DRC enabling us to identify abundance differences that are putatively associated with breast cancer from each of the above categories (higher- and lower-abundance proteins). Taken together, our results extend the use of DRC via hexapeptide libraries for proteomic studies.
EXPERIMENTAL PROCEDURES
Whole saliva collection and processing for glycoprotein analysis
Prior to saliva collection, 14 healthy human volunteers (8 male, 6 female) did not eat or drink for 90 min, and lightly rinsed their mouth with water. Fresh, unstimulated saliva was collected via drooling into 50 mL falcon tubes that were chilled on ice. The saliva was pooled, clarified by centrifugation twice at 12 000 g at 4°C and supplemented with protease inhibitors (Roche Complete EDTA-free). 10 mL of pooled, clarified saliva was boiled in equal volume of 100mM Tris-Cl, pH 6.8, 4% SDS, 10% mercaptoethanol, and 20% glycerol and labeled as “Untreated saliva” prior to being stored at −20°C.
Treatment of whole saliva for DRC prior to glycoprotein identification analysis
Two different methods were used for DRC of ~250 mL of the remaining amount of pooled saliva. In the first method (DRCstd saliva), 100 mL of pooled saliva was incubated with 200 μl pre-washed PM beads (Bio-Rad Inc., Hercules, CA) at room temperature with tumbling for 18 hours. PM beads were washed three times with PBS and flow through fraction was subsequently treated with 200 μl of LIB2 beads (identically as done for PM treatment). LIB2 beads were similarly washed with PBS three times. Protein was eluted from PM and LIB2beads by boiling in elution buffer (100 mM Tris-Cl, pH 6.8, 4% SDS, 10% mercaptoethanol, and 20% glycerol) prior to being stored at −20°C. These eluates were designated as either PMstd or LIB2std. In a second DRC method performed in parallel (DRCpH4_7_9 saliva), pooled saliva was split into three aliquots (45 mL each), and buffered to three different pH values before treatment with hexapeptide libraries. pH of saliva was adjusted by addition of 10X buffer concentrates (1.5M sodium chloride buffered with either 250 mM acetate pH 4.0, 250 mM phosphate pH 7.4 or 250 mM Tris-HCl pH 9.0) to clarified saliva to a final concentration of 1X buffer (and the final pH of saliva was measured at 4.0, 7.4 and 9.0, respectively). Each pool of pH adjusted saliva (50 mL) was separately incubated with 100μL PM beads at room temperature with tumbling for 18 hours. Beads were washed three times with 1X buffer at same pH as the incubation conditions and flow through fractions from each of the PM treatments were subsequently incubated with LIB2 beads. LIB2 incubation was performed identically as for PM treatment and subsequently the beads were washed three times at the same pH as the incubation conditions. Protein elution of each treatment was done as before with boiling of PM or LIB2 beads in elution buffer (100 mM Tris-Cl, pH 6.8, 4% SDS, 10% mercaptoethanol, and 20% glycerol). Eluates from PM beads incubated with saliva at three pH values (4.0, 7.4 and 9.0) were pooled and designated as PMpH4_7_9 saliva, while eluates from LIB2 beads were pooled and designated as LIB2pH4_7_9 saliva and stored at −20°C.
Sample processing for trypsin digestion, SDS-PAGE and glycoprotein/phosphoprotein staining
Untreated saliva, PMstd saliva, LIB2std saliva, PMpH4_7_9 saliva and LIB2pH4_7_9 saliva were acetone precipitated (4 volumes of ice-cold acetone per volume of protein sample) overnight at −20°C, and the protein pellets washed twice in ice-cold acetone prior to dissolving in 50 mM Tris pH 8.0, 5 mM EDTA, 0.5% SDS. Protein amount per sample was determined by BCA protein assay. For trypsin digestion, dissolved protein pellets were diluted 10-fold in 50 mM Tris, pH 8.0, 5 mM EDTA, pH 8.0, without SDS to bring total SDS levels to 0.05%. DTT was added to a final concentration of 5 mM followed by treatment of protein samples at 60°C for 1 hour. After cooling samples to room temperature, trypsin digestion was performed overnight at 37 °C at a 1:25 (w/w) ratio.
For SDS-PAGE analysis, equal volume of 2X SDS sample buffer was added to protein samples and they were boiled at 100°C for 5 minutes, cooled, centrifuged, and equal amount of protein loaded per lane followed by SDS-PAGE analysis. Visualization of salivary glycoproteins was done after ProQ-emerald staining (Invitrogen, CA) and total protein by coomassie staining. ProQ Emerald and Diamond staining were performed as per manufacturer provided instructions.
Glycopeptide enrichment by hydrazide chemistry
Tryptic peptides were purified by sequential steps of MCX (Oasis, Waters) and C18 (Sep-Pak, Waters) cartridge clean-up. Glycopeptide capture was performed using hydrazide chemistry essentially as detailed previously 20, 21. Dried tryptic peptides were dissolved in coupling buffer (100 mM sodium acetate, 150 mM NaCl, pH 5.5) at a concentration of 1.5 mg/500 μL. Undissolved solids were removed by centrifugation, and the supernatant was processed further. To oxidize cis-diol groups of carbohydrates to aldehydes, sodium periodate was added at a final concentration of 10 mM final concentration to the peptide solution and the sample incubated in the dark with rotation at room temperature for 1 h. C18 clean up (Sep-Pak, Waters) was used to remove excess sodium-periodate from the peptide solution. Subsequently, 200 μl (bed volume) of affigel resin (Bio-Rad, CA) was pre-washed thrice with coupling buffer, and coupled with 1.5 mg of peptides redissolved in 800 μl 80% ACN, 0.1% TFA. Coupling was allowed to proceed overnight (18 hours) at room temperature with rotation.
After the coupling reaction, affigel resin was washed extensively to remove non-glycopeptides as follows. Three washes were performed with 1 mL of 1.5 M NaCl each, two washes with 1 mL of 80% (v/v) aqueous ACN each, two washes with 1mL of water each, and three final washes with 1 mL of freshly prepared 100 mM NH4HCO3 each time. All washes were done by rotation at room temperature for 15 minutes. N-Glycopeptides were released by addition of 10 μL of PNGase F (500 units/μL; New England Biolabs, Beverly, MA) in 200 μL of 100 mM NH4HCO3 and overnight incubation at 37 °C. The supernatant, containing released deglycosylated peptides, was collected by centrifugation and combined with the supernatant of an 80% ACN wash, dried and purified by C18 clean up. 10% of the peptides were directly analyzed on an LTQ-Orbitrap mass spectrometer, while 90% of the same sample was reserved for fractionation as described below.
Collection and processing of whole saliva from healthy and metastatic breast cancer patients for mTRAQ labeling
Unstimulated whole saliva was collected via drooling from two groups: female subjects diagnosed with metastatic breast cancer and healthy patients as a control. In each case, saliva was collected with written informed consent using an IRB approved protocol. Subjects with metastatic breast cancer had not undergone any treatment or surgery (including tissue biopsy) at the lesion site prior to saliva collection. Unstimulated whole saliva was obtained using a standard, controlled protocol22 by first having each subject swallow and then expectorate continuously into a 50 mL sterile, polypropylene conical tube for a period of 5 min. This resulted in about 2–5 mL of total saliva from each subject. Following collection, the samples were immediately placed on ice and stored at −70°C until further processing. Subjects included in the study were free of confounding conditions: periodontal/auto-immune disease, a prior history of diseases or current use of potentially confounding medications.
After thawing saliva samples on ice, they were clarified by centrifugation and pooled such that 1.5 mL of saliva was used from each subject to generate a total pooled saliva sample of 15 mL from healthy and breast cancer subjects each. Protein concentration was determined on each sample by BCA assay. 1 mL of pooled, untreated saliva from healthy or breast cancer patients were boiled in SDS sample buffer, acetone precipitated, and redissolved in 50 mM Tris pH 8.0, 5 mM EDTA. From this, 600 μg protein from each pooled sample (healthy or breast cancer patients) was stored at −20°C. This was later used for western blotting confirmation experiments. 100 μg tryptic peptides from the healthy pooled sample and 100 μg tryptic peptides from metastatic breast cancer pooled sample were selected for mTRAQ labeling (Applied Biosystems, CA). The healthy saliva peptide mixture was labeled with the normal mTRAQ, and the breast cancer saliva peptide mixture was labeled with the heavy mTRAQ, as per manufacturer provided instructions. 14 mL (14 mg protein) of pooled saliva from healthy volunteers was processed by DRC at standard pH of saliva using sequential use of 50 μL volume each of PM or LIB2 and pooled (DRCstd saliva). In parallel and identical fashion, DRC was performed on pooled saliva from breast cancer patients. mTRAQ labeling was performed on healthy and breast cancer saliva by labeling 100 μg tryptic peptides from each sample with mTRAQ light and heavy label, respectively.
Strong cation exchange (SCX) HPLC fractionation of glycopeptides and 3D-peptide fractionation of mTRAQ-labeled peptides
SCX HPLC was performed on a Magic 2002 HPLC system coupled with a Magic Variable Splitter set at position R4 (Michrom BioResources, Inc., Auburn, CA) using a Polysulfethyl A column, 150 mm length × 1.0 mm i.d., 5 μm particles, 200 Å pore size (PolyLC, Inc., Columbia, MD). The sample was dissolved in 250 μL of SCX buffer A (20% v/v acetonitrile (ACN), 10 mM KH2PO4, pH 2.7 with phosphoric acid) and loaded onto the column. For glycopeptide fractionation, peptides were eluted at a flow rate of 35 μL/min with a gradient of 0–20% buffer B (20% v/v ACN, 10 mM KH2PO4, pH 3.0, 500 mM KCl) over 15 min followed by a gradient from 20% to 100% buffer B for 6 min. During the chromatography run, absorbance at 215 and 280 nm was monitored, fractions were collected at 3-min intervals, and each fraction vacuum centrifuged to dryness. This fractionation resulted in the collection of 6 fractions. Each SCX fraction was purified by C18 ‘STAGE’ tips23 prior to LTQ-Orbitrap analysis. For the health to breast cancer comparison, 3D-fractionation for mTRAQ labeled peptides was done essentially as described previously6.
Capillary LC-MS/MS
Peptides were dissolved in 5 μL of load buffer (98:2:0.01 water/ACN/formic acid) and processed for LC-MS/MS analysis on an LTQ-Orbitrap mass spectrometer essentially as described previously6 with the following modifications. For analysis of glycopeptides, dissolved peptides were eluted using a gradient of 2–40% ACN in 0.1% formic acid over 60 min with a constant flow of 250 nL/min. For analysis of mTRAQ labeled peptides, they were eluted over the same gradient but over 180 minutes. For the glycopeptide studies, 5 most intense ions from the full scan were selected for fragmentation by collision-induced dissociation (normalized collision energy, 35%) in the LTQ ion trap with automatic gain control settings of 5000 ions or 100 ms concurrent to full-scan acquisition in the orbital trap, while for mTRAQ-labeled peptides, 8 most intense ions were processed similarly.
Protein identification for N-linked glycopeptides and mTRAQ labeled peptides
Data generated from μLC-MS/MS analysis was extracted using ReAdW and searched with Sequest V27.0 against a composite database consisting of the NCBI human database V200806 and its reversed database (70,711 entries which includes reverse database entries). Search parameters used for identification of N-linked glycoproteins were partial tryptic digestion, variable modification of 15.9949 Da on methionine, and 0.984 Da for deamidation of asparagine to aspartic acid induced by PNGase F treatment. As a further filtering, only those peptide sequences having the conserved sequence for N-linked glycosylation, Asparagine-X-Serine/Threonine (where X is not Proline) were considered to be true matches to glycosylated peptides. Search parameters used for identification of mTRAQ labeled peptides were partial tryptic digestion, fixed modification of 140.0949 Da of lysine residues along with N-termini of peptides and fixed modification of 45.9877 Da of cysteine, variable modification of 4.007 Da of lysine residues along with N-termini of peptides, and variable modification of 15.9949 Da on methionine residues.
Sequest output was organized and peptide/protein probabilities were calculated through Peptide/Protein Prophet respectively, using Scaffold (Proteome Software, Inc., Portland, OR). For glycoprotein identification analysis, false discovery rate (FDR) at protein level was determined using the equation, false discovery rate (FDR) % = [nreverse/(nforward + nreverse)] × 100, where nforward equals number of protein matches from the forward database, and nreverse equals number of reversed database matches. As part of the Scaffold software, the reported proteins were subjected to assignment of a protein probability (based on the Protein Prophet program), in order to minimize peptides matching redundantly to proteins and to report the minimal number of unique proteins represented by our data.
Bioinformatic Analysis of N-linked glycoproteins
Proteins were analyzed using Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Inc., Redwood City, CA). Core analysis was performed using direct and indirect relationships. With Fisher’s Exact test, core comparison analysis was used to compare functional diversity of N-linked salivary glycoproteins identified in this study and previously identified saliva proteins6.
Quantification of mTRAQ labeled peptides
The computational proteomics portion of the workflow used for identifying and quantifying proteins after mTRAQ labeling of peptides is similar to a recent report24. Within this workflow, SEQUEST results were used as input to the Trans-Proteomic Pipeline (TPP v4.3 JETSTREAM rev 1, Build 200909091257 (MinGW), http://www.proteomecenter.org) Peptide Prophet and Protein Prophet were used with to select peptides with .05 probability and proteins with .9 probability respectively. XPRESS and ASAPRatio were used for quantification with mass tolerance 1.0 Da, N-terminus and lysine mass difference 4.0, and peak m/z range .5. We used MAYU software25 to generate 1% peptide FDR lists for both Untreated saliva and DRCstd saliva datasets. Resulting Mayu data and Transproteomic Pipeline data (PeptideProphet, ProteinProphet, and ASAPRatio26) were combined using Perl scripting software and stored in an Oracle 11g (Oracle Corporation, Redwood Shores, CA) database enabling novel analyses.
Western blotting for validating breast cancer associated protein abundance changes in pooled saliva
Equal amounts of pooled saliva samples (without DRC treatment) from healthy subjects and metastatic breast cancer subjects were resolved by SDS-PAGE after boiling protein samples in SDS-sample buffer27 and proteins were transferred to PVDF membrane for Western blotting. Antibodies used for CD44a, Kallikrein 13, SCGB2A1 (Secretoglobin family 2A member 1), SCGB2A2 (Secretoglobin family 2A member 2), Neuregulin 3, and Selectin P were purchased from Abcam (Cambridge, MA).
RESULTS
Dynamic range compression (DRC) of saliva by hexapeptide libraries
We first sought to evaluate the effect of DRC using hexapeptide libraries on the detection and identification of salivary glycoproteins. For these studies we pooled saliva from 14 healthy volunteers, setting aside a portion of this saliva for processing without DRC treatment (referred to as Untreated saliva) for glycoprotein analysis.
We analyzed the remaining portion of pooled saliva by DRC using two methods. In both DRC methods, proteins were first treated with PM hexapeptide beads, the flow through collected, and subsequently treated with another set of hexapeptide beads, LIB2. LIB2 is a carboxylated derivative of PM5, which we employed to minimize potential losses of proteins that might bind insufficiently to the PM library. For the first DRC method, DRCstd, salivary proteins were directly applied to the hexapeptide bead libraries at the natural/standard pH of saliva (~6.5). For the second DRC method, DRCpH4_7_9, equal portions of saliva were adjusted to pH 4, 7 or 9 and then applied to the hexapeptide bead libraries. pH adjustment was used because it has been shown to potentially capture a broader set of proteins using hexapeptide libraries 28–30. For DRCpH4_7_9 the proteins bound at each pH value to the PM beads were eluted and combined together prior to glycoprotein analysis; similarly, the proteins bound at each pH valued to the LIB2 beads were eluted and combined together, prior to glycoprotein analysis. Rather than analyze each pH fraction separately, we combined proteins bound and eluted at each pH to potentially capture a larger set of glycoproteins in a single fraction while reducing downstream analysis steps.
Evaluating the effect of DRC on glycoprotein detection
With three different groups of saliva protein samples in hand (Untreated, DRCstd and DRCpH4_7_9) we evaluated the effect of DRC on detection of glycoproteins. We first visualized proteins in the sample groups by standard SDS-PAGE and coomassie protein staining (Figure 1A). For both DRC methods, PM and LIB2 reduced the levels of the most abundant proteins relative to Untreated saliva while apparently capturing distinct proteins. Although generally similar, differences are observable in the protein patterns for DRCstd and DRCpH4_7_9 methods (Figure 1A left panel).
Figure 1. Effect of dynamic range compression (DRC) of saliva on detectability of salivary glycoproteins and strategy used for identification of N-glycoproteins in Untreated saliva versus DRC treated saliva.
(A) Equal amounts of protein from untreated saliva or saliva processed for DRC by two different methods were separated by SDS-PAGE. Total protein was visualized by coomassie staining and glycoproteins visualized by ProQ Emerald staining. For each DRC method, DRCstd or DRCpH4_7_9, proteins were serially treated with PM hexapeptide libraries followed by LIB2 hexapeptide libraries. See text for details. (B) Workflow employed to capture and identify N-linked glycoproteins from Untreated or DRC saliva by mass spectrometric analysis.
We next visualized glycoproteins in each sample using a ProQ Emerald 300 glycoprotein gel stain (Figure 1A right panel). A number of high abundance proteins in Untreated saliva were reactive with the glycoprotein stain and detected, suggesting their modification by glycosylation. Several new bands, notably in the molecular weight region between 10–40 kDA, not detected in the Untreated saliva were detected using both DRC methods. These proteins are likely lower in abundance in Untreated saliva, but enriched by treatment with hexapeptide libraries and thus only detectable after DRC. The samples processed using the DRCpH4_7_9 method generally showed increased glycoprotein staining intensities, for both the PM and LIB2 hexapeptide libraries, indicating that this method may have advantages over the DRCstd method.
Evaluating the effect of DRC on N-linked glycoprotein identification via MS/MS
We next sought to evaluate the effect of DRC on identification of glycoproteins via mass spectrometry. We chose to focus on N-linked glycoproteins given that they account for ~90% of all glycoproteins, are commonly found in secreted fluids such as saliva7, and methods for their enrichment and analysis by MS/MS are well established20, 31, 32.
For each processing method, Untreated saliva, DRCstd saliva and DRCpH4_7_9 saliva, proteins were digested with trypsin and N-linked glycopeptides captured by solid-phase hydrazide chemistry20, 31, 32 (Figure 1B) as described in experimental procedures. Similar to the glycoprotein detection experiments above, proteins captured either by PM and LIB2 for both DRC methods were separately processed for glycopeptide analysis.
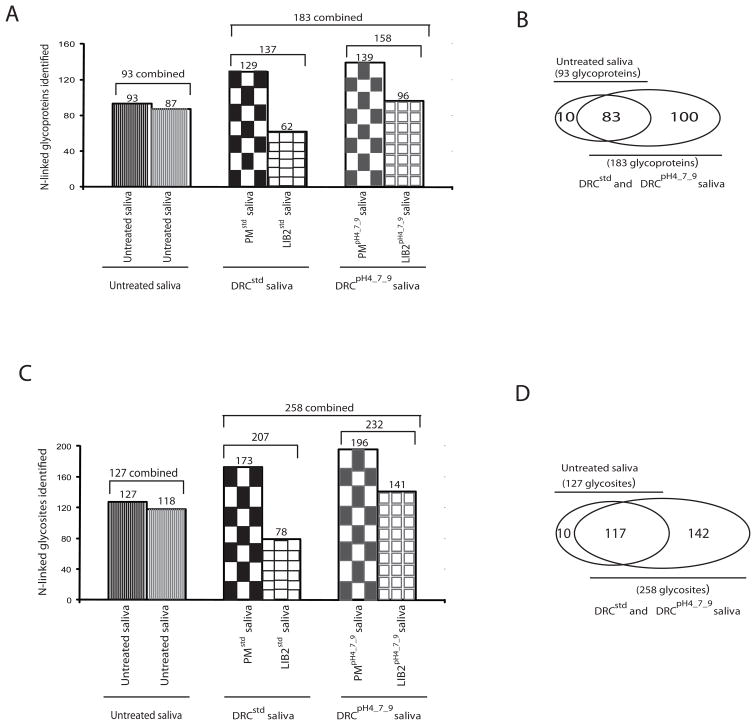
Results from our glycopeptide identification experiments are shown in Figure 2. From two replicate Untreated saliva samples, N-linked glycopeptides derived from 93 distinct glycoproteins were identified (Figure 2A). By contrast, both DRC methods yielded substantially more N-linked glycoproteins. The DRCstd method yielded N-linked glycopeptides derived from 137 distinct glycoproteins, a majority (129/137) of which were identified by analysis of PM treated saliva. LIB2 treatment yielded only 8 distinct glycoproteins that were not identified using PM. The DRCpH4_7_9 method yielded a modest increase in N-linked glycoprotein identifications compared to the DRCstd method, and a majority of the identifications (139/158) were obtained by analysis of PM treated saliva. LIB2 added 19 distinct glycoproteins not identified using PM. The two DRC methods together identified a total of 183 distinct glycoproteins.
Figure 2. Effect of DRC on identification of N-linked glycoproteins and glycosites in saliva.
(A) Total N-linked glycoproteins identified in Untreated saliva versus both DRC methods. (B) Venn diagram illustrating glycoproteins identified in DRC treated saliva and those “missed” by DRC treatment. (C) Total N-glycosites identified in untreated saliva versus both DRC methods. (D) Venn diagram illustrating glycosites identified in DRC treated saliva and those ‘missed’ by DRC treatment.
We next investigated whether or not the use of DRC sacrificed identification of some glycoproteins, ostensibly because these modified proteins do not bind to the hexapeptide libraries. Comparing all distinct glycoprotein identifications from the two DRC methods combined (DRCpH4_7_9 and DRCstd) with those identified in Untreated saliva revealed that 10 glycoproteins were missed when using DRC (Figure 2B).
We also considered the numbers of glycopeptides identified using each different method. Our MS/MS-based identification of glycopeptides provides information not only on the salivary proteins from which they are derived, but also the unambiguous assignment of the exact amino acid site of N-linked glycosylation, which we call “glycosites”. Using this terminology, 258 distinct glycosites were identified by the combined DRC treatments compared to 127 glycosites identified in Untreated saliva (Figure 2C). A total of 10 glycosites were missed with DRC treatment (Figure 2D), corresponding to the 10 missed glycoproteins shown in Figure 2B.
A listing of identified salivary glycoproteins and their corresponding glycosites is available in Supplementary Table 1. Additionally, the entire dataset of glycoproteins, their corresponding glycosites, and annotated spectra are contained in a Scaffold file and may be downloaded from Tranche at ProteomeCommons.org using the hash: dESMiAHFtXsZck1IrrU+W/yxNM/1Jv10dI1av8gwJ8VWo5freKc/ANCPzLBvuCVObR z9dbwoLlQb8otueDw97ZJ3u+kAAAAAAAACkA==. The Scaffold file can be viewed using the freely available Scaffold viewer from Proteome Software, Inc. (http://www.proteomesoftware.com/).
Evaluating a workflow for quantitative proteomics using DRC and differential stable isotope labeling
We next addressed our second question: Can DRC be integrated to a workflow enabling relative abundance measurements via stable isotope labeling? Based on previous studies2, 5, 10, 11, 33, proteins of lower absolute abundance retain their relative abundance levels after treatment with hexapeptide beads, while proteins of higher absolute abundance do not (as they saturate their heaxapeptide binding partner and the portion of unbound protein is lost in the flow-through). Unfortunately, when applying DRC to samples of interest it is not possible to distinguish proteins of low absolute abundance, whose relative abundance ratios are expected to be correct2, 5, 10, 11, from proteins of high absolute abundance, whose relative abundance ratios are expected to be altered2, 5, 10, 11. We reasoned that addressing this central problem required performing two parallel analyses where one portion of the sample is analyzed using DRC and a second portion of the sample is analyzed without DRC. Similar to a previous report5, proteins identified only with the use of DRC are then categorized as low abundance proteins, while proteins identified without DRC are categorized as higher abundance proteins.
We sought to evaluate the proposed proteomic workflow via comparison of saliva from healthy women and women clinically diagnosed with metastatic breast cancer. Using separately pooled saliva from 10 healthy women and 10 metastatic breast cancer women, two portions of each pool were separately prepared, one without DRC (Untreated) and the other using the DRCstd method as described in experimental proedures. Figure 3A details the workflow used, which included peptide labeling with the mTRAQ reagent24 to enable relative abundance measurements.
Figure 3. A DRC-based relative protein abundance profiling workflow applied to whole saliva from healthy women and women with metastatic breast cancer.
(A) mTRAQ reagent stable isotope labeling and steps used for obtaining relative protein abundance ratios. The workflow included identical and parallel analysis of Untreated saliva proteins (filled lines) and DRCstd saliva proteins (dashed lines) obtained from healthy versus cancerous women. For the comparison, equal amounts of saliva protein from 10 healthy women were pooled and compared to equal amounts of saliva protein pooled from 10 women with metastatic breast cancer. (B) Overlap of proteins identified in Untreated saliva samples versus DRCstd saliva samples. The 533 proteins identified only in DRC saliva would be considered low abundance proteins, while the 708 proteins identified without DRC would be considered high abundance proteins. See text for details. (C) Results of Western blotting validation experiments in pooled healthy or pooled metastatic breast cancer saliva for selected proteins showing differential abundance ratios in either Untreated or DRC treated samples.
We identified a total of 708 proteins (1% peptide-level FDR) in the Untreated saliva protein sample (Figure 3B). Among these, a total of 79 proteins showed 2-fold differential abundance between the healthy and metastatic breast cancer pools (Supplementary Table 2). All relevant mass spectrometric information for the entire Untreated dataset is included in Supplementary Table 3. MS/MS scans for single-peptide hits from this dataset are presented in Supplementary Figure 1. For confirmation of our quantitative approach overall we selected for Western blotting three different proteins from the Untreated saliva sample, based on their relative abundance differences and known associations with breast cancer (CD44a36, 37, SCGB2A2/Secretoglobin family 2A member 238, and Kalikrein 1339) (Figure 3C). The relative abundance ratios of these proteins measured by Western blotting were consistent with their mTRAQ relative abundance ratios.
We identified a total of 1032 proteins (1% peptide-level FDR) from DRCstd saliva samples (Figure 3B). Among the identified proteins, a total of 148 proteins showed 2-fold differential abundance between the healthy and metastatic breast cancer DRCstd pools (Supplementary Table 4). All relevant mass spectrometric information for this dataset is included in Supplementary Table 5. MS/MS scans for single and double-peptide hits from this dataset are presented in Supplementary Figure 2.
We next sought to answer an important question: Are at least some of the relative abundance levels measured for proteins after DRC correct? As shown in Figure 3B, about half, 499, of the proteins identified and quantified in the DRC sample were also identified and quantified in the Untreated sample. Based on their identification in the Untreated samples, we categorize them as higher-abundance proteins. We compared the abundance ratios for these 499 common proteins, measured in either the Untreated sample or the DRC treated sample, assuming that the abundance ratios measured from the Untreated sample were the correct ratios. At first look, 317 of the common 499 proteins exhibited homodirectional abundance differences in the Untreated and DRC samples (R-squared value of 0.626 after removing three outlier protein ratios) (Supplementary Table 6). However, upon a closer look at the measured ratios, 131/317 (~41%) homodirectional proteins showed a greater than 25% alteration in their abundance ratio in the DRC-treated sample compared to the Untreated samples, indicating that DRC treatment does introduce significant error in many of these higher abundance proteins.
A second important question then remained: What about relative abundance ratios of putatively lower abundance proteins identified and quantified only after DRC treatment? Based on the analysis above, we reasoned that a majority of the 148 differentially abundant proteins from the DRC dataset should be correct, given that these are from putatively lower abundance proteins whose relative abundance levels should be retained even after DRC. To test this reasoning, we conducted quantitative Western blotting experiments in the pooled healthy and metastatic breast cancer pooled saliva samples without DRC treatment. We compared the Western blot the results to the differential abundance ratios measured by mTRAQ in the mass spectrometer after DRC treatment. Because it was not practical to confirm by Western blotting all of the 148 differentially abundant proteins, we prioritized proteins of interest based on previous studies linking them to cancer and/or breast cancer specifically40–44: Selectin P, Rab6A, Rab27A, Rab3D, Rab5C, SCGB2A1, Neuregulin-3. From these putatively ‘lower-abundance’ salivary proteins, we chose Selectin P, SCGB2A1 and Neuregulin-3 for Western blotting confirmation experiments, but opted against the Rab family of proteins owing to their high homology with each other and the potential for antibody cross-reactivity. As shown in Figure 3C, Western blotting for Selectin P, SCGB2A1, and Neuregulin-3 showed differential abundance levels between the healthy and metastatic saliva pools pre-DRC treatment consistent with the observed mTRAQ abundance ratios obtained after DRC.
DISCUSSION
In this report, we have demonstrated that the enhanced sensitivity offered to mass spectrometry-based shotgun proteomics by hexapeptide libraries extends to two important arenas of proteome research: PTM characterization and profiling protein relative abundance. We used whole human saliva as a representative sample for these studies, given its wide dynamic range of protein abundance, prominence of protein PTMs, especially glycosylation, and potential value for diagnosis of human diseases. Our results confirmed that DRC increases ability to identify glycosylated proteins in whole saliva, and demonstrated a workflow for the successful use of DRC for profiling relative abundance changes in salivary proteins associated with metastatic breast cancer.
DRC increases the detection and identification of post-translationally modified proteins from complex mixtures
To determine the effect of DRC on identification of post-translational modifications, we focused on salivary N-linked glycoproteins. The addition of complex carbohydrate groups introduces a significant chemical change to proteins, which could potentially affect the binding of glycoslyated proteins with their hexapeptide partners, thereby reducing the effectiveness of DRC for this class of proteins. Despite the chemical alterations introduced from glycoslyation, we found that DRC nearly doubled total identifications of salivary N-linked glycoproteins and their corresponding glycosites. These results suggest that glycoslyation does not deter binding to hexapeptide libraries or that existing hexapeptide diversity in the latter provides adequate binding partners for proteins as well as their post-translationally modified variants. Among PTMs, glycosylation is relatively complex and can have profound effects on the physico-chemical properties of targeted proteins. Based on our results, we propose that DRC should serve as a general tool for increasing identification of various protein PTMs of biological interest in complex samples.
Despite the overall increases in glycoproteins identified using DRC, our results suggest that a small proportion of glycoproteins are “missed”. Ten glycoproteins identified in the Untreated saliva sample were not identified using either DRCstd or DRCpH4_7_9 methods. The missed glycoproteins were: apolipoprotein H precursor, calcium activated nucleotidase 1, disulfide isomerase, folate receptor 1 precursor, G-protein-coupled receptor 56 isoform b, LY6/PLAUR domain containing 5 isoform A, lysosomal-associated membrane protein 2 isoform C precursor, sulfatase 2 isoform b precursor, tetraspan 1, transmembrane protease, serine 11E2. The glycopeptides from these proteins were not restricted to any unique SCX fractions analyzed. Analysis of isoelectric point, molecular weight, and gene ontology information did not reveal any conserved features that distinguished the ten “missing” glycoproteins from those identified when using DRC. Even though performing DRC at multiple pH conditions increased overall identifications, it is not sufficient to completely eliminate losses. These losses may result from peptide undersampling during mass spectrometry or a lack of a hexapeptide partner with sufficient binding strength for these proteins. We have observed similar losses of a small percentage of proteins when analyzing total saliva proteins, glycosylated or otherwise6. Similar losses have been observed during mass spectrometric analysis of other samples by other researchers as well 1, 5.
In spite of the relatively small losses when using DRC, we have more than tripled the number of known N-linked glycoproteins and glycosites in saliva, expanding the catalog from 62 glycoproteins found in past studies45, 46 to 193 glycoproteins. Here again, nearly a dozen salivary N-glycoproteins identified in these past studies were not identified by us. It is difficult to know whether these differences are due to losses from DRC, changes in methodology used, or due to biological variability of the samples used in each study. Regardless, it is clear that the use of DRC enabled a significant increase in the identification of glycoslyated proteins from saliva using shotgun proteomics.
We performed initial bioinformatic analyses on our expanded catalog of salivary N-linked glycoproteins using Ingenuity Pathway Analysis (IPA) to better understand characteristics of these proteins. These results are shown in Supplementary Figure 3. Comparing the catalog of N-linked glycoproteins to our previously obtained catalog of over 2000 saliva proteins6, as expected, a higher percentage of salivary N-linked glycoproteins compared to total saliva proteins are located either in the extracellular space or plasma. Additionally, the salivary glycoproteins are distributed across mostly the same functional and disease-related categories as other known salivary proteins. This initial analysis suggests that targeting the N-linked glycoproteome might be a viable alternative to characterizing the entire salivary proteome for disease diagnostic studies.
DRC can be incorporated to a workflow enabling sensitive profiling of relative protein abundance differences
Previous work using ‘spike-in’ of either individual proteins or a complex proteome at known absolute abundance have shown that those of lower absolute abundance preserve their relative abundance ratios after treatment with hexapeptide libraries for DRC5, 10, 11, 33, while those with higher absolute abundance (concentrations of ~1 μM or more) do not2. However, in the practical application of DRC for quantitative proteomic studies (e.g. biomarker discovery studies), it is very difficult, if not impossible, to distinguish between proteins of lower and higher absolute abundance a priori and thus make the determination of which relative abundance ratios are most likely correct.
To address this issue, we presented a workflow, wherein one portion of a complex sample is analyzed without DRC and a second portion of the sample is processed using DRC. Using this workflow, proteins identified and quantified only after DRC and extensive peptide fractionation are classified as putatively low abundance proteins, consistent with terminology used in other descriptions of DRC5; meanwhile those proteins identified without DRC are classified as putatively high abundance proteins. Protein relative abundance ratios of putatively low abundance proteins can be assigned a higher confidence when prioritizing proteins for possible follow-up validation; meanwhile, for protein relative abundance ratios of putatively high abundance proteins (identified both with and without DRC), the ratios measured without DRC should be assigned a higher confidence when prioritizing proteins for possible follow-up validation.
We demonstrated the effectiveness of this workflow by profiling relative protein abundance differences between pooled saliva from healthy women and pooled saliva from women with metastatic breast cancer. We identified and confirmed via Western blotting putatively high abundance proteins (CD44a, Kallikrein 13, and SCGB2A2) and putatively low abundance proteins (SCGB2A1, Neuregulin-3, and Selectin P) showing relative abundance differences between the pooled patient samples. Among the higher abundance salivary proteins, SCGB2A2 (also known as Mammoglobin B), and Keratin 13 have been previously detected in saliva as potential breast cancer biomarkers 34, 35. The use of our workflow enabled the identification several additional proteins with potential as biomarkers (CD44a, SCGB2A1, Neuregulin-3, and Selectin P), with the latter three proteins being detected only with the use of DRC, most likely due to their low abundance in saliva. Although further validations of these proteins are needed, this study provides seminal findings expanding our knowledge of breast-cancer associate proteins that are detectable in whole saliva with possible value as non-invasive biomarkers of this cancer.
In summary, our results should expand the use of DRC with hexapeptide libraries as a general tool for PTM characterization and relative protein abundance profiling using mass spectrometry-based proteomics.
Supplementary Material
Acknowledgments
This research was funded in part by NIH grant 1R01DE017734 and a Seed Grant from the Academic Health Center at the University of Minnesota and University of Minnesota Graduate School Biomedical Informatics and Computational Biology Fellowship awarded to SKV. We are grateful to Dan Wolbrink for assisting with saliva collections from breast cancer patients and Todd Markowski for SCX fractionations. We also thank members of the Center for Mass Spectrometry and Proteomics at the University of Minnesota for instrumental access and support, and the Minnesota Supercomputing Institute for computational proteomics support.
References
- 1.Boschetti E, Righetti PG. The art of observing rare protein species in proteomes with peptide ligand libraries. Proteomics. 2009;9(6):1492–510. doi: 10.1002/pmic.200800389. [DOI] [PubMed] [Google Scholar]
- 2.Mouton-Barbosa E, Roux-Dalvai F, Bouyssie D, Berger F, Schmidt E, Righetti PG, Guerrier L, Boschetti E, Burlet-Schiltz O, Monsarrat B, de Peredo AG. In-depth exploration of cerebrospinal fluid by combining peptide ligand library treatment and label-free protein quantification. Mol Cell Proteomics. 2010;9(5):1006–21. doi: 10.1074/mcp.M900513-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Righetti PG, Boschetti E, Lomas L, Citterio A. Protein Equalizer Technology : the quest for a “democratic proteome”. Proteomics. 2006;6(14):3980–92. doi: 10.1002/pmic.200500904. [DOI] [PubMed] [Google Scholar]
- 4.Thulasiraman V, Lin S, Gheorghiu L, Lathrop J, Lomas L, Hammond D, Boschetti E. Reduction of the concentration difference of proteins in biological liquids using a library of combinatorial ligands. Electrophoresis. 2005;26(18):3561–71. doi: 10.1002/elps.200500147. [DOI] [PubMed] [Google Scholar]
- 5.Roux-Dalvai F, Gonzalez de Peredo A, Simo C, Guerrier L, Bouyssie D, Zanella A, Citterio A, Burlet-Schiltz O, Boschetti E, Righetti PG, Monsarrat B. Extensive analysis of the cytoplasmic proteome of human erythrocytes using the peptide ligand library technology and advanced mass spectrometry. Mol Cell Proteomics. 2008;7(11):2254–69. doi: 10.1074/mcp.M800037-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Bandhakavi S, Stone MD, Onsongo G, Van Riper SK, Griffin TJ. A dynamic range compression and three-dimensional peptide fractionation analysis platform expands proteome coverage and the diagnostic potential of whole saliva. J Proteome Res. 2009;8(12):5590–600. doi: 10.1021/pr900675w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–49. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 8.Lis H, Sharon N. Protein glycosylation. Structural and functional aspects. Eur J Biochem. 1993;218(1):1–27. doi: 10.1111/j.1432-1033.1993.tb18347.x. [DOI] [PubMed] [Google Scholar]
- 9.Peter-Katalinic J. Methods in enzymology: O-glycosylation of proteins. Methods Enzymol. 2005;405:139–71. doi: 10.1016/S0076-6879(05)05007-X. [DOI] [PubMed] [Google Scholar]
- 10.Drabovich AP, Diamandis EP. Combinatorial peptide libraries facilitate development of multiple reaction monitoring assays for low-abundance proteins. J Proteome Res. 2010;9(3):1236–45. doi: 10.1021/pr900729g. [DOI] [PubMed] [Google Scholar]
- 11.Hartwig S, Czibere A, Kotzka J, Passlack W, Haas R, Eckel J, Lehr S. Combinatorial hexapeptide ligand libraries (ProteoMiner): an innovative fractionation tool for differential quantitative clinical proteomics. Arch Physiol Biochem. 2009;115(3):155–60. doi: 10.1080/13813450903154224. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence HP. Salivary markers of systemic disease: noninvasive diagnosis of disease and monitoring of general health. J Can Dent Assoc. 2002;68(3):170–4. [PubMed] [Google Scholar]
- 13.Wong DT. Salivary diagnostics powered by nanotechnologies, proteomics and genomics. J Am Dent Assoc. 2006;137(3):313–21. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]
- 14.Yan W, Apweiler R, Balgley BM, Boontheung P, Bundy JL, Cargile BJ, Cole S, Fang X, Gonzalez-Begne M, Griffin TJ, Hagen F, Hu S, Wolinsky LE, Lee CS, Malamud D, Melvin JE, Menon R, Mueller M, Qiao R, Rhodus NL, Sevinsky JR, States D, Stephenson JL, Than S, Yates JR, Yu W, Xie H, Xie Y, Omenn GS, Loo JA, Wong DT. Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin Appl. 2009;3(1):116–134. doi: 10.1002/prca.200800140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartshorn KL, Ligtenberg A, White MR, Van Eijk M, Hartshorn M, Pemberton L, Holmskov U, Crouch E. Salivary agglutinin and lung scavenger receptor cysteine-rich glycoprotein 340 have broad anti-influenza activities and interactions with surfactant protein D that vary according to donor source and sialylation. Biochem J. 2006;393(Pt 2):545–53. doi: 10.1042/BJ20050695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatton MN, Loomis RE, Levine MJ, Tabak LA. Masticatory lubrication. The role of carbohydrate in the lubricating property of a salivary glycoprotein-albumin complex. Biochem J. 1985;230(3):817–20. doi: 10.1042/bj2300817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loomis RE, Prakobphol A, Levine MJ, Reddy MS, Jones PC. Biochemical and biophysical comparison of two mucins from human submandibular-sublingual saliva. Arch Biochem Biophys. 1987;258(2):452–64. doi: 10.1016/0003-9861(87)90366-3. [DOI] [PubMed] [Google Scholar]
- 18.McBride BC, Gisslow MT. Role of sialic acid in saliva-induced aggregation of Streptococcus sanguis. Infect Immun. 1977;18(1):35–40. doi: 10.1128/iai.18.1.35-40.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prakobphol A, Tangemann K, Rosen SD, Hoover CI, Leffler H, Fisher SJ. Separate oligosaccharide determinants mediate interactions of the low-molecular-weight salivary mucin with neutrophils and bacteria. Biochemistry. 1999;38(21):6817–25. doi: 10.1021/bi990145m. [DOI] [PubMed] [Google Scholar]
- 20.Sun B, Ranish JA, Utleg AG, White JT, Yan X, Lin B, Hood L. Shotgun glycopeptide capture approach coupled with mass spectrometry for comprehensive glycoproteomics. Mol Cell Proteomics. 2007;6(1):141–9. doi: 10.1074/mcp.T600046-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y, Aebersold R, Zhang H. Isolation of N-linked glycopeptides from plasma. Anal Chem. 2007;79(15):5826–37. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- 22.Rhodus NL, Ho V, Miller CS, Myers S, Ondrey F. NF-kappaB dependent cytokine levels in saliva of patients with oral preneoplastic lesions and oral squamous cell carcinoma. Cancer Detect Prev. 2005;29(1):42–5. doi: 10.1016/j.cdp.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663–70. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 24.Kang UB, Yeom J, Kim H, Lee C. Quantitative analysis of mTRAQ-labeled proteome using full MS scans. J Proteome Res. 2010;9(7):3750–8. doi: 10.1021/pr9011014. [DOI] [PubMed] [Google Scholar]
- 25.Reiter L, Claassen M, Schrimpf SP, Jovanovic M, Schmidt A, Buhmann JM, Hengartner MO, Aebersold R. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol Cell Proteomics. 2009;8(11):2405–17. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li XJ, Zhang H, Ranish JA, Aebersold R. Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal Chem. 2003;75(23):6648–57. doi: 10.1021/ac034633i. [DOI] [PubMed] [Google Scholar]
- 27.Bandhakavi S, Xie H, O’Callaghan B, Sakurai H, Kim DH, Griffin TJ. Hsf1 activation inhibits rapamycin resistance and TOR signaling in yeast revealed by combined proteomic and genetic analysis. PLoS One. 2008;3(2):e1598. doi: 10.1371/journal.pone.0001598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fasoli E, Aldini G, Regazzoni L, Kravchuk AV, Citterio A, Righetti PG. Les Maitres de l’Orge: the proteome content of your beer mug. J Proteome Res. 2010;9(10):5262–9. doi: 10.1021/pr100551n. [DOI] [PubMed] [Google Scholar]
- 29.Fasoli E, Farinazzo A, Sun CJ, Kravchuk AV, Guerrier L, Fortis F, Boschetti E, Righetti PG. Interaction among proteins and peptide libraries in proteome analysis: pH involvement for a larger capture of species. J Proteomics. 2010;73(4):733–42. doi: 10.1016/j.jprot.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 30.Righetti PG, Boschetti E, Kravchuk AV, Fasoli E. The proteome buccaneers: how to unearth your treasure chest via combinatorial peptide ligand libraries. Expert Rev Proteomics. 2010;7(3):373–85. doi: 10.1586/epr.10.25. [DOI] [PubMed] [Google Scholar]
- 31.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of N-linked glycopeptides. Nat Protoc. 2007;2(2):334–9. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21(6):660–6. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 33.Frobel J, Hartwig S, Passlack W, Eckel J, Haas R, Czibere A, Lehr S. ProteoMiner() and SELDI-TOF-MS: A robust and highly reproducible combination for biomarker discovery from whole blood serum. Arch Physiol Biochem. 2010 Jul 22; doi: 10.3109/13813455.2010.501082. (epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 34.Streckfus CF, Mayorga-Wark O, Arreola D, Edwards C, Bigler L, Dubinsky WP. Breast cancer related proteins are present in saliva and are modulated secondary to ductal carcinoma in situ of the breast. Cancer Invest. 2008;26(2):159–67. doi: 10.1080/07357900701783883. [DOI] [PubMed] [Google Scholar]
- 35.Streckfus CF, Storthz KA, Bigler L, Dubinsky WP. A Comparison of the Proteomic Expression in Pooled Saliva Specimens from Individuals Diagnosed with Ductal Carcinoma of the Breast with and without Lymph Node Involvement. J Oncol. 2009;2009:737619. doi: 10.1155/2009/737619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klingbeil P, Marhaba R, Jung T, Kirmse R, Ludwig T, Zoller M. CD44 variant isoforms promote metastasis formation by a tumor cell-matrix cross-talk that supports adhesion and apoptosis resistance. Mol Cancer Res. 2009;7(2):168–79. doi: 10.1158/1541-7786.MCR-08-0207. [DOI] [PubMed] [Google Scholar]
- 37.Klingbeil P, Natrajan R, Everitt G, Vatcheva R, Marchio C, Palacios J, Buerger H, Reis-Filho JS, Isacke CM. CD44 is overexpressed in basal-like breast cancers but is not a driver of 11p13 amplification. Breast Cancer Res Treat. 2010;120(1):95–109. doi: 10.1007/s10549-009-0380-7. [DOI] [PubMed] [Google Scholar]
- 38.Zehentner BK, Carter D. Mammaglobin: a candidate diagnostic marker for breast cancer. Clin Biochem. 2004;37(4):249–57. doi: 10.1016/j.clinbiochem.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Obiezu CV, Diamandis EP. Human tissue kallikrein gene family: applications in cancer. Cancer Lett. 2005;224(1):1–22. doi: 10.1016/j.canlet.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 40.Brown NM, Stenzel TT, Friedman PN, Henslee J, Huper G, Marks JR. Evaluation of expression based markers for the detection of breast cancer cells. Breast Cancer Res Treat. 2006;97(1):41–7. doi: 10.1007/s10549-005-9085-8. [DOI] [PubMed] [Google Scholar]
- 41.Cheng KW, Lahad JP, Gray JW, Mills GB. Emerging role of RAB GTPases in cancer and human disease. Cancer Res. 2005;65(7):2516–9. doi: 10.1158/0008-5472.CAN-05-0573. [DOI] [PubMed] [Google Scholar]
- 42.Haas S, Gevensleben H, Rabstein S, Harth V, Pesch B, Bruning T, Justenhoven C, Brauch H, Hamann U, Ko YD, Baisch C, Fischer HP, Buttner R. Expression of heregulin, phosphorylated HER-2, HER-3 and HER-4 in HER-2 negative breast cancers. Oncol Rep. 2009;21(2):299–304. [PubMed] [Google Scholar]
- 43.Laubli H, Borsig L. Selectins promote tumor metastasis. Semin Cancer Biol. 2010;20(3):169–77. doi: 10.1016/j.semcancer.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 44.Ouellette RJ, Richard D, Maicas E. RT-PCR for mammaglobin genes, MGB1 and MGB2, identifies breast cancer micrometastases in sentinel lymph nodes. Am J Clin Pathol. 2004;121(5):637–43. doi: 10.1309/MMAC-TXT5-5L8Q-TKC1. [DOI] [PubMed] [Google Scholar]
- 45.Ramachandran P, Boontheung P, Xie Y, Sondej M, Wong DT, Loo JA. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J Proteome Res. 2006;5(6):1493–503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 46.Ramachandran PBP, Pang E, Yan W, Wong DT, Loo JA. Comparison of N-linked glycoproteins in human whole saliva, parotid, submandibular, and sublingual glandular secretions identified using hydrazide chemistry and mass spectrometry. Clinical Proteomics. 2008;4:80–104. doi: 10.1007/s12014-008-9005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.